Abstract
We have developed the software package Tomato and Potato Assembly Assistance System (TOPAAS), which automates the assembly and scaffolding of contig sequences for low-coverage sequencing projects. The order of contigs predicted by TOPAAS is based on read pair information; alignments between genomic, expressed sequence tags, and bacterial artificial chromosome (BAC) end sequences; and annotated genes. The contig scaffold is used by TOPAAS for automated design of nonredundant sequence gap-flanking PCR primers. We show that TOPAAS builds reliable scaffolds for tomato (Solanum lycopersicum) and potato (Solanum tuberosum) BAC contigs that were assembled from shotgun sequences covering the target at 6- to 8-fold coverage. More than 90% of the gaps are closed by sequence PCR, based on the predicted ordering information. TOPAAS also assists the selection of large genomic insert clones from BAC libraries for walking. For this, tomato BACs are screened by automated BLAST analysis and in parallel, high-density nonselective amplified fragment length polymorphism fingerprinting is used for constructing a high-resolution BAC physical map. BLAST and amplified fragment length polymorphism analysis are then used together to determine the precise overlap. Assembly onto the seed BAC consensus confirms the BACs are properly selected for having an extremely short overlap and largest extending insert. This method will be particularly applicable where related or syntenic genomes are sequenced, as shown here for the Solanaceae, and potentially useful for the monocots Brassicaceae and Leguminosea.
An established strategy to determine the sequence content of target genomes involves large insert clones that are physically mapped into contigs spanning the target of interest, and which are used for shotgun library construction and high-throughput sequencing. Many aspects concerning the clone-by-clone whole-genome sequencing strategy in literature have been addressed, and although much progress has been made in developing this strategy, key steps are the subject of continued evaluation and improvement. Here we present results on the Centre for Biosystems Genomics initiative to sequence tomato chromosome 6 of Solanum lycopersicum cv Heinz 1706 by a clone-by-clone sequencing approach and to establish a resistance gene homolog profiling for the potato (Solanum tuberosum) genome. In this paper we particularly focus on selecting bacterial artificial chromosomes (BACs) for walking and finishing.
The condition of having large insert clones available was fulfilled by Budimann et al. (2000), who constructed a HindIII BAC library for cultivated tomato cv Heinz 1706, covering the target with approximately 15 genome equivalents, and recently with an MboI and an EcoRI BAC library that the United States' part of the International Solanaceae Project (SOL) has made available (Mueller et al., 2005b). A key step in clone-by-clone whole-genome sequencing is determining a reliable minimal-tiling path. This strategy depends on the availability of a high quality physical map. An established approach for map construction involves DNA fingerprinting. With fingerprinting, overlapping clones are identified by determining a pattern of shared bands produced from restriction enzyme analysis, which is indicative for the physical overlap. Owing to its simplicity and low initial costs, often agarose separation and staining is used for detection of bands. A combinatorial comparison of fingerprints through automated physical map assembly software, e.g. FingerPrinted Contigs (FPC), is applied for map construction (Soderlund et al., 1997, 2000). However, low resolution separation, errors in detection and size estimation of separated fragments, uncalibrated FPC parameter settings for size tolerance, and inaccurate probability cutoff scores, cause false negative scoring results, creating gaps in the physical map and resulting in a higher amount of singletons, and false positives creating chimeric contigs (for review, see Meyers et al., 2004). Compared to agarose separation, amplified fragment length polymorphism (AFLP) fingerprinting is a high-resolution separation technique, and this allows for more precise fragment size estimation. Typically 50 to 100 restriction fragments in the range from 50 to 500 nucleotides can be detected (Vos et al., 1995). Budimann et al. (2000) have proposed a sequence-tagged connector (STC) framework for more precise selection of minimally overlapping tomato BAC clones to support whole-genome sequencing of the tomato genome. The selection strategy originally proposed by Venter et al. (1996) involves a fingerprint analysis and BAC end sequencing, which is used in combination with genetically anchored seed BACs that are completely sequenced. Recently a large number of tomato BAC end sequences have been made available by the Solanaceae Genome Network (SGN) for the sequencing community, and these developments make it possible to pursue the STC approach using high-density fingerprints.
Upon selection of fingerprinted BACs, determining the sequence content is the next important step in rebuilding the genomic content of targets. The method most commonly used for genomic DNA sequencing is shotgunning. The sample DNA is randomly sheared into small fragments and cloned into appropriate sequencing vectors. With double-barreled shotgun sequencing, small insert clones are sequenced from both insert ends, producing read pairs or mates. The aim is to cover the target of interest and to reduce the number of sequence gaps between contigs by producing a sufficient amount of sequences from which a reliable consensus can be determined upon assembly. Theoretically, following Poisson distribution rules, the probability for bases not being sequenced leaving sequence gaps reduces with an increase of coverage, as outlined by Lander and Waterman (1988), although cloning bias causes a nonrandom distribution leading to nonsequenced areas regardless of coverage. Uncovered areas are usually rescued by PCR, using custom-designed primers and templates spanning the sequence gap. For tomato and potato BAC sequencing we focus on 6-fold coverage, aiming for a limited and balanced demand of resources. However, low coverage will leave assemblies more incomplete and will demand a dedicated input for the assembly finishing phase. While sequencing and computer technology have facilitated the automated processing and assembly of large amounts of shotgun sequence data, the finishing of contig sequences is a time-consuming process, and needs expert knowledge to evaluate base calls, design primers for gap closure, and untangle complex sequences that obstruct a proper assembly. To compensate for the human input required to finish low-covered BACs, we aim to automate local assembly verification, contig linking, and gap closure.
Several tools for contig linking and gap closure have been presented in the past. Among those, prokaryotic genome assembly assistance system, which was developed to automate contig ordering and gap closure for prokaryotic cyanobacterial genome assembly by finding possible links for Synechococcus contigs with known protein sequences coming from closely related Synechocystis sp. (Yu et al., 2002), using local sequence homology-based searches with BLASTX (Altschul et al., 1990). Finding contig links by BLASTX homology searches depends on gene distribution in the target genome. For tomato, the regions near the centromeric region have the lowest gene density with 15 to 17 kb per gene, while the euchromatin has a gene density of approximately 7 kb. Analysis of sequenced tomato BACs reveal a gene density with an average of 10 kb per gene (Van der Hoeven et al., 2002). Bacterial genomes in general do not contain introns and have a higher gene density compared to eukaryotic plant genomes. Therefore, finding corresponding putative functions on sequences from higher eukaryotic plant origin for gapped assemblies will be more difficult. Additional linkage information might be obtained through comparative genomics. Solanaceae members like tomato and potato share a conserved colinearity between their genomes (Bonierbale et al., 1988). The genomic sequence information from Solanaceae is, however, scarcely available. From studies to analyze gene content and organization though, a large collection of single-pass expressed sequence tags (ESTs) from tomato cDNA have become available (Van der Hoeven et al., 2002) and this opens the possibility for genome-wide comparative studies.
