Abstract
The Arabidopsis (Arabidopsis thaliana) orthologs of Brca2, a protein whose mutations are involved in breast cancer in humans, were previously shown to be essential at meiosis. In an attempt to better understand the Brca2-interacting properties, we examined four partners of the two isoforms of Brca2 identified in Arabidopsis (AtRad51, AtDmc1, and two AtDss1 isoforms). The two Brca2 and the two Dss1 isoforms are named AtBrca2(IV), AtBrca2(V), AtDss1(I), and AtDss1(V) after their chromosomal localization. We first show that both AtBrca2 proteins can interact with either AtRad51 or AtDmc1 in vitro, and that the N-terminal region of AtBrca2 is responsible for these interactions. More specifically, the BRC motifs (so called because iterated in the Brca2 protein) in Brca2 are involved in these interactions: BRC motif number 2 (BRC2) alone can interact with AtDmc1, whereas BRC motif number 4 (BRC4) recognizes AtRad51. The human Rad51 and Dmc1 proteins themselves can interact with either the complete (HsRad51) or a shorter version of AtBrca2 (HsRad51 or HsDmc1) that comprises all four BRC motifs. We also identified two Arabidopsis isoforms of Dss1, another known partner of Brca2 in other organisms. Although all four Brca2 and Dss1 proteins are much conserved, AtBrca2(IV) interacts with only one of these AtDss1 proteins, whereas AtBrca2(V) interacts with both of them. Finally, we show for the first time that an AtBrca2 protein could bind two different partners at the same time: AtRad51 and AtDss1(I), or AtDmc1 and AtDss1(I).
Mutations in the BREAST CANCER 2 (BRCA2) gene are responsible for a hereditary form of breast cancer predisposition in human and are generally associated with genetic instabilities (Wooster et al., 1995; Scully and Livingston, 2000). However, the function of Brca2 remained quite obscure until it was found to interact with Rad51, the eukaryotic RecA homolog, suggesting a role for Brca2 in homologous recombination and DNA repair (Sharan et al., 1997). Consistently, brca2 knockout mice show early embryonic lethality associated with chromosomal rearrangements and breaks (Yu et al., 2000), a phenotype that is reminiscent of rad51 knockout mice (Lim and Hasty, 1996; Tsuzuki et al., 1996), while Rad51 does not organize into foci following DNA damage in Brca2-deficient cells (Yu et al., 2000). A similar defect in Rad51 and in Dmc1 (the meiotic RecA homolog in eukaryotes) foci formation was observed in the meiotically arrested cells of brca2 knockout mice whose viability was restored by a human BRCA2 gene (Sharan et al., 2003). While Brca2 was known to localize on the meiotic chromosomes during the formation of the synaptonemal complex in human (Chen et al., 1998a), the interaction disclosed between Brca2 and Dmc1 in Arabidopsis (Arabidopsis thaliana) helped assign a role for Brca2 at meiosis (Siaud et al., 2004). Another interactant of Brca2, Dss1, was uncovered (Marston et al., 1999), the depletion of which leads to a brca2-like phenotype in Ustilago or mouse (Kojic et al., 2003; Gudmundsdottir et al., 2004). Its yeast (Saccharomyces cerevisiae) counterpart, Sem1, is found associated with the 19S proteasome, but it also participates in the resistance to genotoxic treatment (Funakoshi et al., 2004; Krogan et al., 2004), although no Brca2-like protein can be identified in yeast. These results converge to underline how characterization of its partners was key to better understanding the role of Brca2.
The Brca2 protein thus became subject of intense examination from both a structural and a biochemical point of view, especially in view of its connection with Rad51 and the newly found interactant Dss1 that could be involved in Brca2 stability. Crystallization of Rad51 in the presence of a BRC motif (so called because iterated in the Brca2 protein; Bork et al., 1996) allowed definition of critical amino acids in BRC that mimic the Rad51 interface between Rad51 monomers, suggesting that Brca2 could modulate the association of Rad51 into a nucleofilament (Pellegrini et al., 2002). Yang et al. (2002) demonstrated that a DNA-binding domain is present in the 800-amino acid C-terminal region of Brca2 that can stimulate Replication Protein A (RPA)-dependent strand transfer between homologous DNA molecules by Rad51; this region could only be crystallized in the presence of Dss1, which allowed the authors to define domains in this C-terminal region of Brca2 that were involved in binding Dss1. Work from Galkin et al. (2005) also indicates that the BRC motifs number 3 and number 4 in human Brca2 can form stable complexes with Rad51-DNA nucleoprotein filaments. They also reveal the regions in Rad51 that are involved in contacting the BRC motifs. Finally, a full-length Ustilago maydis Brca2 ortholog (called Brh2) was able to stimulate Rad51-mediated recombination in vitro, acting preferentially at the junction between single-strand and double-strand DNA (Yang et al., 2005). Furthermore, addition of Brh2 to RPA-coated single-stranded regions of double-strand DNA molecules increases the amount of Rad51 incorporation, thus showing that Brh2 overcomes the inhibition of filament formation by RPA (Yang et al., 2005). Recent data show that Dss1 is required for Brh2 to load Rad51 onto RPA-coated single-strand DNA at a duplex junction (Kojic et al., 2005). Dss1 could thus regulate the recombination activity of the Brh2/Rad51 complex in vivo. But no concomitant physical interaction between either all three Brca2, Rad51, and Dss1, or Brca2, Dmc1, and Dss1 was evidenced at this time.
The human BRCA2 gene encodes for a 3,418-amino-acid-long protein, with eight internal iterated BRC motifs that are typical of Brca2 proteins and are responsible for the interaction with Rad51 (Wong et al., 1997; Chen et al., 1998b). Brca2-related proteins can be identified in other eukaryotic genomes that are extremely heterogeneous in their sizes (down to 394 amino acids in Caenorhabditis elegans) and the number of BRC repeats they contain (from one in C. elegans or Ustilago to 15 in Trypanosoma Brca2 proteins; Lo et al., 2003). However, the ability of Brca2 to associate with Rad51 is conserved in Ustilago, Arabidopsis, and C. elegans. In Arabidopsis, two proteins of 1,151/1,155 amino acids, with four BRC repeats each, can be defined: AtBrca2(IV) and AtBrca2(V) that are 94.5% identical (Siaud et al., 2004; Kojic et al., 2005; Martin et al., 2005). Studies considering the role of Brca2 in organisms other than mammals offered interesting clues about its function in vivo, especially since Brca2-depleted Ustilago, Arabidopsis, or Caenorhabditis were viable. Brca2 was thus found to be essential at meiosis in Arabidopsis and C. elegans, while its mutation led to premeiotic arrest and triggered genomic rearrangements in Ustilago (Siaud et al., 2004; Kojic et al., 2005; Martin et al., 2005). An experimental advantage of these heterologous Brca2 proteins is their shorter size, compared with the human Brca2, which facilitates in vitro examination of their activity or potential interactants with a complete protein (Yang et al., 2005).
