Abstract
Diurnal variations in nitrate reductase (NR) activity and nitrogen metabolites were examined in wild-type Nicotiana plumbaginifolia and transformants with various degrees of NR deregulation. In the C1 line, NR was only deregulated at the transcriptional level by placing the NR gene under the control of the cauliflower mosaic virus 35S RNA promoter. In the Del8 and S521D lines, NR was additionally deregulated at the posttranslational level either by a deletion mutation in the N-terminal domain or by a mutation of the regulatory phosphorylation site (serine-521). Posttranslational regulation was essential for pronounced diurnal variations in NR activity. Low nitrate content was related to deregulation of NR, whereas the level of total free amino acids was much higher in plants with fully deregulated NR. Abolishing transcriptional and posttranslational regulation (S521D plants) resulted in an increase of glutamine and asparagine by a factor of 9 and 14, respectively, compared with wild type, whereas abolishing transcriptional regulation (C1 plants) only resulted in increases of glutamine and asparagine by factors <2. Among the minor amino acids, isoleucine and threonine, in particular, showed enhanced levels in S521D. Nitrate uptake rates were the same in S521D and wild type as determined with 15N feeding. Deregulation of NR appears to set the level of certain amino acids, whereas diurnal variations were still determined by light/dark. Generally, deregulation of NR at the transcriptional level did not have much influence on metabolite levels, but additional deregulation at the posttranslational level resulted in profound changes of nitrogen metabolite levels.
In higher plants, nitrate reductase (NR) is regulated at the transcriptional as well as posttranslational level by various external and endogenous signals, and among these signals light has been shown to activate NR at both levels (for review, see Meyer and Stitt, 2001). Indeed, light, through the activation of photosynthesis and the production of sugars, stimulates the NR promoter (Vincentz et al., 1993) and NR activity (for review, see Huber et al., 2002; Lillo et al., 2004). Conversely, darkness generally inhibits NR gene transcription and leads to NR inactivation by phosphorylation and subsequent 14-3-3 binding. Recently, a Nicotiana plumbaginifolia transformant (S521D) with NR deregulated at both these levels was constructed (Lillo et al., 2003; Lea et al., 2004) by expressing a NR mutated for the regulatory phosphorylation site, Ser-521, under the control of the cauliflower mosaic virus (CaMV) 35S promoter. In some systems, Asp has been shown to mimic a phosphorylated Ser residue (Wang et al., 1992). However, this was not the case for mutated Arabidopsis (Arabidopsis thaliana) NR expressed in Pichia (Su et al., 1996), or for cultivated tobacco (Nicotiana tabacum) NR. We have already shown that NR is constitutively active in S521D plants under conditions where wild-type NR would have been rapidly inactivated (Lillo et al., 2003). In this work, we test diurnal variations of NR activity and nitrogen metabolites in wild-type N. plumbaginifolia, S521D, and lines with intermediate degrees of NR deregulation, C1 and Del8. In the C1 line, NR was deregulated at the transcriptional level only by placing the NR coding sequence under the control of the 35S promoter (Vincentz and Caboche, 1991). In the Del8 and S521D lines, NR was also deregulated at the posttranslational level by a deletion mutation in the N-terminal domain (Nussaume et al., 1995) or by mutation of the regulatory phosphorylation site (Ser-521; Lillo et al., 2003), respectively. The results show that posttranslational regulation of NR was apparently much more important than transcriptional regulation for setting the levels of amino acids, ammonium, and nitrate in the plant.
NR mRNA and activity show pronounced diurnal variations in a wide range of higher plant species usually with a peak around the beginning of the day (Galangau et al., 1988; Lillo and Meyer, 2001; Tucker et al., 2004), and downstream products of NR (e.g. Gln, Glu, and Asp) are assumed to exert, in normal conditions, negative feedback on NR expression at the transcriptional level, although the exact signaling mechanism is so far not clear (Langendorfer et al., 1988; Deng et al., 1991; Sivasankar and Oaks, 1995; Scheible et al., 2000; Lillo et al., 2001; Stitt et al., 2002). In C1 plants, circadian rhythmicity of the NR mRNA driven by the 35S promoter is lost, which indicates that this fluctuation is the result of transcriptional control in wild-type plants (Vincentz and Caboche, 1991). The C1 as well as the S521D plants were obtained by transformation of the NR-deficient mutant E23, in which the innate NR gene is inactivated by the insertion of a Tnp2 retrotransposon (Leprince et al., 2001). As a result, a truncated NR mRNA is produced, which is driven by the NR promoter and ends in the first Tnp2 long terminal repeat (Vaucheret et al., 1992). This truncated transcript can thus be used as a reporter gene for the endogenous NR promoter (Vaucheret et al., 1992; Klein et al., 2000). In the S521D plants with fully deregulated NR, diurnal variations and circadian rhythms in NR activity would be expected to be largely wiped out, as was indeed shown in this work. S521D plants were analyzed to test whether the truncated transcript would still show a circadian rhythm, which would indicate that NR is governed by a central biological clock independent of nitrogen metabolism. Our results support the assumption that rhythms in the endogenous NR are dependent on nitrogen metabolism or that the influence by a central oscillator is overridden by the high levels of amino acids in plants with deregulated NR.
RESULTS
Diurnal Variations in NR Activity
When the NR gene is linked to the CaMV 35S promoter, as for C1 plants, the NR gene is constitutively expressed and NR mRNA is high both during the day and night (Vincentz and Caboche, 1991). In early work on C1 plants, experiments with prolonged periods of darkness showed that NR activity changed, although NR mRNA levels were rather constant (Vincentz and Caboche, 1991; Nussaume et al., 1995). Interestingly, diurnal variations of NR activity are still clearly pronounced in leaves of these C1 plants, although the gene is constitutively transcribed (Fig. 1, A and B). The first sample was harvested after 12 h of darkness. The next samples were harvested 2 and 3 h into the light period, and sampling was continued every 3 h. The total amount of NR activity assayed in the presence of EDTA (no Mg2+ available to inhibit the phosphorylated enzyme) increased by 57% during the first 4 h of the light period for C1. This indicates that light may also act positively on the translation process or on the stability of the NR protein. Posttranslational phosphorylation contributed strongly to the diurnal variations of NR activity in wild-type as well as C1 plants, as can be seen when NR activity was measured in the presence of Mg2+ (Fig. 1B). NR tested in the presence of Mg2+ (actual NR activity) is believed to give a better measurement of the in situ NR activity because phosphorylated NR is inactive in the presence of Mg2+ at physiological concentrations (MacKintosh et al., 1995). However, even in the presence of Mg2+, in situ inactivation of NR can be underestimated because some activation by dephosphorylation is likely to take place during extraction and assay of NR for C1 and wild type (MacKintosh, 1992), whereas for mutated NR from S521D plants this is not relevant. Especially in the dark, NR activity in C1 and wild-type plants can thus be even lower, which would explain why these plants behave similarly with respect to accumulation of reduced nitrogen compounds and very differently from S521D, although the Mg2+ assay gave fairly similar measurements for NR extracted from C1 and S521D plants. Actual NR activity increased by 150% to 230% for wild-type and C1 plants in the light, whereas for the S521D plants the increase was very modest (22%). The increase was also modest for Del8 plants (64%) compared with wild-type and C1 plants (Fig. 1B). Activity state (percentage of active NR) was always high (70%–80%) for S521D throughout the day and night, as expected (Fig. 1C). For Del8, activity state was also rather constant throughout the day and night, but lower (45%–60%) compared with S521D. For C1 and wild-type activity, activity state varied between 20% and 70%.
