Abstract
The shoot apical meristem (SAM), a small group of undifferentiated dividing cells, is responsible for the continuous growth of plants. Several genes have been identified that control the development and maintenance of the SAM. Among these, WUSCHEL (WUS) from Arabidopsis (Arabidopsis thaliana) is thought to be required for maintenance of a stem cell pool in the SAM. The MADS-box gene AGAMOUS, in combination with an unknown factor, has been proposed as a possible negative regulator of WUS, leading to the termination of meristematic activity within the floral meristem. Transgenic petunia (Petunia hybrida) plants were produced in which the E-type and D-type MADS-box genes FLORAL BINDING PROTEIN2 (FBP2) and FBP11, respectively, are simultaneously overexpressed. These plants show an early arrest in development at the cotyledon stage. Molecular analysis of these transgenic plants revealed a possible combined action of FBP2 and FBP11 in repressing the petunia WUS homolog, TERMINATOR. Furthermore, the ectopic up-regulation of the C-type and D-type homeotic genes FBP6 and FBP7, respectively, suggests that they may also participate in a complex, which causes the determinacy in transgenic plants. These data support the model that a transcription factor complex consisting of C-, D-, and E-type MADS-box proteins controls the stem cell population in the floral meristem.
Vascular plants continue to grow throughout their life cycle, a condition that does not hold for animals. This indeterminate growth is ensured by two regions of pluripotent, meristematic cells, which are located since early embryogenesis at both ends of the plant body. The root apical meristem gives rise to the underground root system, whereas all the above-ground structures are initiated by the shoot apical meristem (SAM), which originates an orderly sequence of leaves, nodes, and internodes. Upon floral induction, the SAM turns into an inflorescence meristem and/or into a floral meristem (FM), which will eventually cease its meristematic activity to allow the differentiation of a pistil, the formation of gametes, and the completion of the life cycle.
Mutant and genetic analyses in Arabidopsis (Arabidopsis thaliana) have revealed some of the regulatory circuits that perpetuate the delicate balance between stem cell accumulation and organ initiation (for review, see Sharma et al., 2003; Williams and Fletcher, 2005). Meristem activity is promoted by the homeodomain proteins SHOOTMERISTEMLESS (STM) and WUSCHEL (WUS), whose loss-of-function mutants fail to maintain a population of stem cells. In these mutants, the SAM is terminated at the end of embryogenesis and growth stops at the cotyledon stage (Barton and Poethig, 1993; Laux et al., 1996; Long et al., 1996; Mayer et al., 1998). Independent and complementary roles have been assigned to STM and WUS in SAM regulation: A WUS-dependent signal from the organizing center underlying the L1 to L3 layers of the central zone (CZ) specifies the stem cell population in the CZ, whereas STM maintains cell proliferation in the peripheral zone (Long and Barton, 1998; Mayer et al., 1998; Lenhard et al., 2002). The balance between stem cell maintenance and organogenesis in the SAM is tightly controlled by a regulatory circuit that involves CLAVATA (CLV) and WUS proteins. WUS induces stem cell identity in the CZ and subsequent expression of the CLV complex, which in turn activates a signaling pathway that feeds back from the stem cells to repress the transcription of WUS in the center of the meristem (Brand et al., 2000; Schoof et al., 2000). While organogenesis in the shoots can go on indefinitely, only a limited number of floral organs can be formed before the FM ceases to perpetuate itself. Because of its central role in promoting stem cell identity, WUS is likely to be the target of pathways that lead to the termination of the FM. It has recently been proposed that a negative feedback loop involving WUS, the FM identity gene LEAFY (LFY), and the floral organ identity gene AGAMOUS (AG) takes place in the FM and is responsible for its suppression (Lenhard et al., 2001; Lohmann et al., 2001). The combined action of LFY and WUS would activate AG in the FM, which in turn represses WUS in the determinate flower. Furthermore, evidence of AG being a repressor of WUS is based on the opposite phenotypes of wus and ag mutants and on the expression patterns of the respective genes in wild-type and mutant plants. The sporadic flowers that are formed in wus plants lack carpels and most stamens, whereas ag flowers are indeterminate with an indefinite number of perianth organs; furthermore, WUS expression declines after stage 6 of wild-type flowers, but persists in ag flowers. There are, however, some questions that remain to be resolved. First, AG alone is not sufficient to repress WUS because ectopic expression of AG causes termination of the inflorescence meristem, which forms a single terminal flower, but not of the shoot meristem. Second, expression of WUS persists until stage 6 of flower development, while AG mRNA can be detected from stage 3 onward. The presence of another unknown factor, which acts together with AG in the flower to repress WUS, has been proposed as a possible explanation. Finally, it remains to be uncovered how conserved these mechanisms that regulate meristem self maintenance and self termination are among different species.
Here we report about the relation between petunia (Petunia hybrida) MADS-box proteins belonging to the C, D, and E class and the WUS homolog TERMINATOR (TER; Stuurman et al., 2002) and propose a model of how FM determinacy is controlled in petunia.
