Abstract
The regulation of PBC protein function through subcellular distribution is a crucial evolutionarily conserved mechanism for appendage patterning. We investigated the processes controlling PBX1 nuclear export. Here we show that in the absence of MEINOX proteins nuclear export is not a default pathway for PBX1 subcellular localization. In different cell backgrounds, PBX1 can be imported or exported from the nucleus independently of its capacity to interact with MEINOX proteins. The cell context-specific balance between nuclear export and import of PBX1 is controlled by the PBC-B domain, which contains several conserved serine residues corresponding to phosphorylation sites for Ser/Thr kinases. PBX1 subcellular localization correlates with the phosphorylation state of these residues whose dephosphorylation in duces nuclear export. Protein kinase A (PKA) specifically phosphorylates PBX1 at these serines, and stimulation of endogenous PKA activity in vivo blocks PBX1 nuclear export in distal limb mesenchymal cells. Our results reveal a novel mechanism for the control of PBX1 nuclear export in addition to the absence of MEINOX protein, which involves the inhibition of PKA-mediated phosphorylation at specific sites within the PBC-B domain.
Keywords: homeodomain/limb patterning/nuclear export/nuclear import/post-translational regulation
Introduction
The PBC subfamily of TALE (Three-Amino-acid-Loop-Extension) homeodomain proteins includes the products of the vertebrate Pbx1, Pbx2, Pbx3 and Pbx4, and the Drosophila extradenticle (exd) genes (Bürglin, 1997; Wagner et al., 2001). EXD was shown to play important roles in Drosophila development, participating as a cofactor for HOX proteins in the specification of segmental identities (for a review, see Mann and Affolter, 1998). Similarly, vertebrate PBX proteins were shown to cooperate with HOX proteins in pattern formation (Pöpperl et al., 2000; Selleri et al., 2001). PBC proteins act as HOX cofactors by binding cooperatively with HOX proteins to DNA, thus increasing their binding site and promoter activation selectivity (for a review, see Mann and Affolter, 1998).
PBC proteins were also found to form stable heterodimers with MEINOX proteins (Chang et al., 1997; Rieckhof et al., 1997; Berthelsen et al., 1998), which belong to a different subfamily of TALE homeodomain proteins and include the products of the vertebrate Meis and Prep genes and the Drosophila homothorax (hth) genes (Bürglin, 1997; Fognani et al., 2002). Hetero dimerization between PBC and MEINOX proteins requires two conserved regions within the N-terminus of PBC proteins, the PBC-A and PBC-B domains, and two related regions, HR1 and HR2, within the N-terminus of MEINOX proteins (Berthelsen et al., 1998). EXD–HTH heterodimers were shown to be involved in several crucial developmental processes (reviewed in Mann and Affolter, 1998; Morata, 2001).
An intriguing aspect of PBC protein function is its regulation at the post-translational level, a mechanism that plays a crucial role in specific patterning processes (reviewed in Affolter et al., 1999; Vogt and Duboule, 1999). The exd gene, for example, is transcribed and translated in most embryonal cells, but its function is regulated through subcellular localization; the EXD protein is nuclear where the gene is functional and cytoplasmic where its function is not requested. EXD subcellular distribution is regulated by heterodimerization with HTH. In Drosophila leg imaginal discs, EXD is nuclear in proximal regions, which correspond to the HTH expression domain. Conversely, in distal parts, where HTH is not present, EXD is cytoplasmic. Both hth and exd functions are necessary for proximal, but not distal, leg development. Ectopic expression of hth distally induced nuclear localization of EXD and blocked distal development giving rise to truncated appendages (reviewed in Morata, 2001). Truncations were also obtained by mis expression of EXD in the nuclei of distal cells, indicating that EXD can interfere, independently of HTH, with distal leg development (Gonzales-Crespo and Morata, 1996). The correlation between PBC protein subcellular localization and proximodistal limb patterning was shown to be evolutionarily conserved. In mouse and chicken developing limb buds PBX1 is nuclear in proximal cells and cytoplasmic in distal cells, and, as in Drosophila, nuclear PBX1 coincides with the expression of the Meis1 and Meis2 MEINOX genes (Gonzales-Crespo et al., 1998; Capdevilla et al., 1999; Mercader et al., 1999). Misexpression of Meis1 or Meis2 distally in developing chicken limbs leads to proximalization or truncation respectively of distal structures (Capdevilla et al., 1999; Mercader et al., 1999). Moreover, like exd in Drosophila, Pbx1 function is required for the correct development of vertebrate proximal limb structures, as Pbx1-deficient mice display malformations involving only proximal skeletal elements, leaving distal elements unaffected (Selleri et al., 2001).
The mechanisms and the signalling pathways controlling nuclear exclusion of PBX1 and EXD, particularly in distal limb regions, are still poorly understood. It has been reported by us and other workers that the cytoplasmic localization of PBX1 and EXD was due to active leptomycin-B-sensitive nuclear export (Abu-Shaar et al., 1999; Berthelsen et al., 1999). We showed that PBX1 nuclear export, which requires a signal located within the PBC-A domain, is prevented by heterodimerization with the MEINOX proteins PREP1 and HTH (Berthelsen et al., 1999). Hence, nuclear export was assumed to be the default route for subcellular localization of PBC proteins in the absence of MEINOX proteins. In specific cell backgrounds, however, we found PBX and EXD to be exclusively nuclear even without co-expression of MEINOX proteins, suggesting that nuclear export of PBC proteins is not simply triggered by the lack of heterodimerization with MEINOX proteins. In this work, we exploited the differential subcellular localization of PBX1 in different cell contexts to investigate the regulation of its nucleocytoplasmic transport. PBX1 was found to contain two adjacent independent nuclear export signals (NESs) of the leucine-rich type, located within the conserved PBC-A domain, which mediate direct contacts with the CRM1 export receptor. The cell context-specific PBX1 nucleocytoplasmic distribution is controlled by the PBC-B domain, and correlates with the phosphorylation state of specific serine residues. Protein kinase A (PKA) phosphorylates these serine residues in vitro, and PKA activation blocks nuclear export of PBX1 in distal regions of developing chicken limb buds in vivo. Our results indicate that prevention of PKA phosphorylation of the PBC-B domain is the mechanism that triggers nuclear export of PBC proteins, and provide the framework for a possible link between the signalling pathways controlling distal limb development and the active nuclear exclusion of PBX1.
Results
PBX1 contains two independent leucine-rich nuclear export signals
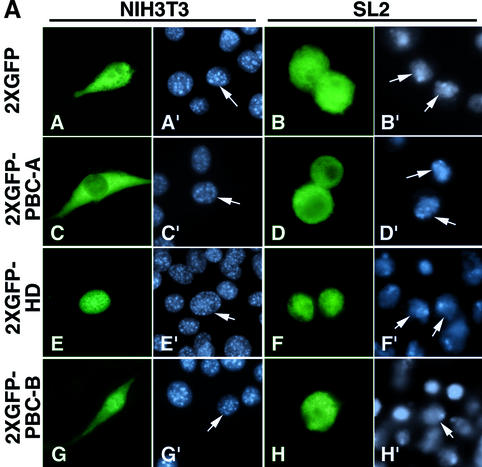
We previously identified a signal required for PBX1 nuclear export within the PBC-A domain (Berthelsen et al., 1999). To characterize this putative NES, we generated a fusion of PBC-A with two molecules of the green fluorescent protein (GFP) reporter. As controls, we also fused to GFP two other domains of PBX1, namely PBC-B and the homeodomain (HD). Different cell contexts were used for transient transfections: NIH 3T3 and COS7 cells where exogenously expressed PBX1 is nuclear, and Drosophila Schneider cells (SL2) where it is exported from the nucleus (Berthelsen et al., 1999). As shown in Figure 1A, panels A and B, the double GFP reporter protein (2XGFP) is uniformly distributed between the cytoplasm and the nucleus in both SL2 and NIH 3T3 cells. In contrast, a PBC-A domain fusion with 2XGFP was found exclusively in the cytoplasm in both cell lines (Figure 1A, panels C and D), and the 2XGFP–HD fusion was found only within the nucleus in both cell contexts (Figure 1A, panels E and F). The 2XGFP–PBC-B fusion behaved instead as 2XGFP alone, being equally distributed in the cytoplasm and the nucleus in both cell lines (Figure 1A, panels G and H). To test whether 2XGFP– PBC-A was exported from the nucleus in NIH 3T3, transfected cells were treated with leptomycin B (LMB), an inhibitor of the CRM1 nuclear export receptor (Wolff et al., 1997). As shown in Figure 1B, panel J, 2XGFP– PBC-A was also present in the nucleus of LMB-treated NIH 3T3. These results demonstrate that, among the three main conserved domains of PBX1, only PBC-A is able to direct nuclear export of a reporter protein. In contrast, the PBC-B domain was unable to selectively direct GFP to the nucleus or the cytoplasm in either cell context.