Figure 1.
Diurnal variations of NR activity in leaves of various N. plumbaginifolia lines. A, EDTA NR (i.e. total NR activity). B, Mg2+ NR (i.e. actual NR activity). C, Activity state, percentage of active NR enzyme out of the total amount of NR enzyme. N. plumbaginifolia lines tested were wild type (circles), C1 (squares), Del8 (triangles), and S521D (inverted triangles). During the photoperiod, light intensity was 150 μmol m−2 s−1 and the temperature was 25°C; during the dark period, temperature was 18°C. Samples were harvested in the morning after 12-h darkness, then 2 and 3 h into the light period, and continued every 3 h. There were four samples for the two first time points, and vertical bars indicate se (when exceeding the size of the symbol). For other time points, there were two samples and the vertical bars indicate the spread.
Diurnal Variations in Content of Nitrate, Ammonium, and Amino Acids
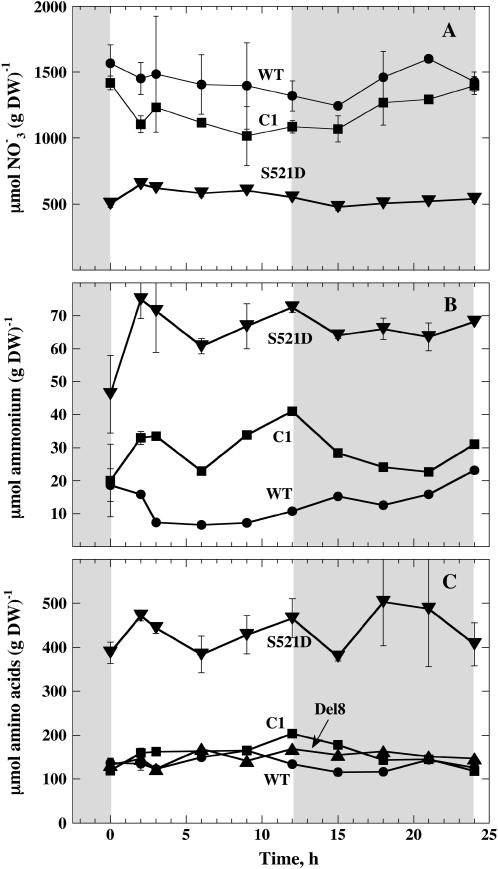
Figure 2A shows diurnal variations in nitrate content of leaves. For wild-type and C1 plants, a small and reproducible decrease in nitrate content was observed during the first 2 h of light, whereas this was not the case for S521D. These findings can be explained by the large increase in actual NR activity in wild-type and C1 plants (Fig. 1B). For wild-type and C1 plants, an increase in nitrate was observed when NR was partly inactivated (during the dark period). Diurnal variations in nitrate content were less pronounced in S521D compared to wild-type and C1. A remarkable difference in the level of nitrate throughout the day and night was observed between S521D and wild type and C1. S521D contained less than one-half the nitrate of wild type and C1. As NR in the S521D mutant is constantly active, a high nitrate reduction is expected and a refill of the nitrate pool may not be possible. Alternatively, uptake of nitrate may also be lower in S521D plants.
Figure 2.
A, Diurnal variations in nitrate content in leaves of various N. plumbaginifolia lines. B, Diurnal variation of ammonium content. C, Diurnal variations in total free amino acids. Samples were harvested as described in Figure 1 and the same symbols are used. For S521D, there were four samples at the two first time points and two samples for the other time points. For other lines, there were two samples for the first two time points and one sample for the other time points. se or spread is indicated by vertical bars.
Figure 2B shows diurnal variations in the ammonium content of leaves. When light was switched on, ammonium content increased for C1 and the S521D mutant, whereas wild-type plants showed a small decrease in ammonium content. On average, ammonium content of S521D was 5 times higher than in wild type and two times higher than in C1.
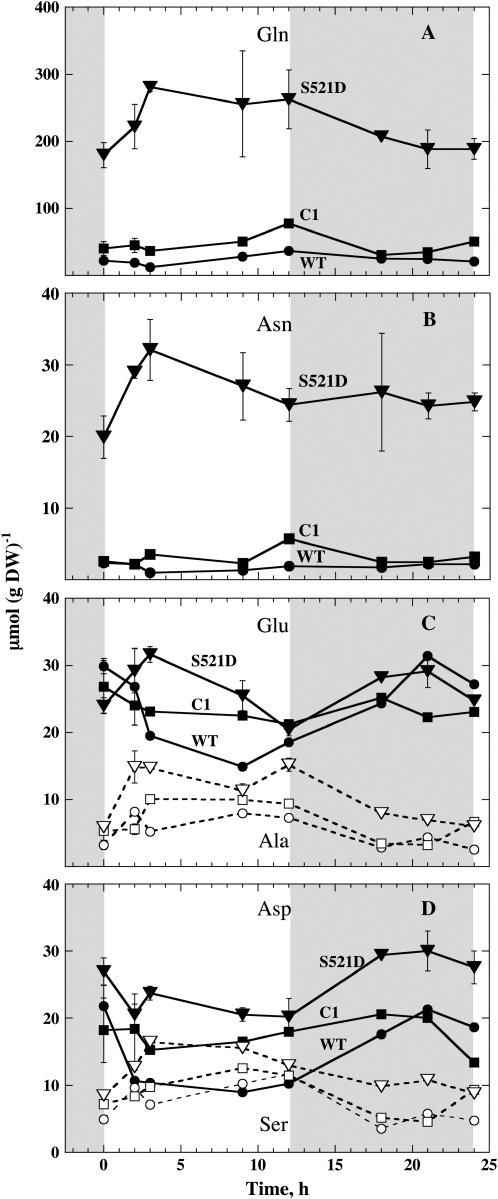
Figure 2C shows total free amino acids. Free amino acids were about 4 times higher in the S521D mutant compared with the other lines. Reduced nitrogen was almost constant throughout the day and night in all lines under these conditions. When concentrations of different amino acids were measured, the most striking difference was a higher percentage of amide amino acids in the S521D mutant. The Gln content was very high in S521D compared with wild-type and C1 plants (Fig. 3A). Indeed, when averaged throughout the day and night, Gln content was 224, 44, and 24 μmol (g dry weight)−1 in S521D, C1, and wild-type plants, respectively. In S521D plants, Gln clearly increased during the first 3 h of the light period, whereas diurnal changes of Gln in wild-type and C1 plants were modest under these conditions. The level of Asn was 14 times higher in S521D compared with wild-type plants when averaged throughout the day and night: 26, 3, and 1.8 μmol (g dry weight)−1 for S521D, C1, and wild type, respectively. For Asn, the concentration also increased significantly during the first 3 h of the light period (Fig. 3B). Concentrations of other amino acids were in the same range for all three lines. For Glu (Fig. 3C) and Asp (Fig. 3D), there was a tendency toward a dip in concentration at the end of the photoperiod and an increase during the first part of the dark period for all lines tested. Ala and Ser increased during the first part of the photoperiod and decreased during the dark period in all lines (Fig. 3, C and D). Concentrations of minor amino acids averaged throughout the day and night are shown in Figure 4. Thr, Ile, and Lys are essential amino acids belonging to the Asp family of amino acids. Interestingly, Thr and Ile, which are also on the same branch in the metabolic reaction pathways, were both elevated in S521D plants, whereas the concentration of Lys was almost unaltered in transgenic plants (Fig. 4). Among the other amino acids, the Pro level was strikingly higher in both S521D and C1 compared with wild type. Concentrations of other minor amino acids were not much different in transgenic plants compared with wild type.