With this work, we establish the ability of the Arabidopsis Brca2 proteins to interact in vitro with not only Rad51 but also Dmc1, a Brca2 partner that was only uncovered in Arabidopsis (Siaud et al., 2004). While the interaction between Brca2 and Rad51 could be inhibited in the presence of a human BRC repeat, this was not the case for Dmc1. However, we could restrict the region of Brca2 involved in the interaction with Dmc1 to one BRC repeat. We also describe the ability of the two Brca2 proteins to differentially bind with the two Dss1 proteins that are present in Arabidopsis. Finally, we show that ternary complexes can form in vitro that comprise either Rad51 and Brca2(V)/Dss1, or Dmc1 and Brca2(V)/Dss1.
RESULTS
In a Coimmunoprecipitation Assay, AtBrca2 Interacts with AtRad51 or AtDmc1
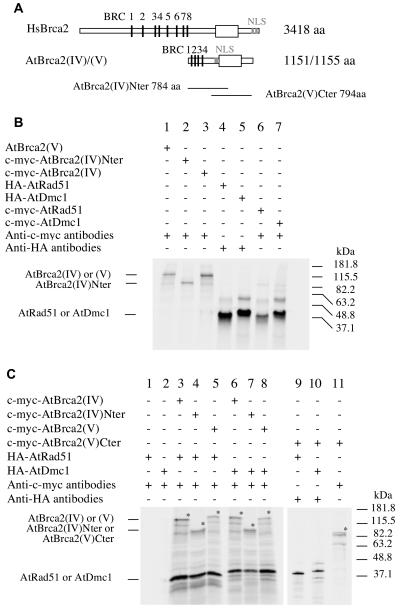
A typical feature of the Brca2 proteins is their ability to interact with Rad51, which was previously observed for both Arabidopsis Brca2(IV) and Brca2(V) protein isoforms in a yeast two-hybrid procedure (Siaud et al., 2004). A truncated 784-amino-acid-long N-terminal version of AtBrca2(IV), AtBrca2(IV)Nter (Fig. 1A; previously called AtBrca2(IV)FS by Siaud et al. [2004]), was also produced in the course of this work. This shorter AtBrca2(IV) comprises all four BRC domains, which was found to be sufficient for it to bind AtRad51 in a yeast two-hybrid assay (Siaud et al., 2004). This property of AtBrca2 was further examined by conducting coimmunoprecipitation assays between the protein products of AtBRCA2 and AtRAD51 in vitro transcribed and translated with adequate tags (Fig. 1B). Thus, we could use specific binding of one or the other target protein. We could then determine whether it bound a potential interacting partner after pairwise in vitro incubation. Following incubation of a c-myc-tagged Brca2 protein in the presence of hemaglutinin (HA)-tagged Rad51 protein, c-myc antibodies retained a complex that could be separated into two bands on a SDS-PAGE gel: one band migrated at the c-myc-AtBrca2 expected size [126.6 kD or 127.2 kD for AtBrca2(IV) and AtBrca2(V), respectively, plus 2.3 kD due to the c-myc tag], while the other band migrated at the HA-AtRad51 expected size (39.3 kD; Fig. 1C, lanes 3 and 5). Incubation of the truncated c-myc-AtBrca2(IV)Nter protein (85.6 kD) with HA-tagged AtRad51 similarly led to coimmunoprecipitation of both proteins in the presence of anti-c-myc antibodies (Fig. 1C, lane 4). Reciprocally, the same result was obtained if HA antibodies were used to bind the resulting complex following in vitro incubation. When c-myc-tagged Brca2 or HA-tagged Rad51 were subjected to, respectively, HA antibodies or c-myc antibodies, no protein was found to be retained following SDS-PAGE migration of the reaction product and autoradiography. Faint bands of smaller sizes, which were not seen after in vitro protein synthesis (Fig. 1B), could also be detected that may correspond to protein degradation occurring in the course of the coimmunoprecipitation experiment. Altogether, our data indicate that each AtBrca2 protein, as well as the truncated AtBrca2(IV)Nter, can coimmunoprecipitate with AtRad51 (Fig. 1B).
Figure 1.
In vitro protein synthesis and coimmunoprecipitation assays of the AtRad51, AtDmc1, and AtBrca2 proteins. A, The AtBrca2 proteins (both share the same structure) compared to the HsBrca2 protein. The BRC motifs are presented as black bars. The box represents a conserved sequence between human and Arabidopsis proteins, and the vertical gray bars indicate the nuclear localization signal (NLS). B, Autoradiography of the products of the coupled transcription-translation reaction. Lane 1, c-myc-AtBrca2(IV); lane 2, c-myc-Nter-AtBrca2(IV); lane 3, c-myc-AtBrca2(V); lane 4, HA-AtRad51; lane 5, HA-AtDmc1; lane 6, c-myc-AtRad51; lane 7, c-myc-AtDmc1. C, Autoradiography after coimmunoprecipitation assays. Lanes 1 and 2, Controls. HA-AtRad51 is not retained when precipitated with anti-c-myc antibodies (lane 1); neither is HA-AtDmc1 (lane 2). Lanes 3 to 5, HA-AtRad51 retained by c-myc-AtBrca2(IV) (lane 3), c-myc-AtBrca2(IV)Nter (lane 4), and c-myc-AtBrca2(V) (lane 5). Lanes 6 to 8, HA-AtDmc1 retained by c-myc-AtBrca2(IV) (lane 6), c-myc-AtBrca2(IV)Nter (lane 7), and c-myc-AtBrca2(V) (lane 8). Lanes 9 to 11, HA-At-Rad51 (lane 9) or HA-AtDmc1 (lane 10) precipitated by anti-HA antibodies does not retain c-myc-AtBrca2(V)Cter (lane 11).
