Abstract
The oxidative cleavage of heme by heme oxygenases (HOs) to form biliverdin IXα (BV) is the committed step in the biosynthesis of the phytochrome (phy) chromophore and thus essential for proper photomorphogenesis in plants. Arabidopsis (Arabidopsis thaliana) contains four possible HO genes (HY1, HO2–4). Genetic analysis of the HY1 locus showed previously that it is the major source of BV with hy1 mutant plants displaying long hypocotyls and decreased chlorophyll accumulation consistent with a substantial deficiency in photochemically active phys. More recent analysis of HO2 suggested that it also plays a role in phy assembly and photomorphogenesis but the ho2 mutant phenotype is more subtle than that of hy1 mutants. Here, we define the functions of HO3 and HO4 in Arabidopsis. Like HY1, the HO3 and HO4 proteins have the capacity to synthesize BV from heme. Through a phenotypic analysis of T-DNA insertion mutants affecting HO3 and HO4 in combination with mutants affecting HY1 or HO2, we demonstrate that both of the encoded proteins also have roles in photomorphogenesis, especially in the absence of HY1. Disruption of HO3 and HO4 in the hy1 background further desensitizes seedlings to red and far-red light and accelerates flowering time, with the triple mutant strongly resembling seedlings deficient in the synthesis of multiple phy apoproteins. The hy1/ho3/ho4 mutant can be rescued phenotypically and for the accumulation of holo-phy by feeding seedlings BV. Taken together, we conclude that multiple members of the Arabidopsis HO family are important for synthesizing the bilin chromophore used to assemble photochemically active phys.
Plants employ a complex web of signaling pathways directed by several photoreceptor families to detect and respond to light (Schafer and Nagy, 2005). Phytochromes (phys) are one of the dominant photoreceptors, directing growth and photomorphogenic responses to red (R) and far-red (FR) light (Smith, 1995; Quail, 2002; Tu and Lagarias, 2005). They are dimeric chromoproteins with each approximately 120-kD monomer containing the linear tetrapyrrole (or bilin) chromophore (3E)-phytochromobilin (PΦB) covalently attached via a thioether bond. Once assembled, phys can assume one of two stable conformers, a R-absorbing Pr form and a FR-absorbing Pfr form, which are repeatedly photointerconvertible by R and FR, respectively. They are initially synthesized as Pr, which is biologically inactive, and only become active upon photoconversion to Pfr. By measuring either the amount of Pfr or the Pr/Pfr ratio, plants use phys as photoreversible switches to assess the direction, intensity, spectral composition, and duration of the ambient light environment. Responses under phy control span the life cycle of plants, including seed germination, chloroplast development, stem and leaf growth, pigmentation, entrainment of the circadian clock, flowering time, and senescence (Smith, 1995; Quail, 2002).
The coordinated action of two spatially separate pathways are required to synthesize photochemically active phys; one in the chloroplast that produces PΦB and another in the cytosol that synthesizes the phy apoproteins, which are encoded by small families of nuclear genes (PHYA–E in Arabidopsis [Arabidopsis thaliana]; Quail, 2002). Photoactive phys are assembled autocatalytically by a bilin lyase activity intrinsic to the polypeptide following transport of PΦB to the cytoplasm. PΦB is synthesized from 5-aminolevulinic acid (ALA) by the same pathway that produces chlorophyll and heme (Terry et al., 2002; Tu and Lagarias, 2005). A key branch point is the cyclic bilin protoporphyrin IX. Protoporphyrin IX is either shunted toward chlorophyll production by Mg+2 chelatase that generates Mg-protoporphyrin IX or toward heme production by ferrochelatase that generates heme (Papenbrock and Grimm, 2001). The committed step in PΦB synthesis is the oxidative cleavage of heme by a heme oxygenase (HO) to form biliverdin IXα (BV; Terry et al., 1993; Ortiz de Montellano and Wilks, 2001). In plants, this reaction requires both O2 and the electron donor, ferredoxin, and releases carbon monoxide (CO) and Fe2+ (Muramoto et al., 2002). BV is further reduced in plants to (3Z)-PΦB by the ferredoxin-dependent PΦB synthase (Frankenberg et al., 2001; Kohchi et al., 2001). Finally, (3Z)-PΦB is isomerized to PΦB; whether this 3Z to 3E conversion is enzymatic or occurs spontaneously remains to be determined. Either or both of the PΦB isomers could be exported to the cytosol for holo-phy assembly. Even though the heme/PΦB and chlorophyll biosynthetic subpathways have different end points, accumulating data suggest that both routes are coordinately regulated, likely by affecting ALA synthesis, which is the rate-limiting step for bilin synthesis (Terry and Kendrick, 1999; Mochizuki et al., 2001; Papenbrock and Grimm, 2001; Cornah et al., 2003). For example, blocking the ferrochelatase/heme branch of the pathway can reduce chlorophyll levels even though there is no direct inhibition of chlorophyll biosynthesis (Cornah et al., 2003).
Molecular genetic approaches have identified a number of genes important for phy assembly. In Arabidopsis for example, null mutants in all five PHY genes have been isolated. Analysis of these mutants showed that each phy isoform has distinct and overlapping roles in light-regulated development (Somers et al., 1991; Parks and Quail, 1993; Aukerman et al., 1997; Devlin et al., 1998; Franklin et al., 2003; Monte et al., 2003). The analysis of mutants that attenuate all phy responses also identified genes responsible for PΦB synthesis. As predicted, these mutants globally reduce the pool of photochemically active phys often without dampening apoprotein accumulation. Examples include the hy1, se5, pcd1, pew1, and yg-2 mutants from Arabidopsis, rice (Oryza sativa), pea (Pisum sativum), tobacco (Nicotiana tabacum), and tomato (Lycopersicon esculentum), respectively, which affect genes encoding HOs that convert heme to BV (Parks and Quail, 1991; Weller et al., 1996; Davis et al., 1999; Muramoto et al., 1999; Izawa et al., 2000; Davis et al., 2001), and the hy2, au, elm1, and pcd2 mutants from Arabidopsis, tomato, maize (Zea mays) and pea, respectively, which affect genes encoding PΦB synthases that convert BV to (3Z)-PΦB (Parks and Quail, 1991; Weller et al., 1997; Kohchi et al., 2001; Sawers et al., 2004; Muramoto et al., 2005).
Whereas subsequent genomic searches in several plant species revealed that PΦB synthase is typically encoded by a single gene (Frankenberg et al., 2001; Kohchi et al., 2001; Sawers et al., 2004), the HOs are often encoded by small gene families (Davis et al., 2001). Arabidopsis for example, contains a single HY2 gene and four possible HO genes that include HO2, 3, and 4 in addition to HY1. The encoded HO proteins fall into two subfamilies based on amino acid sequence alignments. One subfamily includes HY1, HO3, and HO4, which all have the canonical HO active site, and another includes just HO2, which can also be distinguished by the absence of a positionally conserved histidine considered to be an important ligand for heme binding (Davis et al., 2001).
The severe photomorphogenic defects of hy1 mutants coupled with the absence of detectable phy photoactivity revealed that the HY1 protein is responsible for most BV synthesis (Davis et al., 1999; Muramoto et al., 1999). However, the observations that hy1 mutants are often more compromised phenotypically than hy2 mutants (Koornneef et al., 1980; Chory et al., 1989) and that hy1 seedlings are more chlorotic than quadruple mutants lacking phyA, B, D, and E (Franklin et al., 2003b; Monte et al., 2003), implied that loss of HY1 does not solely affect phy assembly in Arabidopsis. Other possible defects include (1) the accumulation of heme to cytotoxic levels; (2) a reduction in chlorophyll accumulation, possibly caused by the stabilization of heme feedback inhibiting ALA synthesis; and/or (3) iron deficiency caused by attenuated Fe2+ recycling (Terry and Kendrick, 1999; Ortiz de Montellano and Wilks, 2001; Papenbrock and Grimm, 2001). Despite the importance of HY1, subsequent genetic analysis suggested that HO2 also has a role in photomorphogenesis and phy assembly. When compared to wild-type seedlings, the ho2-1 mutant is slightly more chlorotic, flowers earlier, and is modestly less responsive to R and FR during hypocotyl growth (Davis et al., 2001). An increased ratio of apo-to-holo phy was also evident, suggesting that loss of HO2 activity limits the availability of PΦB. Whereas hy1 plants regain some of their photomorphogenic responses and accumulate more chlorophyll as the seedlings mature, ho2-1 plants become more chlorotic as they age, suggesting that HY1 and HO2 have distinct developmental roles in bilin synthesis/degradation (Terry and Kendrick, 1999; Davis et al., 2001).
