Abstract
Four starch synthase I (SSI)-deficient rice (Oryza sativa) mutant lines were generated using retrotransposon Tos17 insertion. The mutants exhibited different levels of SSI activities and produced significantly lower amounts of SSI protein ranging from 0% to 20% of the wild type. The mutant endosperm amylopectin showed a decrease in chains with degree of polymerization (DP) 8 to 12 and an increase in chains with DP 6 to 7 and DP 16 to 19. The degree of change in amylopectin chain-length distribution was positively correlated with the extent of decrease in SSI activity in the mutants. The structural changes in the amylopectin increased the gelatinization temperature of endosperm starch. Chain-length analysis of amylopectin in the SSI band excised from native-polyacrylamide gel electrophoresis/SS activity staining gel showed that SSI preferentially synthesized DP 7 to 11 chains by elongating DP 4 to 7 short chains of glycogen or amylopectin. These results show that SSI distinctly generates DP 8 to 12 chains from short DP 6 to 7 chains emerging from the branch point in the A or B1 chain of amylopectin. SSI seemingly functions from the very early through the late stage of endosperm development. Yet, the complete absence of SSI, despite being a major SS isozyme in the developing endosperm, had no effect on the size and shape of seeds and starch granules and the crystallinity of endosperm starch, suggesting that other SS enzymes are probably capable of partly compensating SSI function. In summary, this study strongly suggested that amylopectin chains are synthesized by the coordinated actions of SSI, SSIIa, and SSIIIa isoforms.
Starch biosynthesis in higher plants is catalyzed by four classes of enzymes; ADP-Glc pyrophosphorylase (AGPase), starch synthase (SS), starch branching enzyme (BE), and starch debranching enzyme (DBE; Smith et al., 1997; Myers et al., 2000; Nakamura, 2002). Genome information has shown that many isozymes of each class are involved in starch biosynthesis in higher plants. Amylopectin accounts for about 65% to 85% of storage starch and has a defined structure composed of numerous tandem-linked clusters. The unit cluster size (about 9 nm) is a remarkably constant feature of all plant starches (Jenkins et al., 1993), whereas the fine structure of the cluster varies widely among species, tissues, and genetic backgrounds. This structural variation, which contributes greatly to the differences in the physicochemical properties among starches, is thought to be caused by the differences in the composition and relative activities of the isozymes of SS, BE, and DBE, although the distinct function of each isozyme has not been fully clarified. The analysis of mutants could greatly contribute to understanding the function(s) of individual isozymes involved in starch biosynthesis.
SS (EC 2.4.1.21) elongates glucans by adding Glc residues from ADP-Glc to the glucan nonreducing ends through α-1,4 linkages. The complete purification of SS isozymes from plants is difficult because of the inherent instability of the enzymes. Nevertheless, some biochemical studies of SS isozymes expressed in Escherichia coli (Imparl-Radosevich et al., 1998, 1999; Senoura et al., 2004) and analysis of partially purified SS isozymes from plant tissues (Baba et al., 1993; Mu-Forster et al., 1996; Cao et al., 2000) have been reported. Two SS activity peaks, designated SSI and SSII, have been fractionated by anion-exchange chromatography from soluble extracts of maize (Zea mays; Ozbun et al., 1971; Boyer and Preiss, 1981) and rice (Oryza sativa; Tanaka and Akazawa, 1971) developing endosperm. The enzyme responsible for the SSI activity peak was purified and identified as the product of its cDNA in maize (Mu-Forster et al., 1996; Knight et al., 1998). The SSII activity peak is significantly reduced in dull-1 mutants of maize (Boyer and Preiss, 1981) and it is clear from gene-tagging studies in maize mutants that Du1 codes for SSIII (Gao et al., 1998).
Various SS isoforms have been identified through gene sequences in several plant genomes. In rice, there are 10 SS isoforms separated into five types; two granule-bound starch synthase (GBSS) isoforms (GBSSI and GBSSII) in the GBSS type, one SSI isoform in the SSI type, three SSII isoforms (SSIIa [SSII-3], SSIIb [SSII-2], and SSIIc [SSII-1]) in the SSII type, two SSIII isoforms (SSIIIa [SSIII-2] and SSIIIb [SSIII-1]) in the SSIII type, and two SSIV isoforms (SSIVa [SSIV-1] and SSIVb [SSIV-2]) in the SSIV type (Hirose and Terao, 2004). Mutants defective in GBSSI in several species (waxy rice [Sano, 1984]; waxy maize [Tsai, 1974]; waxy barley [Hordeum vulgare; Eriksson, 1962]; waxy wheat [Triticum aestivum; Nakamura et al., 1995; Triticum monococcum; Fujita et al., 2001]; amf potato [Solanum tuberosum; Hovenkamp-Hermelink et al., 1987]; lam pea [Pisum sativum; Denyer et al., 1995]), SSIII of maize (dull-1; Gao et al., 1998), mutants deficient in SSIIa in rice (most japonica rice varieties are mutants of indica rice varieties [Umemoto et al., 2002; Nakamura et al., 2005]), in barley (sex6; Morell et al., 2003), in wheat (GSP-1 null; Yamamori et al., 2000) and in maize (sugary2; Zhang et al., 2004), and mutants for SSII in pea (rugosus5; Craig et al., 1998) have been identified and analyzed. Most recently, SSI (Delvalle et al., 2005) and SSIII (Zhang et al., 2005) mutants in Arabidopsis (Arabidopsis thaliana) leaves have been isolated. However, mutants for the remaining SS isozymes are yet to be found.
SSI probably plays important role(s) in starch biosynthesis in plants since it has no multiple isoforms, unlike all other SS types having more than two isoforms in rice (SSII, SSIII, SSIV, and GBSS; Hirose and Terao, 2004), maize (SSII; Harn et al., 1998), kidney bean (Phaseolus vulgaris; SSII and GBSS; Matsui et al., 2003; Isono et al., 2004), and wheat (GBSS; Fujita and Taira, 1998; Nakamura et al., 1998). Mu-Forster et al. (1996) reported that more than 85% of the SSI in maize endosperm is associated with starch granules. The N-terminal extension of SSI was thought to be important for its binding with starch granules (Imparl-Radosevich et al., 1998). However, Commuri and Keeling (2001) found that the mobility of both full-length SSI and N-terminally truncated SSI are similarly retarded on native gels containing 2% soluble starch, indicating that the N-terminal extension of SSI is not important for SSI interaction with glucans. To understand the function of SSI, Guan and Keeling (1998) analyzed the chain-length distribution of polyglucans synthesized by different combinations of recombinant maize SS and maize BE isoforms in E. coli and found that SSI preferentially synthesized short chains (degree of polymerization [DP] 6–15). Commuri and Keeling (2001) reported that maize SSI enzyme, despite having the greatest affinity for longer glucans (DP > 20) based on the measurement of dissociation constants, preferentially elongates shorter glucans with DP < 10. They proposed that SSI elongates shorter A and B1 chains during starch biosynthesis in the soluble phase of amyloplast stroma until it becomes entrapped within longer glucans as a relatively inactive protein in the starch granules. Moreover, they speculated that a mutation in SSI would likely have a severe impact on the formation of the crystalline amylopectin matrix, even though mutations in the other SS isozyme genes such as dull-1 (SSIII gene) or sugary2 (SSIIa gene) would affect amylopectin chain length without affecting the basic starch granule structure. The SSI from kidney bean (rPvSSI) expressed in E. coli also has a high elongation performance for extremely short chains of less than DP 6 of glycogen and amylopectin in vitro (Senoura et al., 2004).
Although these reports suggest that monocot or dicot SSIs have a specific role in the elongation of short chains in vitro, there are very few reports regarding their function in vivo. Kossmann et al. (1999) reported that the reduction of potato SSI in antisense plants did not lead to any detectable changes in starch structure in the tuber, since in potato SSI is predominantly expressed in leaves and only to a lower extent in the tuber where SSII and SSIII are the major isozymes expressed. In Arabidopsis mutants defective in SSI (Delvalle et al., 2005), which is one of the major isozymes in its leaves, the chain-length distribution of amylopectin showed a decrease in DP 8 to 12 branches and an increase in DP 17 to 20 branches. Considering these results and the previously described kinetic properties of recombinant maize SSI (Commuri and Keeling, 2001), Delvalle et al. (2005) proposed that SSI is mainly involved in the synthesis of small outer chains during amylopectin cluster synthesis. However, the in vivo function of SSI in amylopectin synthesis in storage starch has not been examined since there are no genetic mutants in the SSI gene in any plants that accumulate starch in storage tissues.
In this study, four allelic mutant lines of rice generated by retrotransposon Tos17 insertion and carrying SSI mutations expressed in endosperm, one containing a null mutation and three exhibiting different levels of SSI activity, were biochemically characterized. Analysis of amylopectin chain-length distribution and starch physicochemical traits of these mutants clarified the in vivo function of rice SSI in amylopectin biosynthesis.