In addition to existing database information, a powerful data source for contig scaffolding and inherent to the double-barreled shotgun sequencing approach, is the assembly position of a sequence read constraint by the assembly position and direction of its mate pair. This information can be used to both relatively position contigs and to solve local assembly problems. Reconstruction of target sequences is often complicated by repeats, resulting in collapsed assemblies. To resolve these phenomena, a tool that reports on violation of direction and size constraints will help to determine contig quality. We report here the development of a Tomato and Potato Assembly Assistance System (TOPAAS) that uses homology-based searches, comparative alignments, read pair information, and high-density AFLP fingerprint data to link contigs, verify assemblies, and select minimal overlapping BACs.
RESULTS
Dataflow and Output
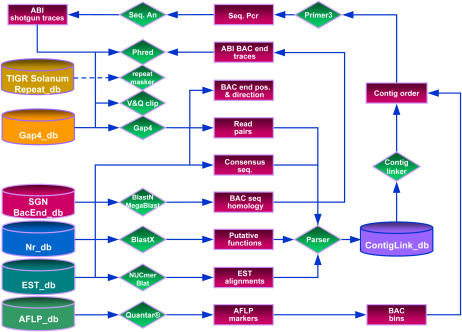
The main purpose of TOPAAS is to automate key steps in the clone-by-clone sequencing approach. Its tasks are to find contig link information for gapped assemblies resulting from low-coverage sequencing, to analyze the assembly integrity, and to assist the selection of overlapping BAC clones for a subsequent sequence walk. To that end we have built a system that extracts read pair information, carries out homology-based searches, and analyzes this information according to user-defined settings. A schematic representation of the TOPAAS pipeline and dataflow is shown in Figure 1. TOPAAS visualizes the link analysis and presents the user with detailed information on type, order, and number of links (see Fig. 2).
Figure 1.
Schematic overview of the dataflow used in this study. Red-colored rectangles represent datasets, databases are depicted as bins, and applications are drawn as green-colored diamonds. Direction of dataflow is indicated by blue-colored connectors. The dashed blue-colored connector represents an additional step that can be included for repeat masking. For processing raw trace data we rely on PREGAP4 of the Staden package (Bonfield et al., 1995), which is flexible in interfacing a diverse set of tools for base calling, vector clipping, repeat masking, and assembly. In this study we have used PHRED base calling and GAP4 assemblies. From the GAP4 database, consensus sequences and assembly positions are extracted, uploaded, and used by TOPAAS for BLASTX, MUMmer, and BLAT analyses. The system also searches a BAC end database with BLASTN or MegaBlast against consensus sequences. To verify quality, overlap, and direction, corresponding BAC end traces are processed and assembled onto contig sequences. Candidate BAC clones are used for AFLP fingerprint analyses. Comigrating fragments are used to deduce the binning of BACs. Read pair information, BLAST scores, EST alignments, and BAC end positions are parsed into the ContigLinkdb. TOPAAS analyzes the data in ContigLinkdb on a project level and predicts contig links and minimal overlapping BAC clones. BAC binning information is then used for extended contig ordering and selection of minimal overlapping BACs. The primer module part designs nonredundant primers, which are then subsequently used for sequence PCR analysis and gap closure.
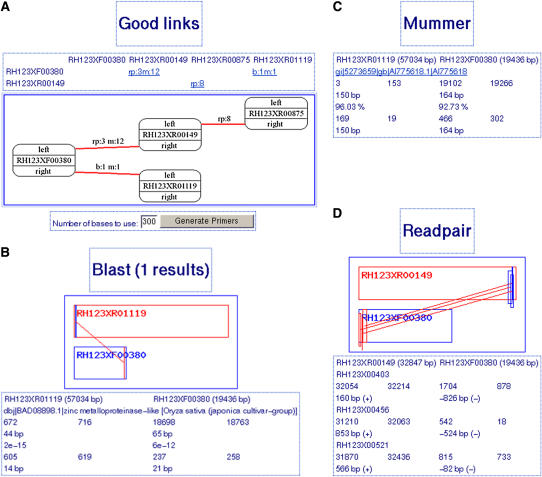
Figure 2.
Typical view of a TOPAAS link analysis output. A, For potato BAC RH123P09 a predicted contig order, gap-flanking read pairs (rp:), gap-spanning MUMs (m:), and contig bridging BLAST alignment (b:) between pairs of contigs are shown and provide a link to more detailed output for BLAST linkage (B), EST alignments (C), and read pairs (D), described in terms of position, length, direction, percentage identity, and E-score. The number of links per link type is indicated behind the colon separator.
TOPAAS provides a web front end in PHP for uploading assembly data and contig sequences, setting alignment constraints and average insert sizes for shotgun libraries. Homology-based alignments can be uploaded manually or provided by TOPAAS via two automated BLASTs. TOPAAS aligns contigs against the nonredundant sequence database from the National Center for Biotechnology Information (NCBI) and against the BAC end sequence database from SGN. The system also carries out a MUMmer (Delcher et al., 2002) or a BLAT (Kent, 2002) alignment against Solanaceae ESTs. Together with the homology-based alignment results, read pair positions and directions are parsed into MySQL tables comprising the TOPAAS database (for an overview of the TOPAAS table scheme, see Supplemental Fig. 1). The actual link analysis is started from a web front end and is carried out by the ContigLinker that queries the TOPAAS database. First the system retrieves and filters hits on cutoff for percentage identity or e-value score. We separated the filtering step from the alignment program filtering options to enable linkage analysis using variable cutoff scores without the need to perform additional homology searches. Next TOPAAS matches identical database accession numbers from EST and BLASTX hits. Subsequently, the system outputs a linkage analysis on the fly rather than storing the analysis. TOPAAS tracks down read pairs both within and between contigs. Violations against direction and spacing constraints point toward possible local assembly problems, and inconsistent read pairs are reported to the editor for extraction and reassembly. Via the web interface primer design constraints can be manipulated and the system will output unique primer pair combinations for sequence gap closing purposes (Supplemental Fig. 2).
The automated BLASTN analysis of contigs against the BAC end sequence database is used for high-throughput screening and rapid preselection of candidate BACs, having a sequence overlap with seed BACs. The single-pass BAC end sequences are reassembled onto the seed BAC consensus. Base pair inconsistencies are edited to exclude high quality base call mismatches and the position of a nearby cloning site upstream of the BAC end sequence start position is verified. When meeting constraints, corresponding BACs are then selected for further analysis with high-density AFLP fingerprinting. The reassembly of BAC ends and AFLP fingerprinting analysis is carried out independently from TOPAAS.
Selection of Tomato BACs for Sequence Walking
Sequence Homology-Based Searches
To examine whether a STC approach with a nonselective AFLP fingerprinting can support the tomato BAC walking, we selected P250I21 and P046G10 from an initial set of tomato seed BACs for sequencing. P250I21 is assembled to full closure, whereas the assembly of P046G10 is gapped (Table I). Different lines of evidence indicate these BACs originate from tomato chromosome 6. Fluorescent in situ hybridization analysis shows P250I21 and P046G10 are located on the short and the long arm of chromosome 6, respectively (for an overview, see http://sgn.cornell.edu/cgi-bin/cview/map.pl?map_id=13). Furthermore, the chromosome 6 known functional gene Mi marker, which has been used as a probe in an overgo plating analysis, shows plausible associations to P250I21. In addition, P112G05 has been associated to the Mi marker and has been assigned to a chromosome 6 FPC contig (for details, see http://www.genome.arizona.edu/fpc/WebAGCoL/tomato/WebFPC/ and http://www.sgn.cornell.edu/cgi-bin/search/direct_search.pl?search=bacs). No FPC data is available for P250I21. However, AFLP mapping shows both BACs coassemble (see also Fig. 5), and upon sequencing we have found a 60-kb overlap between P112G05 and P250I21 (for BAC sequences, see ftp://ftp.sgn.cornell.edu/tomato_genome/bacs/chr06). Gene prediction with Genscan or GlimmerM and subsequent BLASTX analysis reveal the repetitive nature of the P250I21 insert sequence, and five separate putative genes show hits to Mi gene homologs (data not shown). These lines of evidence suggest P112G05 and P250I21 originate from overlapping locations on chromosome 6.