The complex phenotypes of hy1 mutants coupled with the more subtle effects of the ho2-1 mutant raised the possibility that HO3 and HO4 also have important roles in phy assembly and/or heme breakdown. To help define these roles, we report here the analysis of T-DNA insertion mutations affecting the HO3 and HO4 genes alone and in combination with those affecting HY1 or HO2. Although ho3 and ho4 mutants are not phenotypicaly compromised either singly or together, they enhance the photomorphogenic defects of hy1 plants and further depress the level of holo-phy in ho2 seedlings when combined. The light insensitivity of hy1/ho3/ho4 seedlings could be substantially rescued by feeding seedlings BV, suggesting that their photomorphogenic defects are caused primarily by a block in phy assembly and not by perturbing heme/chlorophyll metabolism. Collectively, the data indicate that multiple HOs contribute to the production of PΦB needed to assemble photoactive phys in Arabidopsis.
RESULTS
All Members of the Arabidopsis HO Gene Family Are Expressed
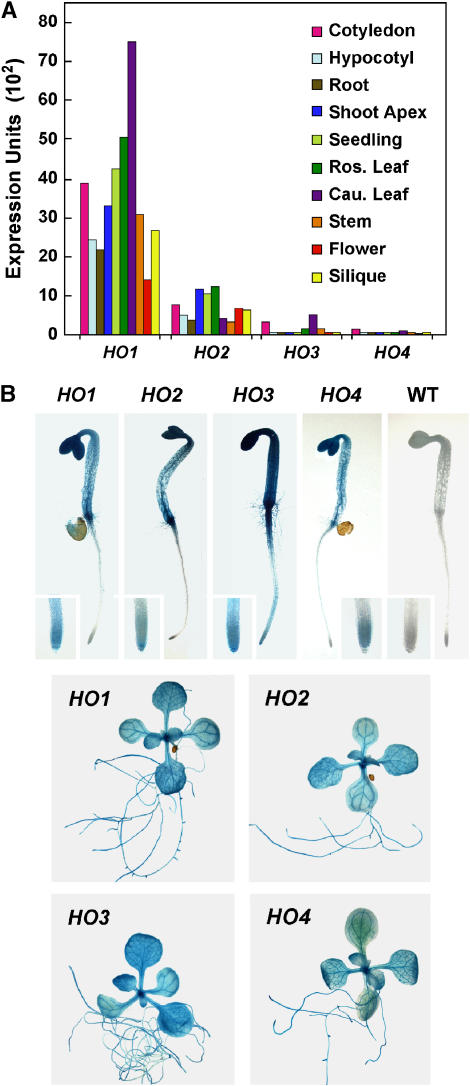
Previous studies by us (Davis et al., 1999, 2001) and others (e.g. Muramoto et al., 1999) showed that the HY1 and HO2 genes are widely expressed in Arabidopsis. To confirm that the HO3 and HO4 genes are also transcriptionally active, we employed several methods to evaluate their expression patterns. First, we subjected RNA isolated from 10-d-old seedlings to reverse transcription (RT)-PCR with gene-specific primers for the four HO loci and then sequenced the products to confirm the presence of each cDNA. The transcript sequences were easily distinguished from their genomic counterparts by the absence of the three intragenic sequences (Davis et al., 2001). In addition to RT-PCR products for HY1 and HO2, products for HO3 and HO4 were generated, indicating that these genes are also transcribed (data not shown). Subsequently, we mined the Genevestigator DNA microarray database (Zimmermann et al., 2004) to compare quantitatively the relative expression of the four HO genes in whole seedlings and various tissues. As shown in Figure 1A, the HY1 mRNA is clearly the most abundant in all tissues examined with HO2 having the second highest amount. Substantially lower but significant transcript levels could be detected for HO3 and HO4. Cotyledons were one of the few organs that had high levels of each of the four HO mRNAs, which is consistent with the high concentration of phys in this tissue (Somers and Quail, 1995; Sharrock and Clack, 2002). Recent data of Matsumoto et al. (2004) and further analysis of the expressed sequence tag (EST; http://www.Arabidopsis.org) and Massively Parallel Signature Sequences (http://mpss.udel.edu/at) databases supported the lower expression levels of HO2 to 4. For example, 42 ESTs were available for HY1, one for HO3, and none for HO2 and HO4. Scans of the various DNA microarray experiments testing various growth and environmental conditions (e.g. oxidative stress and response to various light regimes [http://www.genevestigator.ethz.ch]) failed to find situations that substantially increased or decreased HO mRNA levels, suggesting that the expression of these genes is not dramatically regulated by external signals.
Figure 1.
Expression patterns of the Arabidopsis HO genes. A, Relative expression of the HO genes in various tissues as determined by analysis of the GENEVESTIGATOR DNA microarray dataset (Zimmermann et al., 2004). B, Expression patterns of the HO genes using promoter-GUS fusions. The HY1/HO promoter∷GUS reporters were introduced into wild-type Col-0 Arabidopsis and the plants were stained overnight with the substrate 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid. Representative 6-d-old dark-grown and 21-d-old light-grown seedlings are shown. The insets show a higher magnification of the primary root tip. A wild-type Col-0 seedling (WT) is included as a control.
And finally, we introduced a β-glucuronidase (GUS) reporter gene transcribed under the direction of the HO3 or HO4 promoters into Arabidopsis and compared by GUS histochemical staining the expression patterns of these chimeric genes relative to similar genes containing the HY1 or HO2 promoters. The constructions were generated with genomic fragments starting immediately downstream of the stop codon of the preceding gene and ending immediately upstream of the translation start site for the HO to insure that all 5′ regulatory elements were included. Even though RT-PCR and DNA microarray studies indicated that both HO3 and HO4 genes are expressed at low levels (see Fig. 1A), lines conferring sufficient GUS activity for histochemical staining were generated with the corresponding promoters. Previously, we showed that the HY1 and HO2 promoters are active throughout the plant with the highest staining in the shoot apex, cotyledons, vascular tissue, and hypocotyl/root junction (Davis et al., 2001; Fig. 1B). Analysis of multiple independent transgenic lines harboring the HO3 or HO4∷GUS fusions detected a similar expression pattern (Fig. 1B). For etiolated seedlings, each promoter drove the highest expression in cotyledons and hypocotyls with modest expression in root tips. This pattern extended to light-grown plants as both the HO3 and HO4∷GUS reporters like those for HY1 and HO2 expressed well in cotyledons. The vasculature of leaves, stems, and roots showed strong staining in more mature plants (Fig. 1B). While subtle variations in expression were detected among the four HO∷GUS transgenes (e.g. HO4∷GUS did not express well in root tips of light-grown seedlings and HO2∷GUS plants often did not express well in the apical meristem region), most often the staining patterns were coincident. Taken together, it appears that all four Arabidopsis HO genes are transcriptionally active with substantially overlapping patterns, but that the mRNAs for HO3 and HO4 are present at much lower relative levels.
HY1, HO3, and HO4 Have HO Activity
Whereas the Arabidopsis HY1 protein has been confirmed enzymatically to generate BV from heme in a ferredoxin- and O2-dependent reaction (Muramoto et al., 1999, 2002), this activity has not yet been demonstrated for HO2, 3, and 4. Such confirmation was particularly important for HO2 since it has an arginine in place of the key histidine (position 88) required for heme-iron binding and oxidative cleavage and appears to represent a separate HO type based on phylogenetic analyses (Davis et al., 2001). As a facile way to demonstrate HO activity, we coexpressed in Escherichia coli each HO with the phy apoprotein (BphP) from Deinococcus radiodurans, which naturally employs BV as its chromophore (Bhoo et al., 2001). Because E. coli does not have HO-type activities, the conversion of endogenous heme to BV by each Arabidopsis HO could then be assayed by the formation of a BV-bound D. radiodurans (Dr)BphP, using zinc-induced fluorescence of the complex following SDS-PAGE to confirm covalent ligation and the characteristic R/FR difference spectrum of the BV complex to confirm the nature of the bilin. The DrBphP polypeptide used here was a truncation (amino acids 1–321) that contains just the bilin-binding pocket and the lyase activity capable of autocatalytic assembly (Bhoo et al., 2001; Karniol et al., 2005). For the four HOs, the first 51 to 56 N-terminal amino acids were predicted to encode transit peptides that direct import of the initial translation products into chloroplasts (Muramoto et al., 1999; Davis et al., 2001). These residues were not included in the E. coli-expressed proteins to avoid their possible interference with catalysis (see “Materials and Methods”).