Using the yeast two-hybrid procedure, we previously identified AtDmc1 as a new Brca2-interacting partner. The interaction between the in vitro translated AtBrca2(IV) or (V) and AtDmc1 was similarly confirmed by coimmunoprecipitation. Following incubation of a c-myc-tagged Brca2 protein in the presence of HA-tagged Dmc1 protein, c-myc antibodies retained a complex comprising two proteins that migrated on a SDS-PAGE gel at the sizes predicted for c-myc-AtBrca2 and HA-AtDmc1 (39.5 kD; Fig. 1C, lanes 6 and 8). In a similar assay, AtBrca2(IV)Nter coimmunoprecipitates with AtDmc1, thus restricting the Dmc1-interacting domain of AtBrca2 to its N-terminal region, in which the BRC motifs are located (Fig. 1C, lane 7). Neither AtRad51 nor AtDmc1 interacted with c-myc-AtBrca2(V)Cter, a C-terminal version of AtBrca2(V) in which the BRC motifs region was eliminated (Fig. 1, A and C, lanes 9 and 10). The interactions between AtBrca2 and AtRad51 or between AtBrca2 and AtDmc1 were evidenced in the absence of any other Arabidopsis proteins, and were not affected by a DNase or RNase treatment prior to the incubation step (data not shown).
AtRad51 and AtDmc1 Interact Together or with Their Human Counterparts in Homotypic or Heterotypic Combinations
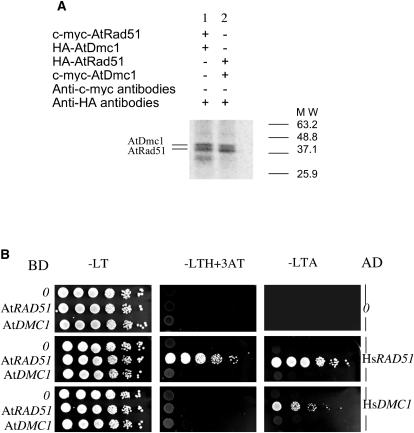
Following incubation of AtRad51 and AtDmc1 in various pairwise combinations, we also established that these proteins could interact in vitro in homotypic or heterotypic combinations. These interactions had previously been observed using the yeast two-hybrid procedure but required further examination (Siaud et al., 2004). Upon binding of the c-myc-tagged AtRad51 onto the Sepharose column, HA-tagged AtRad51 could be coimmunoprecipitated, and, similarly, a c-myc-tagged AtDmc1 allowed coimmunoprecipitation of HA-AtDmc1, evidence of their polymerization properties. After immunoprecipitation and SDS-PAGE analysis of the eluate, only one band was visible around 39 kD. The presence of both tagged proteins in this unique band was ascertained by western blot, using anti-tag antibodies (data not shown). A similar approach was used to analyze the ability of AtRad51 and AtDmc1 to assemble in vitro. HA-tagged Dmc1 retained c-myc-tagged AtRad51 in the presence of anti-HA antibodies, and, reciprocally, c-myc-AtDmc1 retained HA-AtRad51. AtRad51 and AtDmc1 are very similar in term of molecular mass (37.3 kD and 37.5 kD respectively). Nevertheless, their migration patterns can be distinguished, which allowed us to ascertain their assembly in a heterotypic fashion (Fig. 2A).
Figure 2.
A, AtRad51-AtDmc1 heterodimers assembly. Lane 1, c-myc-AtRad51 retained by HA-AtDmc1. Lane 2, c-myc-AtDmc1 retained by HA-AtRad51. B, Yeast two-hybrid assays for interactions between AtRad51 or AtDmc1 and HsRad51 or HsDmc1. −LT, −LTH+3AT, and −LTA correspond to dropout media without L, Leu; T, Trp; H, His; or A, Ade. The −LTH medium was supplemented with 5 mm 3-amino-triazole (3AT). Several dilutions of diploid strains were realized before spotting on media. Yeasts containing both vectors grow on −LT; positive interactions appear as white spots on −LTH+3AT.
A yeast two-hybrid assay was performed to investigate the ability of the human Rad51 and Dmc1 proteins to recognize and interact with the Arabidopsis proteins. Interactions were effective between AtRad51 and either HsRad51 or HsDmc1 (Fig. 2B), suggesting enough conservation among species to allow heterologous interactions.
The Interaction between AtBrca2 and AtRad51 or AtDmc1 Occurs in the BRC Domains Region, and the Interaction between Brca2 and Rad51 Is Conserved among Species
In mammals, the BRC repeats are involved in the interaction of Brca2 with Rad51 (Wong et al., 1997; Chen et al., 1998b; Davies et al., 2001; Galkin et al., 2005). Since an interaction between Brca2 and Dmc1 has never been described in any other organism than Arabidopsis (to our knowledge), there is no clue about the region of Brca2 that is involved in binding to Dmc1. However, the conserved protein structure between Dmc1 and Rad51 (Chen et al., 1999) led us to examine whether Brca2 and Dmc1 could, as in the case of Rad51, interact via the BRC domains region. A 734-bp region was defined between nucleotides (nts) 175 and 909 of the AtBRCA2(IV) cDNA, comprising all four BRC repeats, and cloned in frame with the Gal4 activation domain (AD) and binding domain (BD) of the yeast vectors pGAD424 and pGBT9 (Fig. 3A). In a yeast two-hybrid assay, this BRC repeats-containing region was sufficient to allow binding of not only AtRad51 or AtDmc1, but also HsRad51 and HsDmc1 (Fig. 3D).
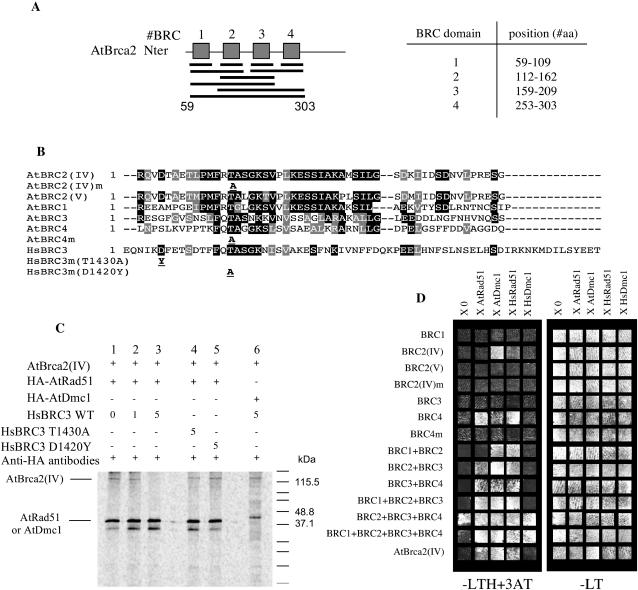
Figure 3.