Table I.
Link analysis by TOPAAS for potato and tomato BACs
Tomato BAC IDs are indicated with a prefix P and potato BACs have a prefix RH or SH. For each BAC the insert size and the amount of contigs remaining after shotgun assembly are shown. Linkage result is represented by the number of contig pairs linked with gap-flanking read pairs (R), gap-bridging BLASTX hits (B), EST gap-spanning alignments (E), and combinations thereof. The gap closure for each link type per BAC is indicated between parentheses. The closing efficiency is shown as the number of closed gaps over the number of predicted contig links per BAC. Link analysis was not determined (n.d.) for P250I21 and P046G10.
| BAC ID
|
Size
|
Contigs
|
TOPAAS Links
|
Gaps/Links
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R | B | E | RB | RE | BE | RBE | ||||
| kb | ||||||||||
| RH123P09 | 131 | 4 | 1 (1) | 0 | 0 | 0 | 1 (1) | 1 (1) | 0 | 3/3 |
| SH196 | 72 | 11 | 10 (9) | 0 | 0 | 0 | 0 | 0 | 0 | 9/10 |
| RH011D17 | 132 | 6 | 2 (2) | 2 (2) | 1 (1) | 0 | 0 | 0 | 0 | 5/5 |
| P073H07 | 130 | 18 | 7 (4) | 0 | 0 | 1 (1) | 1 (1) | 0 | 1 (1) | 7/10 |
| P103N18 | 105 | 6 | 4 (4) | 0 | 0 | 1 (1) | 0 | 0 | 0 | 5/5 |
| P250I21 | 148 | 1 | – | – | – | – | – | – | – | n.d. |
| P046G10 | 90 | 8 | – | – | – | – | – | – | – | n.d. |
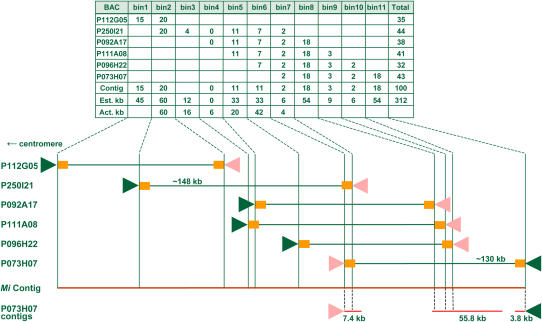
Figure 5.
BAC bins and physical map of the Mi contig. Each bin is defined as a domain in which a set of AFLP markers is shared between BACs. The number of comigrating fragments indicated in the top table is used to estimate the order and size of the overlap. For each shared fragment observed in gel an overlap portion of 3 kb is assumed. The assembly positions of BAC end sequences (orange squares) flanking the T7 (triangle pointing right) and SP6 (triangle pointing left) region on the consensus of P250I2 have been used to calculate actual overlap sizes. BAC end sequences assembled onto P073H07 contigs (horizontal lines) have been used as BAC end sequence tags for extended ordering.
We first searched the contig sequences of P250I21 and P046G10 with TOPAAS against the BAC end database from SGN, containing 75,000 to 126,000 BAC end sequences from a HindIII and an MboI library depending on the time of screening. The raw BLASTN output was converted into html format to provide for a complete overview of hits (Fig. 3; Supplemental Fig. 3). We frequently observe individual seed BAC domains hit by multiple BAC ends. Such can be the result of a repetitive domain within the genome. In addition it may reflect also a redundancy in the BAC library. Indeed, e.g. around the 30-kb position from the start of P250I21, a putative gene predicted by Genscan shows a BLASTX homology against a putative retroelement polyprotein from Arabidopsis (Arabidopsis thaliana) and a hypothetical protein from the wild cabbage (Brassica capitata) transposon Melmoth. Transposable elements account for at least 10% of the Arabidopsis genome and are well represented in other plant genomes as well and most likely also in Solanaceae genomes (Arabidopsis Genome Initiative, 2000). Consistent with this notion is the BLASTX analysis of BAC ends from P005D08, P110K11, P122M05, and P166M18, which hit in the 30-k region of P250I21 and show homology against putative retroelement polyproteins found in potato, Arabidopsis, Solanum demissum, and Oryza sativa. Also we observe single BAC ends hitting with multiple high-scoring pairs. The latter reflects a repetitive sequence present within the seed BAC. For P250I21, Mi homologous sequences around nucleotide positions 95, 110, and 135 kb contribute to this phenomenon. The repetitive nature is confirmed by the fact that BAC end sequence P006L20 shows a BLASTX homology against gene homolog Mi-copy 2 from Solanum esculentum hitting multiple Mi homologous domains in P250I21. By screening the position, direction, and significance level, we preselect for reasonable candidates having a single high-scoring pair against a seed BAC. Although we stringently filter for BAC end hits with a high e-value score, we frequently observe sequence discrepancies to seed BAC consensi, which have in general a lower error rate compared to consensi of single-passed BAC ends. To exclude false-positive scoring, corresponding trace files of BAC ends are examined by assembling them onto seed BAC consensus sequences. For P250I21, four BAC end sequences align consistently. From the BLAST hit and assembly positions an overlap order is deduced (see Fig. 4; Supplemental Fig. 4). Of those alignments, a 768-nucleotide overlap of BAC end sequence P073H07 runs from position 3,996 to 3,228 with a HindIII site starting at P250I21 coordinate 4,016. We find the shortest potential overlap to be 4 kb between P250I21 and P073H07. Taking into account the insert sizes and overlaps, the Mi contig has a spanning distance of approximately 320 kb. For seed BAC P046G10, seven BAC ends align consistently and have been used as sequence tags for ordering purposes. P046G10 contig end sequences adjacent to the T7 and SP6 side of the BAC cloning vector have been identified by assembly of LE_HBa_046G10-SP6 and LE_HBa_046G10-T7 traces and tagged accordingly. An overlap of 720 nucleotides with BAC P103N18 starts at 2,158 nucleotides from the P046G19 insert end, running toward the SP6 region. A HindIII site is positioned 3 nucleotides upstream from the start of the overlap. We determined the minimal potential overlap to be 2.1 kb between P046G10 and P103N18 with a total spanning distance of approximately 205 kb (Supplemental Fig. 5).
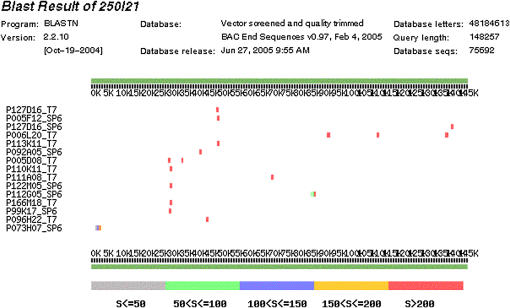
Figure 3.