In accord with previous studies (Bhoo et al., 2001), recombinant DrBphP was neither fluorescent nor R/FR photochemically active when expressed by itself in E. coli even in the presence of heme (Fig. 2, A and B). However, upon adding BV to E. coli extracts containing the DrBphP polypeptide, a chromoprotein was readily assembled with the signature R/FR difference spectrum for BV-bound DrBphP. A photochemically indistinguishable chromoprotein was generated when a HO from Synechocystis PCC6803 (SynHO; Bhoo et al., 2001) was coexpressed with DrBphP, thus validating the assay (Fig. 2, A and B). When each of the Arabidopsis HOs were coexpressed in the same fashion, we found that HY1, HO3, and HO4 could synthesize BV as judged by the formation of a bilin-protein complex that was fluorescent in the presence of zinc and displayed R/FR difference spectra identical to that of authentic BV-DrBphP assembled in vitro (Fig. 2, A and B).
Figure 2.
Assay of Arabidopsis HO proteins for HO activity. The synthesis of BV from heme was determined by a coupled-enzymatic assay using the HO to generate the bilin, which then autocatalytically assembled with the bacteriophytochrome from D. radiodurans (DrBphP). The bilin-binding domain of DrBphP (residues 1–321) was expressed in E. coli by itself or with one of the four HOs from Arabidopsis or a HO from Synechocystis (SynHO). For HO2, both the wild-type sequence and one containing an Arg88 to histidine substitution (R88-H) was tested. DrBphP + BV represents extracts from cells expressing only BphP, which were mixed with purified BV in vitro. The DrBphP proteins were enriched from the crude extracts by nickel-chelate affinity chromatography. A, Samples were subjected to SDS-PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). B, R/FR absorption difference spectra of the samples found in A. The bars on the right represent 0.01 ΔA. The R/FR difference maximum at 696 nm is indicated.
In contrast to the other Arabidopsis HOs, HO2 failed to generate a detectable BV-DrBphP adduct by either assay (Fig. 2, A and B). One obvious problem was that recombinant HO2 expressed poorly with almost all of the protein found in the insoluble fraction. In an attempt to overcome this problem, we attempted to purify the residual recombinant HO2 from the soluble fraction using an appended His6 tag. We then reassayed it for HO activity using an in vitro assay system developed by Muramoto et al. (2002), containing the purified HO2, purified DrBphP, heme, and reduced ferredoxin (the presumed electron donor in planta). Under these assay conditions, we also failed to detect HO activity (data not shown). Given the fact that the predicted active site in HO2 contains an arginine instead of a histidine (Fig. 3; Davis et al., 2001), we examined whether we could generate HO activity by introducing this residue. However, this HO2R88-H mutant (which was also poorly soluble) also failed to display HO activity using either the in vivo or in vitro systems (Fig. 2 and data not shown). Thus, while HY1, HO3, and HO4 are bona fide HOs, we could not confirm a similar enzymatic activity for HO2, possibly because of its poor solubility.
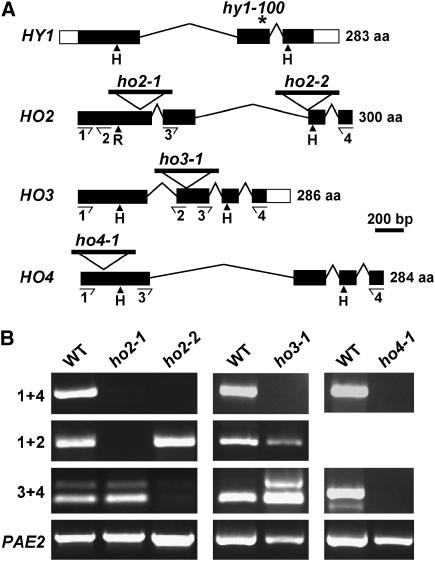
Figure 3.
Description of the Arabidopsis ho3 and ho4 mutants. A, Gene structures of the four HO genes; location of T-DNA insertions in HO2, 3, and 4; and the position of the hy1-100 mutation in HY1. Coding region and 5′ and 3′ untranslated regions are identified by the black and white boxes, respectively; introns are indicated by lines. The histidines (H) required for heme-iron binding and catalysis in animal HOs are located by the arrowheads: In HO2, the N-terminal position contains an arginine (R) instead (Davis et al., 2001). The locations of the primer pairs used to analyze the mutations by RT-PCR in B are indicated by the arrows. B, RT-PCR analysis of the ho2-1, ho2-2, ho3-1, and ho4-1 insertion mutants. Total seedling RNA was reverse transcribed according to “Materials and Methods,” and the RT products were then PCR amplified using the primer pairs 1 + 2, 1 + 4, or 3 + 4. RT-PCR amplification of PAE2 was used as a control.
Genetic Analysis of the HO Gene Family
To help assess the functions of HO3 and HO4 in planta, we identified T-DNA insertion mutants in the Arabidopsis Columbia (Col-0) ecotype that should strongly impair expression of the corresponding genes. To avoid ecotype differences when creating the various ho mutant combinations, we also isolated a second mutant affecting HO2 (ho2-2) in Col-0 background since the first mutation described (ho2-1) is in the Wassilewskija (Ws) background (Davis et al., 2001). Sequence analyses of the regions surrounding the T-DNAs revealed the insert location for all three lines. The T-DNA was at the 5′ splice site of the third exon in ho2-2, within the second exon upstream of nucleotide 556 in ho3-1, and within the first exon upstream of nucleotide 44 in ho4-1 (Fig. 3A). Genomic DNA on either side of the T-DNAs could be amplified by the left-border T-DNA primer in combination with the gene-specific primers, indicating that the T-DNAs are all bracketed by two left T-DNA borders. Sequence analysis of these products showed that the T-DNA insertions did not induce any secondary alterations to the respective loci.
RT-PCR analysis of total seedling RNA demonstrated that the T-DNAs prevented accumulation of full-length mRNAs in all three mutants. Whereas we could detect the expected full-length PCR products using primers that bracketed the entire open reading frame (primers 1 + 4) with wild-type RNA, no full-length products were evident with RNA isolated from the respective mutants (Fig. 3B). A primer pair downstream of the T-DNA insertion site (primer 3 + 4) also failed to amplify a product from the ho4-1 mutant, strongly suggesting that this mutant is a null allele. However for ho2-1, ho2-2, and ho3-1, partial PCR products were amplified using primer pairs hybridizing either upstream (1 + 2) or downstream (3 + 4) of the insertion sites, indicating that truncated transcripts accumulate in these mutants (Fig. 3B). Transcription of the upstream RNA sequences (ho2-2 and ho3-1) was likely driven by the endogenous HO promoters while the downstream RNA sequences (ho2-1 and ho3-1) were possibly driven by promoter elements present within the T-DNA construction used to generate the mutant populations (Sessions et al., 2002; Alonso et al., 2003). Given that all three of these insertions interrupt the coding region spanning the domain required for HO activity (Fig. 3A), we consider it unlikely that these partial transcripts would generate catalytically active proteins even if translated.