Interactions between AtRad51, AtDmc1, and AtBrca2 reside in the BRC motifs. A, Representation of the BRC motif combinations as they were cloned into the yeast two-hybrid vector pGBT9. Position of each BRC domain is indicated on the right. B, Sequence alignment of the AtBrca2 BRC motifs used in the yeast two-hybrid experiments and of the HsBRC3 peptides used for the competition assay. HsBRC3m(T1430A) and HsBRC3m(D1420Y) carry a Thr-to-Ala or a Asp-to-Try mutation; AtBRC2(IV) m and AtBRC4m carry the same Thr-to-Ala mutation as HsBRC3m(T1430A). C, Lanes 1 to 5, Coimmunoprecipitation assay between HA-Rad51 and AtBrca2(IV), using anti-HA antibodies. HA-AtRad51 was preincubated with different amounts of the HsBRC3 domain, 0 μg (lane 1), 1 μg (lane 2), and 5 μg (lane 3), or with 5 μg of the mutated HsBRC3 motifs (D1420Y) (lane 4) or (T1430A) (lane 5). Lane 6, Coimmunoprecipitation assay between HA-AtDmc1 preincubated with 5 μg of HsBRC3 domain and AtBrca2(IV). D, Yeast two-hybrid assays for interactions between combinations of the Arabidopsis BRC motifs and AtRad51, AtDmc1, HsRad51, or HsDmc1. −LT and −LTH+3AT correspond to dropout media without L, Leu; T, Trp; H, His. −LTH medium was supplemented with 5 mm 3-amino-triazole (3AT). Yeasts containing both vectors were grown as patches and replica plated with velvet on −LT and −LTH+3AT. Plates were scanned and the final picture was obtained by assembling squares of each growth area. Regular growth appears as white squares on −LT; positive interactions appear as white squares on −LTH+3AT.
To further extend our understanding of the Brca2-interacting properties, we examined whether a BRC motif peptide could prevent AtBrca2 from interacting with either Rad51 or Dmc1. In human breast cancer cells, expression of BRC motifs can disrupt the formation of Brca2-Rad51 complexes (Chen et al., 1999). Similarly, incubation of the BRC motif number 3 (BRC3), from the human Brca2, with Rad51 prevents nucleoprotein filament formation by Rad51 (Davies et al., 2001). We used this same human BRC3 motif in our experiments (Fig. 3B). The interactions observed between AtBrca2 and HsRad51 or a four-BRC motifs region (Fig. 3D) and either HsRad51 or HsDmc1 suggested that conservation of the BRC motifs allowed their binding to heterologous proteins, and, consequently, that a human BRC motif could eventually perturb the interactions between AtBrca2 and AtRad51 or AtDmc1.
The human BRC3 peptide was thus introduced in the coimmunoprecipitation assays in an attempt to compete out the interaction between AtRad51 or AtDmc1 and AtBrca2. In these experiments, 35S-Met-radiolabeled AtBrca2(IV) was used as a substrate for HA-AtRad51 (or HA-AtDmc1) binding. When AtRad51 was preincubated with no (Fig. 3C, lane 1) or 1 μg (Fig. 3C, lane 2) of the human BRC3 peptide prior to the addition of AtBrca2, we obtained two bands after elution, SDS-PAGE migration, and visualization by autoradiography. These bands corresponded to the AtBrca2(IV) and the HA-tagged AtRad51 predicted sizes. However, when AtRad51 was preincubated with 5 μg of the BRC3 peptide, no AtBrca2 binding could be detected. Only one band at the predicted size for HA-AtRad51 was eluted in this assay, proving that the ability of AtBrca2 to bind AtRad51 was abolished. This amount of BRC3 corresponds to a 10-fold molar excess of peptide over AtRad51 and AtBrca2 (Fig. 3C, lane 3). In human, mutations of BRC3 in key positions prevent its binding with HsRad51 (Davies et al., 2001). Five micrograms of the mutated T1430A and D1420Y BRC3 peptides did not prevent the interaction between AtRad51 and AtBrca2 (Fig. 3C, lanes 4 and 5). In a similar assay, preincubation of HA-AtDmc1 with 5 μg of the wild-type HsBRC3 motif did not affect binding of HA-AtDmc1 to AtBrca2.
We thus examined whether any of the Arabidopsis Brca2 single BRC motifs could bind AtRad51 or AtDmc1 in a yeast two-hybrid assay. As in human, the BRC motifs in AtBrca2 were numbered after their order of occurrence in the Arabidopsis protein (Fig. 3A), which does not mean that their sequence is closer to similarly numbered motifs of the human protein. The two BRC2 motifs are divergent, thus both were tested, while BRC1, BRC3, and BRC4 are identical in AtBrca2(IV) and AtBrca2(V) (Fig. 3B). An interaction was clearly evidenced between BRC2 of AtBrca2(IV) and AtDmc1 (but not AtRad51), or BRC4 and AtRad51. These interactions were eliminated when a Thr-to-Ala amino acid change was introduced at a key position (Fig. 3, B and D). The motifs involved in the fixation of Rad51 (BRC4) or Dmc1 (BRC2) are the minimum sufficient domains we could define for this interaction. When combined with other domains, an interaction is thus also obtained. We examined different combinations of the BRC repeats (Fig. 3D) and evidenced that the BRC1-BRC2 combination binds AtDmc1 and HsRad51. BRC2-BRC3, BRC3-BRC4, and BRC1-BRC2-BRC3 bind AtRad51, AtDmc1, and HsRad51 as well. Autoactivation of the BRC2 + BRC3 + BRC4 combination in the absence of any partner prevents interpretation of data with this particular construct. As previously mentioned, the whole four-motif region of Brca2 (BRC1-BRC2-BRC3-BRC4) is required for the interaction with HsDmc1. Not surprisingly, this last combination also interacts with AtRad51, AtDmc1, and HsRad51 (Fig. 3D).