Schematic representation of a BLASTN analysis of tomato BAC P250I21 against the SGN BAC end sequence database. The linear sequence of P250I21 is represented by horizontal green bars running from position 1 at the left site to position 148,257 at the right site. Each BAC end hit is marked with a tick and positioned according to homologous 250I21 coordinates. The 15 most significant hits are displayed. Ticks are color coded to indicate the level of significance (bottom bar). At the left side the BAC ID is indicated, of which P073H07 is the most left-positioned BAC end hit.
Figure 4.
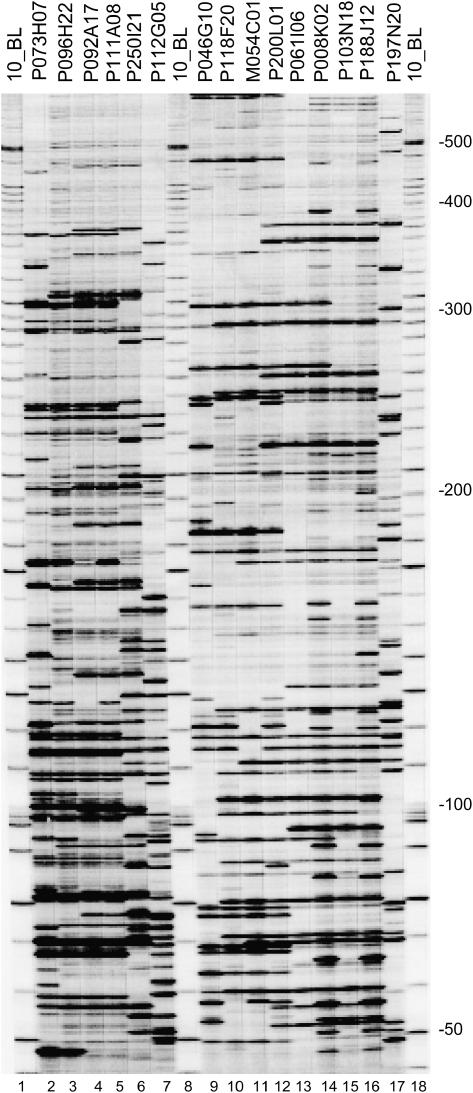
AFLP fingerprints from chromosome 6 tomato BACs. Section I, lanes 2 to 6, contain fingerprints for BACs from the Mi contig. Section II, lanes 9 to 16, contain fingerprints for BACs from contig P103. All fingerprinted BACs originate from a HindIII BAC library except for lane 12, which was pulled from an MboI library. For all fingerprints EcoRI/MseI + 0/+ 0 primer combinations have been used where +0 indicates the absence of selective nucleotides. Lanes 1, 8, and 18 contain a 10-bp size marker. The Mr size range of the fingerprints is between 50 and 500 nucleotides and is indicated at the right side. BACs used for fingerprints are indicated at the top.
High-Density Nonselective AFLP Fingerprinting of Tomato BACs
To investigate the relation between BACs over a larger extent we analyzed AFLP EcoRI/MseI + 0/+ 0 fingerprints by determining the number of comigrating fragments between BACs (Fig. 4), and comparing their sizes with an in silico EcoRI/MseI digest obtained from the seed BAC consensus sequence. From the combinatorial comparison of comigrating fragments, the bins for P250I21 (Fig. 5) and P046G10 (Supplemental Fig. 5) are constructed. In the Mi contig the smallest number of comigrating fragments is shared between P250I21 and P073H07 pointing to a minimal overlap. The other BACs in the Mi contig share a large amount of comigrating fragments, suggesting the overlap size with both P250I21 and P073H07 is considerably larger. The deduced order of BACs overlapping P250I21 is consistent with the BLAST hit positions, although we find a 6-kb extension of P111A8 compared with P092A17 (see Fig. 5). The in silico digest of P250I21 indicates two pairs of consecutive EcoRI/MseI restriction sites are present in this 6-kb domain. However, corresponding comigrating fragments couldn't be scored from gel (Fig. 4, lanes 5 and 6). Several phenomena might account for missing the detection of fragments. We cannot entirely rule out an excessively deviating gel migration behavior. Furthermore, similar sized fragments comigrating as a single band can mask each other and cause ambiguities when scoring fragments in gel. Isolation of fragments from gel and sequencing for positive identification would provide more insight, but is beyond the scope of this study and it will be addressed elsewhere. From experience we assume each fragment observed in gel corresponds to an overlap size of approximately 3 kb. In some instances the estimated overlap size per bin differs from the calculated size. Nevertheless, the overall estimated spanning distance is in agreement with the calculated overlap sizes for bin 1 to bin 5. Taken together these results make it unlikely P250I21 and P073H07 would share a small repeat and suggest the minimal overlap is authentic. Furthermore bin 1, bin 3, and bin 11 contain fragments unique to P112G05, P250I21, and P073H07, respectively, indicating these BACs make up for the largest spanning distance in the Mi contig.
The nature of the overlap is further investigated by shotgun-sequencing P073H07 and 103N18 and assembly onto the consensus of P250I21 and P046G10, respectively. Both P073H07 and P103N18 align without base inconsistencies, and the overlap start position is similar to that determined by BLAST. Furthermore the BAC end assembly positions and directions are in agreement with the mapping results (Supplemental Fig. 4). From these results we conclude to have identified P073H07 as optimal BAC for walking in terms of minimal overlap and largest extending insert. At the time of screening the same did hold true for BAC P103N18. Over time the sequencing community will be provided in total with some 400,000 BAC end sequences obtained from three different libraries (Mueller et al., 2005b). It is likely we will find new BAC candidates with even more favorable features for walking as BAC end sequence data accumulate. This is illustrated by candidate BAC P008K02, which we found later on in the screening process. This BAC has a larger extending insert, but also a larger overlapping portion with seed BAC P046G10 (Supplemental Fig. 5).
Linking of Tomato and Potato Contigs
To analyze the quality of the contig links predicted by TOPAAS, we have constructed an assembly data set from three potato BACs, which were pulled from two different libraries (Rouppe van der Voort et al., 1999; Huang et al., 2005) and two tomato BACs. A total of 21 potato contigs with 18 sequence gaps was obtained for three potato BACs and comprised a contig length of approximately 335 kb. For two tomato BACs P073H07 and P103N18 we obtained 24 contigs with a length of 235 kb. The type, number of links, and references to EST and BLAST matches between tomato and potato contigs was determined by TOPAAS as shown in Table I and Supplemental Table I. All potato BAC contigs have been linked, of which 13 out of 21 contigs are linked by read pairs. For tomato BACs, 17 contigs have been linked. For five contig pairs, 18 gap-spanning EST alignments have been found. P073H07 and RH123P09 have one contig pair, each linked by ESTs from both potato and tomato. One contig pair from RH123P09 has been linked with 12 ESTs from both tomato and potato (see Fig. 2). One contig pair from potato BAC RH11D17 has been linked with two potato ESTs. For six contig pairs, gap-bridging BLASTX hits have been found. One contig pair from RH123P09 showed a gap-spanning BLASTX alignment to a zinc finger-like protein (BAD08898) from O. sativa. Two contig pairs from RH011D17 are linked by BLAST hits against a Pto locus (AF220602) from Lycopersicum pimpinellifolium and a patatin A gene (S51460) from potato, respectively. A P103N18 contig pair shares a hit with a nodulin gene (AAC72337) from Glycine max. BLASTX hits against C3HC4-type zinc finger protein (B84710) from Arabidopsis, and a putative copia-like polyprotein (AAL68851) from Sorghum bicolor links two contig pairs from P073H07. The latter is a known repetitive element in Solanaceae, and TOPAAS may have linked the two contigs from P073H07 incorrectly. However, the contig pairs are also linked by a read pair. Additionally we checked the other contig sequences that were linked by bridging ESTs and BLAST hits against The Institute for Genomic Research Solanaceae Repeat Database at http://www.tigr.org/tdb/e2k1/plant.repeats. No hits were found against the repeat database. These findings suggest incorrect links through alignment to repetitive regions are not likely. For BAC SH196 links only via gap-flanking read pairs are found. In total, six pairs of contigs from RH123P09, P073H07, and P103N18 have the ordering based upon multiple link types, of which one contig pair for P073H07 was linked by a combination of an EST and BLASTX alignment, and a gap-spanning read pair. A typical html output of the linking analysis for RH123P09 by TOPAAS is given in Figure 2.
