Abstract
Over 1,600 genes encoding carbohydrate-active enzymes (CAZymes) in the Populus trichocarpa (Torr. & Gray) genome were identified based on sequence homology, annotated, and grouped into families of glycosyltransferases, glycoside hydrolases, carbohydrate esterases, polysaccharide lyases, and expansins. Poplar (Populus spp.) had approximately 1.6 times more CAZyme genes than Arabidopsis (Arabidopsis thaliana). Whereas most families were proportionally increased, xylan and pectin-related families were underrepresented and the GT1 family of secondary metabolite-glycosylating enzymes was overrepresented in poplar. CAZyme gene expression in poplar was analyzed using a collection of 100,000 expressed sequence tags from 17 different tissues and compared to microarray data for poplar and Arabidopsis. Expression of genes involved in pectin and hemicellulose metabolism was detected in all tissues, indicating a constant maintenance of transcripts encoding enzymes remodeling the cell wall matrix. The most abundant transcripts encoded sucrose synthases that were specifically expressed in wood-forming tissues along with cellulose synthase and homologs of KORRIGAN and ELP1. Woody tissues were the richest source of various other CAZyme transcripts, demonstrating the importance of this group of enzymes for xylogenesis. In contrast, there was little expression of genes related to starch metabolism during wood formation, consistent with the preferential flux of carbon to cell wall biosynthesis. Seasonally dormant meristems of poplar showed a high prevalence of transcripts related to starch metabolism and surprisingly retained transcripts of some cell wall synthesis enzymes. The data showed profound changes in CAZyme transcriptomes in different poplar tissues and pointed to some key differences in CAZyme genes and their regulation between herbaceous and woody plants.
Carbohydrate-active enzymes (CAZymes; http://afmb.cnrs-mrs.fr/CAZY) include families of glycosyltransferases (GTs; Coutinho et al., 2003a) and glycoside hydrolases (GHs; Henrissat, 1991), as well as polysaccharide lyases (PLs) and various carbohydrate esterases (CEs). The multiplicity and complexity of structures that oligosaccharides and polysaccharides can adopt are exploited by living organisms for a multitude of biological functions, including structural aspects, storage of energy, and signaling events. With over 200 genomes from various organisms completely sequenced today, it has become clear that plants have more CAZyme-encoding genes than any other group (Coutinho et al., 2003b). Plants rely on CAZymes to assimilate the products of photosynthesis into sugars and starch, to synthesize cell wall biopolymers such as cellulose, pectin, and xylan, and to create various glycosylated compounds (glycolipids, glycoproteins, lignin precursors, and secondary metabolites). CAZyme genes have been extensively studied at the genomic and transcriptomic levels in Arabidopsis (Arabidopsis thaliana; Henrissat et al., 2001) and other species of plants and microorganisms (Coutinho et al., 2003a). The recently published genome sequence and the extensive tissue-specific expressed sequence tag (EST) library collections available for poplar (Populus spp.), a woody plant species of commercial value and a tree model species, offer unique opportunities to study CAZyme gene diversity and regulation.
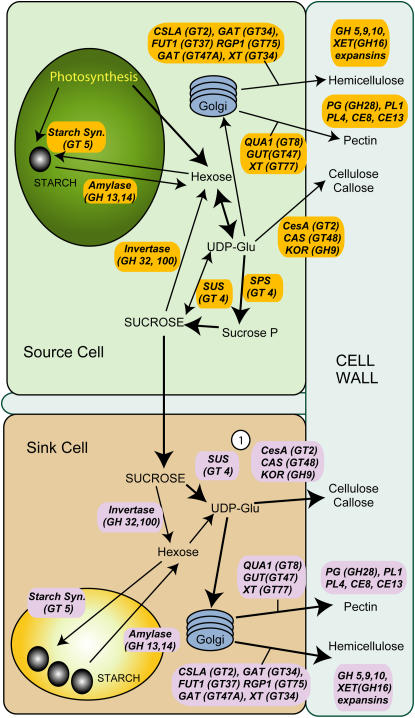
Woody plants are the most important primary producers in terrestrial ecosystems. The biomass accumulated in the secondary walls of trees represents an ecologically essential carbon sink, as well as an important industrial raw material. Trees assimilate carbon in the chlorenchyma (source tissue) situated in leaves and young bark (Kozlowski, 1992). There the carbon is incorporated into triose-P and subsequently used for starch biosynthesis in the chloroplasts (Fig. 1). Carbon from starch and triose-P is used to synthesize hexoses and, finally, Suc in the cytosol of the source tissues. Suc is a major compound transported in the phloem from the source tissues to the nonphotosynthetic (sink) tissues (Konishi et al., 2004). The carbon delivered by Suc transport is then utilized as an energy source and a skeleton for building new molecules during growth and respiration. A major proportion of carbon is used for cambial growth and differentiation of the secondary xylem. Once carbon is allocated to secondary wall cellulose, hemicellulose, or lignin, it is permanently immobilized for the lifetime of the tree. The carbon resources are also stored as starch, either transiently in source tissues, or as seasonal and emergency reserves in storage tissues. These reserves can be mobilized later for growth and development processes or can also be broken down in response to cold stress to produce the cryostabilizing sugars (Kaplan and Guy, 2004).
