Summary
Individual differences in perception are ubiquitous within the chemical senses: taste, smell, and chemical somesthesis [1–4]. A hypothesis of this fact states that polymorphisms in human sensory receptor genes could alter perception by coding for functionally distinct receptor types [1, 5–8]. We have previously reported evidence that sequence variants in a presumptive bitter receptor gene (hTAS2R38) correlate with differences in bitterness recognition of phenylthiocarbamide (PTC) [9–11]. Here, we map individual psychogenomic pathways for bitter taste by testing people with a variety of psychophysical tasks and linking their individual perceptions of the compounds PTC and propylthiouracil (PROP) to the in vitro responses of their TAS2R38 receptor variants. Functional expression studies demonstrate that five different haplotypes from the hTAS2R38 gene code for operatively distinct receptors. The responses of the three haplotypes we also tested in vivo correlate strongly with individuals’ psychophysical bitter sensitivities to a family of compounds. These data provide a direct molecular link between heritable variability in bitter taste perception to functional variations of a single G protein coupled receptor that responds to compounds such as PTC and PROP that contain the N-C═S moiety. The molecular mechanisms of perceived bitterness variability have therapeutic implications, such as helping patients to consume beneficial bitter-tasting compounds—for example, pharmaceuticals and selected phytochemicals.
Results and Discussion
The TAS2R38 Variant from a Sensitive Individual Responds to PTC and PROP, and the Variant from an Insensitive Individual Does Not
The haplotypes of TAS2R38 indicate that this gene, or transcriptionally functional portions of it, determines PTC sensitivity. The three most common polymorphisms observed in TAS2R38 occur at amino acid position 49, where either a proline or an alanine is encoded, at position 262, where either an alanine or a valine is encoded, and at position 296, where either a valine or an isoleucine is encoded, giving rise to two frequent haplotypes, PAV and AVI, plus the less common haplotypes AAI, PVI, and AAV. We cloned PAV and AVI alleles of hTAS2R38 from genomic DNA of two homozygous individuals. Their receptors were functionally expressed in HEK293 cells [9]. Micromolar concentrations of PTC elevated cytosolic [Ca2+]i in cells transiently transfected with the hTAS2R38-PAV variant in a concentration-dependent manner (Figure 1A). Moreover, stimulation of receptor-expressing cells with the related compound PROP resulted in an equally strong response at micromolar concentrations (Figure 1B). hTAS2R38-AVI did not respond to PTC or PROP concentrations as high as 1 mM.
Figure 1.
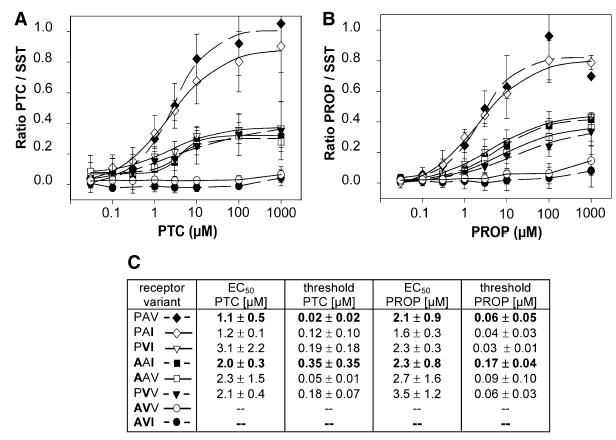
Dose-Response Curves of the Calcium Concentrations in Cells Cotransfected with hTAS2R38 Variants after Stimulation with Increasing PTC and PROP Concentrations
(A) Effects of a PTC concentration series on cells expressing the TAS2R38 variants (descending order): PAV (dashed line), PAI (solid line), PVI (solid line), AAI (dashed line), AAV (solid line), PVV (dashed line), AVV (solid line), or AVI (dashed line). See (C) for symbols key. The amplitudes of PTC (A) and PROP (B) responses have been normalized to those of the peptide hormone somatostatin-14 SST (1 μM), which activates an endogenous receptor. All receptor variants were challenged with PROP and PTC up to 1 mM, at which point PAV responses saturated. Each point represents the mean ± the standard error of the mean of at least three independent experiments carried out in triplicate. See Supplemental Experimental Procedures for methods, and for additional detail see Bufe et al., 2002 [9].
(B) Effects of a PROP concentration series on cells expressing the eight TAS2R38 variants. Symbols are as above.