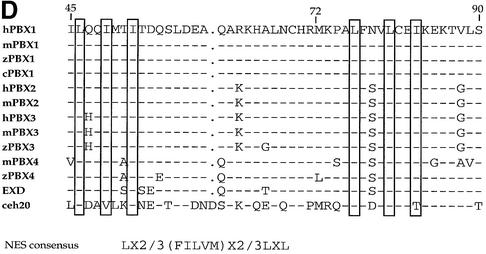
Fig 1. The PBC-A domain of PBX1 contains two independent nuclear export signals. (A) PBC-A drives cytoplasmic localization of 2XGFP, while HD induces nuclear localization (panels C and D, and E and F, respectively). PBC-B does not alter the subcellular localization of 2XGFP (panels G and H). (B) Nuclear export of 2XGFP–PBC-A is sensitive to LMB (panels I and J). Amino acids 73–90 and 45–90 are sufficient for driving LMB-sensitive export of 2XGFP (panels K and L, and M and N respectively). Amino acids 45–72 do not drive export (panel O and P). (C) Amino acids 45–90 are required for PBX1 export. PBX1 is cytoplasmic in SL2 cells (panel Q). Deletion of amino acids 73–90, as well as 45–72, does not affect export in SL2 cells (panels R and T). Deletion of amino acids 45–90 blocks export (panel S). Nuclei were stained with Hoechst (panels labelled with primes). (D) Alignment of the hPBX1 PBC-A domain 45–90 subregion with the corresponding regions from mouse (m), zebra fish (z), chicken (c), Drosophila (EXD) and Caenorhabditis elegans (ceh20) PBC proteins. Dashes indicate conserved amino acids and dots indicate absent ones. Boxed amino acids share homology to the NES consensus (Mattaj and Englmeier, 1998) shown below. Accession numbers are zPBX1 AJ245962, cPBX1 AB043620, zPBX3 AJ245964, mPBX4 AJ300183, zPBX4 AJ245966. hPBX1, mPBX1, hPBX2, mPBX2, hPBX3, mPBX3, EXD and ceh20 (Bürglin, 1997).
A deletion analysis of the PBX1 N-terminus showed that the cytoplasmic localization of PBX1 in SL2 cells requires a region of PBC-A spanning amino acids 73–90 (Berthelsen et al., 1999). We fused this region to 2XGFP (2XGFP PBC-A73–90) and, when expressed in NIH 3T3 cells, the fusion protein was indeed exported in an LMB-sensitive manner (Figure 1B, panels K and L). However, a mutant lacking amino acids 73–90 (PBXΔ73–90) was still cytoplasmic in SL2 cells (Figure 1C, panel R), suggesting that additional sequences mediate nuclear export. There fore we deleted a larger portion of the PBC-A domain, spanning amino acids 45–90 (PBXΔ45–90). As shown in Figure 1C, panel S, PBXΔ45–90 was nuclear in SL2 cells. Conversely, a deletion mutant involving only amino acids 45–72 (PBXΔ45–72), like PBXΔ73–90, was still exported from the nucleus (Figure 1C, panel T). Amino acids 45–90 and 45–72 were also fused to 2XGFP to test for their ability to drive nuclear export. As shown in Figure 1B, panels M and N, region 45–90 was able to drive LMB-sensitive export of GFP, whereas region 45–72 did not (Figure 1B, panels O and P). Identical results were obtained using COS7 in place of NIH 3T3 cells.
These results indicate that the region spanning amino acids 45–90 of PBC-A contains two independent signals, one located between amino acids 45 and 72 and the other between amino acids 73 and 90, both sufficient to mediate PBX1 nuclear export. The sequence between amino acids 45 and 90 (Figure 1D) contains two stretches, one from 45 to 52 (ILQQIMTI) and the other from 75 to 82 (LFNVLCEI), rich in leucine and isoleucine. Their spacing is characteristic of Rev-like leucine-rich consensus NESs, which are recognized by the CRM1 nuclear export receptor (Mattaj and Englmeier, 1998), suggesting that export of PBX1 might be mediated by a direct interaction of CRM1 with these sequences.
The PBC-A domain directly interacts with the CRM1 nuclear export receptor
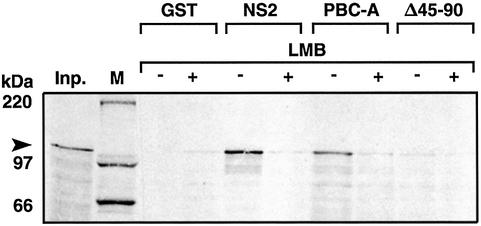
To verify whether PBX1 interacts directly with CRM1, we generated two glutathione S-transferase (GST) fusion proteins, one containing the PBC-A domain and the other its mutant derivative carrying a deletion of amino acids 45–90 (Δ45–90). The GST fusion proteins, expressed in Escherichia coli, and bound to glutathione– Sepharose, were challenged with in vitro translated hCRM1. A GST fusion carrying the minute virus of mice (MVM) NS2 protein NES, which was shown to interact strongly with CRM1 (Askjaer et al., 1999), was used as a positive control. As shown in Figure 2, the GST–PBC-A (PBC-A) fusion significantly retained CRM1 (lane 7), whereas the GST–PBC-A fusion carrying a deletion of amino acids 45–90 did not (lane 9). The addition of LMB strongly impaired the interaction between GST–PBC-A and CRM1 (Figure 2, lane 8). The resin carrying GST alone did not retain CRM1 (Figure 2, lanes 3 and 4), whereas GST–NS2 (NS2) interacted with CRM1 in an LMB-sensitive manner (Figure 2, lanes 5 and 6). Thus the PBC-A domain is capable of specifically establishing LMB-sensitive direct protein–protein contacts with the CRM1 nuclear export receptor, mediated by the NES-containing region.
Fig. 2. The PBC-A domain makes direct protein–protein contacts with CRM1. The MVM NS2 control NES (NS2), the PBX PBC-A domain (PBC-A) or the PBC-A domain deleted of amino acids 45–90, were fused to GST. CRM1 is not retained by GST alone (lane 3), whereas it is by GST–PBC-A (lane 7) and GST–NS2 (lane 5). CRM1 is not retained by the PBC-A Δ45–90 deletion mutant (lane 9). LMB prevents the interaction between CRM1 and PBC-A (lane 8) or NS2 (lane 6). Inp., Input CRM1 protein (∼110 kDa) (lane 1); M, molecular weight marker (weights are in kilodaltons).
The lack of a PBX1–MEINOX interaction is not sufficient to induce PBX1 nuclear export, which is instead controlled by the PBC-B domain
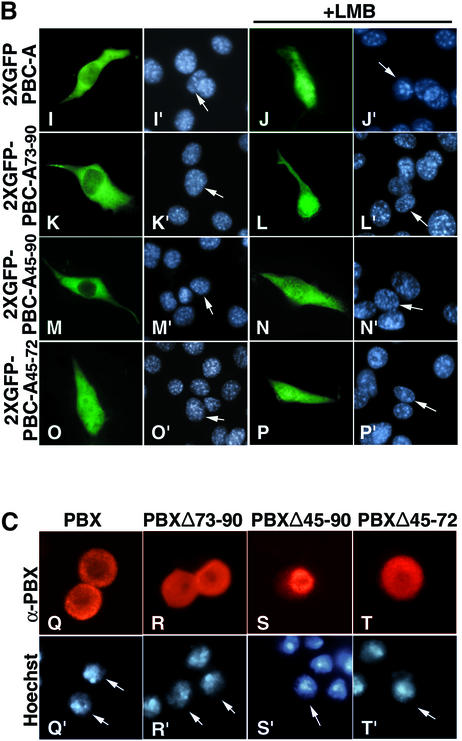
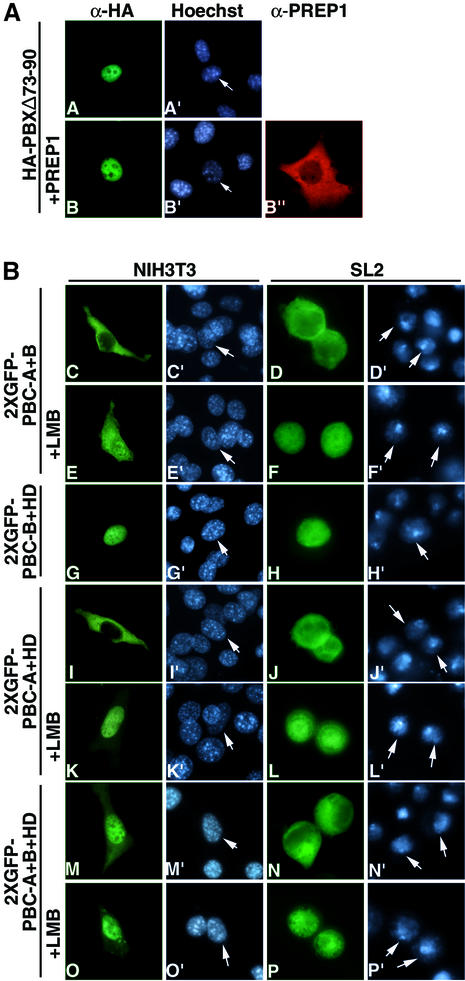
PBX1 subcellular distribution does not exclusively de pend on the presence or absence of MEINOX proteins. Evidence for this was provided by the PBXΔ73-90 mutant, which was not capable of heterodimerizing with MEINOX proteins in either cultured cells (Figure 3A, panel B) or in vitro in electrophoretic mobility shift assays (EMSAs) (data not shown). Despite this, PBXΔ73–90, as wild-type PBX1, was found to be exclusively nuclear in NIH 3T3 or COS7 cells and cytoplasmic in SL2 cells (Figures 3A, panel A and 1C, panel R; data not shown). Thus, PBX1 nuclear import in NIH 3T3 and COS7 cells is independent of interaction with MEINOX proteins and an additional mechanism must be active in specific cell backgrounds to prevent PBX1 nuclear export.