RESULTS
Characterization of SSI in Developing Rice Endosperm
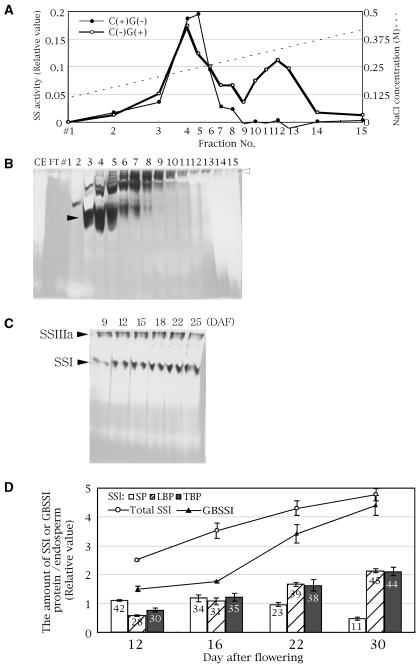
In this study, soluble fraction from rice developing endosperm was separated by anion-exchange (HiTrapQ, Pharmacia) chromatography with a linear gradient of 0 to 0.5 m NaCl. One peak of SS activity that eluted at 0.2 to 0.25 m of NaCl was detected in the presence of 0.5 m citrate and the absence of exogenous primers, whereas two SS activity peaks were detected at about 0.2 to 0.25 m and 0.35 m, in the absence of citrate but in the presence of glycogen primer (Fig. 1A). These results are similar to those obtained from maize fractionation by Q-sepharose (Cao et al., 2000) and it is likely that most of the first peak at 0.2 to 0.25 m of NaCl and the second peak at 0.35 m of NaCl correspond to SSI and SSIII, respectively, in rice developing endosperm. This SSIII should be regarded as SSIIIa, because there are two SSIII isoforms (SSIIIa and SSIIIb) in the rice genome and SSIIIa is specifically expressed in the developing endosperm (Hirose and Terao, 2004; Ohdan et al., 2005). In maize, only the SSI activity peak is detected in the presence of 0.5 m citrate and the absence of exogenous primers, while both SSI and SSIII activity peaks emerge when exogenous primers but no citrate is present in the reaction buffer, indicating that SSIII is dependent on the exogenous primer for activity (Cao et al., 2000). As in maize, SSI in rice exhibited SS activity even in the absence of exogenous primers, while rice SSIIIa activity was dependent on the exogenous primer (Fig. 1A). Native-PAGE/SS activity staining of each fraction from the HitrapQ column in gel containing rice amylopectin indicated that the strong activity middle band found in fraction numbers 3 to 5 corresponds to SSI activity (black arrowhead in Fig. 1B). On the other hand, the slowest migrating SS activity bands in fractions around number 12 seem to be rice SSIIIa (white arrowhead in Fig. 1B). The middle and the slowest migrating bands on these native gels (Fig. 1C) were identified to be SSI and SSIIIa, respectively, using SSI (Fig. 3A) and SSIIIa (N. Fujita, unpublished data) mutants of rice. Other SS activity bands detected between SSI and SSIIIa bands (Fig. 1B) could be either due to other SS isozymes or fragments or oligomers of SSI or SSIIIa.
Figure 1.
Characterization of SSI protein in rice developing endosperm. A, Separation of the SSI and SSIIIa activity peaks of rice developing endosperm using HiTrapQ anion-exchange chromatography. Each fraction was assayed for SS activity under two different conditions, either in a reaction buffer containing 0.5 m citrate and lacking glycogen ([C(+)G(−)], black circles and thin line) or in a reaction buffer lacking citrate and containing 2 mg/mL glycogen ([C(−)G(+)], white circles and bold line). B, Native-PAGE/SS activity staining of each fraction in A. The gel contains 0.1% rice amylopectin. The black and white arrowheads indicate putative SSI and SSIIIa bands, respectively. CE, Crude enzyme extract; FT, flow through fraction; number 1 to approximately 15, fraction numbers as in A. C, Native-PAGE/SS activity staining of rice developing endosperm from DAF 9 to 25. Fifteen percent of pooled SP (see “Materials and Methods”) was loaded on each lane. The gel contains 0.1% rice amylopectin. Black arrowheads indicate putative SSI and SSIIIa bands. These activity bands were dependent on the addition of ADP-Glc in the incubation buffer (data not shown). D, Amount of SSI or GBSSI protein in developing rice endosperm from DAF 12 to 30. Proteins of developing endosperm were divided into three fractions: SP, LBP, and TBP (see “Materials and Methods”). The amount of SSI or GBSSI protein was quantified by immunoblotting using antiserum raised against SSI or GBSSI. The data are the mean ± se of three seeds. The numbers on the graph are the relative SSI amount (%) of each protein fraction in the developing endosperms.
Figure 3.
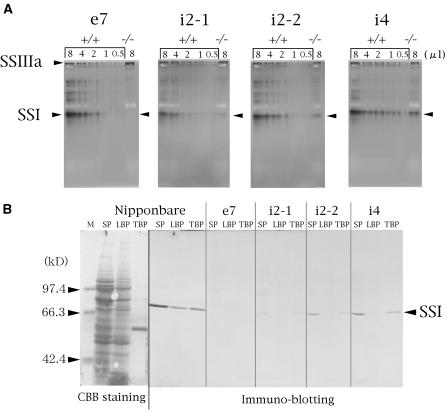
Estimation of SSI activity and amount of SSI protein in rice SSI mutants. A, Native-PAGE/SS activity staining to estimate SSI activity in mutants (−/−) and their respective control lines (+/+). The numbers above the lanes are the volumes (microliters) of crude enzyme extract applied onto each lane. The SSIIIa and SSI activity bands are indicated by arrowheads. B, SDS-PAGE (left section, stained with Coomassie Brilliant Blue) and immunoblotting (right section) of SP, LBP, and TBP from developing endosperm using antiserum raised against rice SSI for quantification of the amount of SSI protein in Nipponbare and the SSI mutant lines. M, Molecular markers.
Mu-Forster et al. (1996) demonstrated in maize that more than 85% of total endosperm SSI is associated with starch granules. To determine the distribution of rice SSI in developing endosperm, proteins in developing rice endosperm (day after flowering [DAF] 12) were separated into three fractions: soluble protein (SP) extracted with aqueous buffer; proteins that are loosely bound to starch granules (loosely bound protein [LBP]) extracted with 2.3% SDS buffer after the removal of SP; and proteins that are tightly bound to starch granules (tightly bound protein [TBP]) extracted by boiling the starch granules in SDS buffer after the removal of LBP (see “Materials and Methods”). These proteins, all approximately 71 kD, were identified to be SSI proteins by immunoblotting using an antiserum raised against SSI from developing endosperm of the wild-type Nipponbare (Fig. 3B). The predicted molecular mass of rice SSI calculated from the DNA sequence of the gene coding for the SSI mature protein is 65 kD (Baba et al., 1993; Jiang et al., 2004), which is smaller than the apparent molecular mass (71 kD) estimated on SDS-PAGE. Similar differences in molecular mass were also reported for SSI proteins from maize (Knight et al., 1998) and potato tubers (Edwards et al., 1995). The relative amounts of SSI protein associated with SP, LBP, and TBP were estimated from the bands in the immunoblots to be roughly 42%, 28%, and 30%, respectively (Fig. 1D; DAF 12), in one endosperm, indicating that the soluble SSI fraction is higher in rice than that reported for maize (12%; DAF 20; Mu-Forster et al., 1996).
SSI and SSIIIa activities of the endosperm SP were detectable from DAF 9 to DAF 25 (dehydration stage of endosperm; Fig. 1C). To analyze the changes in the localization of rice SSI in starch granules during endosperm development, the three protein fractions extracted from developing endosperms at DAF 12 to 30 were subjected to immunoblotting using SSI antiserum, and the amounts of SSI protein were estimated from band densities (Fig. 1D). The amounts of SSI proteins in the endosperm SP per endosperm were high at early stages (DAF 12–16) but gradually diminished at later stages during which LBP and TBP increased (Fig. 1D), while the total amount of SSI protein gradually increased (Fig. 1D). GBSSI was detected in TBP, but not in SP or LBP, and rapidly increased after DAF 16 (Fig. 1D).
Production of SSI-Deficient Mutant Lines
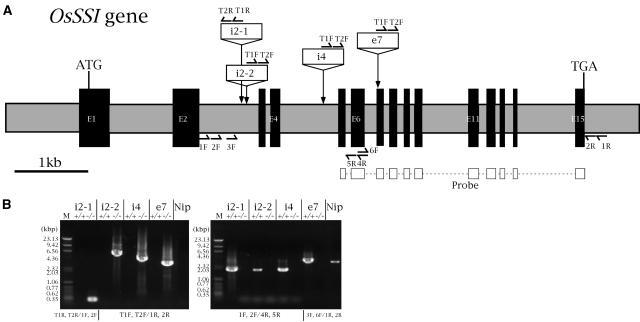
Nine lines containing Tos17 insertion in the rice SSI gene (OsSSI) were isolated by PCR screening of approximately 40,000 Tos17 knockout rice population (see “Materials and Methods”). OsSSI is composed of 15 exons and 14 introns (Fig. 2A). Four lines carrying Tos17 insertion in intron 2 (lines i2-1, i2-2), intron 4 (line i4), or exon 7 (line e7) were chosen for this study (Fig. 2A). The genotype of the lines were determined as either homozygous for Tos17 (−/−) or wild homozygous (+/+) using nested PCR (see “Materials and Methods”; Fig. 2B). For instance, in line i2-1+/+, the PCR reaction using the T1R, T2R/1F, 2F primer pairs had no product (Fig. 2B, left section, lane i2-1+/+), while 1F, 2F/4R, 5R primer pairs generated an approximately 2 kbp band (Fig. 2B, right section, lane i2-1+/+). In line i2-1−/−, the PCR product of the T1R, T2R/1F, 2F primer pairs was an approximately 0.35 kbp band (Fig. 2B, left section, lane i2-1−/−), while the 1F, 2F/4R, 5R primer pairs had no product (Fig. 2B, right section, lane i2-1−/−). The same PCR results were obtained in other lines (Fig. 2B). The −/− lines were used as the mutants and +/+ lines as the controls since the background of the +/+ lines was more similar to those of −/− lines than to the wild-type Nipponbare.
Figure 2.