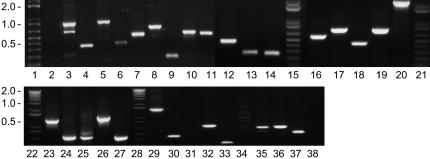
Subsequently, primers designed by TOPAAS on contig ends were used for PCR analysis on BAC template DNA in combinations according to the contig order predicted by TOPAAS. Figure 6 shows 29 out of 33 primer combinations producing amplicons. Amplified products do not exceed a length of 1 kb except for Figure 6, lane 20, which is well within the size limit for bridging. The PCR analysis shows all except one primer pair combination producing single amplicon products, indicating the primer annealing positions are unique and suggesting the primer redundancy check by TOPAAS to be reliable. PCR products have been sequenced and assembled to investigate the gap closure. In all instances, sequences derived from single amplicons (Fig. 6, lanes 4–38) are contig bridging and result in joins between contigs. Multiple amplicons from one primer pair combination were isolated separately, of which the larger product produced a gap-spanning sequence (Fig. 6, lane 3). Four out of 33 primer combinations failed to produce a PCR product, although contig pairs flanking the gaps are linked by read pairs (Fig. 6, lanes 2, 31, 34, and 38). In one instance gap-flanking sequences reveal a potential hairpin structure that probably obstructs a proper PCR (Fig. 6, lane 2). We redesigned PCR oligos at the 3′ site of both arms of the hairpin structure and adapted PCR conditions. The redesigned primers facilitated a proper PCR and produced a gap-closing sequence (data not shown). Thus using the contig ordering information from TOPAAS we are able to efficiently finish the potato BACs to full closure. Also tomato BAC P103N18 was closed, whereas for P073H07 we could not find sufficient links to complete closure. These results indicate the integrity of the contig order predicted by TOPAAS and the sufficient quality of the automatically designed primers for gap closure.
Figure 6.
Gap closure analysis for potato and tomato BAC contigs. Pairs of gap-spanning primers are used for PCR in combinations suggested by TOPAAS on two tomato and three potato BAC templates. Detection of agarose gel separated amplicons is used to determine the bridging efficiency. Lanes 1, 15, 21, 22, and 28: 1 kb + size marker (InvitroGen). PCR products produced from potato BAC templates SH196 (lanes 2–11), RH123P09 (lanes 12–14), RH011D17 (lanes 16–20), and tomato BACs P103N18 (lanes 22–27) and P073H07 (lanes 29–38).
DISCUSSION
Selection of BAC Clones for Sequence Walk
We presented here a software package, TOPAAS, that automates key steps in the selection and finishing of BAC clones. A combination of nonselective AFLP fingerprinting, BLASTN analysis, and assembly of BAC ends supports an accurate physical mapping. The BLASTN search is used for high-throughput screening of BACs and rapid preselection. The selection can be used without laborious screening techniques such as the STS approach (Blake et al., 1996; Marra et al., 1997) or having to fingerprint an entire BAC library. The BAC clones we have screened for building the Mi contig are repetitive for Mi homologous sequences and contain transposable elements, the latter being well represented in plant genomes. Repetitive domains can confound the binning by scoring false overlaps and this also poses a problem for assembly, ordering, and bridging of contigs. By filtering the BLASTN hits, verifying for nearby upstream cloning sites within 50 bps from the start of the overlap on the seed BAC consensus, and manual inspection and curation of base call discrepancies, the screening is made robust enough to discriminate for true BAC end overlaps. An alternative approach to circumvent potential problems caused by alignment to repetitive regions is discussed hereafter.
For screening contigs against BAC ends alternatively MegaBlast might be used. MegaBlast is faster compared to BLASTN and allows for a percentage identity cutoff rather than expected value cutoff. Since e-values depends on the length of the BAC ends and the size of the referenced database, relatively short BAC end sequences with a perfect match might be missed when filtering with a cut-off e-value of 0.0. We have also included the option to screen BAC contig sequences with MegaBlast.
The screening presented here works very efficiently. From a total of 75,000 to 126,000 BACs we have identified four and seven candidates for P250I21 and P046G10, respectively, prior to fingerprinting. The fingerprinting and BLASTN analyses work complementarily in the physical mapping process. With the BAC end sequence homology search we are able to pinpoint the exact start position and direction of the overlap, and the AFLP fingerprinting is used to determine the relationship between overlapping BACs over a larger domain. Whereas the BLASTN hits disclose information on minimal overlap sizes, the multiple BAC comparisons through nonselective AFLP fingerprinting provide vital information for identifying BACs with the largest extending insert. For BAC P073H07, two comigrating fragments with seed BACs P250I21 have been scored (Fig. 4, lanes 2 and 6). For BAC P103N18 one comigrating fragment is scored (Fig. 4, lanes 9 and 15), which alone would be an insufficient number to declare a reliable overlap. Furthermore, AFLP fragments are sometimes not detected from gel reads, causing small overlaps to be missed in the physical mapping process. The BLASTN hit positions and the assembly of BAC ends onto the seed BAC consensus have shown to be able to compensate this shortcoming. By sequencing and assembly of BACs selected for walking, we have confirmed that the overlap of BACs with a few kilobase pair overlap is authentic.
The approach we have taken does not depend on the full closure of a seed BAC. The results for P046G10 show that minimal overlapping BACs can be scored for as well, even when having gapped assemblies, provided the contig ends adjacent to the T7 and SP6 region are identified. Theoretically with this approach it should be possible to identify BACs for walking having only a few hundred base pair overlap. This will depend on the distribution of restriction sites in the tomato genome and the number of BAC clones available to cover the genome. Recently also BAC end sequences from an MboI library have been made available and will be complemented by the United States' part of the SOL initiative with additional sequences coming from an EcoRI library. The use of multiple libraries produced with different restriction enzymes will increase the likelihood of finding BACs with even shorter overlap sizes.