Figure 3.
Diurnal variations in major free amino acids. A, Gln; B, Asn; C, Glu (black symbols) and Ala (white symbols); D, Asp (black symbols) and Ser (white symbols). Samples were harvested as described in Figure 1, and the same symbols were used. For S521D, there were four samples at the two first time points and two samples for the other time points. For other lines, there were two samples for the first two time points and one sample for the other time points. se or spread is indicated.
Figure 4.
Levels of minor amino acids averaged throughout the day and night. Leaves were harvested as described in Figure 1. Black bars, Wild type; gray bars, C1; white bars, S521D. The data are means of 10 different samples, and the se is indicated.
Circadian Rhythms, 30-h Constant Light
We have followed NR expression at the RNA and activity level in an experiment where the different lines were placed in constant light after the normal 12-h night. It has previously been shown that rhythms in NR reach a peak in mRNA level before NR protein and activity in various plants (Lillo, 1991; Vincentz and Caboche, 1991; Scheible et al., 1997; Tucker et al., 2004), as confirmed in these experiments (Fig. 5A). As expected for S521D plants, NR activity was rather constant during the 30-h light period (Fig. 5B). In S521D plants, the endogenous NR gene had been inactivated by a transposon leading to a truncated transcript. This truncated transcript was used as a reporter gene for the endogenous NR promoter. We wished to test whether this transcript would be expressed rhythmically, although the overall rhythm in NR activity was abolished in these plants. Figure 5B shows transcript levels of truncated NR as well as full-length transcripts during 30-h light in the S521D line. The level of truncated NR was low but did not vary significantly. Because the content of free amino acids was high in the S521D plants throughout the day and night (Fig. 2C) or in constant light (C. Meyer, unpublished data), expression of the endogenous NR gene was apparently repressed. Surprisingly, the full-length NR transcript (linked to the constitutive 35S promoter) showed a marked decrease during the light period. There was a small increase after 24 h; however, this increase was not significant according to Student's t test. The decrease in NR transcript linked to a constitutive promoter was unexpected and led us to ask whether the transcript was less stable in the light compared with the dark. Effects of light and dark on transcript levels were therefore tested (Fig. 6) and indeed showed that when plants were exposed to the dark the full-length NR transcript level increased. This result is in agreement with NR transcripts being more rapidly degraded in the light than in the dark. The truncated NR transcript also started to increase in the dark, indicating a difference in stability in light and dark.
Figure 5.
NR activity and relative transcript levels in wild-type and S521D plants during 30 h of continuous light. A, Wild type; B, S521D. For wild type, there were at least three samples for each activity measurement. There was one sample for each wild-type mRNA measurement. For S521D plant activity and mRNA, there were at least three samples for each time point. NR activity is given by black symbols and relative mRNA levels by white symbols (full-length Nia mRNA) or gray symbols (truncated Nia mRNA). mRNA levels are given relative to signals obtained for the EF-1α housekeeping gene. se is indicated by vertical bars.
Figure 6.
Relative transcript levels of truncated and full-length NR in S521D during 12-h-light/24-h-dark cycles. Values of relative mRNA are means of at least three samples of truncated NR and at least four samples of full-length NR. Full-length mRNA is indicated by white symbols and truncated by black symbols. mRNA levels are given relative to signals obtained for the EF-1α housekeeping gene. se is indicated by vertical bars.
Uptake and Assimilation of 15N-NO3− in the Dark
Uptake and assimilation of nitrate was studied in whole plants grown hydroponically with 1 mm nitrate and subsequently supplied with 15N-labeled  at the beginning of a dark period. During 3 h as well as 16 h of darkness, the measured nitrate uptake rate was lower for Del8 but very similar for all other lines tested (Table I). Uptake rates during the first 3 h were almost twice as high as during the latter part of the dark period. The relative decrease in uptake rates was the same in all lines. Plants grown in soil were given 15 mm nitrate in the irrigation solution, whereas plants in hydroponics were exposed to only 1 mm nitrate to facilitate measurement of 15N. This resulted in a difference in nitrate content of leaves for the two different setups. Leaves of wild-type plants grown at 15 mm nitrate had 1,500 μmol
at the beginning of a dark period. During 3 h as well as 16 h of darkness, the measured nitrate uptake rate was lower for Del8 but very similar for all other lines tested (Table I). Uptake rates during the first 3 h were almost twice as high as during the latter part of the dark period. The relative decrease in uptake rates was the same in all lines. Plants grown in soil were given 15 mm nitrate in the irrigation solution, whereas plants in hydroponics were exposed to only 1 mm nitrate to facilitate measurement of 15N. This resulted in a difference in nitrate content of leaves for the two different setups. Leaves of wild-type plants grown at 15 mm nitrate had 1,500 μmol 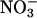 (g dry weight)−1 (Fig. 2), whereas wild type grown at 1 mm nitrate had 803 μmol
(g dry weight)−1 (Fig. 2), whereas wild type grown at 1 mm nitrate had 803 μmol  (g dry weight)−1. In both cases, the absolute difference between nitrate content in wild type and S521D was considerable, 1,000 and 800 μmol
(g dry weight)−1. In both cases, the absolute difference between nitrate content in wild type and S521D was considerable, 1,000 and 800 μmol 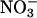 (g dry weight)−1, respectively. At 1 mm nitrate, the relative difference between wild type and S521D was therefore much larger, and S521D plants had only 29 μmol
(g dry weight)−1, respectively. At 1 mm nitrate, the relative difference between wild type and S521D was therefore much larger, and S521D plants had only 29 μmol 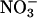 (g dry weight)−1. As seen from Tables I and II, the amount of nitrate was low in both shoots and roots of S521D, as well as in roots of other plants. The nitrate concentrations did not change significantly during the 16-h dark period (data not shown). The
(g dry weight)−1. As seen from Tables I and II, the amount of nitrate was low in both shoots and roots of S521D, as well as in roots of other plants. The nitrate concentrations did not change significantly during the 16-h dark period (data not shown). The  assimilation rate in leaves, roots, and whole plants was estimated during the 16-h labeling period (Table II). The total amount of nitrogen uptake was calculated from the quantity of 15N measured in all organs. Since nitrate concentrations in S521D shoots and roots and in roots of all lines were low, several plant extracts were pooled to measure the quantities of nitrogen stored as
assimilation rate in leaves, roots, and whole plants was estimated during the 16-h labeling period (Table II). The total amount of nitrogen uptake was calculated from the quantity of 15N measured in all organs. Since nitrate concentrations in S521D shoots and roots and in roots of all lines were low, several plant extracts were pooled to measure the quantities of nitrogen stored as  during the labeling experiment. The S521D lines showed a much higher apparent nitrate reduction rate in leaves with almost all nitrate being reduced (Table II). Since the labeling period was relatively long, shoots and roots can exchange both nitrate and reduced nitrogen and thus the value calculated is only a measure of the ratio between nitrate and reduced nitrogen in leaves and roots at the end of the 16-h dark period. It has previously been shown for various lines of wild-type tobacco that nitrate as well as nitrite reduction can take place in the dark in leaves, although at a reduced rate (Reed et al., 1983; Lejay et al., 1997). The values obtained in this work are very similar to those obtained previously for the wild-type and Del lines using detached leaves (Lejay et al., 1997). In roots, the S521D lines again had a much higher apparent nitrate reduction rate (Table II). By adding up the amounts of nitrogen taken up and stored as nitrate during the labeling period in shoots and roots, we calculated the real nitrate reduction rate in the whole plant (Table II). It seems that most of the nitrate taken up in the S521D lines is reduced during the 16-h dark period, whereas the reduction rate in wild type and Del8 is significantly lower.