(C) Half maximal response (EC50) and threshold values of all receptor variants for PROP and PTC. Amino acids identical to those of the AVI variant are listed in bold in the three-letter haplotype name to the left. Responses were normalized to somatostatin responses. EC50 and threshold numerical values for the three haplotypes tested from subjects PAV, AAI, AVI are in bold.
Three less common haplotypes were also characterized. PVI, AAI, and AAV [10] responded to PTC and PROP in the functional expression assay with EC50 values that resembled the sensitivity of the PAV variant (Figure 1). However, these three variants were only activated to approximately 40% of the response of the PAV variant when stimulated with the same concentrations (up to 1 mM). Thus, these data further implicate the common PAV variant as a major determinant of PROP/PTC taster status because it is the most responsive. They also suggest that the AAI, PVI, and AAV receptor variants convey intermediate PROP/PTC response magnitudes and thus confirm previous suggestions that multiple alleles determine PTC sensitivities within the population [10, 12].
The differences in the activity of the functionally expressed receptors could be caused by differences in membrane targeting. However, AVI, AAI, and PAV constructs are seen in the membranes of comparable proportions of HEK293 cells (see Figure S1 in the Supplemental Data available with this article online). The characteristic responses of the different expressed haplotypes are stable and reliable across three replicates of the experiment, so differences are not attributable to random fluctuation in membrane targeting. Alternatively, it is also possible that the intermediate responses are caused by an impaired coupling of receptor with signal-transduction G proteins.
Structure-Function Studies with Haplotype Variants and Diverse Ligands Reveal that hTAS2R38 Responds to N-C--S-Containing Compounds
Next, we determined the sets of ligands that activate these receptor variants, thereby establishing the tuning distribution of each variant. Because PTC sensitivity is also correlated with the bitter taste of other compounds that contain an N-C═S moiety [13], we challenged the hTAS2R38-PAV construct with various structurally related chemicals (Figure S2). Compounds containing the N-C═S group and showing bimodal taste sensitivities in humans [13, 14] activated hTAS2R38-PAV at concentrations that correspond to the bitter thresholds of “tasters” (sensitive subjects) [13]. In contrast, other compounds that are chemically related to PTC and PROP and lack the N-C═S group or do not show bimodal taste sensitivities [13, 14] failed to activate hTAS2R38–PAV (Figure S2). Notably, the artificial sweetener saccharin, which lacks the N-C═S group, failed to activate hTAS2R38 (Figure S2), although a correlation between the perceived bitterness of saccharin and PROP has been reported [15]. This observation, as well as the recent finding that saccharin activates in vitro the bitterness receptors hTAS2R44 and hTAS2R43 from chromosome 12 [16, 17], suggests that hTAS2R38 does not mediate the bitter off-taste of saccharin.
Our results suggest that hTAS2R38-PAV is a bitter taste receptor for chemicals that contain the N-C═S moiety. The AVI receptor, however, did not respond to PTC, PROP (Figures 1A and 1B), or other chemicals containing the N-C═S moiety at any concentration within the functional range of the assay, up to 10 mM (not shown). At the higher concentration ranges, AVI subjects may depend upon an additional TAS2R gene or genes to perceive them. We also tested the PAV haplotype against a battery of diverse bitter compounds and sweeteners, and we found that it only responded to PTC and PROP, further suggesting that hTAS2R38–PAV is a specific detector of anti-thyroid toxins (Figure S2A).
Mutational Analysis Shows that, of the Three Main Polymorphic Sites, the Amino Acids at Positions 49 and 262 Affect Cellular Responding the Most
We constructed receptor types PAI, AVV, and PVV for residue positions 49, 262, or 296; these receptor types do not correspond to known human haplotypes (Figure 1). The presence of alanine at position 49 and valine at position 262 diminishes receptor function, whereas the variation in position 296 had little effect. This is best exemplified by the observation that the AVI and AVV variants do not respond to PTC and PROP, whereas the PAV and PAI variants respond equally strongly. Thus, these results underscore the importance of these two sites and strengthen the correlations between the haplotypes and PTC sensitivity [10]. The mutational analysis conducted on the hTAS2R38 constructs confirmed our earlier observations [10] that amino acid positions 49 and 262 carry greater impact on stimulus binding and cellular activation than does amino acid position 296 (table in Figure 1).