Phenotypic Analysis of ho Mutants
Analysis of homozygous ho3-1 and ho4-1 mutant seedlings either singly or in combination revealed no abnormal growth or developmental phenotypes in light- or dark-grown plants, indicating that the corresponding HO proteins are not essential to Arabidopsis by themselves (Figs. 4 and 5A, and data not shown). The response of hypocotyl elongation to R, chlorophyll accumulation, flowering time, and R suppression of gravitropism were also identical to wild-type plants, implying that most, if not all, phy-controlled responses were unaffected as well (Figs. 5A and 6). The ho2-2 mutation in the Col-0 background displayed subtle growth abnormalities, including slightly early flowering and a diminished inhibition of hypocotyl growth under R and FR (Figs. 4, 5, and 6), consistent with a previous analysis of the ho2-1 mutant in the Ws background (Davis et al., 2001). Both the ho2-1 and ho2-2 mutants were more pale under continuous light or long-day photoperiods (16-h light/8-h dark), especially later in development, due to a slight reduction in chlorophyll (Fig. 6A and data not shown). Given the similarities of the ho2-2 mutant to ho2-1, most subsequent studies were completed with ho2-2 to avoid ecotype differences within the mutant collection.
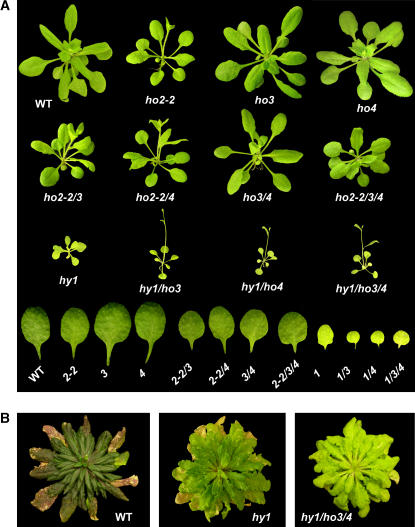
Figure 4.
Phenotype of Arabidopsis plants containing various combinations of ho mutations. A, Plants were grown under continuous white light for 3 weeks. The second true leaf from each line is shown at the bottom. B, Plants were grown for 4.5 months under short days.
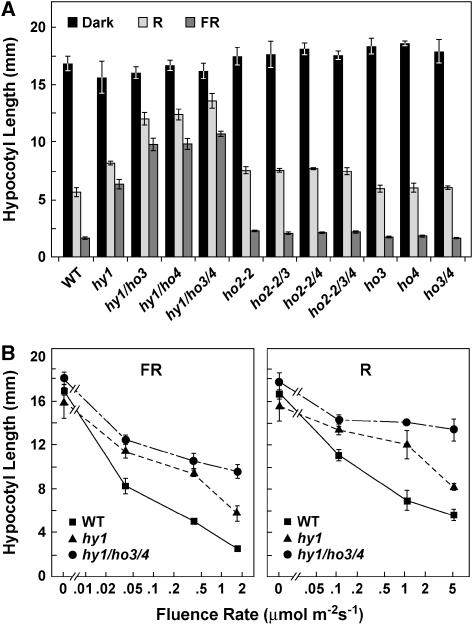
Figure 5.
Effect of various combinations of ho mutations on Arabidopsis hypocotyl growth in R and FR. A, Plants were grown for 6 d in the dark or under continuous R (5.1 μmol m−2 s−1) or FR (1.7 μmol m−2 s−1). B, Plants were exposed to various fluence rates of continuous R and FR. Each bar/point represents the average hypocotyl length of at least 10 seedlings with the error bars reflecting ± 1 sd.
Figure 6.
The effects of various combinations of ho mutants in Arabidopsis on chlorophyll accumulation (A), days to flowering in long days (B), and the gravitropic response (C). Chlorophyll was measured from rosette leaves of 21-d-old plants grown in a long-day photoperiod. Effect on flowering was measured by counting the number of rosette leaves produced before the floral inflorescence. For the gravitropic response, the angle from vertical was measured for seedlings grown for 6 d in continuous 5.1 μmol m−2 s−1 R. Each bar represents the average of at least 10 seedlings with the error bars reflecting ± 1 sd.
To assess genetically the functions of the HO3 and HO4 genes in more detail, we attempted to generate a collection of all possible combinations of double, triple, and quadruple mutants of ho3-1 and ho4-1 with hy1 and ho2-1/ho2-2. The hy1-100 mutant allele used here is a strong phenotypic mutant in the Col background generated by a G to A transition at position 1,079 (Davis et al., 1999). hy1-100 is considered to be a null allele based on the facts that this mutation (1) is predicted to alter both the reading frame and mRNA splicing, (2) dramatically decreases HY1 mRNAs levels, and (3) strongly attenuates holo-phy protein accumulation (see below and Davis et al., 1999). While most mutant combinations could be made, we failed to generate combinations containing hy1-100 and either ho2-1 or ho2-2, and consequently we were unable to assemble the quadruple hy1/ho2/ho3/ho4 mutant, which should be completely absent of HO activity. This failure was caused by the linkage of the HY1 and HO2 loci, which are only 48 kb (or 0.1 cm) apart on chromosome 2 (Davis et al., 1999). For example, despite screening at least 20,000 progeny from hy1-100/ho2-1 double-heterozygote parent, we were unable to identify plants that were homozygous for hy1-100 and at least heterozygous for ho2-1 (as determined by the long hypocotyl phenotype [hy1-100] and kanamycin resistance [ho2-1]) and thus have the two mutations in coupling (data not shown). In the absence of hy1/ho2 double mutants, we generated all other possible combinations, i.e. hy1/ho3, hy1/ho4, hy1/ho3/ho4, ho2/ho3, ho2/ho4, and ho2/ho3/ho4, using the ho2-2 allele. Like the four single ho mutants, none of the double and triple mutants were phenotypically different from wild type when grown in the dark, indicating that inactivation of these HO genes is not detrimental to general metabolism and growth of Arabidopsis (data not shown).
Phenotypic analysis of our ho mutant collection grown in the light revealed that HO3 and HO4 have an additive role in photomorphogenesis, which was evident only when coupled with a mutant inactivating HY1. As shown in Figure 4A, hy1-100 plants were slow growing and produced small chlorotic leaves under continuous light. This phenotype was further exacerbated when combined with the ho3 and ho4 mutations. Consistent with severely reduced levels of photochemically active phys, etiolated hy1-100 seedlings have markedly reduced sensitivity to light, which could be easily seen by the reduced effectiveness of R and FR to repress hypocotyl elongation (Parks and Quail, 1991; Davis et al., 1999; Muramoto et al., 1999; Fig. 5). This insensitivity was further accentuated in the hy1/ho3, hy1/ho4, and hy1/ho3/ho4 mutant combinations, with the triple mutant being the most insensitive (Fig. 5).
Additional hy1 phenotypes enhanced by inactivation of HO3 and HO4 function were (1) flowering time in continuous light and long-day photoperiods, which was accelerated further in the double (hy1/ho3 and hy1/ho4) and triple (hy1/ho3/ho4) mutants; (2) chlorosis, which became more noticeable in the triple hy1/ho3/ho4 mutant; and (3) the R suppression of the gravitropism, which became more severe (Fig. 6, A–C). For example, when grown under continuous light, the hy1/ho3/ho4 triple mutant initiated the floral inflorescence after forming only 4.2 ± 0.2 true leaves, while hy1-100 and wild-type plants made 8.1 ± 0.3 and 18.5 ± 1.8 true leaves before bolting, respectively. One hallmark of hy1 mutants is that they marginally recover with respect to chlorophyll accumulation as the plants mature. This feature was markedly reduced in hy1/ho3/ho4 seedlings, which retained their pale-green/yellow color longer than hy1 seedlings (Fig. 4B). Etiolated wild-type plants have a strong gravitropism response when grown in the dark, causing their hypocotyls to bend and elongate in the opposite direction to the gravity vector. This tropism is substantially dampened in R by a process that requires phyA and phyB (Poppe et al., 1996; Robson and Smith, 1996). Here, we found that this inhibition of graviperception to R is attenuated in the etiolated hy1 seedlings (as measured by the deviation of hypocotyl growth from vertical [Fig. 6C]). Analysis of the hy1/ho3, hy1/ho4, and hy1/ho3/ho4 mutants showed that this inhibition of gravitropism was further compromised upon inactivation of HO3 and HO4 with the triple mutant behaving strongly gravitropic (Fig. 6C).