RESULTS
Expression Domains of C-, D-, and E-Function Genes and TER in the Petunia Flower
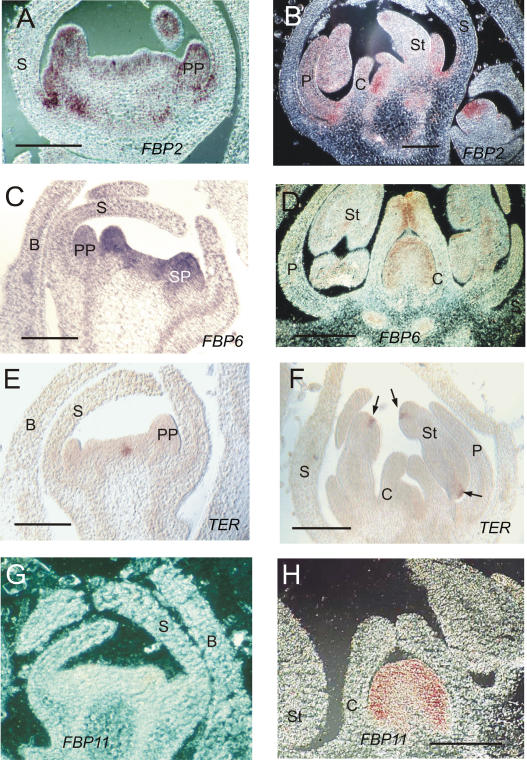
Similar to Arabidopsis, petunia plants with impaired C or E functions show loss of determinacy within the flower, indicating a possible involvement of the corresponding genes in FM determination (Angenent et al., 1994, 2005; Kapoor et al., 2002; Ferrario et al., 2003). The E function in petunia is accomplished by at least two genes, FLORAL BINDING PROTEIN2 (FBP2) and FBP5, as suggested by the floral phenotype of cosuppression plants. When these two genes are simultaneously down-regulated, homeotic conversion of the three inner whorls and loss of determinacy occur within the flower (Angenent et al., 1994; Ferrario et al., 2003). The milder phenotype of the FBP2/FBP5 double knockout mutant, however, indicates the presence of at least one more gene partially redundant in the E function (Vandenbussche et al., 2003). In situ hybridization experiments revealed that FBP2 and FBP5 share a similar expression pattern from the FM stage until complete maturity of the floral bud, where their expression is restricted to the three inner whorls (Ferrario et al., 2003; Fig. 1, A and B).
Figure 1.
In situ localization of petunia gene expression in wild-type flower buds. A and B, FBP2 expression in young (A) and older (B) flower buds. C and D, FBP6 expression in flower buds at the same stage as in A and B. E and F, TER expression is detected in a limited number of cells in the meristematic tissue of young flower buds (E). At a later stage, TER expression appears in the stomium area of the stamens (arrows in F). G and H, FBP11 is only expressed in flower buds at a later stage in the placenta tissue inside the developing pistil; C, E, and F were viewed using bright-field microscopy; others were viewed using dark-field microscopy. B, Bract; C, carpel; P, petal; PP, petal primordium; S, sepal; SP, sepal primordium; St, stamen. Bars = 500 μm.
The fate of reproductive organs is controlled by class C genes belonging to the AG clade, which have been duplicated during evolution in several plant species. In petunia, two genes share high sequence similarity with the Arabidopsis AG gene, pMADS3 and FBP6 (Tsuchimoto et al., 1993; Kater et al., 1998; Kapoor et al., 2002). Similar to AG, FBP6 and pMADS3 transcripts are present in stamens and carpels from the appearance of their primordia until complete maturity of these organs (Fig. 1, C and D; Kapoor et al., 2002). The phenotype of pMADS3 cosuppression plants, where homeotic conversion and loss of determinacy is observed in the third whorl only, support the idea that this gene may act redundantly with FBP6 in the determination of the fourth whorl (Kapoor et al., 2002).
TER is the petunia ortholog of WUS, a key player in meristem determination in Arabidopsis (Stuurman et al., 2002). We compared the expression domains of TER with those of C- and E-class genes in wild-type petunia plants to obtain a preliminary indication about the functional relationships of these factors in the termination of the FM. TER expression, like its Arabidopsis ortholog, is limited to a few cells in the CZ of the SAM, as has been shown by in situ hybridization on shoot apexes of wild-type seedlings (Stuurman et al., 2002; Fig. 2I). In the flower, TER activity is still present in a limited number of cells in the central region at a stage when petal and stamen primordia are emerging (Fig. 1E), but it disappears at a stage during gynoecium development, just prior to the fusion of the carpels (Fig. 1F). At that stage, TER expression remains in an area of the stamen that becomes the stomium.
Figure 2.
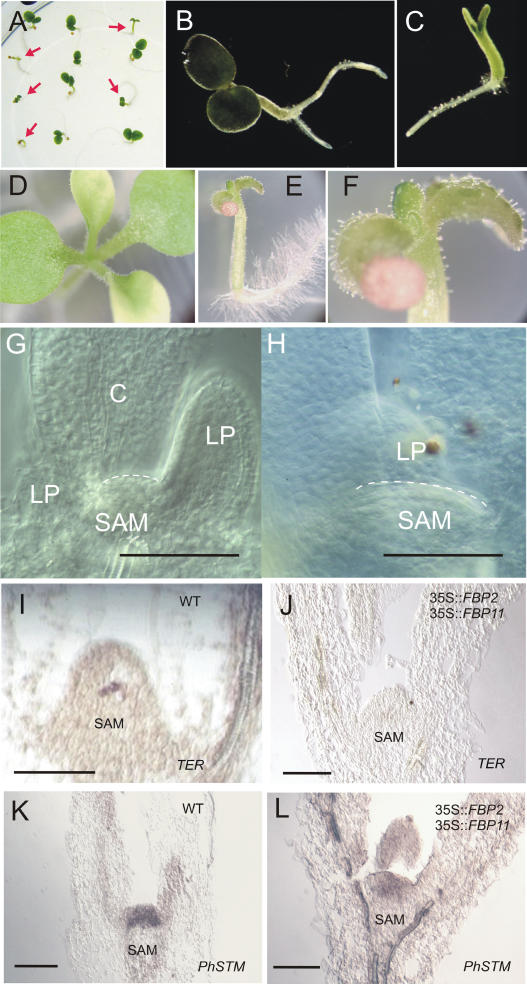
Wild-type and 35S∷FBP2/FBP11 double overexpression seedlings. A, Progeny of a cross between 35S∷FBP2 and 35S∷FBP11 transgenic plants 6 d after germination; the double overexpression seedlings are indicated by the arrows. B and D, Wild-type seedlings 6 and 13 d after germination, respectively. C, E, and F, Double overexpression seedlings 6 and 13 d after germination. D and E were taken with the same magnification, whereas F is a magnification of E. Cleared SAMs of 35S∷FBP2/FBP11 (G) and wild-type (H) seedlings; images were taken with the same magnification. The dome of the SAM is indicated with a broken line. I and K, In situ hybridization of wild-type seedlings using TER and PhSTM as a probe, respectively; the same probes were used to hybridize 35S∷FBP2/FBP11 double overexpression seedlings in J and L, respectively. LP, Leaf primordium; C, cotyledon. Bars = 200 μm.