TAS2R38 Transcripts Are Identified in Human Circumvallate and Fungiform Papillae for Both PAV-Taster and AVI-Nontaster Forms
To determine whether these receptors are within the perceptual bitterness pathway, we looked for their transcripts in taste-receptor cells inside taste buds. We detected TAS2R38 mRNA via RT-PCR in human tongue tissue containing circumvallate and fungiform papillae (Figures 2A and 2B). Moreover, in situ hybridization revealed that hTAS2R38 mRNA is located in a subset of taste-receptor cells within circumvallate taste buds (Figure 2C). We also quantified the amounts of transcript expressed in human fungiform papillae with real-time-quantitative PCR in homozygous AVI and PAV subjects and in two heterozygous subjects. All alleles were expressed (Figure 2B). This observation eliminates the possibility that insensitivity to PTC might be because of failure to express this allele and suggests that the human insensitivity to PTC is explained largely by the in vitro observation that the AVI allele is not activated by PTC over a broad concentration range. The observation that the two heterozygous subjects expressed very different ratios of PAV and AVI alleles may explain the large variability in heterozygotes’ responses to PTC and PROP (Figure 3). Some heterozygous subjects appear PAV-like, and others appear AVI-like.
Figure 2.
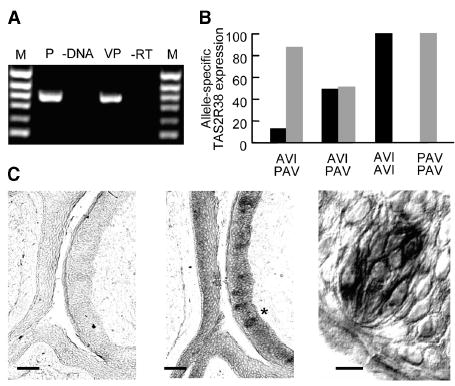
Presence of hTAS2R38 mRNA in Human Taste Papillae
(A) Agarose gel electrophoresis of 765 bp fragments amplified by RT-PCR with primers specific for hTAS2R38 mRNA. (1) 6:100 bp ladder used as size standard; (2) hTAS2R38 plasmid DNA, positive control; (3) negative control, no template present; (4) circumvallate papillae; and (5) reverse transcriptase has been omitted from the reaction to assess the presence of contaminating genomic DNA.
(B) Allele-specific gene expression of hTAS2R38 in human fungiform taste tissue. mRNA levels have been measured by quantitative RT-PCR in relation to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA levels in four subjects of the indicated diplotype: A/P, AVI/PAV; A/A, AVI/AVI; and P/P, PAV/PAV. Grey bars represent mRNA levels for PAV variant, and black bars represent mRNA for the AVI variant.
(C) In situ hybridization of human circumvallate papilla with hTAS2R38 sense (left panel) or antisense cRNA probe (middle and right panels), processed by alkaline phosphatase. Scale bars represent 100 μm (left and middle panels) and 10 μm (right panel) (3 of 11 cells stained in this section). Star denotes the taste bud shown in higher magnification in the right panel.
Figure 3.
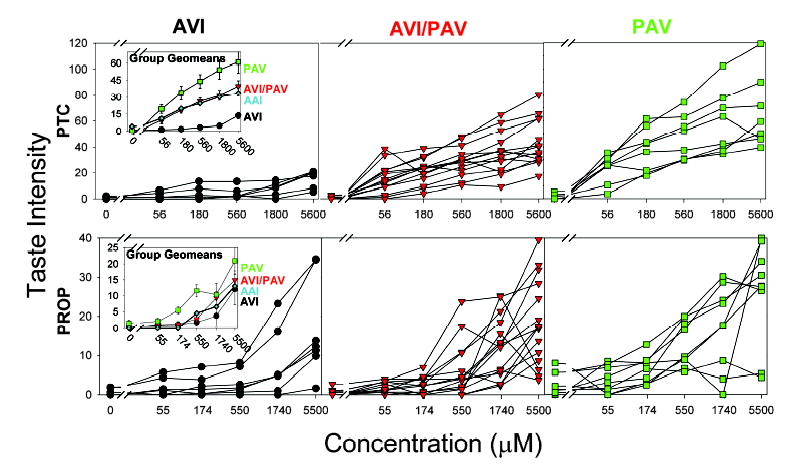
Suprathreshold Concentration-Intensity Functions for PTC and PROP Tasting in 32 Subjects Genotyped for hTAS2R38
The y axis represents standardized perceived intensity as rated on the general labeled magnitude scale. The x axis represents the concentration of the respective compounds tested. Each line represents the average responses over two trials for an individual subject. The top row of panels represents PTC responses, and the bottom row of panels represents PROP responses. The left column represents AVI/AVI subjects, the middle column represents AVI/PAV subjects, and the right column represents PAV/PAV subjects. Symbols identify haplotype groups: black circles, AVI/AVI (left column); red down-triangles, AVI/PAV (middle column); and green squares, PAV/PAV (right column).