Arabidopsis ho2-1 and ho2-2 plants are only weakly compromised in light detection as compared to hy1-100, indicating that the corresponding protein plays a minor role in PΦB synthesis as compared to HY1 (Davis et al., 2001; Fig. 5A). We found that this insensitivity was not made worse when the ho2-2 mutant was combined with either or both ho3 and ho4 mutants. For example, the modest insensitivity of hypocotyl elongation to R or FR for the single ho2-2 mutant was identical to the triple ho2/ho3/ho4 mutant (Fig. 5A). The ho2/ho3/ho4 plants were also phenotypically normal (Fig. 4), and the chlorophyll content, days to flowering, and light-activated gravitropic responses for the ho2/ho3, ho2/ho4, and ho2/ho3/ho4 seedlings were statistically indistinguishable from ho2-2 seedlings (Fig. 6).
Effect of ho Mutations on Holo-Phy Accumulation
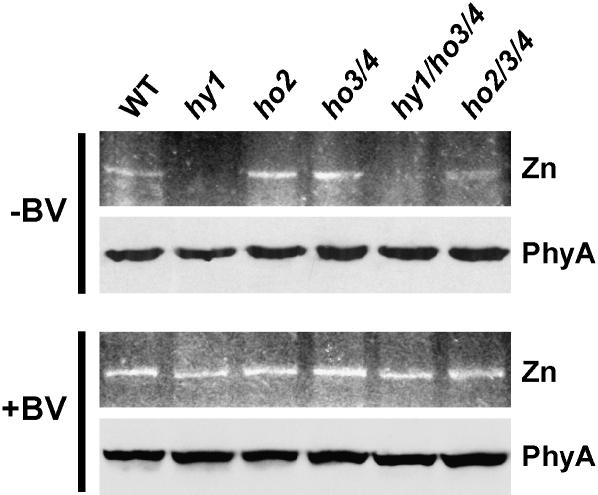
Consistent with a strong suppression of many R and FR light responses, inactivation of the HY1 locus substantially reduces the pool of photochemically active phys without apparent reductions in the abundance of the apoproteins (Parks and Quail, 1991; Davis et al., 1999; Muramoto et al., 1999). A more subtle reduction of holo-phy levels was reported for the ho2-1 mutant in agreement with its weaker effects on photomorphogenesis (Davis et al., 2001). By assessing the levels of phy in the various ho mutant combinations, we found that HO3 and HO4 also contribute to holo-phy assembly. The mutant plants were grown in the dark to avoid Pfr-induced turnover. Crude extracts from these plants were then assayed for the bound holoprotein by zinc-induced fluorescence of the chromophore following SDS-PAGE and for phyA protein levels (which constitutes most of the phy pool in etiolated seedlings [Somers and Quail, 1995; Sharrock and Clack, 2002]) by immunoblot analysis with a monoclonal antibody specific for the phyA isoform.
As shown in Figure 7, none of the various hy1, ho2, ho3, and ho4 mutants either singly or in combination affected accumulation of the phyA apoprotein, further supporting the proposal that phyA polypeptide synthesis is not controlled by bilin availability (Parks and Quail, 1991). Because the levels of holo-phys is already below detection in the hy1-100 mutant, we could not assess whether the ho3-1 and ho4-1 mutations further repress their accumulation when combined in the triple mutant. However, when the two mutants were combined with ho2-2, a decrease in bilin-containing holo-phy levels was observed. As judged by fluorescence intensity of the holo-phys, we found that the ho3/ho4 double mutant had similar levels of photoactive Phys to wild-type and ho2-2 plants. In contrast, this fluorescence was consistently less intense for ho2/ho3/ho4 triple mutant seedlings even though the level of the phyA polypeptide was unaffected (Fig. 7).
Figure 7.
Comparison of holo-phy levels in the collection of ho mutant seedlings. Holo-phys were detected by zinc-induced fluorescence of the chromoproteins following SDS-PAGE (top sections). The phyA polypeptide was detected following SDS-PAGE by immunoblot analysis with the monoclonal anti-phyA antibody O73D (bottom sections). Analysis of crude extracts from plants grown for 6 d in the dark (−BV). Analysis of crude extracts from plants grown for 6 d in the dark on medium supplemented with 100 μm BV (+BV).
Rescue of Phy Responsiveness and Holo-Phy Accumulation with BV
The potential involvement of HO activities in other aspects of heme/chlorophyll metabolism, CO production, and Fe2+ recycling in addition to the synthesis of PΦB implied that some of the phenotypes of hy1 mutants are not solely generated by attenuating holo-phy assembly (Terry and Kendrick, 1999; Ortiz de Montellano and Wilks, 2001; Papenbrock and Grimm, 2001). These role(s) could also explain the strong phenotypic effects of null HY1 mutants as compared to those in HY2 (Koornneef et al., 1980; Chory et al., 1989). To examine this possibility, particularly whether ho mutant phenotypes were generated by a failure to degrade heme and release CO and Fe2+ and not by a block in BV synthesis, we attempted to rescue the hy1, ho2/ho3/ho4, and hy1/ho3/ho4 plants by feeding them BV. Rescue of holo-phy levels was assessed by restoration of the zinc-induced fluorescence of the PΦB-bound holo-phy when etiolated seedlings were grown for 6 d in the dark on medium containing 100 μm BV. This concentration of BV did not appear to have any deleterious effects on etiolated seedling development (data not shown). Similar to the observations of Parks and Quail (1991), we found that feeding BV to etiolated hy1 seedlings restored the fluorescence intensity of the phys to that observed for wild-type seedlings (Fig. 7). A similar rescue in holo-phy levels was observed for the ho2/ho3/ho4 and hy1/ho3/ho4 mutants as well (Fig. 7). Given that phy apoproteins assemble poorly if at all with BV (Elich and Lagarias, 1989; Bhoo et al., 2001), the restoration of holo-phy levels further implied that the exogenous BV was first imported into chloroplasts, converted to PΦB, and the PΦB then exported to the cytoplasm before binding to the polypeptide.
Phenotypic rescue by BV was evaluated by measuring the hypocotyl elongation response of the hy1 and hy1/ho3/ho4 mutants to FR (Parks and Quail, 1991). As described above, continuous FR inhibits hypocotyl elongation of wild-type seedlings; this inhibition is substantially attenuated in the hy1 and hy1/ho3/ho4 mutants with the triple mutant displaying very little response even to high fluences (Fig. 5). Upon feeding the seedlings increasing concentrations of BV, we found that this growth-inhibitory response of FR could be restored in a concentration-dependent manner (Fig. 8). Whereas 1 μm BV did little to rescue this photoresponse, it was partially restored with 3 to 10 μm and completely restored with 100 μm. The rescue of hypocotyl growth inhibition to FR in the hy1/ho3/ho4 mutant was similar to that seen for the hy1 mutant, suggesting that a lack of BV is the sole reason behind the accentuated phenotype of the triple mutant as compared to hy1. When combined with the ability of BV to restore assembly of photoactive phys, we conclude that the photomorphogenic defects of the hy1/ho3/ho4 mutants were caused primarily by a reduction in photochemically active phys and not a failure to either metabolize heme or release CO or Fe2+.
Figure 8.
Rescue of the FR sensitivity to hypocotyl growth by feeding the various ho mutants BV. Wild-type (Col-0), hy1, and hy1/ho3/ho4 seedlings were grown under continuous FR on various concentrations of BV. A, Pictures of representative 6-d-old seedlings grown on 0, 3, and 100 μm BV. B, Hypocotyl length of 6-d-old seedlings grown on various concentrations of BV. Each bar represents the average of at least 10 seedlings with the error bars reflecting ± 1 sd.
DISCUSSION
Genomic analyses indicate that many plant species contain small families of HO-type proteins that are presumably responsible for metabolizing heme and generating the BV precursor needed for synthesis of the phy chromophore PΦB. For example, cursory searches of available EST databases identified multiple HO-type genes in rice (two), maize (six), barley (Hordeum vulgare; two), sorghum (Sorghum bicolor; three), cotton (Gossypium hirsutum; three), tomato (two), and soybean (Glycine max; three; data not shown). Here, we show that all four members of the HO family in Arabidopsis are transcriptionally active with substantially overlapping patterns of expression. Similar to the recent results of Matsumoto et al. (2004) using transcript profiling, we found that HY1 is clearly the highest expressed, followed by HO2, with both HO3 and HO4 expressed at low levels. Moreover, neither our nor their data found substantial changes in HY1, HO2-4 expression by light or other environmental cues. Thus, it appears that most Arabidopsis tissues constitutively express all four genes. By reverse genetic analysis of the four Arabidopsis HO genes either singly or in combination, we provide evidence that that multiple HO isoforms can contribute to synthesizing BV necessary to assemble photoactive phys. Our data reaffirm HY1 as the dominant isoform for holo-phy assembly but show that HO3 and HO4 are also important, especially in the absence of HY1. Moreover, our ability to rescue the photosensitivity of etiolated hy1/ho3/ho4 seedlings to FR by feeding BV argues that that many of the phenotypic defects in these plants can be explained by an inadequate supply of BV.