According to the above-mentioned expression data, C and E gene activities overlap in the center of the flower, in the region where TER is also active. Hence, similar to Arabidopsis, the timing of C and E gene induction does not coincide with TER down-regulation, which occurs at a later stage. It is therefore possible that another factor is responsible for the exact timing of TER down-regulation. Good candidates for this function are the D-type genes FBP7 and FBP11, which determine ovule identity in petunia (Angenent et al., 1995; Colombo et al., 1995). The expression of these D-type genes was analyzed by in situ hybridization using a labeled FBP11 RNA probe, which should also detect the FBP7 transcripts. The hybridization experiments (Fig. 1, G and H) revealed that FBP7/FBP11 transcripts appear approximately at a stage when TER expression is decreasing. Prior to the formation of carpel primordia, FBP7/FBP11 mRNA is absent (Fig. 1G), whereas expression of these MADS-box genes appears in the cells located in the center of the flower and differentiate into placental tissue (Fig. 1H).
Multimeric Complexes among C-, D-, and E-Type Proteins
To investigate the potential combinatorial action of the C-, D-, and E-type proteins in the FM that could coincide with the down-regulation of TER expression, we analyzed the ability of the MADS-box proteins to form multimeric complexes in yeast (Saccharomyces cerevisiae) two- and three-hybrid experiments. Because of the transcriptional activation activity possessed by the C terminus of FBP2 in yeast, a truncated version of the protein (FBP2ΔC) lacking 59 amino acids was used, as described in Ferrario et al. (2003). It has already been shown in previous work that the E-type protein FBP2 interacts in yeast with both C- and D-type proteins (Ferrario et al., 2003), whereas neither the C nor the D types is able to form a heterodimer with each other (Immink et al., 2003). The presence of FBP2, however, is able to bring together the C- and D-type proteins in the same complex, as shown in Table I. The formation of all possible complexes, which included FBP2, could be detected, although with different efficiency, as the interactions between FBP6, FBP7, and FBP2 and between FBP11, FBP7, and FBP2 appeared to be stronger in the yeast system. As a control, the combination of the D-type FBP11, the C-type pMADS3, and the B-type pMADS2 was tested and appeared to be negative in all conditions assayed. The formation of a multimeric complex involving C-, D-, and E-type proteins may also occur in the developing flower, where it may induce repression of the TER gene.
Table I.
Higher order complex formation as determined by a yeast GAL4 three-hybrid assay
Selection for complex formation was performed at room temperature. + + indicates strong interaction; + indicates weak interaction; and − indicates no growth of the yeast cells on the selective medium. Because of the transcriptional activation activity possessed by the C terminus of FBP2 in yeast, a truncated version of the protein (FBP2ΔC) lacking 59 amino acids was used, as described in Ferrario et al. (2003). H, His; L, Leu; T, Thr; 3AT, 3-amino-1,2,4-triazole.
| Protein/Vector Combinations | LTH + 1 mm 3AT Room Temperature |
|---|---|
| pBDGAL4-FBP6 + pADGAL4-FBP7 + pRED-FBP2ΔC | + + |
| pBDGAL4-FBP11 + pADGAL4-FBP6 + pRED-FBP2ΔC | + |
| pBDGAL4-pMADS3 + pADGAL4-FBP7 + pRED-FBP2ΔC | + |
| pBDGAL4-FBP11 + pADGAL4-pMADS3 + pRED-FBP2ΔC | + |
| pBDGAL4-FBP11 + pADGAL4-FBP7 + pRED-FBP2ΔC | + + |
| pBDGAL4-FBP11 + pADGAL4-FBP7 + pTFT1-FBP6 | − |
| pBDGAL4-FBP11 + pADGAL4-FBP7 + pTFT1-pMADS3 | − |
| pBDGAL4-FBP11 + pADGAL4-pMADS3 + pRED-pMADS2 | − |
Simultaneous Ectopic Expression of FBP2 and FBP11 Arrests Seedling Development
To examine whether the simultaneous ectopic expression of the above-mentioned MADS-box genes is sufficient to down-regulate TER expression outside the floral domain, we followed a transgenic approach to combine the constitutive expression of the MADS-box genes. The full-length FBP2 cDNA under the control of the 35S constitutive promoter of the Cauliflower mosaic virus was introduced in petunia plants. Ectopic expression of FBP2 in plants carrying a copy of the transgene, which segregated as a single locus, was confirmed by northern-blot hybridization (data not shown). No visible phenotype was observed in the transgenic plants at any stage of development. Conversely, overexpression of the ovule identity gene FBP11 did cause phenotypic alterations in the flower as previously reported by Colombo et al. (1995). Ectopic ovule formation was observed on the corolla tube and on the inner side of the sepals. Petals were also affected; the corolla at full maturity was nothing more than a tube that barely reached the length of the style. Plants homozygous for the FBP2 gene coming from two independent lines were subsequently crossed with transgenic plants hemizygous for FBP11. When seeds from three independent crosses were germinated in vitro, approximately one-half of the seedlings showed a permanent arrest early in the development at a stage when only the two cotyledons were visible. A clear difference was observed between the two different seedling populations about 6 d after germination (Fig. 2, A–C). Arrested seedlings and seedlings with a wild-type appearance were tested at DNA and mRNA levels. PCR amplification using primers specific for the FBP2 and FBP11 transgenes revealed that all the seedlings contained at least one copy of the FBP2 transgene, whereas only the small ones also carried the FBP11 transgene. The result was confirmed by northern-blot analysis: Ectopic expression of both FBP2 and FBP11 was observed in the arrested seedlings only (data not shown).