Insets (left column) represent geometric mean taste responses with geometric standard errors for PTC and PROP for the various haplotype groups; symbols and colors are the same as with individual curves. The single AAI/AAI subject’s concentration-intensity functions are added to the insets (blue diamonds).
Which of the Perceptual Mechanisms Utilized by Three Complex Psychophysical Tasks Are Explained by the In Vitro Receptor Responses?
To integrate the above results with psychogenomic data, we determined whether the hTAS2R38 receptor variants influence the perception of both PROP and PTC in the population. We studied bitterness perception in groups as well as individual subjects who had been genotyped for hTAS2R38, and we compared these results with the data obtained from the functional expression of their receptors. We measured concentration-intensity functions and recognition thresholds in 32 subjects of known diplotype for TAS2R38: PAV/PAV, PAV/AVI, or AVI/AVI. Three additional individuals, who were genotyped as PAV/PAV, AVI/AVI, and AAI/AAI, were studied more intensively with three psychophysical tests: psychometric detection thresholds, recognition thresholds, and suprathreshold concentration-intensity functions. All three psychophysical tests were necessary to fully characterize these subjects’ bitter responses because each test involves different perceptual mechanisms.
Suprathreshold Measures
hTAS2R38 Haplotypes Account for the Majority of Population Variance for Perceived Suprathreshold PTC Bitterness
Among the PTC concentration-intensity responses, there is clear separation of all the AVI/AVI functions from all the PAV/PAV functions (Figure 3A; compare height of left panel functions with the right panel functions). The large difference in PTC sensitivity of the AVI and PAV diplotype subjects was not caused by general taste insensitivity in AVI carriers because their responses to other bitter and sweet taste stimuli were as strong as those of the PAV homozygotes (data not shown). Therefore, the insensitivity of AVI subjects to PTC is best explained by the observed lack of AVI-receptor response to PTC in the in vitro experiments (Figure 1). Thus, the allele they carry determines, to a large degree, the responses among AVI and PAV subjects (Figure 3).
The heterozygote group’s PTC bitterness functions were intermediate to AVI/AVI and PAV/PAV functions. The heterozygous subjects’ variability encompasses those who appear AVI-like and others who appear PAV-like, with most lying in between. The three haplotypes’ geometric means (Figure 3, inset) show that all responses of the diplotype groups are significantly different from each other at every concentration of PTC (p < 0.001). Clearly, PAV/AVI heterozygotes are PTC and PROP tasters, although their suprathreshold responses are somewhat lower than those of PAV homozygotes [10]. The psychophysical variability (Figure 3) in the heterozygous group may be explained by the varying mRNA proportions for the PAV and AVI transcripts produced in receptor cells (Figure 2B).
hTAS2R38 Haplotypes Predict Less Variance for Perceived PROP Bitterness Than for PTC Bitterness
The PROP concentration-intensity functions did not segregate by haplotype as sharply as the PTC ratings did (Figure 3). The diplotype group geometric means (Figure 3, inset) show that the responses to PROP were only significantly different for the second and third concentrations (170 and 550 μM; p < 0.01). The PAV/PAV group ratings at these concentrations were significantly greater than those of the other two groups. However, the observation that two AVI/AVI subjects responded strongly to PROP (Figure 3, left panel), and two PAV/PAV subjects responded very little (Figure 3, right panel), strongly indicates that suprathreshold sensitivities to PROP are under additional genetic and environmental controls. This is noteworthy because PROP was selected as a test proxy for PTC when it was discovered that the bitterness of the two showed a strong correlation [13]. Because PTC tasting is the oldest studied human chemosensory genetic trait, assuming one could replace PTC with PROP poses a significant change. Our psychogenomic data for PROP indicate that it is not interchangeable with PTC, but at low concentrations shows a strong similarity.