during the labeling experiment. The S521D lines showed a much higher apparent nitrate reduction rate in leaves with almost all nitrate being reduced (Table II). Since the labeling period was relatively long, shoots and roots can exchange both nitrate and reduced nitrogen and thus the value calculated is only a measure of the ratio between nitrate and reduced nitrogen in leaves and roots at the end of the 16-h dark period. It has previously been shown for various lines of wild-type tobacco that nitrate as well as nitrite reduction can take place in the dark in leaves, although at a reduced rate (Reed et al., 1983; Lejay et al., 1997). The values obtained in this work are very similar to those obtained previously for the wild-type and Del lines using detached leaves (Lejay et al., 1997). In roots, the S521D lines again had a much higher apparent nitrate reduction rate (Table II). By adding up the amounts of nitrogen taken up and stored as nitrate during the labeling period in shoots and roots, we calculated the real nitrate reduction rate in the whole plant (Table II). It seems that most of the nitrate taken up in the S521D lines is reduced during the 16-h dark period, whereas the reduction rate in wild type and Del8 is significantly lower.
Table I.
Total nitrate uptake and uptake rates in whole plants during 3 and 16 h of darkness
Hydroponically grown plants were supplied with 1 mm  (approximately 5 atom % 15N). For the 3-h measurements, six plants were originally pooled and analyzed. For the 16-h measurements, three different plants were analyzed separately, and se is indicated. FW, Fresh weight.
(approximately 5 atom % 15N). For the 3-h measurements, six plants were originally pooled and analyzed. For the 16-h measurements, three different plants were analyzed separately, and se is indicated. FW, Fresh weight.
| Plant Tested | Total  in Shoots after 3 h in Darkness in Shoots after 3 h in Darkness |
Total 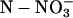 in Roots after 3 h in Darkness in Roots after 3 h in Darkness |
Total 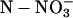 Uptake during 3 h of Darkness per Plant Uptake during 3 h of Darkness per Plant |
Total  Uptake during 16 h of Darkness per Plant Uptake during 16 h of Darkness per Plant |
Uptake Rate of  during 3 h of Darkness during 3 h of Darkness |
Uptake Rate of  during 16 h of Darkness during 16 h of Darkness |
|---|---|---|---|---|---|---|
| μg | μg | μg | μg | μg (g whole plant FW)−1 h−1 | μg (g whole plant FW)−1 h−1 | |
| Wild type | 2,506 | 166 | 152 | 457 ± 27 | 9.4 | 5.8 ± 0.4 |
| C1 | 1,420 | 226 | 180 | – | 10.2 | – |
| Del8 | 1,065 | 113 | 74 | 219 ± 44 | 6.4 | 3.4 ± 0.7 |
| S521D-5 | 68 | 79 | 116 | 351 ± 62 | 9.7 | 5.3 ± 0.9 |
| S521D-7 | 160 | 121 | 138 | 362 ± 50 | 9.3 | 4.8 ± 0.7 |
Table II.
Accumulation and reduction of 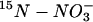 in leaves, roots, and whole plants during 16 h of darkness
in leaves, roots, and whole plants during 16 h of darkness
Values are given as quantities of nitrogen taken up, stored as nitrate, or reduced during the labeling period. Because of the low nitrate content, plants were pooled for measurements of 15N enrichment in the nitrate pool of S521D leaves and in roots of all lines. Total reduced nitrogen corresponds to the quantities of  taken up, which have been reduced during the 16 h of labeling in the whole plants or, apparently, in the leaves and roots. The amounts of nitrogen taken up and stored as nitrate in leaves and roots were added to obtain the values for the whole plants. n = 3. sd is given when calculated.
taken up, which have been reduced during the 16 h of labeling in the whole plants or, apparently, in the leaves and roots. The amounts of nitrogen taken up and stored as nitrate in leaves and roots were added to obtain the values for the whole plants. n = 3. sd is given when calculated.
| Lines | Total Amount of 
|
Total Nitrogen Uptake during 16 h | Total 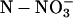 Stored during the Labeling Stored during the Labeling |
Total Reduced Nitrogen | Percentage of  Apparently Reduced during the Labeling Apparently Reduced during the Labeling |
|---|---|---|---|---|---|
| μg | μg | μg | μg | ||
| Leaves | |||||
| Wild type | 2,292 ± 617 | 319 ± 24 | 176 ± 20 | 143 ± 8 | 45 ± 2 |
| Del8 | 1,232 ± 285 | 144 ± 56 | 76 ± 20 | 68 ± 33 | 46 ± 6 |
| S521D-5 | 100 ± 28 | 243 ± 65 | 17 | 226 | 93 |
| S521D-7 | 110 ± 12 | 240 ± 58 | 10 | 230 | 96 |
| Roots | |||||
| Wild type | 232 ± 56 | 138 ± 26 | 83 | 55 | 40 |
| Del8 | 124 ± 16 | 75 ± 19 | 41 | 34 | 45 |
| S521D-5 | 61 ± 30 | 108 ± 45 | 43 | 65 | 60 |
| S521D-7 | 50 ± 18 | 122 ± 46 | 30 | 92 | 76 |
| Whole plants | |||||
| Wild type | 2,524 ± 617 | 457 ± 47 | 259 | 198 | 43 |
| Del8 | 1,356 ± 285 | 219 ± 75 | 117 | 102 | 47 |
| S521D-5 | 161 ± 57 | 351 ± 108 | 60 | 291 | 83 |
| S521D-7 | 160 ± 28 | 362 ± 87 | 40 | 322 | 89 |
DISCUSSION
During the first part of the photoperiod, actual NR activity increased by 22% (S521D) and 64% (Del8) in plants that were deregulated at the transcriptional as well as the posttranslational level. In plants deregulated at the transcriptional level only (C1), actual NR activity increased by 230%, showing that pronounced diurnal variations were revealed unless posttranslational regulation was impaired. NR in C1 plants showed diurnal variations not only due to posttranslational regulation but also due to the amount of total NR (EDTA NR; Fig. 1A). The increase in total NR during the first 3 h of the photoperiod indicates that the translation of NR is stimulated by light. The possibility that the NR level increased in the light due to higher stability of the NR protein has previously been ruled out for N. plumbaginifolia (Lillo et al., 2003), and light stimulation of the translational process is therefore the most likely explanation.