Sites of Tos17 insertion in the OsSSI gene and determination of rice mutant lines genotype by PCR. A, Structure of the OsSSI gene. The exons and introns are depicted as black and gray boxes, respectively. ATG and TGA indicate the translation initiation and stop codons, respectively. The insertion site of Tos17 in the four mutant lines (names in boxes) are indicated by vertical arrows. Horizontal half arrows show the sites of primers for PCR for genotype determination (B) and mutant lines screening. The primers T1R, T2R, T1F, and T2F were designed from the Tos17 sequence while 1F, 2F, 3F, 6F, 1R, 2R, 4R, and 5R were designed from the OsSSI gene sequence. The region used as probe for Southern blotting to screen mutant lines is indicated. B, Determination of genotype (homozygous for Tos17 insertion [−/−, left section] or wild homozygous [+/+, right section]) in the four mutant lines by nested PCR. Primer pairs are indicated below the photographs. T1R, T2R/1F, 2F means that the primer pair T1R/1F was used for the first PCR, and T2R/2F for the second PCR. Nip, Rice cultivar Nipponbare (the wild-type parent of the mutant lines); M, molecular markers.
The expression of the OsSSI gene in mutant lines was confirmed by northern blotting. The 2.9 kb mRNA band previously reported to be the normal mRNA of the OsSSI gene (Tanaka et al., 2004) was detected in wild-type and +/+ lines, and the intensity of the band decreased gradually in the order i4−/− > i2-2−/− > i2-1−/−, and no band was detected in e7−/− (data not shown). The expected higher Mr mRNA band due to the insertion of Tos17 could not be detected because of high background noise. On the other hand, when the 3′ untranslated regions of the OsSSI gene were amplified by semiquantitative reverse transcription (RT)-PCR, the target bands were detected in every mutant line (data not shown), indicating that the long abnormal OsSSI mRNA due to the insertion of Tos17 into the OsSSI gene was expressed in these mutant lines. It is assumed that Tos17 introduced at least one stop codon when inserted into an exon, whereas it interfered with the splicing process when inserted into an intron, both events resulting in the production of truncated and/or nonfunctional SSI proteins.
Characterization of Enzymes Related to Starch Biosynthesis in SSI Mutant Lines
To evaluate the effect of the insertion of Tos17 into the OsSSI gene, SSI activity of the SP from DAF 16 developing endosperm was estimated by native-PAGE/SS activity staining using gels containing oyster glycogen (Fig. 3A). The SSI bands in every mutant line (−/−) were significantly decreased or completely lacking compared to those of control lines (+/+). These bands were identical to the SSI band in Figure 1, B and C. To estimate the amount of remaining SSI activity in these mutant lines, varying amounts of SP from control lines were loaded adjacent to the mutant SP on the gel (Fig. 3A). The SSI activities were calculated to be 0 in e7−/−, roughly 17% in i2-2−/−, 20% in i2-1−/−, and 25% in i4−/−, respectively, of the control lines. In contrast, SSIIIa activity bands in i2-1−/−, i2-2−/−, and i4−/− were comparable to those of their control lines and appeared to be enhanced 2 to 3 times in e7−/− SSI null mutant compared with its e7+/+ control line.
The amount of SSI protein in the three fractions (SP, LBP, and TBP) of DAF 12 developing endosperm detected by immunoblotting using antiserum raised against rice SSI was also significantly decreased in SSI mutant lines and was not detected in any fractions from e7−/− (Fig. 3B). The total amount of the remaining SSI protein in every fraction in lines e7−/−, i2-1−/−, i2-2−/−, and i4−/− was 0%, 3%, 9%, and 20%, respectively, of their control lines, this order being correlated with remaining SSI activity in the respective mutant lines as determined by native-PAGE/SS activity staining (Fig. 3A).
To test whether the reduction in SSI protein has pleiotropic effects, activities of other enzymes involved in starch biosynthesis were measured. The activities of BEI, BEIIa, BEIIb, phosphorylase, isoamylase1, and pullulanase detected by native-PAGE/activity staining and the amount of GBSSI protein detected by SDS-PAGE of TBP showed no obvious differences (data not shown). In contrast, AGPase activity in e7−/− was approximately 1.6 times (10.58 ± 0.20 mmol min−1 endosperm−1) of that of its control (6.59 ± 0.36) and the wild type (6.03 ± 0.37), while AGPase activities in nonnull SSI mutants (i2-1−/−, i2-2−/−, and i4−/−) were not significantly different from their respective control lines (data not shown).
Analysis of Seed Morphology, Starch Structure, and Physicochemical Properties of the SSI Mutant Lines
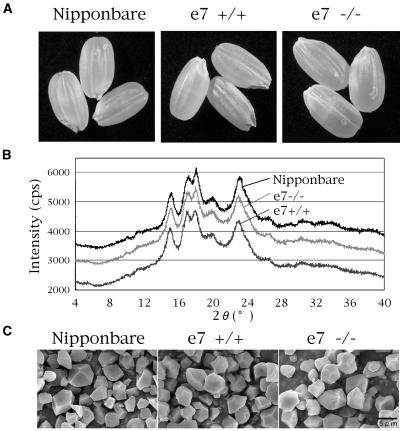
The seed morphology of the SSI null mutant line e7−/− (Fig. 4A) and other lines (data not shown) was similar to that of their respective control lines and the wild type. The seed weights of lines i2-1−/− and e7−/− were not significantly different from those of their control lines and the wild type, while lines i2-2−/− and i4−/− and their control lines had lighter seeds than the wild type (Table I), indicating that these decreases in seed weight could not be caused by SSI deficiency but by the insertion of Tos17 in other genes. The amounts of endosperm starch were similar in SSI mutant and control lines (data not shown). This is one of the reasons for the difficulty in the isolation of SSI-deficient mutants by the screening for seed morphology.
Figure 4.
Characterization of the SSI mutant line e7−/−, its control line e7+/+, and their wild-type parent cultivar Nipponbare. A, Seed morphology. B, X-ray diffraction pattern of endosperm starch. C, SEM of endosperm starch. Bar = 5 μm.
Table I.
Dehulled grain weight of rice SSI mutant lines (−/−), their control lines (+/+), and wild-type rice cultivar Nipponbare
| Lines | Dehulled Grain Weighta |
|---|---|
| mg | |
| Nipponbare | 21.9 ± 0.5 |
| i2-1+/+ | 22.8 ± 0.3 |
| i2-1−/− | 25.0 ± 0.4 |
| i2-2+/+ | 18.7 ± 0.3 |
| i2-2−/− | 18.7 ± 0.3 |
| i4+/+ | 19.9 ± 0.3 |
| i4−/− | 19.0 ± 0.2 |
| e7+/+ | 21.7 ± 0.4 |
| e7−/− | 22.8 ± 0.4 |
Mean ± se of 20 seeds.
X-ray diffraction pattern of the starch granules of Nipponbare, e7+/+, and e7−/− was similar, all exhibiting a typical A type (Fig. 4B), indicating that their starches share similar crystalline properties. Examination of the morphology of the starch granules using scanning electron micrograph (SEM) yielded no significant differences among e7−/−, e7+/+, and the wild-type Nipponbare (Fig. 4C) as well as among i2-1, its control line, and wild type (data not shown).
To examine the effects of SSI deficiency on the amylose content of endosperm starch, the λmax of their starch-iodine complex was measured. The value for e7−/− (566 nm) was only slightly higher than those of e7+/+ (561 nm) and the wild type (557 nm), indicating that SSI deficiency has no significant impact on amylose content of the endosperm starch.
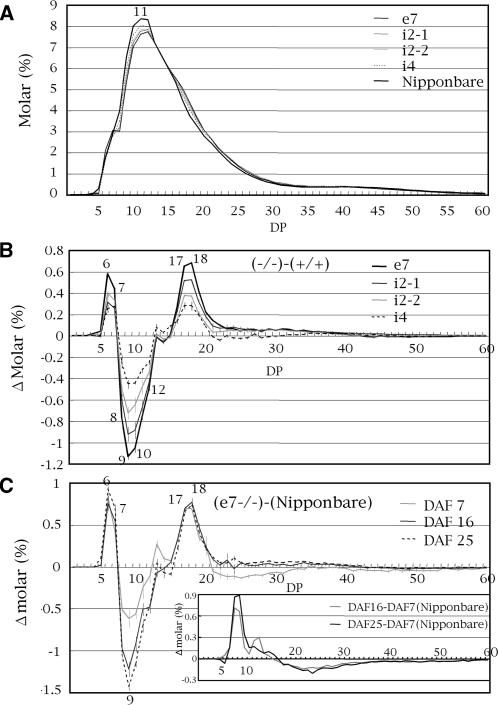
To evaluate the effects of the SSI activity level on the fine structure of the endosperm amylopectin, the chain-length distribution of isoamylolysate of endosperm amylopectin in the four SSI mutant lines was determined by capillary electrophoresis (Fig. 5, A and B). In all mutant lines, chains with DP 6 to 7 and DP 16 to 19 were increased and DP 8 to 12 chains were decreased, while DP 20 to 40 chains were slightly increased, although the change was gradually reduced from DP 20 to DP 40 (Fig. 5B). These chain-length distribution patterns of SSI mutant amylopectin are specific as compared with those of other mutant amylopectin analyzed so far (Nakamura, 2002). It is stressed that as the residual SSI activity decreased in SSI mutant lines, DP 8 to 12 decreased and DP 16 to 19 increased, while DP 6 to 7 level was not correlated with the residual SSI activity (Fig. 5B).
Figure 5.
A, Chain-length distribution patterns of endosperm amylopectin in mature endosperm of mutant lines (−/−) and the wild-type parent cultivar Nipponbare. B and C, Differences in chain-length distribution patterns of endosperm amylopectin between the mature endosperm of mutants (−/−) and their respective control lines (+/+; B), and in developing endosperm at DAF 7, 16, and 25 of mutant line e7−/− and Nipponbare (C). Values for molar % in A and Δ molar % in B and C for each DP are averages of three seeds arbitrarily chosen from a single homozygous plant. Vertical bars in B and C indicate ses. The numbers on the plots are DP values. Inset in C shows differences in chain-length distribution of amylopectin at different endosperm developmental stages in Nipponbare.