The hy1/ho3/ho4 triple mutant is severely attenuated for many phy-controlled responses, including the sensitivity of hypocotyl growth to R and FR, chlorophyll accumulation, flowering time, and enhanced gravitropism by light. In fact, the hy1/ho3/ho4 mutant represents one of the more severely compromised photomorphogenic mutants yet described in Arabidopsis, with phenotypes comparable to those of the quadruple phyA/B/D/E mutant recently described by Franklin et al. (2003a, 2003b). Consequently, this triple mutant should prove useful in defining various responses under phy control. hy1/ho3/ho4 plants still respond to R and FR, albeit weakly. Whether this residual responsiveness is directed by other photoreceptors (e.g. CRY, PHOT) or by low amounts of PΦB-phy holoproteins in which the BV precursor was synthesized nonenzymatically or by HO2 remains to be determined.
While the roles for HO3 and HO4 in photomorphogenesis are clear from HO assays and the phenotype of hy1/ho3/ho4 triple mutant as compared to hy1 single mutant, we cannot yet unequivocally confirm that HO2 functions solely in synthesizing the IXα isoform of BV. Even before this study, HO2 was considered unique within the HO family by its phylogenetic separation from HY1, HO3, and HO4 and by the relatively conservative replacement of an arginine for the histidine that is considered important for the bilin-oxygenase activity of its mammalian orthologs (Davis et al., 2001). Interestingly, for plant species (both monocot and dicot) where sufficient sequence quality is available (rice, barley, sorghum, cotton, tomato, and soybean), we could identify at least one representative of the HO2 subtype by sequence alignments and the presence of this arginine at the predicted ligand binding site, in addition to one or more representatives of the canonical HY1/HO3/HO4 subtype. Such a widespread distribution among plants suggests that the HO2 isoform has a specific role in plant physiology. Under conditions where we could easily confirm HO activity for the other members of the family, we were unable to detect a comparable activity for HO2. Even an attempt to convert the predicted active site of HO2 to better resemble canonical HOs (replacement of Arg88 with a histidine) failed to introduce HO activity. Clearly, a substantial problem was the poor expression and insolubility of recombinant HO2, which could interfere with its activity. Another potential complication could be the requirement by HO2 of unusual enzymatic conditions (e.g. choice of reductant) that were not reproduced here. Alternatively, it is possible that HO2 does not synthesis BV from heme, or more specifically it does not generate the IXα isoform of BV. As an example of the latter, an HO capable of generating BV isoforms besides BV IXα has been recently described in the bacterium Pseudomonas aeruginosa (Wegele et al., 2004). Instead of cleaving between the A and D pyrrole rings, this HO variant cleaves between the A and B rings or the B and C rings to generate a mixture of BV IXβ and BV IXδ isoforms.
Despite the absence of biochemical support, this work and that of Davis et al. (2001) suggest that HO2 does affect BV synthesis. ho2 single mutants have modest but significant effects on photomorphogenesis, evident as a reduced sensitivity of hypocotyls to R and FR and early flowering in continuous light. To further support this role, we attempted to generate various mutant combinations of ho2-2 with hy1-100, ho3-1, and ho4-1 and test for phenotypic additivity. Unfortunately, we were unable to combine the hy1 and ho2 mutations due to genetic linkage, and we failed to exacerbate the ho2 phenotype by introducing the ho3 and ho4 mutants either singly or together. However, we found that disruption of HO3 and HO4 in the HO2 background modestly depressed holo-phy levels without decreasing apoprotein abundance. This effect suggests that BV availability was further limiting in the triple mutant as compared to ho2-2.
Like the Arabidopsis hy1 mutants, the collection of ho mutants in other plant species are not completely R/FR insensitive and sometimes can regain photoresponsiveness later in development. For example, the tomato aurea mutant, like Arabidopsis hy1 mutants, is impaired in several phy-mediated photoresponses and deficient in spectrophotometrically detectable phys early in development (Terry and Kendrick, 1996, 1999). However, as the plants mature, both the levels of photochemically active phys and these photoresponses substantially recover, suggesting that another source of BV becomes available. Likewise, the pea HO mutant pcd1, which is substantially deficient in photochemically active phys and severely compromised in phy-mediated responses in young seedlings, acquires sensitivity to phyB-mediated responses later in development (Weller et al., 1996). While this recovery could suggest that the various ho mutant alleles are leaky or that some BV can be synthesized by nonenzymatic oxidation of heme, a more likely scenario consistent with our data is that other members of the HO family in these species provide a source of BV later in development. The one exception may be the rice se5 mutant defective in its sole representative of the HY1/HO3/HO4 subfamily. This apparent loss-of-function mutant is deficient in many phy-controlled responses that span the life cycle and does not appear to recover as the plants mature (Izawa et al., 2000). Rice has a likely HO2 ortholog, but its presence does not appear to ameliorate the BV deficiency later in development.
It has been proposed that the strong phenotypic defects associated with HO mutants are generated not only by the loss of photochemically active phys but also by changes in heme metabolism. In particular, Terry and Kendrick (1999) provided evidence that the tomato aurea and yellow-green mutants, which affect HO and PΦB synthase activities, respectively, adversely affect chlorophyll synthesis, presumably by an aberrant accumulation of heme. These plants remain chlorotic even when grown on the chlorophyll precursor protoporphyrin IXα and can be mimicked when fed an iron chelator that blocks heme synthesis. One possibility is that heme accumulation feedback inhibits ALA synthesis, which then reduces production of both heme/PΦB and chlorophyll. Another is that some of the phenotypes associated with the ho mutants are generated by the absence of CO and Fe2+ that are also products of heme oxidation by HOs. While we cannot rule out such affects, the lack of obvious phenotypic defects for our Arabidopsis ho mutant collection when grown in the dark argues against the possibilities that these mutants are impaired in general metabolism, accumulate heme to toxic levels, or are deficient in CO or Fe2+. In fact, the ability to phenotypically rescue the Arabidopsis hy1/ho3/ho4 triple mutant by feeding only BV argues that their phenotypic defects are caused primarily by a block in BV synthesis (this study; Parks and Quail, 1991).
Despite our demonstration that the HO2 to 4 isoforms also affect phy assembly in Arabidopsis, it is clear that HY1 is the main source of BV in this plant. Consistent with this role, mRNA levels based on DNA microarrays for HY1 are approximately 3 times higher than that for HO2 and approximately 10 times higher than those for HO3 and HO4. Whereas hy1 mutants are strongly compromised in holo-phy assembly and R and FR light perception (Parks and Quail, 1991; Davis et al., 1999; Muramoto et al., 1999), the ho2 mutants are weakly compromised (Davis et al., 2001) and ho3, ho4, and ho3/ho4 mutants are normal (this study). Consequently, it remains unclear why the HO2, 3, and 4 proteins are necessary. Their overlapping patterns of expression with HY1 suggest that these isoforms do not have tissue-specific roles. Nor is there any evidence to date that one or more of these genes are environmentally regulated. It is possible that HO2, 3, and 4 are mainly responsible for heme degradation but become important for holo-phy assembly only when bilin levels are limiting in the hy1 background. While HY1 has been shown to be chloroplast localized, the intracellular distribution of the other three remains to be demonstrated. Like HY1, HO2, 3, and 4 appear to have an N-terminal transit peptide sequence for chloroplast import, but it remains possible that other destinations exist, including mitochondria that could use the encoded enzymes to metabolize heme-containing proteins found in high concentrations in this compartment. Given the recent evidence that some N-terminal sequences can simultaneously target proteins to both mitochrondria and chloroplasts (Silva-Filho, 2003; Rudhe et al., 2004), one or more Arabidopsis HOs could be directed to both compartments.