Recognition Threshold Measures
hTAS2R38 Haplotypes Predict Recognition Threshold Sensitivity in Individuals and Groups
We measured bitterness recognition thresholds in these same three groups with a modified Harris-Kalmus method because absolute detection thresholds do not convey the quality of taste perceived. Interestingly, unlike the supra-threshold scaling responses of the heterozygote PAV/AVI group, which were intermediate to homozygous PAV and AVI groups, the heterozygote subjects’ recognition thresholds for PTC and PROP resembled those of the homozygous PAV/PAV subjects: They differed by only a factor of ~2 and ~4, respectively (Figure 4; squares on left x axis and table). Recognition thresholds for PTC were significantly different across haplotype groups (F[2,27] = 20.72, p < 0.0001). AVI/AVI PTC recognition thresholds were significantly higher than those of the other two groups (p < 0.001), which did not differ from each other. Recognition thresholds for PROP were also significantly different across haplotypes (F[2,25] = 7.95, p < 0.01). Similar differences between the haplotypes occurred with PROP as with PTC (p < 0.01) (Figure 4; squares on right x axis and table). The bitterness recognition thresholds for PTC and PROP were correlated (r = 0.59, p < 0.01) across all haplotypes, suggesting a common mechanism for tasting PROP and PTC [18] at these lower concentrations.
Figure 4.
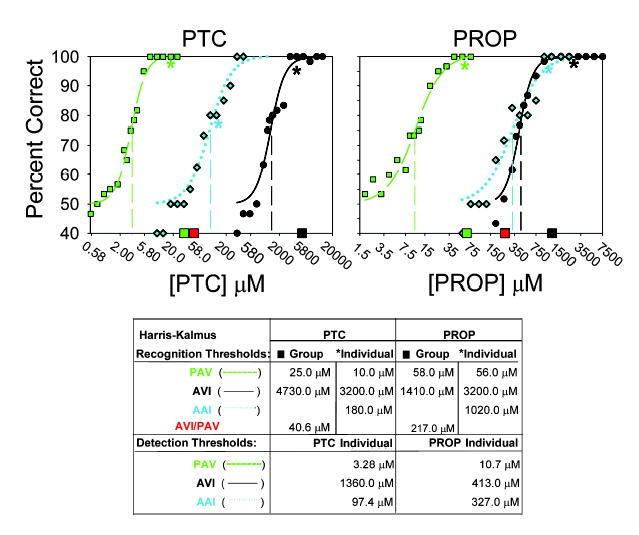
Psychometric Functions for PTC and PROP Tasting in One PAV/PAV Subject, One AVI/AVI Subject, and One AAI/AAI Subject Genotyped for TAS2R38 Variants
The y axis represents percent of trials correct in a two-alternative forced-choice task. The x axis represents the concentrations of the respective compounds that were tested. The dashed, dotted, and solid fitted functions represent the PAV (green), AAI (blue), and AVI (black) subjects’ sensitivities. Asterisks represent the concentrations of the three individuals’ modified Harris-Kalmus (mH-K) bitterness recognition thresholds and are placed on their respective psychometric functions. Large filled squares that lie on the x axis represent the mean mH-K bitterness recognition thresholds for the 32 subjects (from Figure 3), who were tested separately and genotyped as PAV/PAV (green), PAV/AVI (red), or AVI/AVI (black). Vertical dashed lines indicate detection thresholds (~IC50). Numerical threshold values for the three individuals’ and the group means are presented by haplotype in the table beneath the figure.