Starch accumulation in rice endosperm becomes evident after DAF 5 (Sato, 1984; Hirose and Terao, 2004). Prior to this event, the expression of the OsSSI gene starts at DAF 1 (Hirose and Terao, 2004; Ohdan et al., 2005) and is maintained at high levels until DAF 15 (Baba et al., 1993; Hirose and Terao, 2004; Ohdan et al., 2005). The differences in the amylopectin chain-length distribution pattern between e7−/− and the wild type were analyzed during endosperm development (DAF 7–25; Fig. 5C). In the wild type, the chain-length distribution of amylopectin at the middle (DAF 16) or late (DAF 25) stage of endosperm development showed more DP ≤ 17 chains and less DP ≥ 18 chains as compared with those at the early (DAF 7) stage (Fig. 5C, inset). The fact that the alteration of the chain-length pattern in e7−/− was already apparent at DAF 7 (Fig. 5C) indicates that SSI is functional from the very early stage of rice endosperm development. In contrast, the patterns from DAF 16 through seed maturity were the same (Fig. 5, B and C), indicating that the function of SSI does not vary during these periods. The DP 20 to 33 chains decreased (0.13 Δ molar %; Fig. 5C) at DAF 7, although the slight increase in DP ≥ 20 chains (less than 0.1 Δ molar %; Fig. 5, B and C) was evident from DAF 16 through endosperm maturity, suggesting a different physiological role of SSI in the synthesis of amylopectin structure at the very early stage of endosperm development when starch synthesis is being initiated.
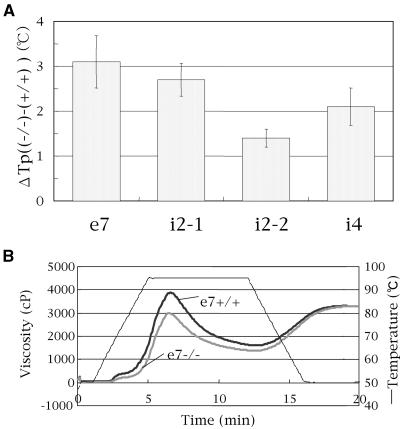
To evaluate the physicochemical properties of endosperm starch in SSI mutant lines, the thermal-gelatinization temperature of endosperm starch was analyzed by differential scanning calorimetry (DSC). The temperatures for the onset (To), peak (Tp), and conclusion (Tc) of gelatinization of endosperm starch in SSI mutant lines were 1°C to approximately 3°C higher than those of control lines (data not shown). The magnitude of Tp change (ΔTp) between the control and SSI mutant lines decreased in the order e7 > i2-1 > i4 > i2-2 (Fig. 6A), which was not correlated with the amount of the residual SSI activity.
Figure 6.
Physicochemical properties of starch granules in mature endosperm of rice SSI mutant lines. A, Differences in peak temperature of thermal gelatinization (°C) as measured by DSC between each mutant line (−/−) and its respective control (+/+). The y axis gives the temperature (°C) difference ([−/−] − [+/+]). The values presented are average ± se of at least three seeds arbitrarily chosen from a single homozygous plant. B, Pasting properties of endosperm starch of the mutant line e7−/− and its control line e7+/+. The viscosity value at each temperature point is the average of three replications. The thin line indicates the change in temperature during measurement by rapid visco analyzer.
The pasting property of the endosperm starch was analyzed by rapid visco analyzer (Fig. 6B). The rate of the rise in the viscosity of e7−/− starch as temperature increased was slower than that of e7+/+, and the peak viscosity of e7−/− starch was 77% of that of e7+/+. The final viscosity of starch in both lines was similar.
Chain-Length Analysis of the SS Activity Band on Native-PAGE/SS Activity Staining Gels
To evaluate the in vitro functions of rice SS isozymes in the wild-type Nipponbare, the changes in chain-length distribution of amylopectin or glycogen in the SS activity band after SS enzymatic reactions in native-PAGE gel were examined. The band corresponding to the SS activity in the gel containing 0.1% of rice amylopectin or 0.8% of oyster glycogen after SS enzymatic reactions for 20 h was excised, and a portion of gel without any activity band at all was also taken as a control. The SS activity band gel segment contained substrate modified by SS isozymes whereas the control gel fragment was assumed to contain unmodified substrate only. The polyglucans in both gel fragments were extracted, processed, and their chain-length distribution patterns were examined and compared.
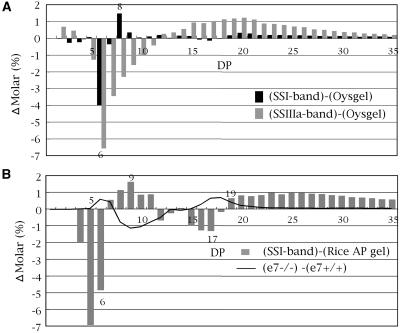
When oyster glycogen was used as the substrate, the polyglucans produced by the SSI activity band on native-PAGE gel had less DP 6 chains and more DP 8 chains compared to the unmodified glycogen (Fig. 7A). In contrast, the chain-length pattern of polyglucans produced by the SSIIIa band showed that shorter glycogen chains with DP ≤ 11 decreased and that longer chains of DP ≥ 12 increased broadly (Fig. 7A). These results imply that SSI specifically elongates short DP 6 chains to DP 8 chains by adding two Glc residues, whereas SSIIIa elongates the short DP ≤ 11 chains to form DP ≥ 12 chains. These results indicate that the function of SSI is quite different from that of SSIIIa.
Figure 7.
Chain-length distribution of polyglucans from SS bands excised from the native-PAGE/SS activity staining gel of the wild-type rice cultivar Nipponbare. Polyglucans were extracted from SSI and SSIIIa activity bands and from a portion without any activity bands. The SS activity band gel segment contained modified substrate whereas the control gel fragment contained unmodified substrate only. A, Chain-length differences between the unmodified substrate (oyster glycogen gel, Oysgel) and the SSI-modified (SSI-band) or SSIIIa-modified (SSIIIa-band) substrate. The SP fraction from developing endosperm was used for native-PAGE/SS activity staining. B, Chain-length differences between the unmodified rice amylopectin (rice amylopectin gel, Rice AP gel) and the SSI-modified rice amylopectin substrate (SSI-band). The partially purified SSI fraction (Fig. 1B, lane 4) was used for native-PAGE/SS activity staining on a gel containing rice amylopectin. Chains with DP 4 to 5 are considered as products of digestion of polyglucans by hydrolytic enzyme(s) included in the SSI fraction during prolonged incubation time (20 h). The superimposed black line shows the difference in amylopectin chain-length distribution between the mature endosperm SSI mutant line e7−/− and its control line e7+/+.
The polyglucans produced by the partially purified SSI activity band (Fig. 1B, SSI band in lane 4) on native-PAGE/SS activity staining rice amylopectin gel had less chains with DP 4 to 6 and DP 12 to 18 but more chains with DP 7 to 11 and DP ≥ 19 as compared to the unmodified substrate (Fig. 7B). It is likely that SSI elongates the very short DP ≤ 6 chains into DP 7 to 11, and converts the intermediate DP 15 to 17 chains to DP ≥ 19 by adding 1 to 5 Glc units. This pattern of changes in the range of DP ≤ 18 chains was almost a mirror image of the differences between the SSI mutants and their respective control lines (for example, [e7−/−] − [e7+/+]; Fig. 7B), indicating that the results of the in vitro analysis of chain preference of SSI on native gel are consistent with those obtained in vivo in SSI mutant lines. Therefore, chain analysis of the SS activity bands excised from native-PAGE/SS activity staining gels is a powerful, yet simple method for dissecting the functions of SS isozymes because it does not require the complete purification of the isozymes to be examined.
DISCUSSION
Isolation and Characterization of Rice SSI Mutant Lines
To date, no mutants deficient in SSI have been characterized in any plant species that accumulate starch in storage tissues, while a lot of mutants defective in GBSSI, SSIIa, and SSIII in a variety of plant species have been examined. The SS isozymes in higher plants such as rice are divided into five isozyme types: SSI, SSII, SSIII, SSIV, and GBSS (Hirose and Terao, 2004). SSI might play a distinct role in starch biosynthesis and its deficiency would induce a direct change in the structure of starch granules because SSI has only a single form, notwithstanding the presence of multiple forms in other SS types (Commuri and Keeling, 2001). In this report, four SSI-deficient mutant lines having different SSI activity levels were isolated through reverse genetics by retrotransposon Tos17 insertion. Surprisingly, seed morphology (Fig. 4A), seed weight (Table I), the amount of endosperm starch (data not shown), and the morphology (Fig. 4C) and crystallinity (Fig. 4B) of endosperm starch granules were not affected in the SSI mutant lines.
On the other hand, the fine structure of endosperm amylopectin was distinctly affected by SSI mutation. Chains with DP 6 to 7 and DP 16 to 19 increased while DP 8 to 12 chains decreased in all SSI mutant lines. These results sharply contrast with the findings in a SSIIa mutant (japonica rice) that DP ≤ 10 chains decreased and DP 12 to 24 chains increased (Nakamura et al., 2002; Umemoto et al., 2002), and in the maize SSIII mutant dull-1 where DP ≥ 37 chains decreased (Jane et al., 1999). Moreover, the magnitude of increase in DP 16 to 19 chains and decrease in DP 8 to 12 chains in all SSI mutant lines were closely related to the levels of the residual SSI activity (Figs. 3A and 5B). It is noteworthy that the maximum change in chain-length pattern even in the SSI null mutant e7−/−, which was within 1.2% (Δ molar %) at DP 9, was much smaller than those of the rice mutants for isoamylase1 (sugary-1) and BEIIb (amylose extender), which reaches up to 5% (Δ molar %) at DP 8 and 4% at DP 9, respectively (Nakamura et al., 1997; Nishi et al., 2001; Nakamura, 2002). This implies that the other SS isozymes at least partially complement SSI deficiency, the uncomplemented function being manifested as alterations in chain length.