The mapping for BACs in AFLP contigs Mi and P103 has revealed some striking differences compared to FPC mapping results. Six BACs coassemble into contig Mi (Fig. 5). FPC data obtained from http://www.genome.arizona.edu/fpc/WebAGCoL/tomato/WebFPC/ show three BACs, P112G05, P111A08, and P096H22, respectively, map into three separate FPC contigs, while for the other three BACs no FPC mapping information could be retrieved. Contig P103 was assembled from eight BACs. For five out of eight BACs, including P250I21, no FPC data was available, whereas only three BACs, P061I06, P008K02, and P188J12, respectively, coassemble into a single FPC contig. BACs like P250I21 that are not assigned to FPC contigs probably represent dropouts. Our mapping results indicate BACs P111A08 and P096H22 from AFLP contig Mi overlap approximately 100 kb and share some 30 comigrating AFLP fragments. This finding is not reflected by the FPC data, and, despite this large overlap, P111A08 and P096H22 have been mapped into two different FPC contigs. The information content used to construct the maps for the AFLP contig Mi and P103 is significantly higher and directly relates to the number of bands produced and detectable size ranges in polyacrylamide and agarose gels (Meyers et al., 2004). For example, the in silico EcoRI/MseI digest for BAC P250I21 show 65 fragments in the size range of 50 to 600 bp, whereas the in silico HindIII digest reveals only 40 fragments in the size range of 600 to 25,000 bp. We conclude the higher information content and the superior resolution power of the AFLP fingerprinting results in more accurate physical maps and a reduced number of contigs, compared to FPC mapping approach.
Other important aspects are cost and labor involved. Recently we have screened 21 seeds from a HindIII library against 350,000 BAC ends. The screening yields 186 BACs from the HindIII library, 126 BACs from the EcoRI library, and 75 BACs from the MboI library (data not shown). Thus on average 18 candidate overlapping BACs have been identified per seed BAC. We can now roughly estimate the total number of BACs to be fingerprinted using the STC approach, and compare this with the classical FPC method. If we follow Batzoglou et al. (1999), the HindIII library with depth d = 15 and an average BAC insert length of λ = 117.5 kb (Budimann et al., 2000) would yield a minimal tilling path with redundant sequencing of 13%. The percentage of redundant sequence will however be closer to 7.1% as a best possible obtainable result, since two additional libraries are available. We estimate the euchromatic part of chromosome 6 with length L to be 20 Mb (http://www.sgn.cornell.edu/help/about/tomato_project_overview.pl). The proportion π, with which 21 seeds from the HindIII library cover chromosome 6, is approximately 2.5 Mb and yields an average gap length ω = (L − π)/π = 7 λ (approximately 819 kb). The number of bidirectional walking steps (κ) to cover 90% of chromosome 6 is roughly equal to the initial mean gap size, and up to 2 κ when covering 98% (Batzoglou et al., 1999). If we consider parallel walking starting from 21 seeds, ignore possible cloning bias and repeat sequences that mask overlaps, and assume all BACs are sequenced at both ends, in total some 2,500 to 5,000 BACs would have to be fingerprinted. A classical map first and sequence second approach like FPC would involve some 350,000 to 400,000 BACs to be fingerprinted.
BAC Finishing
TOPAAS assists the assembly, scaffolding, and finishing of BAC contigs. Read pairs are used commonly for finishing assemblies, and this linking approach has also contributed extensively to the positioning of tomato and potato contigs in this study. The likelihood for finding sequence gap-spanning read pairs depends on the insert sizes used for constructing the shotgun library and the coverage with which the target is sequenced. Approximately 15% of the contigs could not be ordered with gap-spanning mate pairs. This is partly due to the low coverage with which BACs have been sequenced. We have included homology-based searches to increase the chance of finding leads that link contig ends. From the links predicted, approximately 70% belonged to a read pair link type, whereas the remaining 30% were equally divided over BLASTX and ESTs link types.
Multiple factors contribute to the success of the homology-based linking approach. We show here alignments to single-pass ESTs can successfully be used for tomato and potato contig linking. For many plant genomes extensive amounts of ESTs have been produced, and in combination with genomic sequences the approach is feasible for many sequence projects including those from monocots, Brassicaceae, and Leguminosea (http://www.ncbi.nlm.nih.gov/genomes/PLANTS/PlantList.html). The closing efficiency will improve when using unigenes, since the spanning distance in general is larger compared to single-pass EST sequences. Building high quality unigenes requires base calling, accurate preclustering, and assembly, however. Reliable linkage by bridging unigenes will thus depend on the consistency and the overall quality of the build. Some 31,000 for S. lycopersicum and 25,000 unigenes for S. tuberosum have been assembled (Mueller et al., 2005a), each set containing some 38% singletons (http://www.sgn.cornell.edu/search/direct_search.pl?search=unigenes). We have used both unigene sets for alignment against tomato BAC 073H07; however the screening did not yield additional linkages.
MUMmer has been used as the matching algorithm. Its suffix tree-based method is relatively computational inexpensive and is very fast. MUMmer can perform a translated alignment, which is preferable for more distant related genomes. However, it is memory intensive and is originally designed for global rather then local alignments (Delcher et al., 2002; Kurtz et al., 2004). Tools like BLAT are specifically designed for EST-genome alignments. BLAT is also fast but differs from MUMmer in that it uses a hash array. It is very accurate for highly related genomes, but its nucleotide alignment strategy starts to break down when the base identity is below 90%. This makes it less suitable for cross-species alignments that are more distantly related. BLAT can work in translated mode but has limitations for protein alignments with respect to indels (Kent, 2002). We have provided TOPAAS with the option to screen BAC contig sequences with both BLAT and MUMmer.
Both BLAST and EST bridging sequences were checked manually for homology against known Solanum repeats. In one instance we found a contig pair linked by a BLAST hit against a repetitive element. The contig pair also shared a bridging read pair, making an aberrant linkage unlikely. Neither BLAST nor TOPAAS is specifically designed to deal with repetitive sequences. Although not used in this study, we have recently included an automated screen in the assembly phase against The Institute for Genomic Research Solanaceae Repeat Database (http://www.tigr.org/tdb/e2k1/plant.repeats) with RepeatMasker to circumvent potential problems (http://www.repeatmasker.org/RMDownload.html). In a Staden environment RepeatMasker is interfaced by PREGAP4 (Bonfield et al., 1995) and it tags repeats accordingly. Upon assembly, consensus sequences are extracted in which repeats are masked and are being denied from making false overlaps in homology-based alignments and EST alignments.