A more detailed phenotypic analysis of the small seedlings revealed that two to four leaf primordia were emerging from the SAM and the leaves produced always remained very small (Fig. 2, D–F). To analyze the shape and size of the SAM, seedlings of wild-type plants and the double overexpressor were cleared and microscopically studied (Fig. 2, G and H). This revealed that the size of the meristem was reduced in the seedlings of the double overexpressor compared to the wild type. In addition, the SAM size of the single 35S∷FBP2 overexpressor was microscopically analyzed and appeared to be unaffected when compared to the wild-type SAM (data not shown).
Generating plants that simultaneously overexpress either FBP6 or pMADS3 and any of the other MADS-box genes was not possible by crossing because it has been shown previously (Kater et al., 1998) that the 35S∷FBP6 and 35S∷pMADS3 transgenes failed to be transmitted to the progeny.
TER Expression Is Down-Regulated in Seedlings That Are Arrested in Development
The arrest in development that was observed in the double overexpression lines suggested a failure in maintaining the stem cell pool in the shoot meristem, which could be caused by decreased activity of the meristem gene TER (Stuurman et al., 2002). Therefore, we analyzed the expression of TER in the SAM of the seedlings by in situ hybridization on a series of subsequent longitudinal sections. While TER transcripts were present in wild-type seedlings in a small group of cells, we were not able to detect any signal in the CZ of the shoot apex of the arrested transgenic seedlings (Fig. 2, I and J). The petunia ortholog of STM (PhSTM; Stuurman et al., 2002), in contrast, was still expressed in the SAM of the transgenic seedlings (Fig. 2, K and L), which is in line with the independent roles assigned to WUS and STM in determination of the Arabidopsis SAM.
Up-Regulation of MADS-Box Genes in Arrested Seedlings
Although the combined ectopic expression of FBP2 and FBP11 seemed sufficient to induce TER down-regulation, we could not exclude an involvement of the C-type genes. Therefore, we analyzed pMADS3 and FBP6 (Angenent et al., 1993; Tsuchimoto et al., 1993) expression in transgenic seedlings by means of reverse transcription (RT)-PCR. We also checked the expression of both D-type genes, FBP7 and the endogenous FBP11, through the same technique. Although FBP7 and FBP11 share approximately 90% sequence similarity at the protein level, it was possible to design PCR primers in the 3′ part of the sequence that were specific for each endogenous gene (Supplemental Fig. 1).
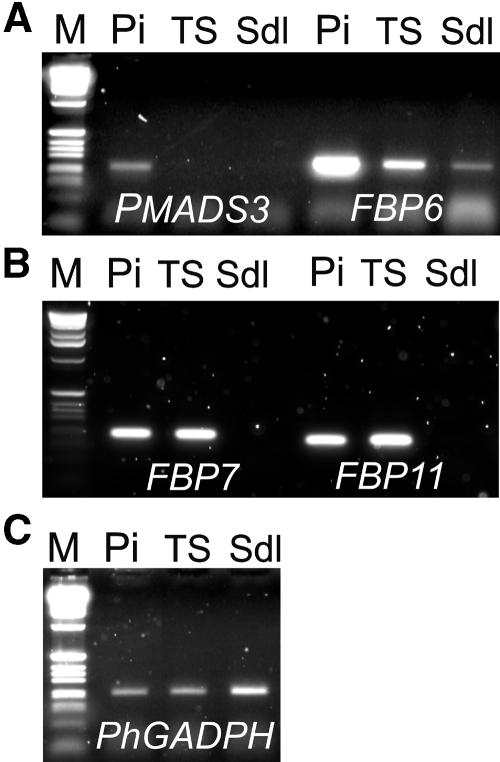
PCR amplifications were performed on cDNA obtained from a single arrested seedling, a single wild-type seedling, and a wild-type pistil. Surprisingly, FBP6, but not pMADS3, was up-regulated to a detectable level in the double overexpression seedling, as shown in Figure 3A, indicating a possible involvement of only one of the two C-type genes in the phenotype observed. It is noteworthy that a basal, although very low, expression of FBP6, but not of pMADS3, was detected in wild-type seedlings as well. Unfortunately, the lack of an fbp6 mutant makes it impossible to assess whether the same arrested phenotype could occur in the absence of the C-type gene product. On the other hand, both the endogenous FBP7 and FBP11 genes appeared to be up-regulated in transgenic seedlings (Fig. 3B) and sequencing of the amplified fragments confirmed the identity of the two genes. Expression of FBP7 or FBP11 was never detected in wild-type seedlings. The petunia housekeeping gene glyceraldehyde-3-P dehydrogenase (PhGADPH), uniformly expressed in all tissues, was used as a control for the cDNA quantity in each tissue used (Fig. 3C).