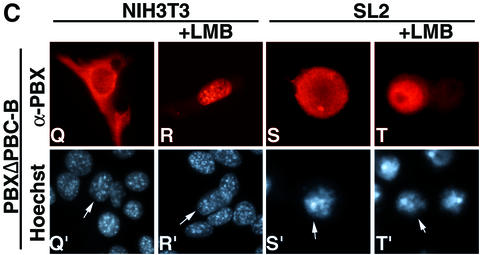
Fig. 3. The PBC-B domain blocks nuclear export according to cell context. (A) PBXΔ73–90 nuclear localization is independent of interaction with MEINOX proteins. PBXΔ73–90 is nuclear in NIH 3T3 cells (panels A and A′) and does not induce nuclear localization of PREP1 (panels B, B′ and B”). (B) A fusion of 2XGFP with PBC-A and PBC-B is cytoplasmic (panels C, C′, D and D′), while a fusion with PBC-B and HD is nuclear in both cell lines (panels G, G′, H and H′). LMB relocalizes 2XGFP–PBC-A+B (panels E, E′, F and F′). A fusion of PBC-A and HD with 2XGFP is cytoplasmic in both cell lines (panels I, I′, J and J′). LMB relocalizes 2XGFP–PBC-A+HD in both cell lines (panels K, K′, L and L′). A 2XGFP fusion containing PBC-A, PBC-B and HD displays cell context-specific subcellular localization (panels M, M′, N and N′). (C) A PBX1 mutant lacking PBC-B is exported in both cell lines (panels Q, Q′, S and S′) in an LMB-sensitive manner (panels R, R′, T and T′). Nuclei were stained with Hoechst.
We speculated that the differential cell context-specific nucleocytoplasmic localization of PBX1 might be due to a modulation of its nuclear export and import signals by other parts of the protein. Therefore we generated GFP fusions containing different combinations of the main functional domains of PBX1. A GFP fusion containing both PBC-A and PBC-B (2XGFP–PBC-A+B; Figure 3B, panels C and D) was cytoplasmic in both NIH 3T3 and SL2 cells, and its relocalization upon LMB treatment showed that it was actively exported (Figure 3B, panels E and F). Conversely, a fusion of GFP with PBC-B and the homeodomain (2XGFP–PBC-B+HD) was nuclear in both cell lines (Figure 3B, panels G and H). A GFP fusion containing PBC-A and the homeodomain (2XGFP–PBC- A+HD) was cytoplasmic in both NIH 3T3 and SL2 cells (Figure 3B, panels I and J), indicating that, if combined outside the PBX1 context, the NESs (PBC-A domain) prevail over the nuclear localization signal (HD) in both cell backgrounds. LMB induced nuclear relocalization of GFP–PBC-A+HD (Figure 3B, panels K and L).
The behaviour of GFP–PBC-A+HD compared with PBX1 suggested that PBC-B could play a role in controlling PBX1 cell context-specific subcellular distribution. Therefore, we generated a GFP fusion containing PBC-A, PBC-B and the homeodomain (2XGFP–PBC- A+B+HD). The GFP–PBC-A+B+HD fusion was nuclear in NIH 3T3 and cytoplasmic in SL2 cells (Figure 3B, panels M and N), thus recapitulating the cell context-specific localization of PBX1. To confirm the role of PBC-B in downregulating PBX1 export in mouse fibro blasts, we generated a PBX1 derivative carrying a dele tion of PBC-B (PBXΔPBC-B). Like wild-type PBX1, PBXΔPBC-B was exported from the nuclei of SL2 cells (Figure 3C, panels S and T). However, unlike wild-type PBX1, in NIH3T3 PBXΔPBC-B was found to be exported from the nucleus in an LMB-sensitive manner (Figure 3C, panels Q and R). Thus the PBC-B domain suppresses PBX1 nuclear export in a cell context-specific manner.
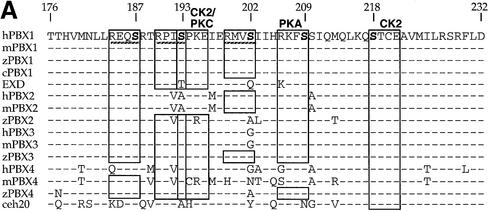
Conserved Ser/Thr phosphorylation sites within the PBC-B domain are required for regulating the subcellular localization of PBX1
To understand how PBC-B controls PBX1 subcellular localization, we scanned the PROSITE database (Falquet et al., 2002) with its peptide sequence to identify potential biologically relevant signals. As shown in Figure 4A, PBC-B contains several potential phosphorylation sites for the cAMP-dependent protein kinase A (PKA), casein kinase 2 (CK2) and protein kinase C (PKC) Ser/Thr kinases. Serines at positions 187, 193, 202, 209 and 218 were predicted with high confidence by the Netphos program (Blom et al., 1999) to be true phosphorylation sites. Significantly, a sequence alignment shows that these serine residues are conserved in the majority of PBC proteins (Figure 4A).
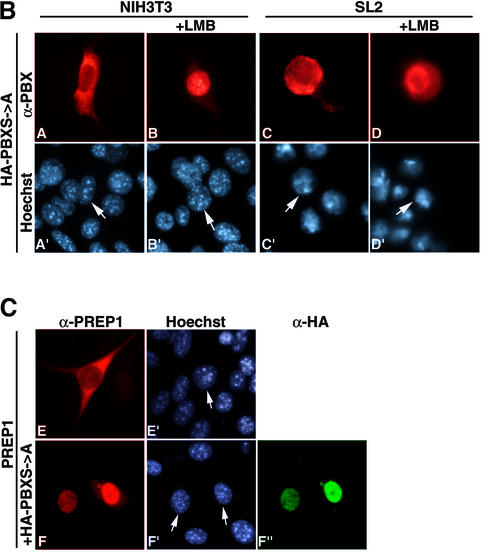
Fig. 4. Conserved Ser/Thr phosphorylation sites within PBC-B are required for regulating the subcellular localization of PBX1. (A) Alignment of part of the PBC protein PBC-B domain from human (h), mouse (m), chicken (c), zebrafish (z), Drosophila (EXD) and C.elegans (ceh20). Dashes indicate identities. Serines predicted to be phosphorylation targets are shown in bold. Numbering corresponds to the hPBX1 sequence. Consensus sites for PKA (Ser 209; R/K(2)-x-S/T Proscan), PKC (Ser 193; S/T-x-R/K Proscan) and CK2 (Ser 193 and 218; S/T-x(2)-D/E Proscan) are indicated. Dashed lines indicate putative PKA target sites (ser 187, 193, 202) showing homology to the looser consensus R-x(2)-S/T (Nethphos). Conserved kinase target sites are boxed. Accession numbers are listed in the caption to Figure 1. (B) Substitution of serines 187, 193, 202 and 209 with alanines (PBXS→A) induces cytoplasmic localization in NIH 3T3 (panels A and A′). Nuclear export in SL2 was not impaired (panels C and C′) and LMB induced nuclear localization in both cell contexts (panels B, B′, D and D′). PBX1S→A was detected with a polyclonal anti-PBX antibody (red). (C) Serine to alanine substitution does not impair heterodimerization with MEINOX proteins. In NIH 3T3, PREP1 is cytoplasmic (panels E and E′) but translocates to the nucleus as a heterodimer with PBXS→A (panels F, F′ and F”).