Two Isoforms of the Dss1 Protein Are Encoded in the Arabidopsis Genome That Interact in a Specific Manner with Each AtBrca2 Proteins
Two potential Dss1-encoding sequences were identified in the Arabidopsis genome named AtDSS1(I) (At1g64750) and AtDSS1(V) (At5g45010) after their chromosomal localization. Both AtDSS1 genomic DNA sequences comprise two exons and one 550-nt-long intron. Using specific primers, two different AtDSS1 cDNAs were isolated via reverse transcription-PCR that correspond to either the chromosome I (222 nt) or the chromosome V (219 nt) DSS1 cDNAs, thus revealing that both genomic sequences are transcriptionally active. These cDNAs share 90.7% identity and encode, respectively, for 74-amino-acid- (I) and 73-amino-acid-long (V) putative proteins, which are 88% identical to each other and show high conservation with their human, fungus, and yeast counterparts (Fig. 4A).
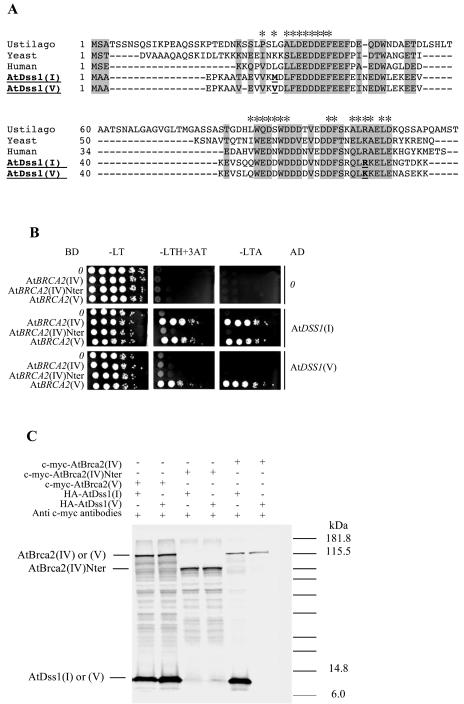
Figure 4.
Interactions between the AtBrca2 and the AtDss1 proteins. A, Alignment of the Dss1 orthologs. Stars indicate amino acids that were defined to contact Brca2 (Yang et al., 2002). Amino acids positions conserved in three or more of the five sequences aligned are in gray. The only two different key amino acids positions between AtDss1(I) and (V) are underlined. B, Yeast two-hybrid assay with BRCA2(IV), AtBRCA2(IV)Nter, and BRCA2(V) fused to the BD of pGBT9, and DSS1(I) or DSS1(V) bound to the AD of pGAD424. Serial dilutions of yeast diploid strains containing combinations of the indicated AD and BD protein fusions where spotted on −LT, −LTH, and −LTA (dropout media without L, Leu; T, Trp; H, His; or A, Ade). C, Coimmunoprecipitation assay between the AtBrca2 and AtDss1 proteins. Lanes 1 to 2, HA-AtDss1(I) (lane 1) and (V) (lane 2) retained by c-myc-AtBrca2(V). Lanes 3 to 4, HA-AtDss1(I) (lane 3) and (V) (lane 4) retained by c-myc- AtBrca2(IV)Nter. Lanes 5 to 6, HA-AtDss1(I) (lane 5) and (V) (lane 6) retained by and c-myc-AtBrca2(IV).
The AtDSS1 coding sequences were fused in frame to the AD and BD of the yeast two-hybrid vectors pGADT7 and pGBKT7. Potential interactions were examined between the two AtBrca2 and AtDss1 coding sequences following their introduction in pairwise combinations into the same yeast reporter strains. The assay indicated potential interactions between AtBrca2(V) and AtDss1(I) or AtDss1(V), and also between AtBrca2(IV) and AtDss1(I). No interaction was observed between AtBrca2(IV) and AtDss1(V), or between Nter-AtBrca2(IV) and AtDss1(I) or AtDss1(V) (Fig. 4B). We analyzed the in vitro interactions between the corresponding proteins in a coimmunoprecipitation assay (Fig. 4C). When coimmunoprecipitation was realized with c-myc-AtBrca2(V) and HA-AtDss1(I) or (V) in the presence of anti-c-myc antibodies (Fig. 4C, lanes 1 and 2), two bands were eluted, one at the predicted size for c-myc-AtBrca2(V) and one at 11 kD [either HA-AtDss1(I) or (V)]. When we immunoprecipitated c-myc-AtBrca2(IV) and HA-AtDss1(I) in the presence of anti-c-myc antibodies, we eluted two bands around 129 and 11 kD (Fig. 4C, lane 5), whereas we obtained only one band around 129 kD when we immunoprecipitated c-myc-AtBrca2(IV) and HA-AtDss1(V) in the presence of anti-c-myc antibodies (Fig. 4C, lane 6). When the assay was performed with c-myc-AtBrca2(IV)Nter and HA-AtDss1(I) or (V) in the presence of anti-c-myc antibodies, only one band at 88 kD [c-myc-AtBrca2(IV)Nter] was obtained (Fig. 4C, lanes 3 and 4). These results indicate that AtBrca2(V) is able to interact with both AtDss1(I) and (V), whereas AtBrca2(IV) only interacts with AtDss1(V) in vitro. The Nter-truncated version of AtBrca2(IV) is unable to interact with AtDss1(V), indicating that amino acids that are key to binding Dss1 must be present beyond amino acid 784 in AtBrca2.
AtBrca2(V) Is Able to Form a Three-Partner Complex
A coimmunoprecipitation assay was performed after incubating together all three in vitro-translated proteins: AtBrca2(V), HA-AtRad51, and AtDss1(I). HA antibodies were used to bind the interacting complex. SDS-PAGE analysis of the reaction products indicated three well defined bands that migrated at the expected sizes for AtBrca2(V), HA-tagged AtRad51, and AtDss1(I) (Fig. 5, lane 1). Since the AtRad51 protein alone had proven unable to bind AtDss1(I) (Fig. 5, lane 3), AtBrca2(V) and AtDss1(I) were thus both retained by HA-AtRad51 binding in this particular assay. Following the same experimental strategy, we also obtained evidence that AtBrca2(V) is able to bind both AtDmc1 and AtDss1(I) (Fig. 5, lane 2). AtDss1 and AtDmc1 cannot directly interact (as shown in Fig. 5, lane 4). Our results establish that AtBrca2(V) can interact with both Rad51 and Dss1(I) or both Dmc1 and Dss1(I) in a tripartite complex.
Figure 5.