Figure 3.
RT-PCR amplification products of different genes in wild-type and transgenic tissues. A, Up-regulation of the C-type gene FBP6 but not of pMADS3 in 35S∷FBP2/FBP11 transgenic seedlings (TS). B, Up-regulation of the D-type genes FBP7 and FBP11 in 35S∷FBP2/FBP11 TS. C, PhGADPH RT-PCR products in wild-type and transgenic tissues. Pi, Pistil; TS, 35S∷FBP2/FBP11 transgenic seedling; Sdl, wild-type seedling; M, 1-kb DNA size marker.
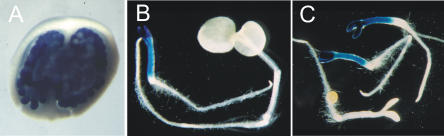
Another indication that FBP7 is indeed activated by simultaneous overexpression of FBP2 and FBP11 came from the analysis of arrested seedlings in which the β-glucuronidase (GUS) reporter gene was expressed under the control of an FBP7 promoter fragment. GUS staining in transgenic petunia plants from line T51013 was specifically observed in ovules (Fig. 4A), as was previously described by Colombo and colleagues (Colombo et al., 1997). This line was used in crosses with 35S∷FBP2 plants, and the progeny plants, selected for the presence of both constructs, were crossed again with 35S∷FBP11 plants. The progeny of this second cross segregated arrested and wild-type seedlings, which were tested for GUS activity. All the wild-type-looking seedlings, which also included transgenic seedlings containing only one of the overexpression constructs and pFBP7∷GUS, did not show any GUS activity. On the other hand, all small seedlings containing the pFBP7∷GUS construct showed clear GUS activity. The results are illustrated in Figure 4, B and C, and indicate that increased expression of both FBP2 and FBP11 is necessary to activate the FBP7 promoter.
Figure 4.
FBP7∷GUS expression in wild-type and transgenic tissues. A, Cross section through an FBP7∷GUS ovary showing GUS staining in the ovules. B, GUS staining of a 35S∷FBP2/FBP11, FBP7∷GUS transgenic (left), and wild-type (right) seedling. C, GUS staining of 35S∷FBP2/FBP11 seedlings segregating for the FBP7∷GUS locus.
Gene Activation and Repression in the Flower of 35S∷FBP11 Transgenic Plants
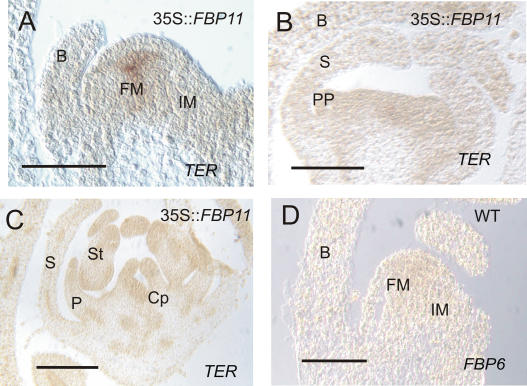
Strong activation of FBP2 and FBP11 genes seems to be sufficient to trigger the repression of TER, as has been shown in the double overexpressor seedlings. However, this occurs in the SAM, where these genes normally are not expressed. Therefore, we wondered whether TER expression was also affected in the FM of 35S∷FBP11 plants, where both FBP2 (endogenous) and FBP11 (35S∷FBP11) are strongly active. Indeed, in situ experiments performed on flower buds confirmed the down-regulation of TER expression in 35S∷FBP11 FMs at a stage when it is normally still active in a wild-type flower (compare Figs. 1E and 5B). TER expression could still be detected in a transgenic FM at a very early stage of flower development, when floral organ primordia are not yet apparent (Fig. 5A) and FBP6 transcripts are still absent (Fig. 5D). TER repression occurs only later in development, after petal primordia have appeared, at a stage when the C-type gene FBP6 is also strongly activated (Fig. 1C). The precocious down-regulation of TER in the transgenic FM, when FBP2 (Fig. 1A), FBP11 (35S∷FBP11), and FBP6 (Fig. 1C) are simultaneously highly expressed, strongly suggests that the three gene activities together are required for TER repression. The hypothesis that FBP11 may take part in TER down-regulation is also indicated by the lack of TER expression in the stomium area of the transgenic stamens (Fig. 5C). In wild-type flowers, TER expression could still be detected in stamens where normally FBP2 and FBP6 are also present (Fig. 1F, arrows), but its expression is abolished in the 35S∷FBP11 stamens. Although a precocious down-regulation of TER occurs in 35S∷FBP11 plants, transgenic flowers can still develop a complete pistil (Colombo et al., 1995). This could be explained by the fact that TER down-regulation in these plants occurs at a stage when sepals are formed, petal primordia are arising, and the number of meristematic cells present is most likely sufficient to ensure the completion of the remaining inner floral organs.
Figure 5.
In situ hybridization of 35S∷FBP11 (A–C), wild-type (D), and flower buds. A to C, Hybridized with a TER-specific probe. An FBP6 probe was used for D. B, Bract; Cp, carpel; IM, inflorescence meristem; P, petal; PP, petal primordium; S, sepal; St, stamen. Bars = 200 μm in A and D; 500 μm in B and C.