We generated a PBX mutant derivative (PBXS→A) in which serine residues 187, 193, 202 and 209 were substituted with alanines. In SL2 cells, PBXS→A behaved like wild-type PBX1, being exported in an LMB-sensitive manner (Figure 4B, panels C and D). In contrast, in NIH3T3 cells, unlike wild-type PBX1, PBXS→A was cytoplasmic in ∼33% of expressing cells (Figure 4B, panel A). LMB treatment induced nuclear localization of PBXS→A (Figure 4B, panel B), indicating that it is exported from the nucleus. To exclude the possibility that PBXS→A nuclear export is due to loss of interaction with endogenous MEINOX proteins, we tested its ability to induce PREP1 nuclear localization. PREP1 is cytoplasmic in NIH 3T3 (Figure 4C, panel E) and is imported into the nucleus upon heterodimerization with PBX1 (Berthelsen et al., 1999). When co-expressed with PBXS→A in NIH 3T3 cells, PREP1 was found to be nuclear (Figure 4C, panels F and F”), demonstrating that interaction with MEINOX proteins was not impaired. Heterodimerization and DNA-binding of PBXS→A with PREP1 were also tested in EMSAs; they were as efficient as those of wild-type PBX1 (data not shown). These data indicated that phosphorylation of PBC-B is the mechanism by which PBX1 nuclear export and import are modulated.
PBX1 is differentially phosphorylated according to cell context
We next investigated whether the differential subcellular distribution in SL2 compared with NIH 3T3 or COS7 cells correlated with the phosphorylation state of PBX1. To this end, since phosphorylation alters the isoelectric points (pI) of proteins adding negative charges, we used two-dimensional electrophoresis and immunoblotting (2D IEF). Protein-A-tagged PBX1 (ProtA-PBX), expressed and immunopurified from SL2 or COS7 cells, was first sub jected to IEF, allowing the separation of differentially charged isoforms, subsequently to SDS–PAGE, which separates according to molecular weight, and finally detected by immunoblotting. ProtA-PBX1 isolated from COS7 cells was separated into several isoelectric isoforms, one of which migrated into the more acidic region of the gel (Figure 5, panel A). PBX1 isolated from SL2 cells also separated into several isoforms, but none was present in the more acidic region of the gel (Figure 5, panel B). Treatment of COS7-derived PBX1 with alkaline phosphatase caused the disappearance of the more acidic isoform (data not shown). We also analysed the PBXS→A mutant carrying substitutions of serines 187, 193, 202, and 209 with alanines. PBXS→A isolated from COS7 cells (Figure 5, panel C), displayed only the pattern of less acidic isoforms. Significantly, the isoform pattern of PBXS→A co-migrated with that of SL2-cell-derived wild-type PBX1 (Figure 5, compare panels C and B). PBXS→A purified from SL2 cells showed the same isoform pattern as wild-type PBX1 (Figure 5, compare panels B and D). Thus, PBX1 displays a differential cell context-specific phosphorylation which correlates with its subcellular localization.
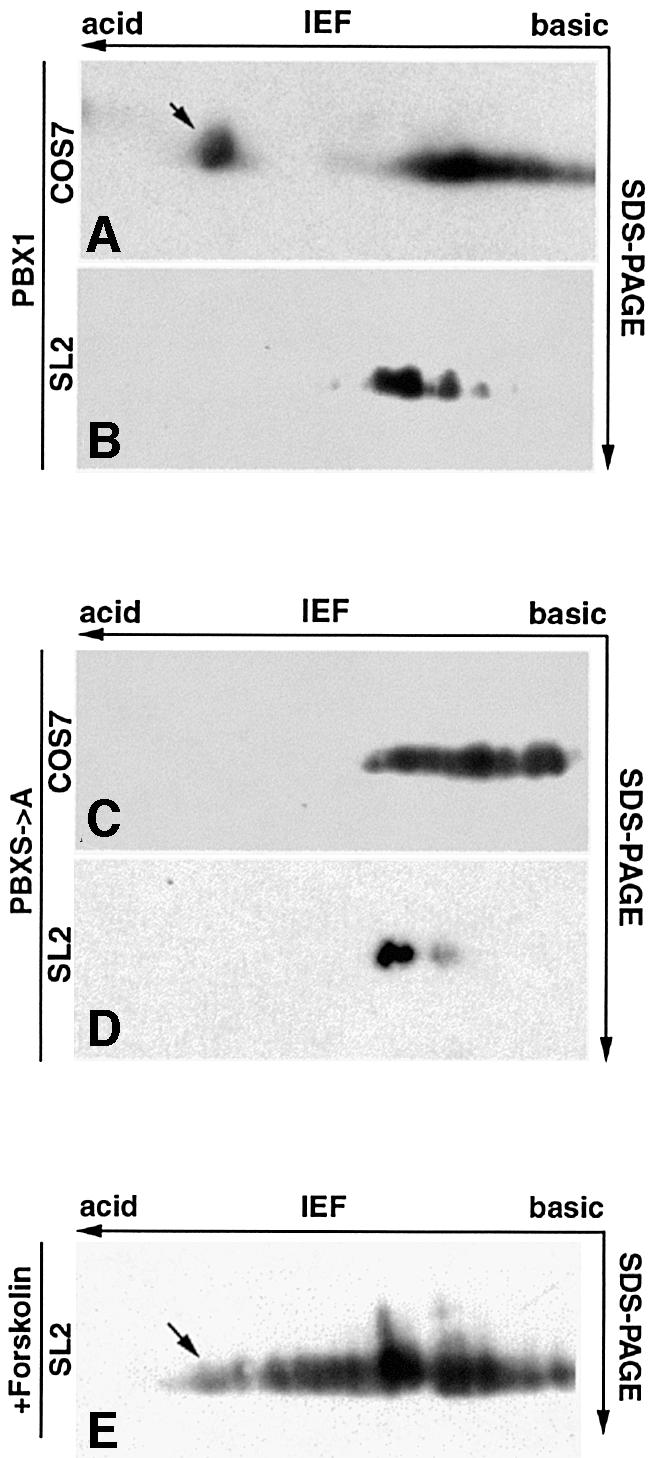
Fig. 5. PBX1 is differentially phosphorylated in COS7 and SL2 cells. (A) PBX1 purified from COS7 cells is separated into one more acidic isoelectric isoform (apparent pI 4) and several less acidic isoforms (pI ranging between 5 and 5.6) (B) PBX1 immunopurified from SL2 cells displays only the less acidic isoforms. (C) Similarly, PBXS→A purified from COS7 cells displays only the less acidic and not the more acidic isoform. (D) PBXS→A isolated from SL2 cells displays the same isoform pattern as wild-type PBX1. (E) PBX1 isolated from forskolin-treated SL2 cells shows the appearance of more acidic isoforms. The acidic isoelectric isoform of PBX1 is indicated by an arrow.
PKA specifically phosphorylates the PBC-B domain of PBX1 in vitro
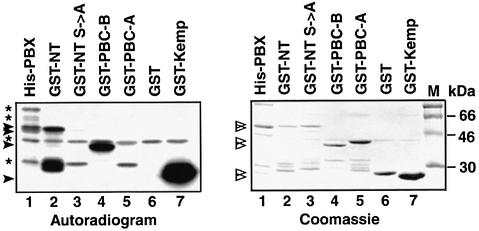
The four serine residues required for downregulating nuclear export of PBX1 represent potential targets for phosphorylation by the PKA, PKC and CK2 kinases. We performed in vitro phosphorylation experiments to test whether any of these kinases was able to phosphorylate PBC-B. As targets for in vitro phosphorylation, we used partially purified bacterially produced proteins. As shown in Figure 6, full-length PBX1 (His-PBX, lane 1) was phosphorylated by purified PKA. GST fusions containing the N-terminus of PBX1, including the PBC-A and PBC-B domains (GST–NT, lane 2) or the PBC-B domain alone (GST–PBC-B, lane 4), were also efficiently phosphorylated by PKA, whereas a mutant derivative of the PBX-NT region (GST–NT S→A), bearing substitutions of serines 187, 193, 202 and 209 with alanines, was not phosphorylated (Figure 6, lane 3). No phosphorylation was detected with GST–PBC-A or with GST alone (Figure 6, lanes 5 and 6). The Kemptide, a substrate of PKA, was also efficiently phosphorylated (Figure 6, lane 7). Neither PKC nor CK2 were able to phosphorylate PBX1 in vitro (data not shown). Thus, of the three Ser/Thr kinases tested, only PKA is capable of modifying PBC-B.