In wild-type and C1 plants, nitrate content decreased slightly during the first 2 h of the photoperiod, and reduced nitrogen increased slightly during the photoperiod (Fig. 2). This is the same pattern found previously in N. tabacum (Scheible et al., 2000), but the extent of increase/decrease here is much smaller in N. plumbaginifolia. The reason for smaller changes in N. plumbaginifolia is probably a lower NR activity level because, generally, low NR activity (low gene dosage) was found to dampen diurnal variations (Scheible et al., 2000). Use of a natural day/night regimen (i.e. higher day temperature than night temperature; Figs. 1–3) is also likely to contribute to dampening of the diurnal variations as previously found in other plants (Lillo, 1984). Furthermore, a higher light intensity (600 μmol photons m−2 s−1) was used for N. tabacum (Scheible et al., 2000) compared with N. plumbaginifolia (120 μmol photons m−2 s−1) in this work; the higher light intensity is also expected to contribute to more pronounced diurnal variations. A higher rate of nitrate decrease was also observed during the day in nitrogen-starved wild-type tobacco plants by Lejay et al. (1997).
The steady-state levels of nitrate, ammonium, and free amino acids were shifted toward the reduced form of nitrogen in plants with deregulated NR; lower nitrate content and higher content of ammonium and amino acids were found in the S521D plants compared with wild-type or C1 plants. It has been shown for barley (Hordeum vulgare), N. plumbaginifolia, and N. tabacum that mutants or transformants with decreased NR levels contain more nitrate and less Gln than wild type, and that transformants with increased NR have less nitrate and more Gln than the wild type (Warner and Kleinhofs, 1981; Foyer et al., 1994; Lejay et al., 1997; Scheible et al., 1997; Gojon et al., 1998). For instance, constitutive expression of NR in C1 plants has previously been found to increase the concentration of total free amino acids (Foyer et al., 1994). This accumulation of amino acids was mainly due to an increase in Gln levels by a factor of 2 on average (Foyer et al., 1994; Quilleré et al., 1994). The same doubling in Gln concentrations was observed in transgenic tobacco expressing NR under the control of the 35S promoter (Gojon et al., 1998). The effect of constitutive expression of NR was, however, very modest compared with the astonishing effect abolishing posttranslational regulation had on the content of nitrogen metabolites (Figs. 2 and 3). Abolishing posttranslational regulation resulted in an increase of Gln and Asn by a factor of 9 and 14, respectively, compared with wild type (averaged throughout the day and night), whereas abolishing transcription regulation (C1) only resulted in increases of Gln and Asn by factors <2 (data from Fig. 3) as already observed by Foyer et al. (1994).
Although a general increase in content of amino acids is often found in different plants during the photoperiod, this depends on growth conditions, especially light intensity and day length (Matt et al., 1998). The level of various amino acids must depend on carbon precursors, available reduced nitrogen, as well as flux into further metabolism. Light in the morning was followed by an increase in both Gln and Asn in S521D plants (Fig. 3), showing that the levels of the two amides were probably restricted by (products of) photosynthesis. In wild type and C1, other factors (e.g. reduced nitrogen) could be limiting for synthesis of these amino acids. Indeed, ammonium content was much higher in S521D compared with wild type and C1 (Fig. 2B).
Glu and Asp have been found to decrease or increase during the photoperiod depending on day length (Matt et al., 1998). In this work, a general dip toward the end of the photoperiod was found (Fig. 3, C and D). Concentrations of the pyruvate-derived amino acids Ala and Ser increased after onset of light in all lines tested (Fig. 3, C and D), confirming that these amino acids always increase during the photoperiod under very different conditions (Matt et al., 1998). Ala and Ser are made from three-carbon skeletons upstream of most other carbon skeletons important for synthesis of different amino acids, and replenishing of the carbon skeleton pool by onset of the Calvin cycle can be expected to be most pronounced in these two amino acids.
Although the levels of Gln and Asn concentrations were much higher in the S521D mutant, the diurnal variations in concentrations of the various amino acids in the S521D mutant were very similar to what is seen in other lines. These results can be interpreted as NR activity restricting the level of some amino acids, with other factors (photosynthesis) being the driving force for the diurnal variations observed. The level of Thr was 2.5 times higher in S521D compared with wild type (Fig. 4). This is of special interest regarding possible practical implications for deregulation of NR because this is one of the essential amino acids that can limit growth of domestic animals.
Transcript level of the endogenous, truncated NR gene in S521D plants was low in continuous light, and no significant rhythm was observed (Fig. 5B). Circadian rhythmicity in the level of the NR truncated transcript was also found to be lost in the E23 NR-deficient mutant, but was regained in an E23 × wild-type cross (Vaucheret et al., 1992). However, the abundance of this truncated transcript was much higher in the E23 mutant than in the other plants (Vaucheret et al., 1992). This suggests that high or low levels of Gln disrupt the diurnal rhythmicity of the (truncated) NR transcript with opposite effects on the abundance of this transcript. The results with S521D are in agreement with a suggested model that the rhythm in transcription directed by the NR promoter is influenced by activity of the NR enzyme and downstream products thereof (Deng et al., 1991; Lillo et al., 2001). Apparently, expression of the endogenous NR transcript is not under the control of a central oscillator, or at least regulation by a central oscillator is overridden in plants with deregulated NR activity because of the high level of amino acids.
The 35S promoter is widely used as a strong promoter in dicot species and is known to confer constitutive expression on the adjacent gene. Search for any specific control of the 35S promoter in Nicotiana species excluded light influence, but some evidence for tissue-specific factors was found (Lam and Chua, 1989; Gilmartin et al., 1990). The full-length NR transcript governed by the 35S promoter in S521D plants decreased during continuous light (Fig. 5B), and, when plants were transferred back into the dark, transcript levels then increased again (Fig. 6). Also, the truncated NR transcript increased in the dark, which suggests that the stability of NR mRNA is mediated by cis-acting elements present in the 5′-untranslated leader or in the NR first exon. This could also point to an involvement of the NR mRNA translation in the regulation of its degradation rate. Because transcription rate is generally found to be constant from the 35S promoter, these observations are best explained by NR mRNA being less stable in the light than in the dark in S521D plants. A decline in NR mRNA during the photoperiod was found previously for another line having NR constitutively expressed by the 35S promoter (i.e. line 30.313; Vincentz and Caboche, 1991). The C1 transformant (also named 30.C), which also has NR constitutively expressed by the 35S promoter, did not show any decrease of NR mRNA during the light period (Vincentz and Caboche, 1991). NR mRNA levels were higher in the C1 compared with the 30.313 transformant, and this could possibly make it more difficult to detect changes in the mRNA level in C1. It should also be stressed that different techniques were used to measure mRNA levels in the work by Vincentz and Caboche (northern) and this work (reverse transcription [RT]-PCR). Generally, RT-PCR is a more sensitive method. The possible involvement of high amino acid levels or the mechanism behind lower mRNA stability in the light compared with the dark is unknown.