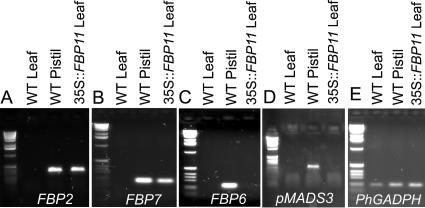
We also tested whether ectopic expression of FBP11 alone could trigger the up-regulation of the set of genes that was observed in the double overexpressors. RT-PCR performed on wild-type leaves and carpels and on transgenic leaves with different sets of gene-specific primers showed the activation of FBP2 and FBP7 genes only, whereas transcripts of the C-type genes FBP6 and pMADS3 were never detected (Fig. 6). The lack of FBP6 up-regulation in transgenic leaves could be explained by the absence of an FBP6 basal expression in these organs. The absence of an arrested phenotype in 35S∷FBP11 plants, despite the up-regulation of FBP2, could be due either to a lack of FBP6 or to an insufficient amount of FBP2 protein produced by FBP2 activation.
Figure 6.
RT-PCR amplification products of different genes in wild-type and 35S∷FBP11 transgenic tissues. A, Up-regulation of FBP2 in 35S∷FBP11 transgenic leaves. B, Up-regulation of FBP7 in 35S∷FBP11 transgenic leaves. C and D, FBP6 and pMADS3 transcripts were not detected by RT-PCR in 35S∷FBP11 transgenic leaves. E, PhGADPH RT-PCR products in wild-type and transgenic tissues.
DISCUSSION
In contrast to the SAM, the FM has to be terminated to allow the determinate structure of the flower to be completed. This occurs in Arabidopsis via a feedback loop involving the C-type gene AG, which is activated by WUS and, in turn, promotes its suppression (Lenhard et al., 2001; Lohmann et al., 2001). Mutant and expression analyses on both genes, however, indicate that at least one other unknown factor is necessary to down-regulate WUS together with AG. With the evidence produced in this study, we suggest that the C-type gene in petunia acts together with the E- and D-type genes in the down-regulation of the WUS ortholog and the termination of the FM.
The involvement of E-type genes in FM identity has been well documented by mutant studies in petunia, tomato (Lycopersicon esculentum), gerbera (Gerbera hybrida), and Arabidopsis (Angenent et al., 1994; Pnueli et al., 1994; Pelaz et al., 2000; Ditta et al., 2004; Uimari et al., 2004). In all mutants in which the E function was impaired, floral determinacy was affected, demonstrating its involvement in the termination of the flower. However, the petunia C- and E-type genes FBP6/pMADS3 and FBP2 (as well as their Arabidopsis counterparts AG and SEPALLATA) are activated within the flower at a stage when the TER transcript is still detectable, suggesting the presence of another factor whose later appearance is finally responsible for TER down-regulation. Possible candidates for this role are the D-type genes FBP7 and FBP11, whose expression coincides with the down-regulation of WUS-like genes, which ultimately leads to loss of meristematic activity. Furthermore, at later developmental stages, WUS is activated in nucellar cells of the ovule, where expression of the D-type gene is lacking (Angenent et al., 1995; Gross-Hardt et al., 2002). Although no direct evidence is available about the mode of action of the above-mentioned MADS-box genes, the yeast three-hybrid experiments we performed lead us to suggest that the corresponding proteins, which interact in yeast, may form a multimeric complex responsible for TER repression within the flower.
Thus, involvement of FBP2, FBP11, and FBP6 in blocking meristematic activity via TER down-regulation is also indicated by the arrested phenotype of petunia seedlings in which FBP2 and FBP11 were overexpressed. Ectopic expression of the two genes caused a significant up-regulation of FBP6, bringing together in the same cell the three components of the hypothetical complex that acts in the flower. Also, the endogenous D-function MADS-box genes FBP7 and FBP11 themselves were strongly up-regulated; however, unlike FBP6, no transcripts were ever detected in wild-type seedlings. Whether these feedback loops are controlled by dimers or higher order protein complexes containing the target gene remains to be solved. In a similar way, the B-type genes PISTILLATA (PI) and APETALA3 (AP3) from Arabidopsis and DEFICIENS (DEF) and GLOBOSA (GLO) from Antirrhinum are autoregulated by complexes containing the AP3-PI or DEF-GLO heterodimers, respectively (Schwarz-Sommer et al., 1992; Tröbner et al., 1992; Goto and Meyerowitz, 1994; Zachgo et al., 1995; Krizek and Meyerowitz, 1996; Samach et al., 1997). Expression of two other MADS-box genes, AGL15 and AG, in two separate studies has recently been shown to respond to the accumulation of their own product (Gómez-Mena et al., 2005; Zhu and Perry, 2005). Although direct evidence is at present still limited, there are indeed several indications that MADS-box genes are regulated by members of the same family and that, for some of them, a mechanism of autoregulation may be responsible for the maintenance of their own expression (deFolter et al., 2005).
There are, however, some discrepancies between the double overexpressor phenotype and a ter knockout mutant. Like wus mutants, ter plants are not completely arrested in development; they form adventitious meristems that terminate prematurely, leading to the characteristic bushy phenotype (Laux et al., 1996; Stuurman et al., 2002). Therefore, it has been postulated that other factors may act redundantly with WUS to promote stem cell activity (Fletcher, 2002). Furthermore, recent work in Arabidopsis revealed that WUS may not be necessary for stem cell maintenance in the shoot apex (Green et al., 2005). Therefore, we hypothesize that, in our double overexpressor plants, the high concentration of transgenic proteins may also interfere with the pathway that acts in parallel with WUS/TER, leading to complete arrest of any meristematic activity.