Fig. 6. PKA specifically phosphorylates the PBC-B domain in vitro. His-PBX1 (lane 1), a GST fusion containing both PBC-A and PBC-B (GST–NT, lane 2) and a GST fusion with PBC-B (GST–PBC-B, lane 4) were all phosphorylated by PKA. The corresponding phosphorylated bands are indicated by solid arrowheads. Conversely, GST (lane 6), a GST fusion of the PBX1 N-terminus bearing alanine substitutions at serine residues 187, 193, 202 and 209 (GST–NT S→A, lane 3) and a GST fusion with PBC-A (lane 5) were not phosphorylated. A GST–Kemptide fusion was used as positive control (lane 7). Phosphorylated bacterially derived contaminant proteins are indicated by asterisks. Open arrowheads in the Coomassie Blue staining indicate the recombinant PBX1-derived fusion proteins.
PKA activity blocks nuclear export of PBX1 in SL2 cells
To test whether PKA activity could affect the subcellular localization of PBX1, we co-expressed PBX1 and the catalytic subunit of PKA (PKAcat) in SL2 cells. As shown in Figure 7A, panel A, co-expression of PKAcat induced nuclear relocalization of PBX1 in 90% of the cells. Nuclear PBX1 was observed only in cells that also expressed PKAcat (Figure 7A, panel A′). We also tested the effect of PKAcat expression on two PBX1 mu tants, PBXΔPBC-B (Figure 7A, panel B) and PBXS→A (Figure 7A, panel C), lacking the serines phosphorylated in vitro by PKA. Significantly, the cytoplasmic localization of neither of these two mutants was affected by PKAcat expression (Figure 7A, panels B and C). We next tested whether stimulation of endogenous PKA activity in SL2 cells could block PBX1 nuclear export by treating cells with the adenylate cyclase stimulator forskolin. SL2 cells treated with 60 µM forskolin for 2 h displayed a nuclear accumulation of PBX in 90% of expressing cells (Figure 7B, panel D). In a time-course experiment, signi ficant nuclear accumulation of PBX1 was already present after 1 h of forskolin treatment (data not shown). Forskolin-induced relocalization of PBX1 requires serine residues 187, 193, 202 and 209, because neither the PBXΔPBC-B nor the PBXS→A mutants showed any change in localization upon forskolin treatment (Figure 7B, panels E and F). Furthermore, forskolin treatment caused the appearance of more acidic PBX1 isoforms in 2D IEF, indicating an induction of PBX1 phosphorylation in SL2 cells (Figure 5, panel E). These results indicate that induction of PKA activity in SL2 cells is sufficient to block nuclear export of PBX1.
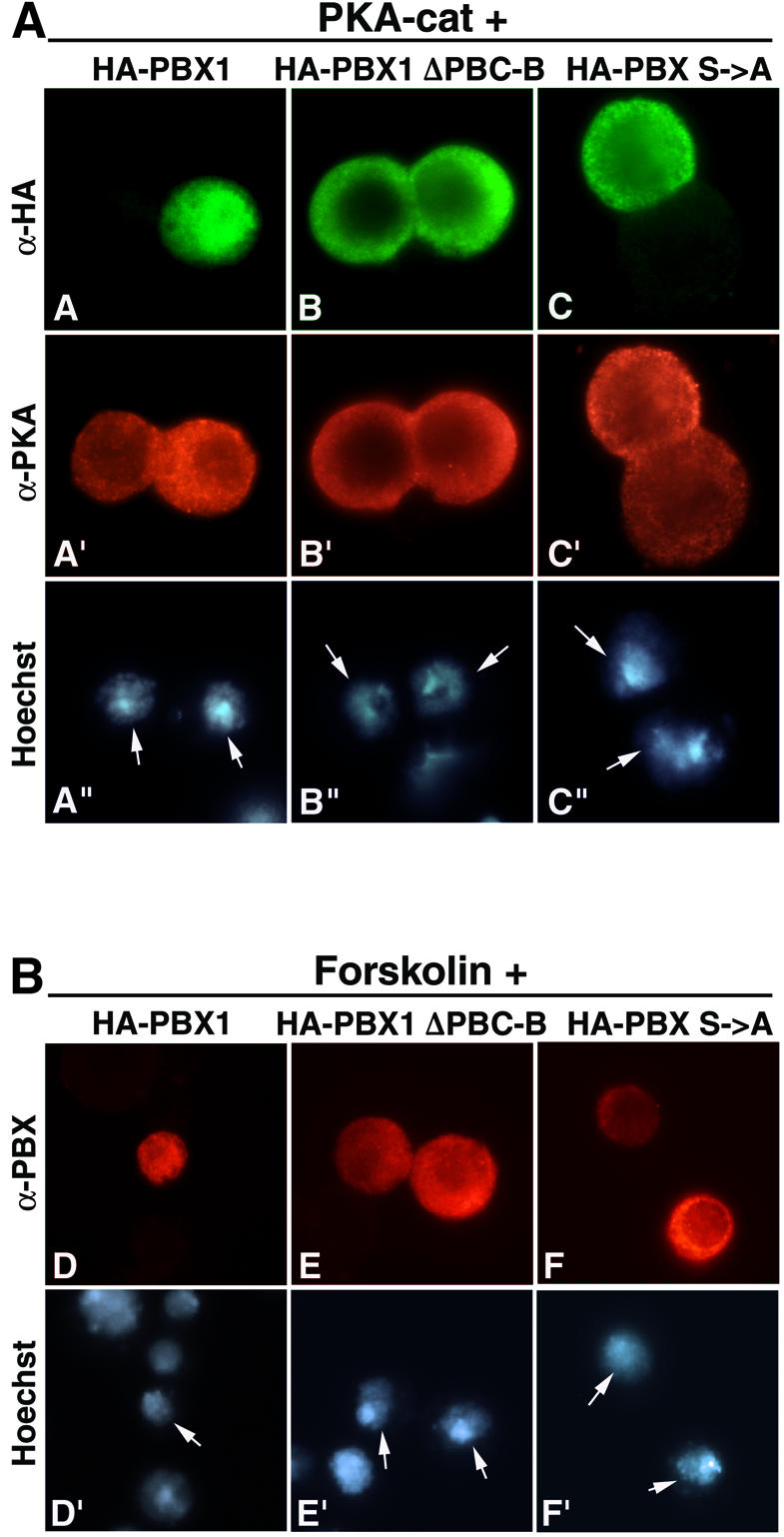
Fig. 7. PKA activity induces PBX1 nuclear translocation in SL2 cells. (A) PBX1 is relocalized to the nucleus in SL2 cells co-expressing PKAcat (panels A, A′ and A”). PKAcat does not trigger nuclear translocation of PBXΔPBC-B (panels B, B′ and B”) and PBXS→A (panels C, C′ and C”). (B) Activation of endogenous PKA in SL2 cells causes nuclear accumulation of PBX1. Nuclear accumulation of PBX1 (panels D and D′) was observed after treatment for 2 h with 60 µM forskolin, whereas PBXΔPBC-B (panels E and E′) and PBXS→A (panels F and F′) were not relocalized to the nucleus. PBX1 in forskolin-treated cells was detected with a polyclonal anti-PBX antibody.
PKA activation induces nuclear localization of PBX1 in developing chicken limbs
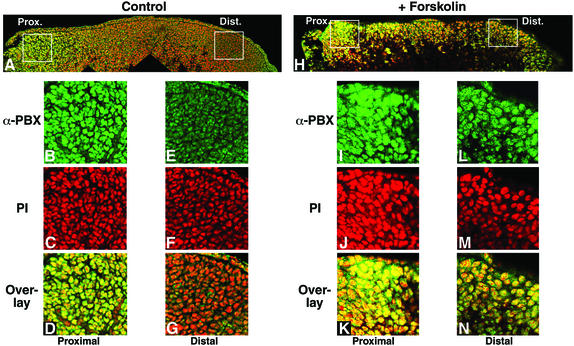
We next wanted to verify whether, in vertebrate distal limb mesenchymal cells in vivo, cytoplasmic PBX1 correlates with its dephosphorylated state, and whether stimulation of endogenous PKA activity would induce nuclear relocalization of endogenous PBX1. We dissected chicken limb buds at stage 20–21 (Hamburger and Hamilton, 1992), cultured them for 2 or 4 h, as described previously (Cusella-De Angelis et al., 1994), in the presence or absence of 60 µM forskolin, and stained them using anti-PBX antibodies. In control limbs, PBX is nuclear in mesenchymal cells only proximally in the limb bud (Figure 8A, panel A). In contrast, in limb buds treated for 4 h with forskolin, mesenchymal cells displayed PBX nuclear localization along the entire proximodistal axis (Figure 8B, panel H). The same degree of PBX relocalization was observed after only 2 h of treatment (data not shown). To exclude the possibility that relocalization of PBX distally was due to ectopic activation of MEIS proteins, we stained limb buds, treated for 2 or 4 h with 60 µM forskolin, with anti-MEIS1 antibodies. No change in MEIS1 protein distribution was observed, which was nuclear and restricted to proximal mesenchymal cells in both untreated and forskolin-treated limb buds (data not shown). This finding confirms that PBX cytoplasmic localization in chicken distal limb mesenchymal cells can also be reversed by inducing PKA activity, and that in vivo PBX nuclear export correlates with its dephosphorylated state.