Three-partner interaction between AtRad51 or AtDmc1 and AtBrca2/AtDss1. Lane 1, c-myc-AtDss1(I) and c-myc-AtBrca2(V) retained by HA-AtRad51. Lane 2, c-myc-AtDss1(I) and c-myc-AtBrca2(V) retained by HA-AtDmc1. Lane 3, c-myc-AtDss1(I) is not retained by HA-AtRad51 alone. Lane 4, c-myc-AtDss1(I) is not retained by HA-AtDmc1. Lane 5, c-myc-AtBrca2(V)Cter is retained by HA-AtDss1(I). Lane 6, c-myc-AtBrca2(V)Cter is retained by HA-AtDss1(V). Lane 7, Neither c-myc-AtDss1(I) nor c-myc-AtBrca2(V)Cter is retained by HA-AtRad51 alone. Lane 8, Neither c-myc-AtDss1(I) nor c-myc-AtBrca2(V)Cter is retained by HA-AtDmc1 alone.
This tripartite interaction is not observed in the presence of the truncated AtBrca2(V)Cter form. When HA-AtRad51, c-myc-AtDss1(I), and c-myc-AtBrca2(V)Cter were immunoprecipitated using anti-HA-antibodies, only one band around 37 kD was eluted (Fig. 5, lane 7). Similar results were obtained with HA-Dmc1, c-myc-AtDss1(I), and c-myc-AtBrca2(V)Cter (Fig. 5, lane 8). We previously observed that AtBrca2(V)Cter can bind AtDss1(I) or (V) (Fig. 5, lanes 5 and 6), thus indicating that this truncated version of AtBrca2 is still partially functional. These experiments clearly show that AtRad51 or AtDmc1 cannot directly interact with AtDss1, but that a three-partner complex can exist in the presence of a full-length AtBrca2 protein.
DISCUSSION
In this work, we have examined in vitro the interactions of the Arabidopsis Brca2 proteins with four of their key partners: Rad51, Dmc1, and Dss1 (two isoforms). We first confirmed that AtBrca2 is able to bind AtRad51 and AtDmc1 efficiently in vitro. The interaction between AtBrca2 and AtDmc1, in particular, which has never been described in any other species than Arabidopsis, had only been observed in a yeast two-hybrid assay and thus remained to be established at the protein level (Siaud et al., 2004). The role of Brca2 at meiosis, as it has now been unveiled in mouse, Arabidopsis, and Caenorhabditis (Chen et al., 1998; Sharan et al., 2003; Siaud et al., 2004), could be considered the only result of its interaction with Rad51, since this protein not only plays a role in somatic DNA double-strand break repair but also at meiosis. However, its interaction with Dmc1, whose function is strictly restricted to meiotic recombination (Bishop et al., 1992; Couteau et al., 1999), definitely points to a specific involvement of Brca2 in this specialized division where elaborate chromosome transactions take place. Brca2 thus appears as a general organizer in all homologous recombination events that take place in somatic or meiotic cells, Rad51 and Dmc1 being the operators that will drive homologous recombination into specific outcomes.
In a yeast two-hybrid assay, we could establish that Brca2 binds to AtDmc1 via the same four-BRC-motifs-containing region that is involved in AtRad51 binding. While we determined that the BRC3 motif of human Brca2 was able to compete the binding of AtRad51 to AtBrca2 and that, like in human, a single mutation at a key position of HsBRC3 suffices to prevent this effect, the Brca2/Dmc1 interaction was not similarly affected. It was only after the examination of every single Arabidopsis BRC motif in a genetic assay that we could determine that the BRC2 motif of AtBrca2(IV) is specifically involved in an interaction with AtDmc1 but not with AtRad51. Our data thus establish that this activity of the BRC motifs, i.e. binding of recombination proteins, is altogether largely conserved, since a human motif can be responsible for destabilizing the interaction between the two Arabidopsis Brca2 and Rad51 proteins, but also specific, since the same AtBrca2 BRC motif that interacts with AtDmc1 does not recognize AtRad51. The C-terminal region of the human Brca2 protein was also recently shown to bind Rad51 in a manner that is regulated through cyclin-dependent kinase (CDK)-dependent phosphorylation (Esashi et al., 2005). In our hands, the C-terminal region of in vitro-synthesized AtBrca2 did not seem to bind AtRad51 or AtDmc1, but one cannot exclude potential in vivo regulations that would affect the binding abilities of AtBrca2. The AtBrca2 proteins align very poorly with this region of HsBrca2. Nevertheless, S/TP potential CDK target motifs and cyclin-binding motifs can be identified that are scattered in the C-terminal region of both AtBrca2 proteins, which may thus be subject to the same regulation.
In human, while BRC3 interacts specifically with HsRad51, no interaction could be detected between BRC3 and Dmc1 or any of the Rad51-like proteins (Xrcc2, Xrcc3, Rad51B, Rad51C, and Rad51D; Davies et al., 2001). Yet, the interaction observed between the four-BRC motifs region of AtBrca2 and the human Dmc1 suggests that the human Brca2 could be involved in Dmc1 binding, although in a manner that would be more species specific than in the case of Rad51 binding. Since eight BRC motifs are present in the human protein, of which six were shown to bind Rad51 (Wong et al., 1997), it is possible that the remaining two, or combinations of the eight BRC repeats, are involved in binding another partner such as Dmc1, or that some motifs could bind both proteins. It has been reported that, in the absence of Brca2, Rad51 and Dmc1 do not organize into chromosome-associated foci in mouse meiocytes at the prophase I stage. A sole role for Rad51 to bring Dmc1 into the meiotic apparatus in eukaryotic meiosis cannot be experimentally examined in the absence of viable rad51 mutants in mouse, or considering how heavily affected early meiosis is in Arabidopsis rad51 mutants (Lim and Hasty, 1996; Li et al., 2004). However, the mouse data possibly indicates a requirement for Brca2 to directly drive not only Rad51 but also Dmc1 into the recombination apparatus at meiosis in mammals (Sharan et al., 2003). Whether the mammalian Brca2 and Dmc1 proteins do interact will thus merit closer examination. In particular, some of the sterilities in human could be related to Brca2 mutations that are not involved into hereditary breast cancer pathologies, if they were to only affect the ability of Brca2 to bind Dmc1.