Absolute Threshold Measures
hTAS2R38 Haplotypes Predict Absolute Subject Sensitivity with High Correspondence to In Vitro Sensitivity Measures
This is an important measure because the psychometric curves for PTC and PROP are most closely analogous to the in vitro dose-response curves and thresholds of their receptors. Unlike the suprathreshold ratings, these psychometric thresholds in homozygous PAV and AVI subjects showed very clear and parallel differences in sensitivity to both PTC and PROP. The threshold values, defined as 75% performance (the inflection point) on the psychometric functions, for PAV/PAV and AVI/AVI differed over 400-fold for PTC but only 40-fold for PROP (Figure 4, vertical dashed lines). Note that AVI subjects are not blind to PTC and PROP but, rather, show a rightward shift of their psychometric functions by approximately two to three orders of magnitude (Figure 4). The lowest concentrations that the PAV and AVI homozygotes could statistically distinguish from water, defined as performance above the 61% level (p < 0.05 via the chi-square statistic), were 2.4 μM PTC and 4.2 μM PROP (PAV) and 1000 μM PTC and 320 μM PROP (AVI). Interestingly, the PAV subject’s lowest distinguishable concentration from water correlates remarkably well with the in vitro EC50 values of the subject’s expressed PAV-receptor for PTC (1.1 μM) and PROP (2.1 μM). A similar comparison cannot be made for the AVI-receptor because it did not respond to PTC or PROP in vitro. Recognition thresholds in these same two individuals were near the upper asymptotes of the psychometric functions, where detectability approaches perfect performance and differed 320-fold for PTC but only 57-fold for PROP (Figure 4, asterisks). The data for these two individuals are representative of their haplotype groups (Figure 4; compare with squares along x axis) and show that the differences in perception of PTC are more pronounced between the different haplotypes than are the differences in perception of PROP.
We also intensively tested another individual subject who is homozygous for the less common AAI haplotype [19]. The AAI subject’s absolute detection and recognition thresholds for PTC were intermediate; they fell between the PAV/PAV and the AVI/AVI mean values (Figure 4). The AAI subject’s values for PROP were more similar to the AVI values than to the PAV values. The discordance between the PTC and PROP sensitivities was also evident in this subject’s suprathreshold responses to these stimuli. The AAI subject’s concentration-intensity function for PTC was intermediate to the AVI and PAV values (Figure 3, insets). At higher PTC concentrations, the AAI function was higher than the highest AVI subject’s response, and it was at the same level as the lowest PAV subject’s response (Figure 3). At the highest concentration of PROP, the AAI rating was very similar to the mean response of the AVI subjects (Figure 3). These observations support the idea that different haplotypes code for receptors that vary in their sensitivities to PTC and PROP semi-independently. Here, we see that genetic variants of TAS2R38 do not always cause perception of PTC and PROP to be scaled in parallel. The observation that the less common AAI haplotype is also a PTC taster but is less sensitive than are carriers of PAV is consistent with the reduced function of the AAI receptor variant in the in vitro studies.
Conclusion
We demonstrate here that the alleles of hTAS2R38 code for receptors that form three broad categories: those that are sensitive to PTC and PROP (PAV), those that have little or no sensitivity to them within the limited response range possible in the expression assay (AVI), and those with intermediate sensitivity (AAI). Thus, the hTAS2R38 polymorphisms that differ on chromosomes within an individual and between individuals code for functionally distinct receptor types that directly affect bitterness perception of N-C═S-containing compounds. Because different TAS2R alleles can code for different receptors, across multiple humans we may not have only 25 bitter taste receptors, but could have as many bitter taste receptors as there are TAS2R alleles [19].
The observation that TAS2R38 receptors are tuned to many chemicals with the N-C═S moiety has important implications. Overingestion of N-C═S-containing compounds, like isothiocyanates, in geographical regions of low iodine is associated with thyroid disease and goiter [20], and ingestion of isothiocyanates from Brassica vegetables [13, 14], such as broccoli and brussel sprouts, is associated with potent anti-cancer effects [21]. Thus, the observed polymorphisms in the TAS2R38 gene may be due to evolutionary pressures that foster variability at the receptor to enhance bitterness detection and rejection of these compounds and, alternatively, to allow them to be ingested without inducing aversive taste experiences [19].
Supplementary Material
Acknowledgments
The authors thank Dr. J.-D. Raguse (Berlin) for providing the lingual tissue for in situ analysis, Dr. Roy Feldman for providing fungiform biopsy for quantitative polymerase chain reaction (QPCR) analysis, Dr. Tauf Huque for assistance with cDNA synthesis, Dr. Joel Levine for critical comments on the manuscript, and Ms. Ellen Schöley-Pohl for her expert technical assistance. This work was made possible by a grant from the German Science Foundation (DFG) to W.M. and grants from the National Institutes of Health to P.A.S.B. (DC02995) and D.R.R. (DC0004698). The authors declare that they have no competing financial interests, except for J.P.S., owing to his employment by Givaudan Flavors Corporation.