On the other hand, the maximum change in amylopectin chain length in SSIIa mutant was 2.3% (Δ molar %) at DP 8 to 9 (Nakamura, 2002; Umemoto et al., 2002), which is larger than that for any SSI mutant. This suggests that the function of SSIIa might be hardly complemented by other SS isozymes including SSI.
Characterization of SSI in Developing Rice Endosperm
The SS activity of the SP in rice developing endosperm obtained by anion-exchange chromatography in this study exhibited two peaks, designated as SSI and SSIIIa (Fig. 1A), consistent with the results obtained in maize endosperm (Ozbun et al., 1971; Cao et al., 2000) and rice endosperm (Tanaka and Akazawa, 1971). The primer dependence of these rice SS isozymes is identical to that of maize (Ozbun et al., 1971; Cao et al., 2000). SSI and SSIIIa are major enzymes in developing rice endosperm and the activity of SSI is higher than that of SSIIIa (Fig. 1, A and B), consistent with the fact that maize SSI and SSIII account for 61% to 66% and 28%, respectively, of the total SS activity in the soluble fraction of developing maize endosperm (Cao et al., 1999). SSI and SSIII are also the main enzyme components of developing wheat endosperm (Li et al., 2000). By contrast, SSII and SSIII account for the major SS activities in potato tuber (Abel et al., 1996; Marshall et al., 1996) and pea embryos (Tomlinson et al., 1998).
Based on the measurement of dissociation constants, Commuri and Keeling (2001) argued that SSI is not able to elongate polyglucan chains after having been entrapped into the long DP > 20 chains. Their speculation is consistent with the process of SSI binding to starch granules in this study. The SSI protein in SP appeared to move into the granule-bound fractions (LBP or TBP) during endosperm development (Fig. 1D). Based on results of Commuri and Keeling (2001) and this study, we further speculate that the active SSI proteins in SP, but not in LBP or TBP even at the late stage of endosperm development, act on the short chains and elongate them just after branch formation by BE(s). The SSI proteins trapped in the long DP > 20 chains might be the proteins bound to starch granules as LBP or TBP. Further elongation of DP > 20 chains should be a function of other SS isozymes after elongation by SSI.
The decrease in DP > 20 chains was not evident in SSI mutant lines compared to their control lines after DAF 16 (Fig. 5C) and in the mature endosperm (Fig. 5B) in vivo. Probably, SSI is not able to elongate DP > 20 chains due to its entrapment by DP > 20 chains after starch concentration becomes high in vivo. In contrast, DP > 20 chains on native-PAGE/SS activity gels increased (Fig. 7B), indicating that SSI has an inherent ability to elongate DP > 20 chains provided it is free from binding by long polyglucans when starch concentration is low as in gels containing rice amylopectin. Increasing the in vitro reaction time for the SSI activity magnified the rate of increase in longer chains produced by the SSI activity band on native-PAGE gel (data not shown).
The Function of Rice SSI
The activities (or Vmax) of maize SSI in developing endosperm (Ozbun et al., 1971; Boyer and Preiss, 1981; Cao et al., 2000) and the recombinant SSI of Arabidopsis (Delvalle et al., 2005), kidney bean (Senoura et al., 2004), and maize (Imparl-Radosevich et al., 1998; Commuri and Keeling, 2001) are higher when glycogen is used as the primer rather than amylopectin, although the Km for glycogen is higher. The predominant polyglucan chains produced by recombinant maize SSI are DP < 10 chains when glycogen (average chain length of DP 6–7) is used as the primer (Commuri and Keeling, 2001). SSI preferentially synthesizes short chains (DP 6–15) when maize SSI and BE are expressed in E. coli (Guan and Keeling, 1998). Recombinant kidney bean SSI predominantly elongates chains around DP 15 and DP < 10 and around 10 and DP < 6 when amylopectin and glycogen are used as the primer, respectively, while SSII produces chains around DP 15 regardless of the primer used (Senoura et al., 2004). These reports suggest that in these plants, SSI has a distinct role of elongating very short chains in vitro.
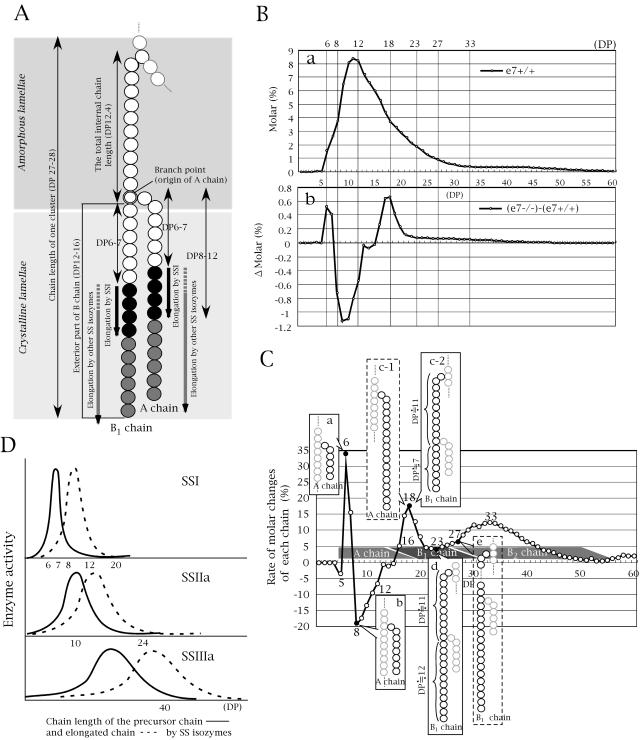
This study is the in vivo attempt to analyze SSI function in storage tissues. The schematic representation of a partial structure of amylopectin is shown in Figure 8A. More than 90% of the chains in rice or maize amylopectin are composed of A chains (that carry no other chains and are linked to the other chains at their reducing ends) and B1 chains (that carry one or more chains belonging to only one cluster; Hizukuri, 1986; Bertoft, 1991). The rate of molar change of each chain (Δ molar %/molar % × 100) in DP 5 to 60 of rice SSI mutant (e7−/−; Fig. 8C) was calculated from the chain-length distribution of endosperm amylopectin in e7+/+ (molar %; Fig. 8B, a) and the differences between the chain-length distribution of e7−/− and e7+/+ (Δ molar %; Fig. 8B, b). We assume that the basic structure of amylopectin such as the proportion of A to B chains and the location of the branch point, is almost the same for the SSI mutant and its wild-type parent as indicated by their similar crystalline type and the crystallinity of their starches (Fig. 4B). Based on this assumption, the variations in chain length and the branching pattern in the SSI mutant from those in the wild type may be explained as follows, as depicted in Figure 8C. As shown in section a, DP 6 and DP 7 chains increased because they were not elongated to DP 8 to 12 due to SSI deficiency (Fig. 8C, inset a). As shown in section b, DP 8 and DP 9 to 12 chains decreased because their precursors DP 6 to 7 chains were not elongated (Fig. 8B, inset b) and most of the available DP 8 to 12 chains were converted into longer chains by other SS isozyme(s) (probably SSIIa and/or SSIIIa). As shown in section c, the increases in DP 18 and DP 16 to 19 chains are mainly attributed to the increase in B1 chains. In maize, the total internal chain length of the B1 chain, which is defined as the length of a B1 chain without its external chain, is estimated to be DP 12.4 (Bertoft, 1991, 2004; see Fig. 8A). If this value holds true for rice amylopectin, the total length of the B1 chain would be defined by the exterior part of the B1 chain (from the branch point connecting the A chain or other B chain to the nonreduced end; Fig. 8A) plus 11. Thus DP 18 chains (B1 chains; Fig. 8C, inset c-2) are predicted to have an exterior portion (A chain) composed of seven Glc units (DP 7), in agreement with the observed length of A chains (DP 6 and 7), which increased in the mutant amylopectin (Fig. 8C, inset a). As shown in section d, the magnitude of the increase in DP 23 chains was significantly lower compared to those in chains with around DP 18 (Fig. 8C). It is likely that the shortage of B1 chains with DP 20 to 23 was due to the nonelongation of the B1 chains having exterior chain lengths with DP 8 to 12 in the absence of SSI (Fig. 8C, inset d).
Figure 8.