MATERIALS AND METHODS
Analysis of HO Activity
Full-length HY1/HO2-4 cDNAs were amplified from total RNA extracted from 10-d-old liquid-grown Arabidopsis (Arabidopsis thaliana) ecotype Col-0 seedlings using the Trizol protocol (Invitrogen). Each first-strand cDNA reaction was performed using a gene-specific 3′ primer (see below), a PAE2-specific primer (Downes et al., 2003), 3 μg of RNA, and SuperScript III reverse transcriptase (Invitrogen). The first-strand cDNA was then PCR amplified using the same 3′ primer and an upstream 5′ primer for each HO gene designed to introduce BamH1 and Xho1 restriction sites at the 5′ and 3′ ends, respectively, and a Met codon followed by the HO-coding region without the predicted plastid transit sequence. The resulting constructions were missing the first 51, 56, 55, and 56 codons of HY1, HO2, HO3, and HO4, respectively. The primer sets were: HY1-5′, GGCGGGATCCATGGCGGCTACTACTGCGGCAGAGAAGCAG; HY1-3′, CGGCCTCGAGTCAGGACAATATGAGACGAAGTATCTC; HO2-5′, GGCGGGATCCATGGCCTCACAGAGGAAGAGAACAAGGTAC; HO2-3′, CCGCCTCGAGTTACAAGATGATCAAGCGAACTATCTGA; HO3-5′, GGCGGGATCCATGGCGGCGGCTATAACGGAGAAGCAAC; HO3-3′, CCGCCTCGAGTCAGGACAATATGAGTCGAAGAATCTCC; HO4-5′, GGCGGGATCCATGGTTGTGGCAGCAGCAGGGGAGAAGC; and HO4-3′, CCGCCTCGAGTCAGGACAATAAGTAACGAAAAATCTCCC.
The HO cDNAs were inserted into pET24a for expression in Escherichia coli strain BL21 Rossetta pLys (Novagen). Each HO was coexpressed with the N-terminal 321 amino acids of DrBphP, which also included a C-terminal His6 tag (Bhoo et al., 2001). The HO2R88-H mutant was generated by the QuickChange protocol (Stratagene) with appropriate mutagenic primers. The Synechocystis HO cDNA was described previously (Bhoo et al., 2001). Coexpression of the HO and DrBphP cDNAs was induced for 5 h in 1-L log-phase cultures following the addition of 1 mm isopropyl-β-d-thiogalactoside. Recombinant DrBphP was purified by nickel chelate affinity chromatography according to manufacturer's recommendations (Novagen). Assembly of a photochemically active DrBphP was detected by zinc-induced fluorescence of the adduct following SDS-PAGE and by R/FR absorption difference spectra as described (Bhoo et al., 2001). For in vitro assembly of DrBphP with purified BV, the E. coli extracts were incubated with 2 μm BV IXα hydrochloride (Porphyrin Products) for 30 min before purification.
In vitro activity assays for Arabidopsis HO2 employed the method of Muramoto et al. (2002) coupled with BV-DrBphP assembly. The HO2 cDNA was cloned into pET24a to append a C-terminal His6 tag and expressed in E. coli strain BL21 Rossetta pLys. Following disruption of the cells, the insoluble fraction was removed by centrifugation, and the residual HO2 protein was purified from the soluble fraction by nickel-chelate chromatography and concentrated into 100 mm HEPES-NaOH (pH 7.2) by ultrafiltration using a Centricon column (Millipore). HO2 activity was assayed in a reaction containing 0.15 mg/mL bovine serum albumin, 10 μm hemin, 4.2 μm ferredoxin, 0.025 units/mL ferredoxin-NADPH reductase, 5 mm ascorbate, 2 mm EDTA, 100 μm NADPH, approximately 0.5 μg recombinant HO2, and approximately 50 μg DrBphP in 100 mm HEPES-NaOH buffer (pH 7.2). After 30 min at 25°C, the reaction was quenched by mixing with SDS-PAGE sample buffer. Detection of the BV-DrBphP adduct was attempted by zinc-induced fluorescence following SDS-PAGE (Bhoo et al., 2001).
Identification and Analysis of Arabidopsis HO T-DNA Mutations
The hy1-100 (Col-0) and ho2-1 (Ws) mutants were described previously (Davis et al., 1999, 2001). The ho2-2 (Salk_025840) and ho4-1 (Salk_044934) T-DNA insertion lines were identified in the SIGNAL T-DNA collection (Alonso et al., 2003) and obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The ho3-1 line was identified in the Syngenta SAIL T-DNA collection (Sessions et al., 2002). Gene-specific primers: HO2-5′, ACAGGTCTGGTGCCGGAAACT; HO2-3′, TTCACACATTGCTGGTGGTCA; HO3-5′, CCCTAATTTCGATATAACTTGA; HO3-3', GTTCCACGAAACTAGCAGAAGG; HO4-5′, CACAAACCCATTCGGTTCTCG; and HO4-3′, TCACAACAAACCGGAAAAAGGT were used to amplify the wild-type genes. T-DNA products were amplified using the gene-specific 5′ primer in combination with either the SIGNAL left-border primer TGGTTCACGTAGTGGGGCCATCG or the SAIL left-border primer GCCTTTTCAGAAATGGATAAATAGCC. The exact positions of the T-DNA insertions were identified by DNA sequencing the PCR products generated with the left-border primer in combination with the gene-specific primer. For ho3-1, kanamycin resistance associated with the T-DNA segregated in a 3:1 pattern, consistent with the presence of a single T-DNA insertion site. For ho2-2 and ho4-1, kanamycin resistance was too weak to be reliable, a characteristic found in other insert mutants within the SIGNAL T-DNA collection (Alonso et al., 2003). The various ho mutant combinations were generated by crossing homozygous individuals and then selecting the appropriate mutant lines by PCR in segregating F2 populations. Both the ho3-1 and ho4-1 lines were backcrossed once to wild-type Col-0 before generating the homozygous mutant populations.
For RT-PCR, total RNA was isolated as described above and subjected to first-strand cDNA synthesis and PCR using a set of four gene-specific primers (see Fig. 3). The HO2 primers are: primer 1, CTCCATTATCTCTACACCACG; primer 2, AGAAACAAAGGAGCACTTTCT; primer 3, GTGAGAAGCTTAACGTGCTT; and primer 4, TTACAAGATGATCAAGCGAAC. HO3 primers are: primer 1, CATCTTCCTTCATCTTTCTTATTCA; primer 2, AAGGTTGGGAAATTGGATCCA; primer 3, CGGTGGCCAGATGATTGGA; and primer 4, TAGCAGAAGGCTTTATTGACTATTG. HO4 primers are: primer 1, ATGGCTACATCAAGACTTAATGCC; primer 3, CCTCTTGCCGCTTTCCTGCAA; and primer 4, GGACAATAAGTAACGAAAAATCTCC. Total seedling RNA was reverse transcribed using primers 2 or 4. The RT products from primer 2 reactions were then PCR amplified using the 1 + 2 primer combinations, and the RT products from primer 4 reactions were then PCR amplified with primer pairs 1 + 4 or 3 + 4.
Plant Growth Conditions
For visual assessment of plant phenotypes, plants were germinated on solid one-half times Murashige and Skoog medium containing 2.5 mm MES (pH 5.7) and stratified for 3 d at 4°C. The seedlings were grown at 22°C for 10 d under long days and then transplanted to soil if necessary. Soil-grown plants were grown at approximately 22°C in either 8-h light/16-h dark (short days), 16-h light/8-h dark (long day), or 24-h light (continuous day) photoperiods. Flowering time was assessed based on the number of rosette leaves at bolting when grown under 25 μmol m−2 s−1 white light in long days. Total chlorophyll content was determined spectrophotometrically from the rosette leaves of 21-d-old plants following extraction in 96% ethanol (1 mL/100 μg fresh weight; Davis et al., 2001).