We examined the interaction between AtBrca2 and another of its essential partners, Dss1. Key Dss1-interacting domains were defined in the C-terminal region of the human Brca2 that are found mostly conserved in the AtBrca2 proteins (Yang et al., 2002). Our data indicate that while AtBrca2(V) interacts with both AtDss1(I) and (V), AtBrca2(IV) only interacts with AtDss1(I). This result suggests a high specificity of this latter pair of partners: Not only are the two Arabidopsis Brca2 proteins very conserved (94.2% identities) but also the two Dss1 proteins that diverge by nine residues, of which only four are nonconservative changes (Fig. 4A). Moreover, of the 25 amino acids of Dss1 that were determined to be involved in contacting Brca2 (Yang et al., 2002), only two differ between AtDss1(I) and AtDss1(V). Consistently, while Brca2 is able to bind Rad51 proteins from other species (our data; see also Yang et al., 2005), this is not true for Dss1 recognition: No heterologous interaction could be observed between the yeast or the mouse Dss1 orthologs and the two AtBrca2 proteins (data not shown). Thus, in the absence of AtDss1, no heterologous Dss1 proteins were involved in stabilizing AtBrca2 in our experiments, whether in yeast two-hybrid assays or during in vitro synthesis in reticulocyte lysates from rabbit. The 50 amino acids that are theoretically required for the human Brca2 protein to bind HsDss1 are more or less conserved but strictly identical in the two Arabidopsis forms of Brca2, thus suggesting that the specificity of the AtBrca2/AtDss1 interactions also resides in other as yet undefined residues.
Finally, although the ability of Brca2 to associate with either Rad51 or Dss1 has been documented in different organisms and is clearly conserved in Arabidopsis, we also establish that AtBrca2 can exist in a three-partner complex, interacting simultaneously with AtRad51 and AtDss1, or with AtDmc1 and AtDss1. This result indicates that AtBrca2, AtRad51 or AtDmc1, and AtDss1 could interact in vivo to form a ternary complex, whether it be transitory or not, in which AtBrca2 is either stabilized or activated by Dss1 (as shown in Kojic et al., 2005), possibly at the same time as it binds AtRad51 or AtDmc1. Experiments by Kojic et al. (2005) indicate that the interaction with Dss1 is required to ensure Brh2 function. In mammalian cells, following DNA damage, Brca2 and Rad51 colocalize into nuclear foci that are thought to represent sites of DNA repair by homologous recombination. Upon Dss1 depletion, these Rad51 foci no longer assemble, which indicates a role for Dss1 in enabling Brca2 to bring Rad51 to the sites of DNA lesions (Gudmundsdottir et al., 2004). Our in vitro evidence of a ternary Brca2/Dss1/Rad51 complex supports this view. Actually, in Ustilago, the Dss1 requirement could be bypassed upon deletion of the Brh2 C-terminal region or its replacement by a DNA-binding domain from RPA70, suggesting that this region of Brh2 negatively regulates its function (Kojic et al., 2005). However, establishing the exact sequence of events that govern the organization of the complex formed by Brca2 and its partners (stoichiometry, dynamics of the complex assembly, and precise regulation of each partner) will need further study. Our data indicate that these interactions do not necessarily occur sequentially but may also occur concomitantly.
MATERIALS AND METHODS
Dss1 and BRC Repeats cDNA Cloning
One microgram of total RNA extracted from a 2-d freshly subcultured cell suspension of Arabidopsis (Arabidopsis thaliana) was reverse transcribed by the MMLV reverse transcriptase with oligo(dT) primers and in the presence of dNTPs in a final volume of 20 μL. PCR was performed, using Dss1(I)- or (V)-specific primers (I-Nter ATCGTGAATTCATGGCGGCAGAACCGAAGGCAGCG; I-Cter GCTTAGTCGACTTATTTCTTGTCAGTACCATTCTC; V-Nter ATCGTGAATTCATGGCGGCAGAACCCAAGGCAGCT; V-Cter GCTTAGTCGACTCATTTCTTCTCACTAGCATTC) with Pfx DNA polymerase (Invitrogen). PCR products were cloned in frame with the BD and AD domains of pGBKT7 (EcoRI and SmaI sites) and pGADT7 (EcoRI and SalI sites; CLONTECH). The BRC motifs, in single or multiple combinations, were isolated by performing PCR reactions with different sets of primers using BRCA2(IV) or BRCA2(V) cDNAs as a matrix (Siaud et al., 2004): BRC1 (B1F GTGGAATTCCGGGAAGAAGCGATGCCTGGG × B1R GTGCTGCAGGGGAATTGAGCAATTTGTGTT), BRC2 (B2F GTGGAATTCAGACAAGTGGATACAGCTGAA × B2R GTGCTGCAGTCCACTCTCCCTGGGAAGAACGTT), BRC3 (B3F GTGGAATTCAGGGAGAGTGGATTTGGAGTT × B3R GTGCTGCAGAGACGACTGGTTCACATGGTT), BRC4 (B4F GTGGAATTCTTAAATCCATCTTTAAAGGTG × GTGCTGCAGTTGATCACCTCCAGCTACATC), BRC1 + BRC2 (B1F × B2R), BRC2 + BRC3 (B2F × B3R), BRC3 + BRC4 (B3F + B4R), BRC1 + BRC2 + BRC3 (B1F × B3R), BRC2 + BRC3 + BRC4 (B2F × B4R), and BRC1 + BRC2 + BRC3 + BRC4 (B1F × B4R). Mutations were introduced in motifs BRC2 and BRC4 using the following primers: B2F(m) GTGGAATTCAGACAAGTGGATACAGCTGAAACTTTGCCAATGTTCAGGGCGGCCT and B4F(m) GTGGAATTCTTAAATCCATCTTTAAAGGTGCCTCCAACAAAGTTTCAGGCTGCTGGT, which turned a consensus Thr into Ala.
Yeast Two-Hybrid Assay
PCR products previously described were cloned in frame with the DB and AD of pGBT9 (EcoRI and PstI sites) and verified by sequencing. HsRad51 and HsDmc1 were provided by S. West (Masson et al., 1999). AtRAD51, AtDMC1, AtBRCA2(IV), AtBRCA2(IV)Nter, and AtBRCA2(V) cDNAs sequences were introduced in pGAD424 and pGBT9 (CLONTECH), and yeast two-hybrid assays were conducted as described (Siaud et al., 2004).