Ordering contig ends with BLASTX depends on the gene distribution in the tomato and potato genome. In this study we have finished BACs containing inserts of the euchromatic part of tomato chromosome 6. The genes are not evenly distributed in the tomato and potato genome (Van der Hoeven et al., 2002), and the likelihood of linking contigs in regions with few genes, e.g. in the heterochromatic parts of the genome, will be lower compared to the euchromatic domains of the genome. In addition, information on Solanaceae (putative) protein sequences are only scarcely available, and finding relationships depends on the availability of more distantly related (putative) protein sequences. The results show four out of six contig-bridging BLASTX alignments having a homology against non-Solanaceous protein sequences. Furthermore, coding regions in higher eukaryotes like tomato and potato contain introns, and this further decreases the chance to find contig ends matching the same protein sequence. We have included comparative alignments between tomato and potato ESTs and genomic sequences in the link analysis. The alignments between genomic and EST sequences show both species-specific and tomato-potato alignments that provide useful linking leads. Even more linking information could be obtained by comparative alignments to non-Solanaceae ESTs. A computational comparison of some 120,000 ESTs against tomato BACs from tomato cv Heinz 1706 and the Arabidopsis genome revealed 70% of the tomato unigenes having identifiable homologs in the Arabidopsis genome. Furthermore a comparison of gene repertoires indicates a set of highly conserved genes (17%) is shared between Arabidopsis, S. esculentum, and Medicago truncatula (Van der Hoeven et al., 2002). Therefore, alignments between, for example, full-length cDNAs or At-ESTs coming from studies to verify transcription units within the Arabidopsis genome (Yamada et al., 2003) to tomato and potato genomic sequences seems a promising possibility. Yet, caution should be taken to use sources from more distantly related species in comparative studies. Where genome rearrangements have occurred in evolution between species, changes on a microsyntheny level might lead to inaccurate projection and false ordering information. Nevertheless, the chances for finding ordering leads based on comparative alignments will surely increase with the rapidly expanding number of genome sequences and EST data sets from closely related species. We will continue to explore data sets and new linking approaches for the BAC finishing process. In this respect we are currently investigating whether matching AFLP gel fingerprints to in silico AFLP fingerprints can be used effectively for automated scaffolding purposes.
The TOPAAS software is available for nonprofit, academic, and personal use. Please contact http://www.cbsg.nl for nonexclusive commercial licenses. The software can be downloaded from http://www.appliedbioinformatics.wur.nl.
MATERIALS AND METHODS
Sequencing and PCR Analysis
BAC DNA was isolated with the Qiagen large construct kit, sized by hydro shearing, fractionated by gel electrophoresis, and 2-kb sized fragments were cloned into the dephosphorylated EcoRV site of pBlueScriptSK (Stratagene) or pGEM-TEasy (Promega). Shotgun templates were prepared from XL2 transformants (Stratagene) and sequenced using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready reaction kit with FS AmpliTaq DNA polymerase (Perkin Elmer) or the DYEnamic ET Terminator Cycle Sequencing kit (Amersham).
For gap closure, PCR products were amplified with custom-made primers using a regular PCR protocol. Typically a 10-μL PCR reaction contained 1 μL 5 μm forward and 1 μL 5 μm reversed custom primer, 1 μL 2.5 mm dNTPs, 2 μL 25 mm MgCl2, 2 μL 10× sequence buffer (200 mm Tris-HCl pH 9.0, 5 mm MgCl2), 0.2 μL 5 units/μL Goldstar (Eurogentec) polymerase, and 1 μL 10 μg/μL BAC template DNA. PCR products were analyzed on agarose gel, purified using QIAquick gel extraction kit (Qiagen) as described by the manufacturer, and diluted into 30 μL. Sequence PCR was carried out in 10 μL reaction mixture with 2 μL Amerdye (Amersham), 1 μL sequence primer, 2 μL sequence buffer (200 mm Tris-HCl pH 9.0, 10 mm MgCl2), and 5 μL template DNA. Sequence PCRs were analyzed on a 3730 XL DNA analyzer (Applied Biosystems).
Assembly
Using the PREGAP4 interface of the Staden package 2004, raw trace data was processed into assembly ready sequences. Sequences were base called by the PHRED base caller (Ewing and Green, 1998; Ewing et al., 1998). Clipping was performed to remove sequencing vector, cloning vector, and bad quality sequences. Processed sequences were subsequently assembled with GAP4, with a sequence percentage mismatch threshold of 8%, and parsed into the GAP4 assembly database. The GAP4 contig editor interface was used for editing and finishing. Consensus calculations with a quality cutoff score of 40 were performed from within GAP4 using a probabilistic consensus algorithm based on the expected error rates output by PHRED.
Software Dependencies
To manage the sequence, assembly, and scaffolding data we developed TOPAAS with components that are available as open-source components or with an academic user license. In particular we use MySQL as a database management system (http://www.mysql.com/downloads). Perl (http://www.perl.org) and PHP (http://www.phpmyadmin.net) are used for scripting purposes, and Apache (http://www.apache.org) is used for web hosting. Graphical output relies on the use of the graphics draw library (http://www.sunfreeware.com, or http://www.boutell.com/gd). The core program for primer design is built upon Primer3 (http://www-genome.wi.mit.edu/genome_software/other/primer3.html), though additional scripting has been used to manipulate Primer3 to automated primer design for sequence gap closure. The software also includes scripts to build a local database of contig sequences for redundancy check purposes of primer sequences using BLASTN. To find matching putative functions that can be attributed to contig sequences we rely on BLASTX hits. We have adopted the prokaryotic genome assembly assistance system approach, but we use our own implementation to screen for identical accession ID. We have extensively revised the table structure so that storage of datasets for multiple projects is supported. The software does not cover the implementation of a local BLAST facility and a proper environment to run BLAST. This should be implemented by the user (for details, see http://www.ncbi.nih.nlm.gov/BLAST). For multiple alignment viewing of BLASTX matches we rely on Mview (http://mathbio.nimr.mrc.ac.uk/∼nbrown). Base calling is carried out using PHRED (http://www.phrap.org). GAP4 assemblies were carried out using the Staden package 2004 (http://staden.sourceforge.net). The MUMmer package was used for sequence alignments between contig sequences and ESTs (http://www.tigr.org/software/mummer; http://mummer.sourceforge.net). Alternatively BLAT (http://www.cse.ucsc.edu/∼kent/) can be used for EST alignments. The software is implemented on a UNIX platform and tested on a SUN V440 server running Solaris 2.9.
Data Manipulation
Consensus sequences of contig ends were cured with the GAP4 assembly viewer using a PHRED quality threshold of 40 over a length of 1 kb for both ends of a contig. Assembly information was extracted from the GAP4 assembly database and parsed into the ContigLink database with TOPAAS. Subsequently, read pairs were evaluated with respect to direction and size constraints that underlie the shotgun library properties. Bridging read pairs are considered valid when positioned on different contig ends, pointing toward each other with respect to their sequencing direction, and meeting size constraints. For gap-flanking read pairs we calculate the sequence-spanning distance, excluding the size of the gap itself. The left distance, dleft, is taken from position 1 at the 5′-end of the first mate pair to the end position of the contig it is assembled in, running in the direction similar to the sequence direction of the first mate pair. The right distance, dright, is taken from the start position of the second contig to the 5′ end coordinate of the second mate pair running opposite to the sequence direction of the second mate pair. The total spanning distance is calculated as dtot = dleft + dright. The size constraint dtot for read pairs can be set to a value related to the average insert size used to construct a shotgun library. In this study dtot is set to 2.5 kb.
To align tomato (Solanum lycopersicum) and potato (Solanum tuberosum) EST sequences to contig sequences, we use an extension of the MUMmer package, designated NUCmer, using mummer2 as the matching algorithm. Consensus sequences in multi-fasta format from assembled contigs are used as a reference, and multi-fasta formatted potato and tomato EST sequences derived from NCBI are used as a query data set. An EST is considered contig bridging when aligning to different contig end sequences, with its domains aligned in a consecutive order, and with a minimal sequence identity threshold of 90% for each aligned domain.