Footnotes
Supplemental Data
Detailed Experimental Procedures, as well as two supplemental figures, are available at http://www.current-biology.com/cgi/content/full/15/4/▪▪▪/DC1/.
References
- 1.Menashe I, Man O, Lancet D, Gilad Y. Different noses for different people. Nat Genet. 2003;34:143–144. doi: 10.1038/ng1160. [DOI] [PubMed] [Google Scholar]
- 2.Fischer, A., Gilad, Y., Man, O., and Paabo, S. (2004). Evolution of bitter taste receptors in humans and apes. Mol. Biol. Evol. 21, in press. Published online October 20, 2004. 10.1093/molbev/msi027 [DOI] [PubMed]
- 3.Drewnowski A. The science and complexity of bitter taste. Nutr Rev. 2001;59:163–169. doi: 10.1111/j.1753-4887.2001.tb07007.x. [DOI] [PubMed] [Google Scholar]
- 4.Delwiche JF, Buletic Z, Breslin PAS. Covariation in individuals’ sensitivities to bitter compounds: Evidence supporting multiple receptor/transduction mechanisms. Percept Psychophys. 2001;63:761–776. doi: 10.3758/bf03194436. [DOI] [PubMed] [Google Scholar]
- 5.Ueda T, Ugawa S, Ishida Y, Shibata Y, Murakami S, Shimada S. Identification of coding single-nucleotide polymorphisms in human taste receptor genes involving bitter tasting. Biochem Biophys Res Commun. 2001;285:147–151. doi: 10.1006/bbrc.2001.5136. [DOI] [PubMed] [Google Scholar]
- 6.Kim UK, Larsen JK, Drayna D. Worldwide genetic variation in human bitter taste receptor genes. Am J Hum Genet. 2003;73 (supplement):378. [Google Scholar]
- 7.Inoue M, Reed DR, Li X, Tordoff MG, Beauchamp GK, Bachmanov AA. Allelic variation of the Tas1r3 taste receptor gene selectively affects behavioral and neural taste responses to sweeteners in the F2 hybrids between C57BL/6ByJ and 129P3/J mice. J Neurosci. 2004;24:2296–2303. doi: 10.1523/JNEUROSCI.4439-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 9.Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 10.Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- 11.Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, Reed DR, Snyder DJ, Bartoshuk LM. Bitterness receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin Exp Res. 2004;28:1629–1637. doi: 10.1097/01.ALC.0000145789.55183.D4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blakeslee AF. Genetics of sensory thresholds: Taste for phenyl thio carbamide. Proc Nat Acad Sc. 1931;18:128–130. doi: 10.1073/pnas.18.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnicot NA, Harris H, Kalmus H. Taste thresholds of further eighteen compounds and their correlation with PTC thresholds. Ann Eugen. 1951;16:119–128. doi: 10.1111/j.1469-1809.1951.tb02464.x. [DOI] [PubMed] [Google Scholar]
- 14.Harris H, Kalmus H. Chemical specificity in genetical differences of taste sensitivity. Ann Eugen. 1949;15:32–45. doi: 10.1111/j.1469-1809.1949.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 15.Bartoshuk LM. Bitter taste of saccharin related to the genetic ability to taste the bitter substance 6-n-propylthiouracil. Science. 1979;205:934–935. doi: 10.1126/science.472717. [DOI] [PubMed] [Google Scholar]
- 16.Pronin AN, Tang H, Connor J, Keung W. Identification of ligands for two human bitter T2R receptors. Chem Senses. 2004;29:583–593. doi: 10.1093/chemse/bjh064. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. . J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo SW, Reed DR. The genetics of phenylthiocarbamide perception. Ann Hum Biol. 2001;28:111–142. doi: 10.1080/03014460151056310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wooding S, Kim UK, Bamshad MJ, Larsen JK, Jorde LB, Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VanEtten, C.H. (1969). Goitrogens. In Toxic Constituents of Plant Foodstuffs, I.E. Liener, ed. (New York: Academic Press), pp. 103–142.
- 21.Fahey, J.W., Stephenson, K.K., and Talalay, P. (1997). Glucosinolates, myrosinase, and isothiocyanates: Three reasons for eating Brassica vegetables. In Functional Foods for Disease Prevention I: Fruits, Vegetables, and Teas, Volume ACS Symposium Series 701, T Shibamoto, J Terao, and T Osawa, eds (Washington, DC: American Chemical Society), pp 16–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