For hypocotyl growth assays, the germinating seedlings were irradiated continuously in the vertical position for 6 d with R or FR (Davis et al., 2001). Hypocotyl length and angles from vertical were measured from computer-scanned images. For BV rescue, the seedlings were grown in the same manner under FR light on solid one-half times Murashige and Skoog medium containing 2.5 mm MES (pH 5.7) and supplemented with various concentrations of BV IXα hydrochloride aliquoted from a 10 mm stock solution in dimethylsulfoxide.
Detection of Phy Protein
Arabidopsis seedlings were grown for 4 d in the dark on solid one-half times Murashige and Skoog medium containing 2.5 mm MES (pH 5.7) and frozen to liquid-nitrogen temperatures. Tissue was homogenized in SDS-PAGE sample buffer, heated and clarified, and the supernatant was subjected to SDS-PAGE. Chromophore-bound phys and the apo-phy polypeptides were detected by zinc-induced fluorescence (Bhoo et al., 2001) and by immunoblot analysis using the monoclonal anti-phy antibody O73D (Jordan et al., 1997), respectively.
HO Expression Analysis
Relative transcript abundance among the four HO genes was approximated using the GENEVESTIGATOR DNA microarray dataset described by Zimmermann et al. (2004). For expression studies using fusions of the HO promoters to the GUS coding region from the uidA gene, the 5′-flanking sequence immediately upstream of the translation start site of Arabidopsis HO3 or HO4 (657 bp and 1,950 bp, respectively) were cloned into pPZP211 (Hajdukiewicz et al., 1994). The HY1∷GUS and HO2∷GUS fusions were as previously described (Davis et al., 2001). The HO∷GUS reporters were introduced into Arabidopsis Col-0 plants by the floral-dip method using the Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998). T1 seeds were selected on Gamborg's B5 medium (Sigma) supplemented with 50 mg/L kanamycin. GUS activity was assayed using the substrate 2-mm 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid dissolved in 100 mm sodium phosphate (pH 7.0), 10 mm EDTA, and 0.15 (v/v) Triton X-100. Seedlings and tissues at various developmental ages were stained for 24 to 36 h at room temperature, and then cleared using an ethanol-gradient series. The progenies of at least three independent transformants for each of the four HO genes were examined to avoid positional effects on the transgenes.
Acknowledgments
We thank Dr. Seth Davis for providing the HY1 and HO2∷GUS reporter lines and Jeremiah Wagner for assistance with the HO activity assays.
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–88ER13968), the National Science Foundation (grant no. MCB–0424062), and the Research Division of the University of Wisconsin College of Agriculture and Life Sciences (Hatch grant no. 142–E443).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard D. Vierstra (vierstra@wisc.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074211.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD (2001) Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414: 776–779 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cornah JE, Terry MJ, Smith AG (2003) Green or red: What stops the traffic in the tetrapyrrole pathway? Trends Plant Sci 8: 224–230 [DOI] [PubMed] [Google Scholar]
- Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD (2001) The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol 126: 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD (1999) The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA 96: 6541–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD (2003) The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35: 729–742 [DOI] [PubMed] [Google Scholar]
- Elich TD, Lagarias JC (1989) Formation of a photoreversible phycocyanobilin-apophytochrome adduct in vitro. J Biol Chem 264: 12902–12908 [PubMed] [Google Scholar]
- Frankenberg N, Mukougawa K, Kohchi T, Lagarias JC (2001) Functional genomic analysis of the hy2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13: 965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC (2003. a) Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15: 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC (2003. b) Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131: 1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile Ppzp family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22: 391–399 [DOI] [PubMed] [Google Scholar]
- Jordan ET, Marita JM, Clough RC, Vierstra RD (1997) Characterization of regions within the N-terminal 6-kilodalton domain of phytochrome A that modulate its biological activity. Plant Physiol 115: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Wagner JR, Walker JM, Vierstra RD (2005) Phylogenetic analysis of the phytochrome superfamily reveals distinct subfamilies of microbial photoreceptors. Biochem J 392: 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC (2001) The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-induced hypocotyl elongation in Arabidopsis thaliana L Heynh. Z. Pflanzenphysiol 100: 147–160 [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K-I, Masuda T (2004) Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol 135: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (gun5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Alonso JM, Ecker JR, Zhang YL, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15: 1962–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T, Kami C, Kataoka H, Iwata N, Linley PJ, Mukougawa K, Yokota A, Kohchi T (2005) The tomato photomorphogenetic mutant, aurea, is deficient in phytochromobilin synthase for phytochrome chromophore biosynthesis. Plant Cell Physiol 46: 661–665 [DOI] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM (1999) The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T, Tsurui N, Terry MJ, Yokota A, Kohchi T (2002) Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol 130: 1958–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz de Montellano PR, Wilks A (2001) Heme oxygenase structure and mechanism. Adv Inorg Chem 51: 359–407 [Google Scholar]
- Papenbrock J, Grimm B (2001) Regulatory network of tetrapyrrole biosynthesis: studies of intracellular signalling involved in metabolic and developmental control of plastids. Planta 213: 667–681 [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH (1991) Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3: 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH (1993) hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell 5: 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C, Hangarter RP, Sharrock RA, Nagy F, Schafer E (1996) The light-induced reduction of the gravitropic growth-orientation of seedlings of Arabidopsis thaliana (L.) Heynh. is a photomorphogenic response mediated synergistically by the far-red-absorbing forms of phytochromes A and B. Planta 199: 511–514 [DOI] [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Robson PR, Smith H (1996) Genetic and transgenic evidence that phytochromes A and B act to modulate the gravitropic orientation of Arabidopsis thaliana hypocotyls. Plant Physiol 110: 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudhe C, Clifton R, Chew O, Zemam K, Richter S, Lamppa G, Whelan J, Glaser E (2004) Processing of the dual targeted precursor protein of glutathione reductase in mitochondria and chloroplasts. J Mol Biol 343: 639–647 [DOI] [PubMed] [Google Scholar]
- Sawers RJH, Linley PJ, Gutierrez-Marcos JF, Delli-Bovi T, Farmer PR, Kohchi T, Terry MJ, Brutnell TP (2004) The Elm1 (ZmHy2) gene of maize encodes a phytochromobilin synthase. Plant Physiol 136: 2771–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer E, Nagy F (2005) Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms, Ed 3. Springer, Dordrecht, The Netherlands
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Filho MC (2003) One ticket for multiple destinations: dual targeting of proteins to distinct subcellular locations. Curr Opin Plant Biol 6: 589–595 [DOI] [PubMed] [Google Scholar]
- Smith H (1995) Physiological and ecological functions with the phytochrome family. Annu Rev Plant Physiol Plant Mol Biol 46: 269–315 [Google Scholar]
- Somers DE, Quail PH (1995) Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J 7: 413–427 [DOI] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH (1991) The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3: 1263–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE (1996) The aurea and yellow-green-2 mutants of tomato are deficient in phytochrome chromophore synthesis. J Biol Chem 271: 21681–21686 [DOI] [PubMed] [Google Scholar]
- Terry MJ, Kendrick RE (1999) Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-greeen-2 mutants of tomato. Plant Physiol 119: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Linley PJ, Kohchi T (2002) Making light of it: the role of plant haem oxygenases in phytochrome chromophore synthesis. Biochem Soc Trans 30: 604–609 [DOI] [PubMed] [Google Scholar]
- Terry MJ, Wahleithner JA, Lagarias JC (1993) Biosynthesis of the plant photoreceptor phytochrome. Arch Biochem Biophys 306: 1–15 [DOI] [PubMed] [Google Scholar]
- Tu S-L, Lagarias JC (2005) The phytochromes. In WR Briggs, JL Spudich, eds, Handbook of Photosensory Receptors. Wiley-VCH, Weinheim, Germany, pp 121–149
- Wegele R, Tasler R, Zeng YH, Rivera M, Frankenberg-Dinkel N (2004) The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J Biol Chem 279: 45791–45802 [DOI] [PubMed] [Google Scholar]
- Weller JL, Terry MJ, Rameau C, Reid JB, Kendrick RE (1996) The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IX alpha. Plant Cell 8: 55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Terry MJ, Reid JB, Kendrick RE (1997) The phytochrome-deficient pcd2 mutant of pea is unable to convert biliverdin IX alpha to 3(Z)-phytochromobilin. Plant J 11: 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]