NR is known to be degraded in the light with a half-life of approximately 20 h (Lillo et al., 2003), and it is therefore clear that a new enzyme is being formed in continuous light in the experiment presented in Figure 5. Although transcript levels decreased markedly during 12 h of light, apparently the transcript level was still sufficiently high to support a high level of NR enzymes because NR activity did not decrease in prolonged light. Because NR activity in the S521D mutant is not down-regulated in the dark, it could be expected that nitrate assimilation would be higher in S521D plants compared with wild type, especially in the dark. This was confirmed by following uptake and reduction of  and almost all the nitrate taken up during the 16-h labeling period in the dark by the S521D lines is reduced (Tables I and II). These results demonstrate that inactivation of NR in the dark is the major controlling step for the nitrate assimilation pathway. The value that we found for the percentage of nitrate taken up being reduced in wild type (45%) is very close to the one found by Gojon et al. (1998) for N. plumbaginifolia whole plants (46.6%). It was previously shown that constitutive transcription of NR (C1 plants) did not lead to higher assimilation rates (Lejay et al., 1997), and this seems logical because posttranslational inactivation of NR would still take place in these plants and assure low NR activity during darkness even though the NR protein level was high.
and almost all the nitrate taken up during the 16-h labeling period in the dark by the S521D lines is reduced (Tables I and II). These results demonstrate that inactivation of NR in the dark is the major controlling step for the nitrate assimilation pathway. The value that we found for the percentage of nitrate taken up being reduced in wild type (45%) is very close to the one found by Gojon et al. (1998) for N. plumbaginifolia whole plants (46.6%). It was previously shown that constitutive transcription of NR (C1 plants) did not lead to higher assimilation rates (Lejay et al., 1997), and this seems logical because posttranslational inactivation of NR would still take place in these plants and assure low NR activity during darkness even though the NR protein level was high.
S521D plants grow and develop normally, although NR is constitutively active. Apparently, plants have various ways to assure that nitrate is not reduced in excess under conditions (e.g. in the dark) where toxic levels of nitrite or side products of the NR reaction might accumulate due to reduced metabolism of nitrite (Meyer et al., 2005). At 1 mm external nitrate, the uptake rate was apparently balanced with the assimilation rate, and S521D then reduced almost all the nitrate in the leaves. At higher concentrations of nitrate (15 mm) used in the experiments on diurnal variations, S521D plants also started to store nitrate. Probably nitrate in the storage pool is not available for reduction by the NR enzyme and thereby storage acts to restrict the flux of nitrogen through the assimilation pathway, but not sufficiently to avoid shifting the balance in nitrogen compounds toward reduced nitrogen. Compartmentation as another level of regulation of nitrate reduction was also confirmed when a high concentration (50 mm) of  was forced upon cut leaves, which resulted in nitrite excretion from S521D leaves, but not from wild type or plants with intermediate deregulated NR (Lea et al., 2004; Lillo et al., 2004).
was forced upon cut leaves, which resulted in nitrite excretion from S521D leaves, but not from wild type or plants with intermediate deregulated NR (Lea et al., 2004; Lillo et al., 2004).
Generally, deregulation of NR at the transcriptional level did not have much influence on metabolite levels, but (additional) deregulation at the posttranslational level resulted in profound changes of nitrogen metabolite levels.
MATERIALS AND METHODS
Plant Material
Plants were in the rosette stage and had approximately seven leaves when leaf samples were harvested. Seeds were provided by Unité de Nutrition Azotée des Plantes (UNAP), Institut National de la Recherche Agronomique (INRA), Versailles, France. Plants tested were Nicotiana plumbaginifolia (var. Viviani) and transgenic N. plumbaginifolia expressing the full-length coding sequence of the tobacco (Nicotiana tabacum) Nia2 gene (C1; Vincentz and Caboche, 1991). Del8 plants were similar to C1 with the exception that the expressed tobacco NR had a deletion of 168 bp in the 5′-end of the coding sequence (Nussaume et al., 1995), and S521D clones 5 and 7 had the tobacco Nia2 gene with Ser-521 mutated into Asp (Lillo et al., 2003). Generally, no difference was found between clones 5 and 7, and in the figures mean values are presented. In C1, Del8, and S521D, NR was constitutively expressed using the CaMV 35S promoter. All the NR activity detected in these transgenic plants is derived from transgene expression, as the innate NR gene is inactivated by a retrotransposon insertion (Leprince et al., 2001). Plants were grown in a 12-h photoperiod (150 μmol m−2 s−1) at 25°C and 12-h dark period at 18°C. In the experiments for testing circadian rhythms, plants were grown at 20°C with a 12-h photoperiod at 80 μmol m−2 s−1, before being kept in constant conditions at the same temperature and light intensity. Plants were irrigated with Hoagland solution containing 15 mm KNO3 three times a week.
Plants used in 15N-labeling experiments were germinated and first grown in vitro as described previously (Vaucheret et al., 1992). When seedlings had two to three leaves, they were first transferred on sand and grown later in hydroponic conditions in a complete nutrient solution (Quilleré et al., 1994; Lejay et al., 1997) and 8 h of light and 16 h of dark (light intensity 150 μmol m−2 s−1). The nutrient solution was replaced every 2 or 3 d. Air was bubbled through the solution to ensure aerobic conditions. Three weeks after the transfer to hydroponic conditions, plants were selected for uniformity. Plants were used approximately when the seven-leaf rosette stage was reached.
Nitrate Measurements
Aliquots of 10 mg of freeze-dried powder were extracted successively with 400 μL of 80% ethanol (v/v in water), 200 μL of 80% ethanol, 400 μL of 50% ethanol, and finally with 400 μL of water at 80°C for 15 min. After centrifugation at 15,000 rpm for 5 min, the four successive extractions were pooled and homogenized. An aliquot (1 mL) of the supernatant was then evaporated in a Speed-Vac and resuspended in 0.5 mL of water for nitrate determination. Nitrate concentrations were determined by HPLC (Dionex analyzer, DX-120, HPLC AS14 column, 3.5 mm Na2CO3, and 1 mm NaHCO3 eluant). Peaks were identified and quantified in an integrator (peak-net station) by comparison with standard nitrate concentrations (10 μm to 1 mm).