A role of the D-type genes FBP7/11 in determinacy is in contrast to the FBP7/11 cosuppression phenotype because a knock down of these genes did not cause indeterminacy in the flower (Angenent et al., 1995). However, in this study, we demonstrated that when FBP11 is activated in floral tissues where normally only FBP2 and FBP6 are present, a premature TER down-regulation occurs. In fact, ectopic expression of FBP11 alone caused TER repression in the FM and stomium area of the transgenic stamens only at a stage when both FBP2 and FBP6 were highly active. The lack of indeterminacy in FBP7/11 cosuppression plants could possibly be explained by the presence of another MADS-box gene that acts redundantly with FBP7/11 in TER down-regulation or by a residual amount of FBP7/FBP11 protein in the cosuppression plants not detectable by northern-blot analysis, but still sufficient to cause TER repression.
Although overexpression phenotypes should be considered cautiously because of the high level of transgenic protein produced, the phenotypes generated in the double overexpressors are consistent with the results obtained in the single overexpressors and are supported by the expression data of several endogenous genes in wild-type and transgenic plants.
In conclusion, our study with simultaneous activation of different MADS-box genes in petunia and consequent repression of the meristem gene TER suggests the requirement of C-, D-, and E-type genes in FM determination, most likely through the formation of an active multimeric complex involving SEP-, AG-, and SEEDSTICK (STK)-like proteins. A similar regulation of floral determinacy might occur in Arabidopsis as well, where genetic and molecular interaction among SEP, AG, and STK were also documented (Favaro et al., 2003).
MATERIALS AND METHODS
Plant Material
Petunia (Petunia hybrida) lines W115 and W138 and transgenic plants were grown under normal greenhouse conditions (22°C, 14-h light/10-h dark).
In vitro germination was performed after surface sterilization of petunia seeds by chlorine gas. Seeds were kept 3 h in a desiccator with a mixture of 100 mL commercial bleach and 3 mL concentrated hydrochloric acid in a fume hood. Sterilized seeds were grown on one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) containing 0.5% (w/v) Suc and 0.8% agarose at 22°C in 16-h light/8-h dark.
The transgenic plant T46008 carrying the 35S∷FBP11 transgene was used in this study (Colombo et al., 1995).
In Situ Hybridization
In situ hybridizations were performed as described by Cañas et al. (1994). Digoxigenin-labeled RNA probes were synthesized by T7 polymerase-driven in vitro transcription from the PCR fragments containing part of the TER open reading frame (ORF), the PhSTM ORF, and the FBP6 ORF, excluding the MADS box, according to the instructions of Boehringer Mannheim. The amplification products were obtained with the following forward and reverse primers: TERfw, 5′-TGGAGAAGAGCTTTAGGG-3′ and TERrev, 5′-TAATACGACTCACTATAGGGGATACGTAGTACATGGCC-3′; PhSTMfw, 5′-GGCAAAGATTATGGCTCATCC-3′ and PhSTMrev, 5′-TAATACGACTCACTATAGGGCATATCTTCGGATGGTTTCC-3′; FBP6fw, 5′-AGGTACAAGAAACACCATGCCG-3′ and FBP6rev, 5′-TAATACGACTCACTATAGGGCATCAGACAAGCTGTAGAGCAG-3′; the T7 polymerase promoter site is underlined. The FBP11 probe was prepared as described by Angenent et al. (1995).
Nomarski Microscopy
Seedlings were cleared for 2 to 16 h in a drop of Hoyer's solution (7.5 g gum arabic, 100 g chloral hydrate, 5 mL glycerol in 30 mL water) on a microscope slide. Cleared seedlings were examined using a Nikon Optiphot microscope equipped with Nomarski optics.
RT-PCR Analyses
Total RNA was extracted from single seedlings, pistils, young leaves, and roots using the RNeasy plant mini kit (Qiagen). cDNA was obtained from 1 μg of total RNA in a 50-μL reaction using the TaqMan reverse transcription kit (Applied Biosystems). Prior addition of the reverse transcriptase, 1 μL of DNase (1 unit/μL; Invitrogen) was added and the reaction was carried out for 30 min at 37°C. After heat inactivation of the DNase at 75°C for 5 min, 10 μL were removed from the reaction and used in the PCR amplification step as a control for the absence of genomic DNA. One microliter of reverse transcriptase was added to the remaining 40 μL and the reaction continued for 10 min at 25°C, followed by 30 min at 37°C, and 5 min at 95°C to inactivate the enzyme.
RT-PCR amplification products were obtained using the following gene-specific forward and reverse primers: FBP2fw, 5′-GCAAAGAACTTGAATCACTTGAAAGGC-3′ and FBP2rev, 5′-GCTTTCAAGGCAACCAGCC-3′; pMADS3fw, 5′-CTGAATCTCAGAGATCTGAGG-3′ and pMADS3rev, 5′-CAAGGTCATAGCTAGAACTCC-3′; FBP6fw, 5′-GTACAGGATCTGTTTCTGAAGC-3′ and FBP6rev, 5′-AATCTCCCTCTTTTGCATGAGC-3′; FBP7fw, 5′-CCCAAGTAAGTCCTCACATCG-3′ and FBP7rev, 5′-GCAAGAAAGCTTGAACAAACACC-3′; FBP11fw, 5′-GTAAATTTATTGGGCGCTGG-3′ and FBP11rev, 5′-GAGAAAAGCTGACGAGTTCACC-3′; and PhGADPHfw, 5′-GGTTGGAGAAAGAAGCCACC-3′ and PhGADPHrev, 5′-CGTTGTCGTACCAAGACACG-3′.