Fig. 8. PKA activation induces nuclear accumulation of endogenous PBX in developing chicken limbs. (A) Low magnification confocal image of a control limb bud stained for PBX (green). The limb bud was sectioned along its proximodistal axis and perpendicular to its anteroposterior axis (proximal is left and distal right). Two white squares indicate the regions shown magnified in (B)–(D) and (E)–(G). PBX is nuclear only in the proximal part of the bud (B) and cytoplasmic in distal regions (E). Nuclei are stained with propidium iodide (PI) (C and F). Co-localization of nuclear and PBX staining appears in yellow (D and G). (H) Confocal image of a limb bud cultured in the presence of 60 µM forskolin for 4 h, sectioned and immunostained as above. White squares indicate the regions shown magnified in (I)–(K) and (L)–(N). Upon forskolin treatment PBX is located in the cell nuclei proximally (I and K), as well as distally (L and N) of the limb bud. Co-localization of nuclear and PBX staining appears in yellow (K and N).
Discussion
The control of PBC protein function through differential nucleocytoplasmic distribution is a crucial evolutionarily conserved mechanism for the proximodistal patterning of appendages (reviewed in Vogt and Duboule, 1999). In prospective distal territories of both vertebrate and Drosophila limbs, PBX1 and EXD are functionally inactivated by exclusion from the cell nucleus. If PBX or EXD are artificially driven into the nucleus by overexpression or by distal misexpression of MEINOX proteins, distal limb development is impaired in both vertebrates and flies (Abu-Shaar and Mann, 1998; Casares and Mann, 1998; Capdevilla et al., 1999; Mercader et al., 1999). It has been reported previously by us and other workers that PBX1 and EXD subcellular localization is controlled by competing nuclear export and import signals (Abu-Shaar et al., 1999; Berthelsen et al., 1999). However, it still remained unclear whether interaction with MEINOX proteins is the sole mechanism by which these opposing signals are modulated or whether there are additional controls of PBC protein nuclear export. Taking advantage of the differential subcellular localization of PBX1 in various cell lines, which we used as models for studying PBX1 nucleocytoplasmic transport, we found that in the absence of MEINOX proteins PBX1 nuclear export is not active by default, but is regulated by post-translational modification of the protein.
Two independent leucine-rich nuclear export signals mediate nuclear export of PBX1
Nuclear export of PBX1 requires sequences containing a putative NES located within the PBC-A domain, a conserved region that is required for heterodimerization with MEINOX proteins (Berthelsen et al., 1999). In this work, we used fusions with GFP and deletion analysis to characterize this signal and to verify whether other conserved domains of PBX1 contain additional signals mediating export. We found that a region spanning amino acids 45–90 within PBC-A is sufficient to drive nuclear export of GFP. Deletion analysis showed that this region contains two separable signals, one spanning amino acids 45–52 and the other spanning amino acids 75–82, each capable of driving LMB-sensitive export of PBX1. As expected for NESs recognized by the CRM1 nuclear export receptor, both signals are composed of leucine and isoleucine hydrophobic amino acids, with a spacing similar to that of the consensus sequence for Rev-like leu-rich NESs (Mattaj and Englmeier, 1998). Moreover, like typical Rev-like NESs, amino acids 45–90 mediate direct LMB-sensitive interactions with CRM1. The two Leu/Ile-rich signals found within PBC-A do not constitute a bipartite NES but represent two independently working NESs, since a deletion of either of the two in the context of PBX1 did not impair nuclear export. Thus PBX1 represents one of the few reported cases of multiple independent leu-rich NESs within a protein.
The NES of the PBX1-orthologue EXD has been mapped within a region spanning amino acids 178–220 of the PBC-B domain (Abu-Shaar et al., 1999). However, we did not observe nuclear export of a GFP–PBC-B fusion or nuclear relocalization of a PBC-B deletion mutant. Moreover, while an LMB-sensitive export was demonstrated for EXD, no sequence resembling a canonical Rev-like NES was identified within PBC-B (Abu-Shaar et al., 1999), whereas the two Rev-like NESs, which we map within the PBC-A domain of PBX1, are perfectly con served in EXD. At present, we have no explanation for the discrepancy between our results and those obtained with EXD other than the possible existence of non-obvious structural differences between the two orthologues.
The two NESs we found are located within one of the domains required for heterodimerization with MEINOX proteins. Interestingly, deletion of either of the two NESs impairs interaction with MEINOX proteins (this work; C.K.Nielsen and V.Zappavigna, unpublished data), indi cating that the contact surfaces with CRM1 and with MEINOX proteins actually overlap, and supporting a model by which heterodimerization with MEINOX proteins favours nuclear import of PBC proteins through masking of the NESs.
Phosphorylation of the PBC-B domain controls the balance between nuclear export and import of PBX1
In the absence of co-expressed MEINOX proteins, PBX1 nuclear export is not active by default. The exogenous expression of PBX1 in several cell lines including NIH 3T3, COS7, HeLa and P19 (this work; C.K.Nielsen and V.Zappavigna, unpublished data) shows that the protein is completely imported into their nuclei. This nuclear localization is not due to endogenous MEINOX proteins because the PBXΔ73–90 mutant, which cannot heterodimerize with MEINOX proteins, is still nuclear in these cells. Rather, the differential subcellular localization of PBX1 according to cell background, i.e. nuclear in NIH 3T3 and other cell lines, and cytoplasmic in SL2 cells, is due to a cell context-specific modulation of its two competing nuclear export and import signals. We found that the PBC-B domain plays a crucial role in this process. An internal deletion of PBC-B causes PBX1 to be exported from the nuclei of NIH 3T3 and COS7 cells, showing that PBC-B suppresses export of PBX1 in these cells.
We identified several potential phosphorylation sites for Ser/Thr kinases within PBC-B, which are conserved among other members of the PBC protein family. Four serines (187, 193, 202 and 209), which were predicted with high confidence to be bona fide phosphorylation sites, were substituted with alanines and the resulting mutant (PBXS→A) was found to be exported in NIH 3T3 cells in an LMB-sensitive manner. Using 2D IEF/SDS–PAGE, we found that the PBX1 phosphorylation state correlates with its cell background-specific subcellular localization. In COS7 cells, PBX1 is phosphorylated and nuclear, whereas in SL2 cells PBX1 is not phosphorylated and cytoplasmic. This differential phosphorylation involves serines 187, 193, 202 and 209 since the PBXS→A mutant displayed no cell context-dependent difference in phosphorylation.
Subcellular localization of a variety of proteins was shown to be regulated by phosphorylation, for example cyclin B1 (reviewed in Mattaj and Englmeier, 1998). CRM1 interaction with cyclin B1 is negatively regulated by phosphorylation of a specific serine within the NES, probably causing steric hindrance of CRM1 binding. In the case of PBX1, it is unlikely that phosphorylation directly influences binding of PBC-A to CRM1, since a GFP fusion containing both PBC-A and PBC-B is exported in SL2, as well as in NIH 3T3 cells where PBC-B is phosphorylated. Phosphorylation could instead induce a conformational change of PBX1, favouring nuclear import. Further experiments will be required to clarify this point. The phosphorylation state of PBX1 does not affect its heterodimerization capacity with MEINOX proteins, because the PBXS→A mutant could still interact with PREP1 both in vitro and in cultured cells. Therefore PBX1 phosphorylation can be considered as another block to nuclear export, in addition to heterodimerization with MEINOX proteins; the two mechanisms are not mutually exclusive. Accordingly, the absence of MEINOX proteins may be regarded as a permissive step for nuclear export of PBC proteins, which requires in addition dephosphorylation of PBC-B. This dual post-translational control of PBC protein function is analogous to the regulation of the Prospero homeodomain protein subcellular localization, which was shown to be regulated by the cell context-dependent activity of competing nuclear export and import signals, whose function is modulated by an NES-masking region containing several potential phosphorylation sites (Demidenko et al., 2001).
PKA activity controls the subcellular localization of PBX1 both in cell culture and in vivo
Serines 187, 193, 202, and 209 within PBC-B are potential phosphorylation sites of several Ser/Thr kinases, including PKC, CK2 and cAMP-dependent PKA. However, we found that among these only PKA was capable of efficiently phosphorylating PBC-B in vitro. Therefore we investigated whether PKA activity could play a role in controlling PBX1 nucleocytoplasmic localization. We found that an increase in PKA activity blocks PBX1 nuclear export in cultured cells. Co-expression of the PKA catalytic subunit with PBX1, or stimulation of endogenous PKA activity, in SL2 cells induces nuclear relocalization of PBX1, which requires serines 187, 193, 202 and 209. Most interestingly, stimulation of endogenous PKA activity leads to nuclear relocalization of PBX1 in distal limb mesenchymal cells also, indicating that cytoplasmic PBX1 in these cells is in a dephosphorylated state. Therefore antagonization of PKA basal activity in distal mesenchymal cells may be the mechanism by which cytoplasmic localization of PBC proteins is triggered during limb development.