15N-NO3− Measurements
Plants were transferred to a nutrient solution containing 1 mm 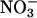 1 week before the 15N-labeling experiment started. 15N labeling was initiated at the light-to-dark transition by transferring the plants to a nutrient solution obtained by the addition of an aliquot of 10 mm K15NO3 (99 atom % 15N) to the solution containing
1 week before the 15N-labeling experiment started. 15N labeling was initiated at the light-to-dark transition by transferring the plants to a nutrient solution obtained by the addition of an aliquot of 10 mm K15NO3 (99 atom % 15N) to the solution containing  , giving a final concentration of 1 mm
, giving a final concentration of 1 mm  . The exact 15N enrichment of
. The exact 15N enrichment of 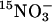 in the nutrition solution (about 5 atom % 15N) was determined from an aliquot sampled 15 min after the addition of K15NO3. Plants were harvested 3 and 16 h into the dark period. Leaves and roots were weighed before freezing. Plant material was freeze dried and ground, and 15N analysis of total nitrogen and
in the nutrition solution (about 5 atom % 15N) was determined from an aliquot sampled 15 min after the addition of K15NO3. Plants were harvested 3 and 16 h into the dark period. Leaves and roots were weighed before freezing. Plant material was freeze dried and ground, and 15N analysis of total nitrogen and  pools was performed as previously described (Lejay et al., 1997). The amount of nitrate reduced was calculated by subtracting the amount of 15N stored as nitrate from the total amount of 15N taken up.
pools was performed as previously described (Lejay et al., 1997). The amount of nitrate reduced was calculated by subtracting the amount of 15N stored as nitrate from the total amount of 15N taken up.
Extraction and Assay of NR Activity
Leaves (1 g) were homogenized with 4 mL of 0.1 m HEPES-KOH, pH 7.5, 3% (w/v) polyvinylpolypyrrolidone, 1 mm EDTA, and 7 mm Cys. The assay mixture contained 50 mm HEPES-KOH, pH 7.5, 100 μm NADH, 5 mm KNO3 with 2 mm EDTA or 6 mm MgCl2. The assay volume was 0.70 mL. Activity was measured in crude extracts by determining 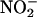 formation by the addition of 1% sulfanilamide and 0.02% N-(1-naphtyl)-ethylene-diamine dihydrochloride in 1.5 m HCl. Activity state is defined as NR assayed in the presence of Mg2+ (and 14-3-3 proteins) as percentage of NR activity measured in the presence of EDTA, and reflects how much of the enzyme is in the nonphosphorylated active form. Assays were run at 25°C.
formation by the addition of 1% sulfanilamide and 0.02% N-(1-naphtyl)-ethylene-diamine dihydrochloride in 1.5 m HCl. Activity state is defined as NR assayed in the presence of Mg2+ (and 14-3-3 proteins) as percentage of NR activity measured in the presence of EDTA, and reflects how much of the enzyme is in the nonphosphorylated active form. Assays were run at 25°C.
Amino Acid and Ammonium Analyses
The concentrations of amino acids were measured on a JEOL Aminotac JLC-500/V as recommended by the manufacturer using water-ethanol extraction and after ninhydrin derivatization. Free  was extracted in a 2% solution of 5-sulfosalicylic acid (50 mg fresh weight mL−1) and determined by the phenol hyperchlorite colorimetric method (Berthelot reaction) using (NH4)2SO4 as a reference.
was extracted in a 2% solution of 5-sulfosalicylic acid (50 mg fresh weight mL−1) and determined by the phenol hyperchlorite colorimetric method (Berthelot reaction) using (NH4)2SO4 as a reference.
RT-PCR
Total RNA was isolated by the RNeasy kit (Qiagen). DNA impurities in the isolated RNA were digested before synthesizing the cDNA by adding DNase (2 μL RQ Dnase; Promega) to 1 μg RNA, 2.5 μL RQ1 buffer, and 0.5 μL rRNAsin ribonuclease inhibitor (Promega) in a total volume of 23 μL and incubated for 30 min at 37°C. DNase was then inactivated by adding 2 μL of 25 mm EDTA and incubated for 10 min at 65°C before diluted 1:1. Right primers (0.4 μm) were added to 20 and 40 ng RNA, heated for 5 min at 70°C, and immediately transferred to ice. Reverse transcriptase (Moloney murine leukemia virus; reverse transcriptase; Promega) 200 units, rRNAsin 0.5 μL, nucleotides, and buffer according to the manufacturer's instructions (Promega) were added to a total of 25 μL. The mixture was incubated at 42°C for 60 min. The synthesized cDNA was diluted 1:1 and frozen at 20°C. cDNA, 2.5 and 5.0 μL, was added into a volume of 25 μL with 0.5 units Hot Star Taq polymerase (Qiagen), and specific primers picking up the tobacco (Nia-2; right, GAGTCAGAGGTGTAACCAGT; left, CCCTCAATACTCAACCCGAGA), truncated (right, GACAATTTTCTTCTGCACACC; left, ACGACGATGATGATGACGAA), and elongation factor-1α (EF-1α; right, AAGAGCTTCGTGGTGCATCT; left, GATGCTACCACCCCCAAGTA) genes were added at 0.2 μm and amplified according to the manufacturer's instructions. PCR primers amplified products of 498, 334, and 404 bp, respectively. Annealing temperature was 50°C. After 28 and 32 PCR cycles, the products were separated on 1% agarose gel containing 0.5 μg mL−1 ethidium bromide, and digital in-gel analysis was made with a GelDoc 2000 (Bio-Rad) using Quantity One 4.0.3 software. Data presented are based on nonsaturating conditions for amplification of the transcripts and presented relative to the control gene EF-1α.
Acknowledgments
We thank Pascal Tillard and Alain Gojon for advice and for performing the mass spectrophotometric measurements of 15N.
This work was supported by the Norwegian Research Council and the Marie Curie host fellowship QLK–3–CT–2002–60058 at the Unité de Nutrition Azotée des Plantes (to U.S.L.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Christian Meyer (meyer@versailles.inra.fr) and Cathrine Lillo (cathrine.lillo@uis.no).
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074633.