The annealing temperature was 59°C for all the primer combinations used. PhGADPH PCR products were visualized on a 2% agarose gel after 25 cycles of amplification. Thirty cycles were used in the PCR profile of all the MADS-box genes.
Three-Hybrid Analysis of Petunia Protein-Protein Interactions
The full-length pMADS3 and FBP6 coding regions were cloned into the pTFT1 vector (Egea-Cortines et al., 1999). The three-hybrid analyses were performed in the modified GAL4 yeast (Saccharomyces cerevisiae) two-hybrid system as described by Ferrario et al. (2003). Growth of yeast and hence complex formation has been scored after a 10-d incubation at room temperature (20°C) on a selective medium lacking His and supplemented with 1 mm 3-amino-1,2,4-triazole (Sigma).
Supplementary Material
This work was supported by the Dutch Organization for Research (VENI grant to R.G.H.I.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gerco C. Angenent (gerco.angenent@wur.nl).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.072660.
References
- Angenent GC, Franken J, Busscher M, Colombo L, van Tunen AJ (1993) Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J 4: 101–112 [DOI] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, van Dijken A, van Went JL, Dons HJ, van Tunen AJ (1995) A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7: 1569–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, Weiss D, van Tunen AJ (1994) Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant J 5: 33–44 [DOI] [PubMed] [Google Scholar]
- Angenent GC, Stuurman J, Snowden KC, Koes K (2005) Use of petunia to unravel plant meristem functioning. Trends Plant Sci: 243–250 [DOI] [PubMed]
- Barton MK, Poethig RS (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Cañas LA, Busscher M, Angenent GC, Beltran JP, Van Tunen AJ (1994) Nuclear localization of the petunia MADS box protein FBP1. Plant J 6: 597–604 [Google Scholar]
- Colombo L, Franken J, Koetje E, Van Went J, Dons HJM, Angenent GC, Van Tunen A (1995) The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo L, Franken J, Van der Krol AR, Wittich PE, Dons HJM, Angenent GC (1997) Down-regulation of ovule-specific MADS box genes from petunia results in maternally controlled defects in seed development. Plant Cell 9: 703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deFolter S, Immink RGH, Kieffer M, Pařenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L (2003) MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario S, Immink RG, Shchennikova A, Busscher-Lange J, Angenent GC (2003) The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15: 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC (2002) Shoot and floral meristem maintenance in Arabidopsis. Annu Rev Plant Biol 53: 45–66 [DOI] [PubMed] [Google Scholar]
- Gómez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R (2005) Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438 [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8: 1545–1560 [DOI] [PubMed] [Google Scholar]
- Green KA, Prigge MJ, Katzman RB, Clark SE (2005) CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17: 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M, Laux T (2002) WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev 16: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RGH, Ferrario S, Busscher-Lange J, Kooiker M, Busscher M, Angenent GC (2003) Analysis of the petunia MADS-box transcription factor family. Mol Genet Genomics 268: 598–606 [DOI] [PubMed] [Google Scholar]
- Kapoor M, Tsuda S, Tanaka Y, Mayama T, Okuyama Y, Tsuchimoto S, Takatsuji H (2002) Role of petunia pMADS3 in determination of floral organ and meristem identity, as revealed by its loss of function. Plant J 32: 115–127 [DOI] [PubMed] [Google Scholar]
- Kater MM, Colombo L, Franken J, Busscher M, Masiero S, Van Lookeren Campagne MM, Angenent GC (1998) Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Meyerowitz EM (1996) The Arabidopsis homeotic genes APETALA3 and PISTlLLATA are sufficient to provide the B class organ identity function. Development 122: 11–22 [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Jurgens G, Laux T (2002) The WUSCHEL and SHOOTMERISTEMLESS genes fulfill complementary roles in Arabidopsis shoot meristem regulation. Development 129: 3195–3206 [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Long JA, Barton MK (1998) The development of apical embryonic pattern in Arabidopsis. Development 125: 3027–3035 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford J, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the SHOOTMERISTEMLESS gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–479 [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hareven D, Broday L, Hurwitz C, Lifschitz E (1994) The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Kohalmi SE, Motte P, Datla R, Haughn GW (1997) Divergence of function and regulation of class B floral organ identity genes. Plant Cell 9: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lonnig WE, Saedler H, Sommer H (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Carles C, Fletcher JC (2003) Maintenance of stem cell populations in plants. Proc Natl Acad Sci USA 100: 11823–11829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman J, Jaggi F, Kuhlemeier C (2002) Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev 16: 2213–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig WE, Saedler H, Sommer H, Schwarz-Sommer Z (1992) A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J 11: 4693–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S, van der Krol A, Chua N-H (1993) Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell 5: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uimari A, Kotilainen M, Elomaa P, Yu D, Albert VA, Teeri TH (2004) Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc Natl Acad Sci USA 101: 15817–15822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Souer E, Koes R, Tornielli GB, Pezzotti M, Ferrario S, Angenent GC, Gerats T (2003) Towards the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C and D floral identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15: 2680–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Fletcher JC (2005) Stem cell regulation in the Arabidopsis shoot apical meristem. Curr Opin Plant Biol 8: 1–5 [DOI] [PubMed] [Google Scholar]
- Zachgo S, de Silva EA, Motte P, Trobner W, Saedler H, Schwarz-Sommer Z (1995) Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121: 2861–2875 [DOI] [PubMed] [Google Scholar]
- Zhu C, Perry SE (2005) Control of expression and autoregulation of AGL15, a member of the MADS-box family. Plant J 41: 583–594 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.