A, Schematic representation of the proposed model for the elongation of rice amylopectin glucan chains by SSI and other SS isozymes. Circles represent Glc residues. In the wild type, A and B1 chains grow through the addition of two to six Glc residues (black circles) by SSI. Black and gray circles in A and B1 chains are elongated by other SS isozymes when SSI is deficient. The double circle marks the point in the B1 chain where a branch (A chain) emerges. The A chains and the exterior parts of B chains (from nonreduced end to branch point), both ranging from DP 12 to DP 16 in length (Hizukuri, 1986), compose the crystalline domain of amylopectin clusters. The length of one cluster of amylopectin corresponds to DP 27 to 28 (Hizukuri, 1986). In waxy maize, Bertoft (1991, 2004) estimated the total internal chain length of amylopectin to be DP 12.4. If this value holds true for rice amylopectin, the length of the B1 chain would be the combined length of the exterior part plus DP 11. The partially broken arrows labeled “elongation by other SS isozymes” indicate compensatory function of other SS isozymes when SSI is deficient. B, The chain-length distribution (molar %) of endosperm amylopectin of control line (e7+/+; section a) and the difference in chain-length distribution (Δ molar %) of endosperm amylopectin between the SSI mutant (e7−/−) and the control line (e7+/+; section b). These data are from Figure 5B. C, The rate of molar changes of each chain relative to the amount of its chain (Δ molar %/molar % × 100) as calculated from Figure 8B sections a and b, for DP 5 to 60 amylopectin chains of SSI mutant (e7−/−). The changes in DP 6 to 7 and DP ≥ 23 chain regions where molar percentages are low (Fig. 8B, a) are emphasized. The zones for the A, B1, and B2 chain were estimated along with the x axis from the result of rice amylopectin described in Hizukuri (1986). Insets are the schematic representation of the chain structure in the amylopectin molecule. D, Schematic representation of the proposed range of glucan chains acted upon (solid line) and synthesized (broken line) by SSI, SSIIa, and SSIIIa in developing rice endosperm based on our present results on SSI, previous data on SSIIa (Nakamura et al., 2005), and our present results and those of Jane et al. (1999) on SSIIIa, respectively. Regions of overlaps define the range of glucan chains that could possibly be synthesized by SSIIa and/or SSIIIa in compensation for SSI deficiency.
The crystalline domains of starch granules appear to be composed of A chains and the exterior parts of B chains of amylopectin, their average length being seemingly in the range of DP 12 to 16 (Hizukuri, 1986). The length of these chains is thought to affect gelatinization temperature; the abundance and shortage of DP ≥ 12 to 16 exterior chains (long chains) result in increase and decrease of the starch gelatinization temperature, respectively, while the abundance and shortage of DP < 12 to 16 exterior chains (short chains) result in decrease and increase of the starch gelatinization temperature, respectively. According to our hypothesis described above, the increases in short A chains with DP 6 to 7 (Fig. 8C, inset a) and B1 chains with DP 16 to 19 (Fig. 8C, inset c-2) lower the gelatinization temperature while the decrease in short A chains with DP 8 to 12 (Fig. 8C, inset b) elevates it. The fact that the total increase in the molar percent of DP 6 to 7 plus DP 16 to 19 chains was almost the same as that of the decrease in DP 8 to 12 chains (Fig. 8B, b), tempts us to predict that the gelatinization temperature of the starch in rice SSI mutant and its control line would be the same. However, the Tp of the starch in the rice SSI mutant (e7−/−) was about 3°C higher than that of its control line (Fig. 6A). This inconsistency could be explained by assuming that the increased DP 16 to 19 chains must contain long A chain with DP 16 to 19 elongated by the other SS isozymes (Fig. 8C, inset c-1) as well as nonelongated B1 chains (Fig. 8C, inset c-2) due to SSI deficiency.
The broad increase of about 5% to 12% in chains around DP 27 to 40 might have resulted from the elongation of the exterior portions of B1 and B2 chains by other SS isozyme(s) (Fig. 8C, inset e). The elongation of the exterior parts of B chains or A chains with DP ≥ 12 to 16 by other SS isozyme(s) when SSI is deficient seems to elevate the gelatinization temperature of starch.
Results of the analysis of polyglucans produced in vitro by the SSI activity band on native-PAGE/SS activity staining lend additional support for the current hypothesis. The changes in distribution patterns of amylopectin chains with DP ≤ 18 in the SSI activity band were almost mirror images of Δ molar % patterns of SSI mutant lines with respect to their control lines in vivo (Fig. 7B). The decrease in DP 4 to 6 and DP 15 to 17 chains and the increase in DP 7 to 11 and DP ≥ 19 chains of the polyglucan produced from rice amylopectin on the native-PAGE gel (Fig. 7B) were considered to be consequences of the elongation of A and B1 chains, respectively. These results strongly suggest that SSI distinctly prefers to elongate the short exterior DP 6 to 7 and DP 8 to 12 chains of A and B1 chains of amylopectin.
Results of the comparison of the chain-length distribution of amylopectin between SSIIa mutants and their respective parents in rice (japonica rice; Nakamura et al., 2002; Umemoto et al., 2002), maize (sugary2; Zhang et al., 2004), barley (sex6; Morell et al., 2003), wheat (GSP-1 null; Yamamori et al., 2000), and antisense-SSII potato (Edwards et al., 1999) suggest that SSIIa preferentially elongates DP ≤ 10 chains to DP 12 to 24 chains in vivo. Moreover, Nakamura et al. (2005) showed that indica-type SSIIa expressed in E. coli elongates short DP 6 to 11 chains to DP 13 to 28 chains in japonica rice amylopectin in vitro. The range of the amylopectin chains that SSIIa can use as substrate (DP 6–11) and that of its products (DP 13–28) are broader than those of SSI, which elongates DP 6 to 7 chains to form DP 8 to 12 chains (Fig. 8D). The analyses of the starch structure of maize dull-1 mutant (Inouchi et al., 1987; Jane et al., 1999), antisense-SSIII potato (Lloyd et al., 1999), and SSIII mutant of Arabidopsis (Zhang et al., 2004) indicate that SSIII in these plants produces the long B2 and B3 chains of amylopectin in vivo. The chain analysis of polyglucans generated by the SSIIIa active band on native-PAGE/SS activity staining gel in this study revealed that the shorter DP ≤ 11 chains decreased and longer DP ≥ 12 chains increased (Fig. 7A) when glycogen was used as the primer. We assume that the number of Glc residues added by SSIIIa is more than that added by SSIIa (Fig. 8D).
SSIIIa activity bands in e7−/− SSI null mutant appeared to be enhanced compared with its e7+/+ control line (Fig. 3A). In view of the report that SSI activity in maize SSIII mutant (dull-1) was higher than that of the wild type (Cao et al., 1999), we consider these two findings as related and indicative of the interaction among SS enzymes. This interaction could be a component of a compensatory mechanism for overcoming the deleterious effects of SS isozyme(s) deficiency, a topic that needs immediate attention. Detailed analyses of mutants of other SS isozymes are prerequisites to the full clarification of this compensatory mechanism.
In summary, in wild-type rice, the short DP 6 to 7 chains of A or B1 chain produced by BE are initially elongated by SSI to DP 8 to 12 chains. Further elongation to form DP > 20 chains should be performed by other SS isozymes including SSIIa and SSIIIa. SSI specifically elongates the narrow range of short chains with DP 6 to 7 and adds a limited number of Glc residues (DP 2–6) as compared with SSIIa and SSIIIa (Fig. 8D).
In vivo analysis of amylopectin chain-length distribution pattern was first attempted on leaves of Arabidopsis SSI mutant (Delvalle et al., 2005). Two lines of Arabidopsis SSI mutants showed a significant decrease in DP 8 to 12 chains and a significant increase in DP 17 to 20 chains, which is similar to our current results in rice. Delvalle et al. (2005) attributed the increase in the amounts of medium-length chains in Arabidopsis SSI mutants to the redirection of the activity of other SS isoforms, whereas we think that the increase of DP 16 to 19 in rice SSI mutants is at least partly due to a failure to elongate short B1 chain. However, the increase in DP 6 to 7 in rice SSI mutants was not evident in Arabidopsis SSI mutants, and the decrease in DP 26 to 34 or DP 33 to 38 in Arabidopsis SSI mutants was absent in rice SSI mutants. These discrepancies might be partly attributed to the differences between Arabidopsis SSI and rice SSI in gene structure (57% amino acid sequence similarity), the contribution of SSI relative to other SS isozymes in the overall SS proteins function(s), or pleiotropic effects of SSI deficiency.
More detailed analysis of amylopectin structure in rice SSI mutants, including a more precise estimation of the lengths of A and B chains is required to substantiate the current hypothesis.
MATERIALS AND METHODS
Plant Materials
The wild-type parental rice (Oryza sativa) cultivar Nipponbare, its four mutant lines (−/−) containing Tos17 insertion at the OsSSI gene, and their control plants (+/+; Tos17 not inserted on the SSI gene but have a genetic background common to all mutant lines) were used in the study. Rice plants were grown during the summer months in an experimental paddy field under natural environmental conditions.
Tos17 Mutagenesis and Screening for SSI-Deficient Mutant Lines by PCR
Mutagenesis with Tos17 and pool sampling were performed as described previously (Hirochika, 2001; Kumar and Hirochika, 2001). To screen for SSI-deficient mutant lines, DNA fragments carrying the Tos17 transposon from DNA pools constructed using the three-dimensional sampling method from approximately 40,000 Tos17-containing plants were subjected to nested PCR using the transposon-specific primers T1F (for first PCR), T2F (for second PCR), T1R (for first PCR), and T2R (for second PCR; Fig. 2A) and the OsSSI specific primers 1R (for first PCR) and 2R (for second PCR; Fig. 2A). The PCR products were hybridized with the 1.2 kb OsSSI cDNA NdeI fragment probe (Fig. 2A), and positive products were gel purified and sequenced to identify those containing the real OsSSI sequence and the location of Tos17 in the OsSSI gene.
Preparation of Enzyme from Developing Rice Endosperm and Fractionation of Crude Enzyme Extract Using Anion-Exchange Column
Developing rice grains at the midmilky stage (12 g fresh weight) were hand homogenized using a prechilled mortar and pestle on ice in 30 mL of grinding solution (GS) containing 50 mm imidazole-HCl (pH 7.4), 8 mm MgCl2, 500 mm 2-mercaptoethanol, and 12.5% (v/v) glycerol. The homogenate was squeezed through two layers of gauze and the filtrate was centrifuged at 10,000g at 4°C for 20 min. The supernatant was once more centrifuged at the same conditions and the supernatant was used as the crude enzyme extract.