Figure 1.
CAZyme-encoding genes involved in carbon flow to the cell wall. General scheme of carbon flow from photosynthesis (green chloroplast in source cell) or mobilization of starch reserves (yellow amyloplast in sink cell) to the production of cell wall carbohydrates (blue). Major CAZyme families are identified, including GTs, GHs, PLs, and CEs. Note that some functions only involve a subset of family members (e.g. GT4, GT2). 1, SUS and CesA complexes are known to associate at the plasma membrane for direct conversion of Suc to UDP-Glc for cellulose biosynthesis.
Given the ecological as well as economical value of woody plants, it would be very useful to achieve a comprehensive characterization of CAZymes in at least one model system for woody plants. Detailed comparison of the transcriptional regulation of CAZymes in a woody plant and Arabidopsis is also a means toward new insight into genetic differences that make trees and herbs utilize carbon so differently. Wood as structural and transporting tissue and the turnover of carbohydrate reserves are essential for trees during their long life. Therefore, it is likely that trees invest more in specific CAZymes, both in terms of genetic diversity and expression levels, than herbaceous plant species. Publication of the Populus trichocarpa (Torr. & Gray) genome (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) made it possible for the first time to examine the full spectrum of CAZymes in a woody plant species. We identified all members of different CAZyme families and closely related expansins and compared their genetic diversity between P. trichocarpa and Arabidopsis. The Populus EST database (http://www.populus.db.umu.se/index.html), containing over 100,000 ESTs from 17 different tissue libraries (Sterky et al., 2004), was used to determine the tissue-specific regulation of highly expressed genes. In addition, our in-house poplar microarray expression data, together with Arabidopsis microarray data available in the public domain, were analyzed to determine the global pattern of CAZyme expression in the two species, focusing on sugar/starch metabolism and cell wall carbohydrate biosynthesis. The results revealed the dynamics of the major carbon fluxes among tissues and organs and during the annual cycle of activity and dormancy in a tree at the transcription level. Comparison of CAZyme diversity and expression at the genomic scale between an annual herbaceous (Arabidopsis) and a woody (poplar) species allowed for unique insights into metabolic differences in the gene expression level of the carbon utilization pathways between these developmental habits.
RESULTS AND DISCUSSION
Identification of 1,647 CAZymes and Expansins in Poplar
Candidate CAZymes in poplar were identified by comparing the approximately 55,000 P. trichocarpa gene models (version 1.0) downloaded from the U.S. Department of Energy (DOE) Joint Genome Institute (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) to the approximately 33,000 curated entries of the CAZyme database (CAZy at http://afmb.cnrs-mrs.fr/CAZy; December 2004 version) supplemented with the sequences of all known expansins. CAZymes are typically modular proteins containing one or more catalytic modules associated with a variable number of independent substrate binding or other accessory modules (for review, see Henrissat and Davies, 2000). To avoid false positives due to this frequent modularity, only the models displaying similarity to the catalytic module of CAZymes were analyzed further. In addition to 42 expansin-related sequences, 1,603 CAZyme gene models were identified (including >600 GHs, approximately 840 GTs, approximately 40 PLs, and >100 CEs). All identified genes are deposited in Supplemental Table I. Because several poplar gene models are likely to be fragments requiring refinement of the assembly, these figures might decrease slightly with subsequent versions of the genome. However, our manual sequence alignment analyses of selected gene families did not reveal major discrepancies in the number of genes (data not shown; N. Nishikubo, J. Takahashi, K. Pien, M. Hertzberg, T.T. Teeri, B. Sundberg, and E.J. Mellerowicz, unpublished data; J. Takahashi, E. Master, U. Rudsander, T.T. Teeri, B. Sundberg, and E.J. Mellerowicz, unpublished data).