To find related putative gene functions, contig sequences were queried against the nonredundant sequence database from NCBI with BLASTX. A link is considered valid when hitting against protein sequences with the same accession ID. A threshold for the expected value was set to 1 × 10−5 to avoid low similarity matches.
Primers are automatically designed on contig end sequences, using Primer3 as a core primer design program. Maximum distance of primer positions to contig ends is set to 500 bp. Additional custom scripting is applied to prefer primer sequences pointing outward with respect to the contig end positions and positioned nearest to a contig end. An automated redundancy check is used by aligning the primer sequence against the consensus sequence of the contigs using BLASTN. The expected value threshold for reporting primers as redundant was set to 0.1. Possible mispriming that could give rise to ambiguous PCR results is output by the program and described in terms of position, number of aligned bases, and alternative melting temperature.
To identify minimal overlapping BAC clones for walking, we use tomato BAC end sequences from the SOL Genomics Network available at ftp://ftp.sgn.cornell.edu/tomato_genome, and perform a BLASTN analysis against assembled tomato contigs. Position and direction of overlap were verified, and candidate BAC clones were preselected setting a threshold expected value to 0.0. When meeting constraints, corresponding ABI traces were subsequently assembled onto BAC contig sequences to which the BLAST hit was found and verified at nucleotide level for integrity. Assembled BAC end sequences showing high quality base call differences compared to contig consensus sequences, or showing its assembly start more than 50 bp downstream from a candidate HindIII or MboI cloning site are rejected. Remaining candidate BAC clones are further analyzed by fingerprint analysis.
AFLP Fingerprinting and BAC Insert Sizes
BAC DNA was isolated by standard alkaline lysis method (Sambrook et al., 1989) and EcoRI/MseI, HindIII/MseI, and PstI/MseI AFLP templates were prepared as described by Vos et al. (1995). Five microliters of the restriction ligation mix was diluted 10-fold in 10 mm Tris-HCl pH 7.5, 0.1 mm EDTA buffer. A nonselective amplification with [γ-33]ATP-labeled EcoRI + 0 and a MseI + 0 primers was performed in a total volume of 20 μL (Vos et al., 1995). Typically a 30-s DNA denaturing step at 94°C, a 1-min annealing step at 56°C, and a 1-min extension step at 72°C for 35 cycles was performed. For the HindIII/MseI and PstI/MseI templates, respectively, the HindIII + 0 and PstI + 0 [γ-33]ATP-labeled primers were used in combination with the MseI + 0 primer. All amplification reactions were performed in a PE-9700 thermocycler (Perkin Elmer). After the amplification step electrophoretic gel analysis of the reaction mix was carried out (Vos et al., 1995) and the fingerprint patterns were visualized using a Fuji BAS-2000 phosphoimaging analysis system (Fuji Photo Film). Band sizes were calculated relatively to a 10-bp size ladder with AFLP-Quantar fingerprint analysis software, and comigrating bands were scored by visual inspection. AFLP-Quantar fingerprint analysis software (http://www.keygene.com/technologies/technologies_keymaps.htm) is distributed by KeyGene and is not part of TOPAAS. For insert size determination BAC DNA was prepared by a standard alkaline lysis method (Sambrook et al., 1989) from a 3-mL overnight culture. BAC DNA was digested with NotI (New England Biolabs) to completion and separated by field inversion gel electrophoresis (Bio-Rad FIGE MAPPER) on a 1% agarose gel in 0.5× Tris-borate/EDTA, with a linear run time, forward (3–30 s) reverse (1–10 s), 14 h and 160 V, along with a mid-range PFGE marker I (New England Biolabs).
Supplementary Material
Acknowledgments
We thank Joyce van Eck for providing us with the MboI and EcoRI library from tomato cv Heinz 1706, and Andy Pereira and Roeland van Ham for reading the manuscript and for advice.
This work was supported by the research program of the Centre of BioSystems Genomics, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sander A. Peters (sander.peters@wur.nl).
The online version of this article contains Web-only data.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Batzoglou S, Berger B, Mesirov J, Lander ES (1999) Sequencing a genome by walking with clone-end sequences: a mathematical analysis. Genome Res 9: 1163–1174 [DOI] [PubMed] [Google Scholar]
- Blake TK, Kadyrzhanova D, Shepherd KW, Islam AKMR, Langridge PL, McDonald CL, Erpelding J, Larson S, Blake NK, Talbert LE (1996) STS-PCR markers appropriate for wheat-barley introgression. Theor Appl Genet 93: 826–832 [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Smith KF, Staden R (1995) A new DNA sequence assembly program. Nucleic Acids Res 23: 4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonierbale MW, Plaisted RL, Tangsley SD (1988) RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120: 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budimann MA, Mao L, Wood TC, Wing RA (2000) A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res 10: 129–136 [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL (2002) Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res 30: 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Basecalling of automated sequencer traces using PHRED. II. Error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Basecalling of automated sequencer traces using PHRED. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Huang S, van der Vossen EAG, Kuang H, Vleeshouwers VGAA, Ningwen Z, Borm TJA, van Eck HJ, Baker B, Jacobsen E, Visser RGF (2005) Comparative genomics enabled the isolations of the R3a late blight resistance gene in potato. Plant J 42: 251–261 [DOI] [PubMed] [Google Scholar]
- Kent JW (2002) The BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software to compare large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Waterman MS (1988) Genomics mapping by fingerprinting random clones: a mathematical analysis. Genomics 2: 231–239 [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH (1997) High throughput fingerprint analysis of large-insert clones. Genome Res 7: 1072–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BB, Scalabrin S, Morgante M (2004) Mapping and sequencing complex genomes. Nat Genet 5: 578–588 [DOI] [PubMed] [Google Scholar]
- Mueller AL, Solow TH, Taylor N, Skwarecki B, Buels R, Bins J, Lin C, Wright MH, Ahrens R, Wang Y, et al (2005. a) The SOL genomics network: a comparative resource for Solanaceae biology and beyond. Plant Physiol 138: 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AL, Tanksley SD, Giovannoni JJ, van Eck J, Stack S, Choi D, Kim BD, Chen M, Cheng Z, Li C, et al (2005. b) The tomato sequencing project, the first cornerstone of the international Solanaceae project (SOL). Comp Funct Genomics 6: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouppe van der Voort JR, Kanyuka K, van der Vossen E, Bendahmane A, Mooijman P, Klein-Lankhorst R, Stiekema W, Balcombe D, Bakker J (1999) Tight physical linkage of the nematode resistance gene Gpa2 and the virus resistance gene Rx on a single segment introgressed from wild species Solanum tuberosum subsp. andigena CPC1673 into cultivated potato. Mol Plant Microbe Interact 12: 197–206 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Soderlund C, Humphray S, Dunham A, French L (2000) Contigs built with fingerprints, markers, and FPC V4.7. Genome Res 10: 1772–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund C, Longdon I, Mott R (1997) FPC: a system for building contigs from restriction fingerprinted clones. Comput Appl Biosci 13: 523–535 [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Rijans M, Van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S (2002) Deductions about the number, organization and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Smith HO, Hood I (1996) A new strategy for genome walking. Nature 381: 364–366 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yu Z, Zhao J, Luo J (2002) PGAAS: a prokaryotic genome assembly assistance system. Bioinformatics 18: 661–665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.