The crude enzyme preparation (about 30 mL) was applied to a HiTrapQ HP column (5 mL, Amersham Bioscience) equilibrated with Solution A (50 mm imidazole-HCl [pH 7.4], 8 mm MgCl2, 50 mm 2-mercaptoethanol). The column was washed with Solution A, followed by a linear gradient of 0 to 0.5 m NaCl in Solution A for 30 min at a flow rate of 1.0 mL min−1.
Enzyme Assay
The assay of SS for each fraction from the HiTrapQ column was conducted in a reaction mixture containing citrate and glycogen following the method of Nishi et al. (2001). The assay for AGPase was performed using the methods of Nakamura et al. (1989).
Native-PAGE/Activity Staining
Native-PAGE/activity staining of DBE and BE was performed using the methods of Fujita et al. (1999) and Yamanouchi and Nakamura (1992), respectively. SS activity staining was performed on 7.5% and 6.0% (w/v) acrylamide slab gel containing 0.8% (w/v) oyster glycogen (Sigma) and 0.1% rice amylopectin purified from rice waxy mutant line (EM-21), respectively, according to Nishi et al. (2001) with the modification that 0.5 m citrate was included in the reaction mixture for oyster gel but not for the rice amylopectin gel.
Extraction of Proteins from the Developing Endosperm
One developing endosperm each from DAF 9 to 25 of Nipponbare, DAF 12 of SSI-deficient mutant lines, and their controls were individually homogenized using a plastic pestle in 3 volumes of cold GS. The homogenate was centrifuged at 20,000g at 4°C for 10 min and supernatant was set aside. The pellet was washed twice with 2 volumes of cold GS and the pooled supernatants (about 80 μL) were used as the SP fraction. The residual pellet was homogenized in 3 volumes of cold SDS solution containing 55 mm Tris-HCl (pH 6.8), 2.3% SDS, 5% 2-mercaptoethanol, and 10% glycerol. The homogenate was centrifuged at 20,000g at 4°C for 10 min and the supernatant was set aside. The pellet was washed twice with 2 volumes of cold SDS solution and the pooled supernatants (about 80 μL) were used as the LBP fraction. The residual pellet (starch granules) was washed with 1 mL of distilled water and twice with 1 mL of acetone and dried under pressure. The starch granules (about 4 mg) were suspended with 10 volumes of SDS solution and boiled for 7 min. After cooling, 10 volumes of SDS solution were added while stirring. The slurry was centrifuged at 20,000g for 10 min at 4°C. The supernatant was set aside and the pellet was resuspended in 10 volumes of SDS solution and recentrifuged. The pooled supernatants (about 80 μL) were used as the TBP fraction.
Preparation of Antibody against SSI and GBSSI of Rice and Immunoblotting
Polyclonal antibodies for rice SSI and GBSSI were prepared from SDS-PAGE purified granule-bound proteins. The SP and LBP were sequentially removed from 10 g of starch granules isolated from developing rice seeds. The remaining TBP was suspended in 24 volumes of SDS solution and boiled for 10 min. After cooling, the slurry was homogenized and centrifuged at 10,000g for 30 min at 4°C. The supernatant was set aside and the pellet was resuspended in 40 volumes of SDS solution and recentrifuged. An equal volume of 30% tricholoroacetic acid was added to the pooled supernatants (about 640 mL) and mixed. The solution was cooled on ice for 1 h and centrifuged at 40,000g for 15 min at 4°C. The pellet was washed with 50 mL of acetone and dried under pressure. The dried pellet was solubilized with 8 mL of SDS solution and analyzed on a 7.5% acrylamide SDS-PAGE gel (145 mm × 100 mm, 1 mm thick). The SSI (71 kD) and GBSSI (60 kD) proteins were excised and the gel slices containing 30 mg SSI and 200 mg GBSSI protein were homogenized with Freund's incomplete adjuvant and injected into rabbits. Four injections were administered at 2-week intervals until the titer of polyclonal antibodies against the rice SSI and GBSSI were fully elevated. The serum was pooled and used as a source of polyclonal antibodies. Immunoblotting was performed according to the methods of Fujita et al. (1999).
Chain-Length Distribution of Polyglucans from Endosperm and SS Bands on Native-PAGE Gels
Extraction of starch from mature and developing rice endosperm for chain-length distribution was performed according to the methods of Fujita et al. (2001).
Native-PAGE/SS activity staining for analysis of modified α-polyglucans from gels were performed as described above. The SSI and SSIIIa bands, 100 mg each, were excised and homogenized with plastic pestles in 1 mL of distilled water (DW) and centrifuged at 20,000g for 1 min at 4°C to remove the supernatant. DW (185 μL) was added to the gels to a total volume of 285 μL, followed by 15 μL of 5 n NaOH and mixed. The homogenate was boiled for 5 min and subjected to debranching of the polyglucans according to Fujita et al. (2001).
The chain-length distributions of α-polyglucans from endosperm and SS bands from the native-PAGE gels were analyzed using the capillary electrophoresis methods of O'Shea and Morell (1996) and Fujita et al. (2001) in a P/ACE MDQ carbohydrate system (Beckman Coulters).
Northern Blotting and RT-PCR
Northern blotting and RT-PCR of SSI gene using the primer pair 5′-gggccttcatggatcaacc-3′ and 5′-ccgcttcaagcatcctcatc-3′ were performed according to the methods of Tanaka et al. (2004) and Ohdan et al. (2005), respectively.
Analysis of Starch Granules of Endosperm
Pasting properties of endosperm starch measured by rapid visco analyzer, x-ray diffraction measurement of endosperm starch, and observation of endosperm starch granules by SEM were performed as described previously (Fujita et al., 2003). For the measurement of thermal properties of endosperm starch by DSC, dried rice grain was dehulled, crushed with pliers, and hand homogenized using mortar and pestle. The weighed starch (about 3 mg) was placed in an aluminum sample cup (SSC000C009, Seiko Instrument) mixed with 9 μL of DW and sealed. Gelatinization properties of the starch were analyzed by a differential scanning calorimeter (DSC-6100, Seiko Instrument). The heating rate was 3°C min−1 over a temperature range of 5°C to 90°C. For measurement of λmax of the iodine-starch complex, a dehulled rice seed was powdered using a mortar and pestle and about 7.5 mg of powder was homogenized with 600 μL of DW and 150 μL of 5 n NaOH and incubated overnight at 4°C. After incubation, the homogenate was boiled for 5 min and neutralized with 750 μL of 1 n HCl. A 5 μL aliquot of 10 times diluted solution was stained with 45 μL of 0.5% KI/0.05% I2 solution. The iodine-starch complex was scanned from 450 to 650 nm using a spectrophotometer (DU7400, Beckman Coulter).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number D38221.
Acknowledgments
The authors are grateful to Professor Hikaru Satoh (Kyushu University) for kindly providing us the waxy mutant line EM-21, to Dr. Perigio B. Francisco Jr. (Japan Science and Technology-Core Research for Evolutional Science and Technology, Akita) for reading the manuscript, and to Professor Jay-Lin Jane (Iowa State University) for helpful discussions.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Naoko Fujita (naokof@akita-pu.ac.jp).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071845.