PKA plays a crucial role in mediating responses to a variety of extracellular signals (reviewed in Taylor et al., 1990). Several studies identified PKA as a negative regulator in the hedgehog/Sonic hedgehog (Hh/Shh) signalling pathway in both Drosophila and vertebrates (reviewed in Ingham and McMahon, 2001). In Drosophila, a PKA loss-of-function mutation causes ectopic activation of Hh target genes, mimicking ectopic Hh signalling. PKA inhibits the Hh pathway by phosphorylating Cubitus-Interruptus (Ci), the transcriptional mediator of Hh signalling. In vertebrates, PKA promotes proteolytic cleavage of Gli3, a Ci orthologue, and negatively regulates transcriptional activation of Gli1, indicating that this antagonistic relationship is evolutionarily conserved. The mechanism by which Hh/Shh antagonizes PKA activity is largely unknown. While the control of PKA activity through modulation of cAMP levels has been excluded, some evidence was obtained that Hh signalling may activate a specific phosphatase (reviewed in Ingham and McMahon, 2001).
The Hh/Shh pathway is required for the distal outgrowth of appendages in both vertebrates and Drosophila. In mice, targeted inactivation of Shh causes truncation of distal limb elements, sparing proximal structures (Chiang et al., 1996). Hh-deficient flies also display truncation of all distal leg structures, whereas proximal structures are unaffected (Diaz-Benjumea et al., 1994). exd/hth function was shown to inhibit Hh target genes proximally, and the inactivation of Exd distally, by exclusion from the nucleus, allows the Hh function to be fully effective (Gonzales-Crespo et al., 1998). This antagonism, required for limb proximodistal axis regionalization, appears to be conserved in vertebrates because misexpression of Meis2 in distal chicken limb regions leads to a strong repres sion of genes involved in the Shh/FGF regulatory loop, and ectopic application of BMP2, a component of this regulatory loop, represses proximal Meis2 expression (Capdevilla et al., 1999).
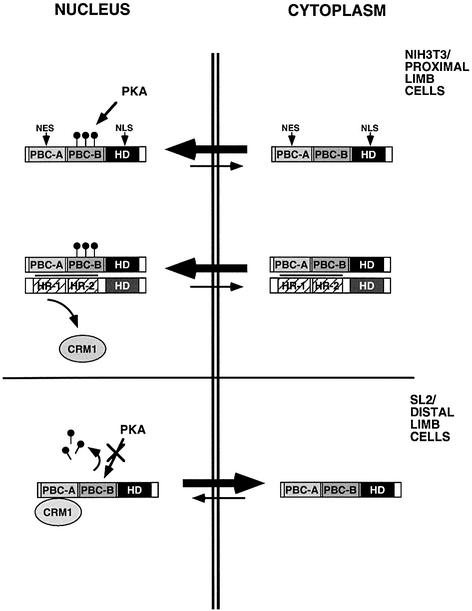
Hh/Shh is believed to prevent PBC protein nuclear localization distally by repressing, through the action of the wg/Wnt and dpp/Bmp pathways, distal expression of the MEINOX genes (reviewed in Vogt and Duboule, 1999). Our results suggest that the Hh/Shh pathway might play an additional role to counteract PBC/ MEINOX proximalizing functions. In addition to restricting MEINOX gene expression proximally, thereby preventing PBC/MEINOX heterodimerization, it would trigger PBC protein nuclear export by counteracting their PKA-mediated phosphorylation. Based on our results, we propose a model (Figure 9) for the control of PBC protein subcellular localization. In cells that co-express MEINOX proteins, PBC proteins heterodimerize with these and PBC–MEINOX complexes are found in the nucleus. In cells that do not express MEINOX proteins, PBC proteins are not exported unless the PBC-B domain is in a dephosphorylated state. PKA basal activity is probably sufficient to phosphorylate PBC proteins in most cell types, thus favouring, independently of heterodimerization with MEINOX proteins, their nuclear import. Conversely, PKA phosphorylation of PBC-B is either blocked or is counteracted by a phosphatase in cells in which nuclear export of PBC proteins is active.
Fig. 9. A model for the regulation of PBC-B protein subcellular distribution (see text for a detailed description). Dark rectangles represent the homeodomain (HD). Grey and striped rectangles represent conserved N-terminal regions (PBC-A, PBC-B, HR-1 and HR-2) within PBC and MEINOX proteins respectively. CRM1, nuclear export receptor; NES nuclear export signal; NLS, nuclear import signal. Lollipops indicate phosphorylated serine residues.
Materials and methods
Expression constructs
Mammalian expression vectors for PBX1, HA-PBX1 and PREP1 have been described previously (Berthelsen et al., 1998, 1999). The PBX1ΔPBC-B and PBX S→A mutants, with deleted amino acids 142–232 of PBC-B and alanine substitutions of serines 187, 193, 202 and 209, respectively, were generated by PCR mutagenesis, cloned in-frame with a hemagglutinin (HA) tag into the pSG5 expression vector (Stratagene). The p2XGFP vector containing two GFP molecules was generated by cloning a BamHI–XbaI fragment containing GFP from pEGFP-N2 into pEGFP-C2 (Clontech). GFP fusions containing different domains of PBX1 were generated by cloning the corresponding PCR-amplified sequences into an EcoRI site of p2XGFP. 2XGFP–PBC-A contains amino acids (aa) 1–127 of PBX1; 2XGFP–PBC-B contains aa 141–231; 2XGFP–HD contains aa 231–295; 2XGFP–PBC-A45–72 contains aa 45–72; 2XGFP–PBC-A73–90 contains aa 73–90; 2XGFP– PBC-A45–90 contains aa 45–90; 2XGFP–PBC-A+HD contains aa 1–127+231–295; 2XGFP–PBC-A+B contains aa 1–231; 2XGFP–PBC- B+HD contains aa 141–295; 2XGFP–PBC-A+B+HD contains aa 1–295. Protein-A-tagged PBX1 was cloned into pSG5. GFP fusions were expressed in SL2 cells from the pPAC5C expression vector. All PBX1 mutant derivatives were expressed in SL2 cells using the pRmHa3 Cu(SO4)2 inducible expression vector. GST expression constructs containing the indicated PBX1 domains were generated by cloning PCR-amplified sequences into the pGEX-4T-1 vector (Pharmacia). All constructs generated by PCR were verified by sequencing.
Transfections, LMB and forskolin treatments
NIH 3T3 mouse fibroblast cells and COS7 monkey kidney cells were grown in Dulbecco’s modified Eagle′s medium (DMEM) supplemented with 10% newborn calf serum and antibiotics. Drosophila Schneider cells (SL2) were grown in Schneider’s Drosophila medium (GIBCO) supplemented with 10% fetal calf serum and antibiotics. Cells were transfected as described previously (Berthelsen et al., 1999). Inducible expression from the pRmHa3-based expression vectors in SL2 cells was obtained by treating cells with 0.7 mM Cu(SO4)2 24 h post-transfection. LMB treatment of Drosophila SL2 cells was performed with 100 nM LMB for 6 h prior to fixation. Forskolin treatment was performed with 60 µM forskolin (Sigma) for 1–2 h prior to fixation.
Antibodies and immunocytochemistry
The primary antibodies used for immunohistochemistry were rabbit anti-PBX (C-20, Santa Cruz Biotechnology, CA), rabbit anti-PREP1 (N-15, Santa Cruz), rabbit anti-PKA (C-20, Santa Cruz) and mouse monoclonal anti-HA (F-7, Santa Cruz). They were used according to the manufacturer. FITC- or TRITC-conjugated anti-rabbit and anti-mouse secondary antibodies (Jackson ImmunoResearch) were used according to the manufacturer. Nuclei were visualized by Hoechst 33258 (Sigma) stain ing. Immunocytochemistry was performed as described previously (Berthelsen et al., 1999). The subcellular localization of GFP fusion proteins was analysed in living cells by fluorescent microscopy on an Olympus IX50 microscope; subsequently cells were fixed and mounted for examination with a Zeiss Axiophot microscope.