In Vitro Synthesis of the Proteins
The BRCA2(IV), BRCA2(V), and AtBRCA2(IV)Nter cDNA sequences in plasmids pGAD424 and pGBT9 (CLONTECH) were used as matrix to in vitro transcribe and translate the relevant proteins. Primer pairs were designed to PCR amplify a fragment containing the expression domain of the vectors, adding a HA tag (pGAD-HA-up AAAATTGTAATACGACTCACTATAGGGCGAGCCGCCACCATGTACCCATACGACGTTCCAGATTACGCTCCACCAAACCCAAAAAAAGAG and pGAD-HA-do ACTTGCGGGGTTTTTCAGTATCTACGAT) or a c-myc tag (pGBT9-c-myc-up AAAATTGTAATACGACTCACTATAGGGCGAGCCGCCACCATGGAGGAGCAGAAGCTGATCTCAGAGGAGGACCTGGGTCAAAGACAGTTGACTGTATCG and pGBT9-c-myc-do TACCTGAGAAAGCAACCTGACCTACAGG) 3′ to the cDNAs. Primer was designed to amplify BRCA2 cDNA from the 1,070th bp to the end, to delete the N-terminal region containing the BRC domains (pGBT9-cmyc C-ter: AAAATTGTAATACGACTCACTATAGGGCGAGCCGCCACCATGGAGGAGCAGAAGCTGATCTCAGAGGAGGACCTGAACTTGGAGAACCTAGCTTCAGGGGGT). When used with the pGBT9-c-myc-do oligonucleotides, the amplified fragment contains the expression domain of pGBT9 and a c-myc tag in frame with the truncated BRCA2 cDNA. RAD51, DMC1, DSS1(I) and (V), and each AtBRCA2 cDNA sequence cloned into plasmids pGADT7 and pGBKT7 (CLONTECH) were used as matrix to in vitro produce the proteins. Primer pairs were designed to amplify the cDNAs (pGADT7do, AAGTGAACTTGCGGGGTTTTTCAGTATCTACG, and pGBKT7do, CATAAGAAATTCGCCCGG, plus the left border of the vector [pGADT7up, TATTCGATGATGAAGATACCCCACAAACC, or pGBKT7up, TCATCGGAAGAGAGTAG]). PCR was conducted with Pfx DNA polymerase (Invitrogen). Coupled transcription/translation was performed for each protein with the PCR products as a matrix in the presence of 35S Met (15.1 μCi/ reaction; Redivue, Amersham; TNT T7 Quick for PCR DNA kit, Promega). Translation products were analyzed by SDS-PAGE. To eliminate DNA or RNA, 5 μL of the translation products were treated with DNaseI (Invitrogen) or RnaseA (Qiagen).
Coimmunoprecipitation Assays
Coimmunoprecipitation was conducted with the Matchmaker kit (CLONTECH), using anti-c-myc or anti-HA antibodies and protein A beads, as recommended. Results were analyzed after SDS-PAGE on a 12% acrylamide gel by autoradiography performed with a Molecular Imager Fx Pro (Bio-Rad).
In Vitro Competition Assay
Human BRC3 wild-type and mutant peptides were provided by S. West (Davies et al., 2001). The lyophilized peptides were resuspended in fresh phosphate-buffered saline, and their concentration was estimated after hot NaOH hydrolysis. In vitro-translated HA-AtRad51 was preincubated for 45 min at room temperature with 0, 1, 3, or 5 μg of human BRC3 peptide. Then AtBrca2 was added, and the mix was further incubated at room temperature for 1 h. Coimmunoprecipitation was then performed using the HA antibodies as recommended by CLONTECH. The assay results were analyzed by SDS-PAGE and autoradiography.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ619975 [for Dss1(I)] and AJ619976 [for Dss1(V)].
Acknowledgments
We thank Dr. Steve West for providing HsRad51 and HsDmc1 cloned into yeast two-hybrid vectors, and for providing the HsBRC3 peptide. We are grateful to F. Ambart and J. Vidal for useful comments in the course of our experiments, and to C. Mézard and S. Lemaire for their critical reading and useful comments on our manuscript.
This work was supported in part by the Association pour la Recherche contre le Cancer. E. Dray was supported by a Ministère de l'éducation Nationale, de la Recherche et de la Technologie fellowship, and N.S. was a recipient of a Ligue Nationale contre le Cancer fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Marie-Pascale Doutriaux (doutriau@ibp.u-psud.fr).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.075838.
References
- Bishop DK, Park D, Xu L, Kleckner N (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456 [DOI] [PubMed] [Google Scholar]
- Bork P, Blomberg N, Nilges M (1996) Internal repeats in the BRCA2 protein sequence. Nat Genet 13: 22–23 [DOI] [PubMed] [Google Scholar]
- Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH (1999) Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem 274: 32931–32935 [DOI] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, et al (1998. a) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2: 317–328 [DOI] [PubMed] [Google Scholar]
- Chzen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH (1998. b) The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci USA 95: 5287–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux M-P (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC (2001) Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell 7: 273–282 [DOI] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC (2005) CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434: 598–604 [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Li X, Velichutina I, Hochstrasser M, Kobayashi H (2004) Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J Cell Sci 117: 6447–6454 [DOI] [PubMed] [Google Scholar]
- Galkin VE, Esashi F, Yu X, Yang S, Wes SC, Egelman EH (2005) BRCA2 BRC motifs bind RAD51-DNA filaments. Proc Natl Acad Sci USA 102: 8537–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A (2004) DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep 5: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M, Yang H, Kostrub CF, Pavletich NP, Holloman WK (2003) The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell 12: 1043–1049 [DOI] [PubMed] [Google Scholar]
- Kojic M, Zhou Q, Lisby M, Holloman WK (2005) Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol Cell Biol 25: 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, et al (2004) Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell 16: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DS, Hasty P (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol 16: 7133–7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T, Pellegrini L, Venkitaraman AR, Blundell TL (2003) Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair (Amst) 2: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A (1999) Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol 19: 4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ (2005) RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol 25: 3127–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, Stasiak A, West SC (1999) The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J 18: 6552–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420: 287–293 [DOI] [PubMed] [Google Scholar]
- Scully R, Livingston DM (2000) In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408: 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A (1997) Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804–810 [DOI] [PubMed] [Google Scholar]
- Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, Swing D, Martin BK, Tessarollo L, Evans JP, et al (2003) BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131: 131–142 [DOI] [PubMed] [Google Scholar]
- Siaud N, Dray E, Gy I, Takvorian N, Gérard E, Doutriaux MP (2004) Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J 23: 1392–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA 93: 6236–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL (1997) RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem 272: 31941–31944 [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–792 [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP (2002) BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297: 1837–1848 [DOI] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP (2005) The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433: 653–657 [DOI] [PubMed] [Google Scholar]
- Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR (2000) Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev 14: 1400–1406 [PMC free article] [PubMed] [Google Scholar]