References
- Deng M-D, Moureaux T, Cherel I, Boutin J-P, Caboche M (1991) Effects of nitrogen metabolites on the regulation and circadian expression of tobacco nitrate reductase. Plant Physiol Biochem 29: 239–247 [Google Scholar]
- Foyer CH, Lescure J-C, Lefebvre C, Morot-Gaudry J-F, Vincentz M, Vaucheret H (1994) Adaptation of photosynthetic electron transport, carbon assimilation, and carbon partitioning in transgenic Nicotiana plumbaginifolia plants to changes in nitrate reductase activity. Plant Physiol 104: 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galangau F, Daniel-Vedele F, Moureaux T, Dorbe M-F, Leydecker M-T, Caboche M (1988) Expression of leaf nitrate reductase genes from tomato and tobacco in relation to light-dark regimes and nitrate supply. Physiol Plant 88: 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua N-H (1990) Molecular light switches for plant genes. Plant Cell 2: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Gojon A, Dapoigny L, Lejay L, Tillard P, Rufty TW (1998) Effects of genetic modifications of nitrate reductase expression on
 uptake and reduction in Nicotiana plants. Plant Cell Environ 21: 43–53 [Google Scholar]
uptake and reduction in Nicotiana plants. Plant Cell Environ 21: 43–53 [Google Scholar] - Huber SC, MacKintosh C, Kaiser WM (2002) Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol Biol 50: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Klein D, Morcuende R, Stitt M, Krapp A (2000) Regulation of nitrate reductase expression in leaves by nitrate and nitrogen metabolism is completely overridden when sugars fall below a critical level. Plant Cell Environ 23: 863–871 [Google Scholar]
- Lam E, Chua N-H (1989) ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langendorfer RL, Watters MT, Smarrelli J Jr (1988) Metabolite control of squash nitrate reductase. Plant Sci 57: 119–125 [Google Scholar]
- Lea US, ten Hoopen F, Provan F, Kaiser WM, Meyer C, Lillo C (2004) Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in high nitrite excretion and NO emission from leaf and roots tissue. Planta 219: 59–65 [DOI] [PubMed] [Google Scholar]
-
Lejay L, Quilleré I, Roux Y, Tillard P, Cliquet J-B, Meyer C, Morot-Gaudry J-F, Gojon A (1997) Abolition of post-transcriptional regulation of nitrate reductase partially prevents the decrease in leaf
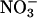 reduction when photosynthesis is inhibited by CO2 deprivation, but not in darkness. Plant Physiol 115: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
reduction when photosynthesis is inhibited by CO2 deprivation, but not in darkness. Plant Physiol 115: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar] - Leprince A-S, Grandbastien M-A, Meyer C (2001) Retrotransposons of the Tnt1B family are mobile in Nicotiana plumbaginifolia and can induce alternative splicing of the host gene upon insertion. Plant Mol Biol 47: 533–541 [DOI] [PubMed] [Google Scholar]
- Lillo C (1984) Optimization of assay conditions and growing temperature in studies of diurnal variations in the nitrogen metabolism of barley leaves. Meldinger fra Norges landbrukshøgskole 63: 1–17 [Google Scholar]
- Lillo C (1991) Diurnal variations of corn leaf nitrate reductase: an experimental distinction between transcriptional and post-transcriptional control. Plant Sci 73: 149–154 [Google Scholar]
- Lillo C, Lea US, Leydecker M-T, Meyer C (2003) Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in constitutive activation of the enzyme in vivo and nitrite accumulation. Plant J 35: 566–573 [DOI] [PubMed] [Google Scholar]
- Lillo C, Meyer C (2001) Biological clocks and the nitrate reductase oscillating system. Biol Rhythm Res 32: 489–500 [Google Scholar]
- Lillo C, Meyer C, Lea US, Provan F, Oltedal S (2004) Mechanism and importance of post-translational regulation of nitrate reductase. J Exp Bot 55: 1275–1282 [DOI] [PubMed] [Google Scholar]
- Lillo C, Meyer C, Ruoff P (2001) The nitrate reductase circadian system. The central clock dogma contra multiple oscillatory feedback loops. Plant Physiol 125: 1554–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh C (1992) Regulation of spinach-leaf nitrate reductase by reversible phosphorylation. Biochim Biophys Acta 1137: 121–126 [DOI] [PubMed] [Google Scholar]
- MacKintosh C, Douglas P, Lillo C (1995) Identification of a protein that inhibits the phosphorylated form of nitrate reductase from spinach (Spinach oleracea) leaves. Plant Physiol 107: 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt P, Schurr U, Klein D, Krapp A, Stitt M (1998) Growth of tobacco in short-day conditions leads to high starch, low sugars, altered diurnal changes in the Nia transcript and low nitrate reductase activity, and inhibition of amino acid synthesis. Planta 207: 27–41 [DOI] [PubMed] [Google Scholar]
- Meyer C, Lea US, Provan F, Kaiser WM, Lillo C (2005) Is nitrate a major player in the plant NO (nitric oxide) game? Photosynth Res 83: 181–189 [DOI] [PubMed] [Google Scholar]
- Meyer C, Stitt M (2001) Nitrate reduction and signalling. In PJ Lea, JF Morot-Gaudry, eds, Plant Nitrogen. Springer-Verlag, Berlin, pp 39–59
- Nussaume L, Vincentz M, Meyer C, Boutin J-P, Caboche M (1995) Post-transcriptional regulation of nitrate reductase by light is abolished by an N-terminal deletion. Plant Cell 7: 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilleré I, Dufossé C, Roux Y, Foyer CH, Caboche M, Morot-Gaudry JF (1994) The effects of deregulation of NR gene expression on growth and nitrogen metabolism of Nicotiana plumbaginifolia plants. J Exp Bot 45: 1205–1211 [Google Scholar]
- Reed AJ, Canvin DT, Sherrard JH, Hageman RH (1983) Assimilation of [15N]nitrate and [15N]nitrite in leaves of five plant species under light and dark conditions. Plant Physiol 71: 291–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar S, Oaks A (1995) Regulation of nitrate reductase during early seedling growth. Plant Physiol 107: 1225–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W-R, Gonzáles-Fontes A, Morcuende R, Lauerer M, Geiger M, Glaab J, Gojon A, Shculze E-D, Stitt M (1997) Tobacco mutants with a decreased number of functional nia genes compensate by modifying the diurnal regulation of transcription, post-translational modification and turnover of nitrate reductase. Planta 203: 304–319 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Krapp A, Stitt M (2000) Reciprocal diurnal changes of phosphoenolpyruvate carboxylase expression and cytosolic pyruvate kinase, citrate synthase and NADP-isocitrate dehydrogenase expression regulate organic acid metabolism during nitrate assimilation in tobacco leaves. Plant Cell Environ 23: 1155–1167 [Google Scholar]
- Stitt M, Müller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible W-R, Krapp A (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Su W, Huber SC, Crawford NM (1996) Identification in vitro of a post-translational regulatory site in the hinge 1 region of Arabidopsis nitrate reductase. Plant Cell 8: 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker DE, Allen DJ, Ort DR (2004) Control of nitrate reductase by circadian and diurnal rhythms in tomato. Planta 219: 277–285 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Marion-Poll A, Meyer C, Faure J-D, Marin E, Caboche M (1992) Interest in and limits to the utilization of reporter genes for the analysis of transcriptional regulation of nitrate reductase. Mol Gen Genet 235: 259–268 [DOI] [PubMed] [Google Scholar]
- Vincentz M, Caboche M (1991) Constitutive expression of nitrate reductase allows normal growth and development of Nicotiana plumbaginifolia plants. EMBO J 10: 1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz M, Moureaux T, Leydecker M-T, Vaucheret H, Caboche M (1993) Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J 3: 315–324 [DOI] [PubMed] [Google Scholar]
- Wang YH, Duff SMG, Lepiniec L, Cretin C, Sarath G, Condon SA, Vidal J, Gadal P, Chollet R (1992) Site-directed mutagenesis of the phosphorylated serine (Ser 8) in C4-phosphoenolpyruvate carboxylase from sorghum—the effect of negative charge at position-8. J Biol Chem 267: 16759–16762 [PubMed] [Google Scholar]
- Warner RL, Kleinhofs A (1981) Nitrate utilization by nitrate reductase-deficient barley mutants. Plant Physiol 67: 740–743 [DOI] [PMC free article] [PubMed] [Google Scholar]