References
- Abel GJW, Springer F, Willmitzer L, Kossman J (1996) Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.). Plant J 10: 981–991 [DOI] [PubMed] [Google Scholar]
- Baba T, Nishihara M, Mizuno K, Kawasaki T, Shimada H, Kobayashi E, Ohnishi S, Tanaka K-I, Arai Y (1993) Identification, cDNA cloning, gene expression of soluble starch synthase in rice (Oryza sativa L.) immature seeds. Plant Physiol 103: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoft E (1991) Investigation of the fine structure of alpha-dextrins derived from amylopectin and their relation to the structure of waxy-maize starch. Carbohydr Res 212: 229–244 [DOI] [PubMed] [Google Scholar]
- Bertoft E (2004) On the nature of categories of chains in amylopectin and their connection to the super helix model. Carbohydr Polym 57: 211–224 [Google Scholar]
- Boyer CD, Preiss J (1981) Evidence for independent genetic control of the multiple forms of maize endosperm branching enzymes and starch synthases. Plant Physiol 67: 1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Imparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM (1999) Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 120: 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, James MG, Myers AM (2000) Purification and characterization of soluble starch synthases from maize endosperm. Arch Biochem Biophys 373: 135–146 [DOI] [PubMed] [Google Scholar]
- Commuri PD, Keeling PL (2001) Chain-length specificities of maize starch synthase I enzyme: studies of glucan affinity and catalytic properties. Plant J 25: 475–486 [DOI] [PubMed] [Google Scholar]
- Craig J, Lloyd JR, Tomlinson K, Barber L, Edwards A, Wang TL, Martin C, Hedley CL, Smith AM (1998) Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell 10: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvalle D, Dumez S, Wattebled F, Roldan I, Planchot V, Berbezy P, Colonna P, Vyas D, Chatterjee M, Ball S, et al (2005) Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J 43: 398–412 [DOI] [PubMed] [Google Scholar]
- Denyer K, Barber LM, Burton R, Hedley CL, Hylton CM, Johnson S, Jones DA, Marshall J, Smith AM, Tatge H, et al (1995) The isolation and characterization of novel low-amylose mutants of Pisum sativum L. Plant Cell Environ 18: 1019–1026 [Google Scholar]
- Edwards A, Fulton DC, Hylton CM, Jobling SA, Gidley M, Rossner U, Martin C, Smith AM (1999) A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J 17: 251–261 [Google Scholar]
- Edwards A, Marshall J, Sidebottom C, Visser RGF, Smith AM, Martin C (1995) Biochemical and molecular characterization of a novel starch synthase from potato tubers. Plant J 8: 283–294 [DOI] [PubMed] [Google Scholar]
- Eriksson G (1962) Radiation induced reversions of a waxy allele in barley. Radiat Bot 2: 35–39 [Google Scholar]
- Fujita N, Hasegawa H, Taira T (2001) The isolation and characterization of a waxy mutant of diploid wheat (Triticum monococcum L.). Plant Sci 160: 595–602 [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Francisco PB Jr, Nakakita M, Harada K, Minaka N, Nakamura Y (1999) Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta 208: 283–293 [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Suh S-D, Wong K-S, Jane J-L, Ozawa K, Takaiwa F, Inaba Y, Nakamura Y (2003) Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiol 44: 607–618 [DOI] [PubMed] [Google Scholar]
- Fujita N, Taira T (1998) A 56-kDa protein is a novel granule-bound starch synthase existing in the pericarps, aleurone layers, and embryos of immature seed in diploid wheat (Triticum monococcum L.). Planta 207: 125–132 [DOI] [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM (1998) Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell 10: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Keeling PL (1998) Starch biosynthesis: understanding the functions and interactions of multiple isozymes of starch synthase and branching enzyme. Trends Glycosci Glycotechnol 10: 307–319 [Google Scholar]
- Harn C, Knight M, Ramakrishnan A, Guan H, Keeling PL, Wasserman BP (1998) Isolation and characterization of the zSSIIa and zSSIIb starch synthase cDNA clones from maize endosperm. Plant Mol Biol 37: 639–649 [DOI] [PubMed] [Google Scholar]
- Hirochika H (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4: 118–122 [DOI] [PubMed] [Google Scholar]
- Hirose T, Terao T (2004) A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220: 9–16 [DOI] [PubMed] [Google Scholar]
- Hizukuri S (1986) Polymodal distribution of the chain lengths of amylopectins, and its significance. Carbohydr Res 147: 342–347 [Google Scholar]
- Hovenkamp-Hermelink JHM, Jacobsen E, Ponstein AS, Visser RGF, Vos-Scheperkeuter GH, Bijmolt EW, de Vries JN, Witholt B, Feenstra WJ (1987) Isolation of an amylose-free starch mutant of the potato (Solanum tuberosum L.). Theor Appl Genet 75: 217–221 [Google Scholar]
- Imparl-Radosevich JM, Li P, Zhang L, McKean AL, Keeling PL, Guan H (1998) Purification and characterization of maize starch synthase I and its truncated forms. Arch Biochem Biophys 353: 64–72 [DOI] [PubMed] [Google Scholar]
- Imparl-Radosevich JM, Nichols DJ, Li P, McKean AL, Keeling PL, Guan H (1999) Analysis of purified maize starch synthase IIa and IIb: SS isoforms can be distinguished based on their kinetic properties. Arch Biochem Biophys 362: 131–138 [DOI] [PubMed] [Google Scholar]
- Inouchi N, Glover DV, Fuwa H (1987) Chain length distribution of amylopectins of several single mutants and the normal counterpart, and sugary-1 phytoglycogen in maize (Zea mays L.). Starch 39: 259–266 [Google Scholar]
- Isono N, Senoura T, Yoshikawa M, Sakurai Y, Watanabe K, Ito H, Matsui H (2004) Occurrence of multiple forms for starch synthase II isozyme in developing seeds of kidney bean. J Appl Glycosci (1999) 51: 101–107 [Google Scholar]
- Jane J-L, Chen YY, Lee LF, McPherson AE, Wong K-S, Radosavljevic M, Kasemsuwan T (1999) Effects of amylopectin branch chain length and amylose content on the gelatinization and pasting properties of starch. Cereal Chem 76: 629–637 [Google Scholar]
- Jenkins PJ, Cameron RE, Donald AM (1993) A universal feature in the structure of starch granules from different botanical sources. Starch 45: 417–420 [Google Scholar]
- Jiang H, Dian W, Liu F, Wu P (2004) Molecular cloning and expression analysis of three genes encoding starch synthase II in rice. Planta 218: 1062–1070 [DOI] [PubMed] [Google Scholar]
- Knight ME, Harn C, Lilley CER, Guan H, Singletary GW, Mu-Forster C, Wasserman BP, Keeling PL (1998) Molecular cloning of starch synthase I from maize (W64) endosperm and expression in Escherichia coli. Plant J 14: 613–622 [DOI] [PubMed] [Google Scholar]
- Kossmann J, Abel GJW, Springer F, Lloyd JR, Willmitzer L (1999) Cloning and functional analysis of a cDNA encoding a starch synthase from potato (Solanum tuberosum L.) that is predominantly expressed in leaf tissue. Planta 208: 503–511 [DOI] [PubMed] [Google Scholar]
- Kumar A, Hirochika H (2001) Applications of retrotransposons as genetic tools in plant biology. Trends Plant Sci 6: 127–134 [DOI] [PubMed] [Google Scholar]
- Li Z, Mouille G, Kosar-Hashemi B, Rahman S, Clarke B, Gale KR, Appels R, Morell MK (2000) The structure and expression of the wheat starch synthase III gene: Motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol 123: 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Landshutze V, Kossmann J (1999) Simultaneous antisense inhibition of two starch-synthase isoforms in potato tubers leads to accumulation of grossly modified amylopectin. Biochem J 228: 515–521 [PMC free article] [PubMed] [Google Scholar]
- Marshall J, Sidebottom C, Debet M, Martin C, Smith AM (1996) Identification of the major starch synthase in the soluble fraction of potato tubers. Plant Cell 8: 1121–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Senoura T, Isono N, Sakurai Y, Hamada S, Ito H (2003) Granule-bound starch synthase I isozyme localized predominantly in kidney bean leaves. J Appl Glycosci (1999) 50: 493–497 [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z (2003) Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J 34: 173–185 [DOI] [PubMed] [Google Scholar]
- Mu-Forster C, Huang R, Powers JR, Harriman RW, Knight M, Singletary GW, Keeling PL, Wasserman BP (1996) Physical association of starch biosynthetic enzymes with starch granules of maize endosperm. Plant Physiol 111: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG (2000) Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol 122: 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Vrinten P, Hayakawa K, Ikeda J (1998) Characterization of a granule-bound starch synthase isoform found in the pericarp of wheat. Plant Physiol 118: 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Yamamori M, Hirano H, Hidaka S, Nagamine T (1995) Production of waxy (amylose-free) wheats. Mol Gen Genet 248: 243–259 [DOI] [PubMed] [Google Scholar]
- Nakamura Y (2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol 43: 718–725 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Francisco PB Jr, Hosaka Y, Satoh A, Sawada T, Kubo A, Fujita N (2005) Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol 58: 213–227 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kubo A, Shimamune T, Matsuda T, Harada K, Satoh H (1997) Correlation between activities of starch debranching enzyme and α-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J 12: 143–153 [Google Scholar]
- Nakamura Y, Sakurai A, Inaba Y, Kimura K, Iwasawa N, Nagamine T (2002) The fine structure of amylopectin in endosperm from Asian cultivated rice can be largely classified into two classes. Starch 54: 117–131 [Google Scholar]
- Nakamura Y, Yuki K, Park SY, Ohya T (1989) Carbohydrate metabolism in the developing endosperm of rice grains. Plant Cell Physiol 30: 833–839 [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H (2001) Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol 127: 459–472 [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56: 3229–3244 [DOI] [PubMed] [Google Scholar]
- O'Shea MG, Morell MK (1996) High resolution slab gel electrophoresis of 8-amino-1,3, 6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis 17: 681–688 [DOI] [PubMed] [Google Scholar]
- Ozbun JL, Hawker JS, Preiss J (1971) Adenosine diphosphoglucose-starch glucosyltransferases from developing kernels of waxy maize. Plant Physiol 48: 765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y (1984) Differential regulation of waxy gene expression in rice endosperm. Theor Appl Genet 68: 467–473 [DOI] [PubMed] [Google Scholar]
- Sato K (1984) Starch granules in tissues of rice plants and their changes in relation to plant growth. JARQ 18: 78–86 [Google Scholar]
- Senoura T, Isono N, Yoshikawa M, Asao A, Hamada S, Watanabe K, Ito H, Matsui H (2004) Characterization of starch synthase I and II expressed in early developing seeds of kidney bean (Phaseolus vulgaris L.). Biosci Biotechnol Biochem 68: 1949–1960 [DOI] [PubMed] [Google Scholar]
- Smith AM, Denyer K, Martin C (1997) The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol 48: 67–87 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Fujita N, Nishi A, Satoh H, Hosaka Y, Ugaki M, Kawasaki S, Nakamura Y (2004) The structure of starch can be manipulated by changing the expression levels of starch branching enzymes IIb in rice endosperm. Plant Biotechnol J 2: 507–516 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Akazawa T (1971) Enzymic mechanism of starch synthesis in ripening rice grains. VI. Isozymes of starch synthetase. Plant Cell Physiol 12: 493–505 [Google Scholar]
- Tomlinson K, Craig J, Smith AM (1998) Major differences in isoform composition of starch synthase between leaves and embryos of pea. Planta 204: 86–92 [Google Scholar]
- Tsai CY (1974) The function of waxy locus in starch synthesis in maize endosperm. Biochem Genet 11: 83–96 [DOI] [PubMed] [Google Scholar]
- Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y (2002) Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet 104: 1–8 [DOI] [PubMed] [Google Scholar]
- Yamamori M, Fujita S, Hayakawa K, Matsuki J, Yasui T (2000) Genetic elimination of a starch granule protein, SGP-1, of wheat generates an altered starch with apparent high amylose. Theor Appl Genet 101: 21–29 [Google Scholar]
- Yamanouchi H, Nakamura Y (1992) Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant Cell Physiol 33: 985–991 [Google Scholar]
- Zhang X, Colleoni C, Ratushna V, Sirghie-Colleoni M, James MG, Myers AM (2004) Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa. Plant Mol Biol 54: 865–879 [DOI] [PubMed] [Google Scholar]
- Zhang X, Myers AM, James MG (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol 138: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]