Bacterial expression and GST pull-down experiments
His-tagged PBX1 was purified from BL21(DE3) cells using a Ni-NTA column (Qiagen), and GST fusions were purified from DH5α cells using glutathione–Sepharose 4B (Amersham), both according to the manufacturer. Thirty picomoles of GST fusion proteins bound to the resin were mixed with 10 µl in vitro translated 35S-labelled hCRM, and incubated as described previously (Askjaer et al., 1999). LMB was added to a final concentration of 400 nM. Washes were performed as described previously (Askjaer et al., 1999) with the addition of 0.1% NP40. Beads were resuspended in sample buffer and loaded on an SDS–PAGE gel.
In vitro phosphorylation and two-dimensional isoelectric focusing/SDS–PAGE
One to three micrograms of purified bacterially expressed proteins were incubated in 20 µl of 50 mM Tris–HCl (pH 7.5), 5 m M MgCl2, 5 µM Mg–ATP and 5 µCi γ-[32P]ATP for 5 min at 30°C; 2.5 U of purified PKA (Roche) were added, and the reaction was incubated for 30 min at 30°C and stopped by denaturation in loading buffer. Proteins were resolved by SDS–PAGE, stained with Coomassie Blue and detected by autoradiography. Total extracts for 2D IEF were prepared as described previously (Berthelsen et al., 1998). Protein-A-tagged PBX was purified on IgG–Sepharose 6 Fast Flow resin (Pharmacia) according to the manufacturer. 2D IEF/SDS–PAGE was performed using 7 cm Immobiline DryStrips (Pharmacia) with a non-linear 3–10 pH range on a Multiphor apparatus according to the manufacturer. Protein-A-tagged PBX was detected by western blotting using the polyclonal anti-PBX antibody (C-20, Santa Cruz).
Chicken limb bud culture
Limbs were excised from the flank of stage 20–21 chick embryos and cultured for 2–4 h in DMEM containing 10% FCS, 2% chicken serum and antibiotics in the absence or presence of 60 µM forskolin. Limb buds were fixed in 4% paraformaldehyde for 2 h at 4°C and included in OCT resin. Cryostat sections were cut parallel to the proximodistal and dorsoventral axes, and perpendicular to the anteroposterior axis. Endogenous PBX was detected, after antigen exposure by boiling for 5 min in 10 mM citric acid pH 6, with polyclonal anti-PBX (C-20, Santa Cruz, CA) and Alexa™ 488 (Molecular Probes, The Netherlands) secondary antibody. Nuclei were stained with propidium iodide (Sigma). The sections were analysed and images generated with a Bio-Rad MRC1024 laser scan confocal microscope.
Acknowledgments
Acknowledgements
We thank Sarah M.Smolik for providing the pPac5C–PKAcat expression construct, M.Fornerod for providing the hCRM1 expression vector and Barbara Wolff (Novartis) for the kind gift of LMB. We also thank Fulvio Mavilio, Francesco Blasi and Arturo Verrotti for comments on the manuscript. Confocal imaging experiments were performed within Alembic (Advanced Light and Electron Microscopy Bio-Imaging Center) at the San Raffaele Scientific Institute. This work was supported by grants from the Italian Association for Cancer Research (AIRC) to V.Z., from the Italian Telethon to V.Z., from the Danish Research Academy to C.K.N. and from the MIUR Center of Excellence in Physiopathology of Cell Differentiation Program to M.A.
References
- Abu-Shaar M. and Mann,R.S. (1998) Generation of multiple antagonistic domains along the proximodistal axis during Drosophila development. Development, 125, 3821–3830. [DOI] [PubMed] [Google Scholar]
- Abu-Shaar M., Ryoo,H.D. and Mann,R.S. (1999) Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev., 13, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affolter M., Marty,T. and Vigano,M.A. (1999) Balancing import and export in development. Genes Dev., 13, 913–915. [DOI] [PubMed] [Google Scholar]
- Askjaer P. et al. (1999) RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol., 19, 6276–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J., Zappavigna,V., Mavilio,F. and Blasi,F. (1998) Prep1, a novel functional partner of Pbx proteins. EMBO J., 17, 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J., Kilstrup-Nielsen,C., Blasi,F., Mavilio,F. and Zappavigna, V. (1999) The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev., 13, 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Gammeltoft,S. and Brunak,S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol., 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- Bürglin T.R. (1997) Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved betwen plants and animals. Nucleic Acids Res., 25, 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevilla J., Tsukui,T., Rodrigues Esteban,C., Zappavigna,V. and Izpisua Belmonte,J.C. (1999) Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol. Cell, 4, 839–849. [DOI] [PubMed] [Google Scholar]
- Casares F. and Mann,R.S. (1998) Control of antennal versus leg development in Drosophila. Nature, 392, 723–726. [DOI] [PubMed] [Google Scholar]
- Chang C.-P., Jacobs,Y., Nakamura,T., Jenkins,N.A., Copeland,N.G. and Cleary,M.L. (1997) Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol. Cell. Biol., 17, 5679–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C., Litingtung,Y., Lee,E., Young,K.E., Corden,J.L., Westphal,H. and Beachy,P.A. (1996) Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature, 383, 407–413. [DOI] [PubMed] [Google Scholar]
- Cusella-De Angelis M.G., Molinari,S., Le Donne,A., Coletta,M., Vivarelli,E., Bouche,M., Molinaro,M., Ferrari,S. and Cossu,G. (1994) Differential response of embryonic and fetal myoblasts to TGF β: a possible regulatory mechanism of skeletal muscle histo genesis. Development, 120, 925–933. [DOI] [PubMed] [Google Scholar]
- Demidenko Z., Badenhorst,P., Jones,T., Bi,X. and Mortin,M.A. (2001) Regulated nuclear export of the homeodomain transcription factor Prospero. Development, 128, 1359–1367. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea F.J., Cohen,B. and Cohen,S.M. (1994) Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs. Nature, 372, 175–179. [DOI] [PubMed] [Google Scholar]
- Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fognani C., Kilstrup-Nielsen,C., Berthelsen,J., Ferretti,E., Zappavigna,V. and Blasi,F. (2002) Characterization of PREP2, a paralog of PREP1, which defines a novel sub-family of the MEINOX TALE homeodomain transcription factors. Nucleic Acids Res., 30, 2043–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Crespo S. and Morata,G. (1996) Genetic evidence for the subdivision of the arthropod limb into coxopodite and telopodite. Development, 122, 3921–3928. [DOI] [PubMed] [Google Scholar]
- Gonzales-Crespo S., Abu-Shaar,M., Torres,M., Martinez-A.,C., Mann,R.S. and Morata,G. (1998) Antagonism between extradenticle function and Hedgehog signalling in the developing limb. Nature, 394, 196–200. [DOI] [PubMed] [Google Scholar]
- Hamburger V. and Hamilton,H.L. (1992) A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn., 195, 231–272. [DOI] [PubMed] [Google Scholar]
- Ingham P.W. and McMahon,A.P. (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev., 15, 3059–3087. [DOI] [PubMed] [Google Scholar]
- Mann R.S. and Affolter,M. (1998) Hox proteins meet more partners. Curr. Opin. Genet. Dev., 8, 423–429. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Mercader N., Leonardo,E., Azpiazu,N., Serrano,A., Morata,G., Martinez-A,C. and Torres,M. (1999) Conserved regulation of proximodistal limb axis development by Meis1/HTH. Nature, 402, 425–429. [DOI] [PubMed] [Google Scholar]
- Morata G. (2001) How Drosophila appendages develop. Nat. Rev. Mol. Cell Biol., 2, 89–97. [DOI] [PubMed] [Google Scholar]
- Pöpperl H., Rikhof,H., Chang,H., Haffter,P., Kimmel,C.B. and Moens,C.B. (2000) Iazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol. Cell, 6, 255–267. [DOI] [PubMed] [Google Scholar]
- Rieckhof G.E., Casares,F., Ryoo,H.D., Abu-Shaar,M. and Mann,R.S. (1997) Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell, 91, 171–183. [DOI] [PubMed] [Google Scholar]
- Selleri L., Depew,M.J., Jacobs,Y., Chanda,S.K., Tsang,K.Y., Cheah, K.S., Rubenstein,J.L., O’Gorman,S. and Cleary,M.L. (2001) Require ment for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development, 128, 3543–3557. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Buechler,J.A. and Yonemoto,W. (1990) cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem., 59, 971–1005. [DOI] [PubMed] [Google Scholar]
- Vogt T.F. and Duboule,D. (1999) Antagonists go out on a limb. Cell, 99, 563–566. [DOI] [PubMed] [Google Scholar]
- Wagner K., Mincheva,A., Korn,B., Lichter,P. and Pöpperl,H. (2001) Pbx4, a new Pbx family member on mouse chromosome 8, is expressed during spermatogenesis. Mech. Dev., 103, 127–131. [DOI] [PubMed] [Google Scholar]
- Wolff B., Sanglier,J.J. and Wang,Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol., 4, 139–147. [DOI] [PubMed] [Google Scholar]