Genetic Diversity of CAZyme Gene Families in Poplar and Arabidopsis
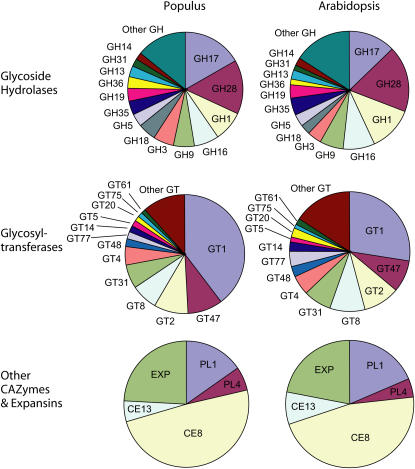
This analysis shows that the P. trichocarpa genome contains approximately 1.6 times the number of CAZyme and expansin genes identified in the Arabidopsis genome (Henrissat et al., 2001; http://afmb.cnrs-mrs.fr/CAZy; http://www.bio.psu.edu/expansins), which is the largest number of genes encoding these enzymes observed among fully sequenced genomes. The large number of gene copies is apparently the consequence of ancient polyploidization events during the history of the Populus genus (Djerbi et al., 2005). Comparison of the number of genes in specific CAZyme families between the two species (Fig. 2) demonstrated that, indeed, most gene families are proportionally increased in P. trichocarpa, indicating chromosome duplication events. Interestingly, a few gene families did not follow this general trend. The families that were disproportionately smaller in P. trichocarpa than in Arabidopsis include xylanases (GH10 family) and pectin-related enzyme families, such as pectate lyases (PL1 family) and rhamnogalacturonan (RG) II xylosyl transferases (GT77 family; Egelund et al., 2004). In contrast, the GT1 family, encompassing inverting, single sugar-adding transferases responsible for glycosylation of secondary metabolites, was larger than expected in poplar. Similarly, the cell wall-related genes have been shown to be underrepresented in the rice (Oryza sativa) genome when compared to the Arabidopsis genome (Yokoyama and Nishitani, 2004). This is a good example of how gene diversification may serve to shape the genomes toward purposeful functional diversification.
Figure 2.
Comparison of the genetic diversity of CAZymes and expansins in poplar (Populus) and Arabidopsis. In the P. trichocarpa sequenced genome, genes were identified by similarity to known CAZymes and compared to the genes found in the Arabidopsis genome. A total of 1,647 poplar genes and 969 Arabidopsis genes were identified. Pie charts show proportional size of each gene family, GT, GH, and other CAZyme families (PL and CE), and expansins (EXPs).
Global CAZyme Expression Analysis
Validation of EST Sources from Different Poplar Species
Transcription is an important level at which the activity of genes and their products is regulated. Recent large-scale transcript-profiling studies revealed that many genes that act together are transcriptionally coregulated (e.g. Andersson-Gunnerås et al., 2006), suggesting that evolutionary change involves modifications of transcriptional programs. Thus, the information on transcriptional regulation of CAZymes in poplar is an essential step toward elucidating the regulation of carbon metabolism in trees and its evolution.
The collection of over 100,000 ESTs from well-defined tissues/organs of several poplar species (http://www.Populus.db.umu.se/index.html; Sterky et al., 2004) constitutes a valuable resource for in silico gene expression analyses (Aspeborg et al., 2005; Moreau et al., 2005). To validate using P. trichocarpa gene models for assigning to them ESTs from this EST collection, we examined the degree of interspecific sequence variation within the genus. Table I compares the full-length nucleotide sequences of CAZyme genes of Populus alba × Populus tremula, P. trichocarpa, P. alba, P. tremula × Populus tremuloides, and P. tremuloides available in the public domain. On average, about 2% of sequence divergence was observed between P. trichocarpa and other poplar species examined. This level of divergence was taken into account when assigning ESTs from various poplar species to the P. trichocarpa gene models.
Table I.
Comparison of coding sequences from different poplar species with coding sequences of P. trichocarpa
| Other Poplar Species Used for EST Libraries | Populus trichocarpa Nucleotide Sequence Divergencea |
|---|---|
| % | |
| P. alba (Pal) | 2.37 ± 0.9 |
| P. alba × P. tremula (Pat) | 2.51 ± 1.59 |
| P. tremuloides (Ptl) | 1.81 ± 0.47 |
| P. tremula × P. tremuloides (Ptt) | 2.11 ± 1.0 |
Full-length EST nucleotide sequences aligned and average percent difference calculated. N = 4, 2, 9, and 26, respectively, by row.
CAZyme Families Expressed in Different Tissues and Seasons
ESTs were assigned to the P. trichocarpa gene models using a stringent criterion of 95% identity to be able to distinguish among the closely related members of gene families. This allowed about 2% of intraspecific sequence divergence (Table I) and 3% of sequencing error. In all, we mapped 2,449 ESTs to the CAZyme gene models (Table II; Supplemental Table I). Over 1,100 ESTs represented GHs, 970 represented GTs, and the rest represented the remaining CAZyme families and expansins. There was a striking difference in the abundance of the expressed CAZymes in different, approximately equally sized, cDNA libraries (Table II). Wood-forming tissues, including the cambial region (cambium plus developing xylem and phloem), cambium, and developing tension wood (TW), were the richest sources of CAZyme transcripts. Developing wood tissues were also a rich source of genes encoding GTs, GHs, PLs, and expansins. However, the highest number of expressed genes for CEs was found in petioles and cold-treated leaves. Expansins showed a stark contrast in EST levels between active and dormant stages, being completely absent in dormant tissues.
Table II.
The EST library distribution and number of expressed genes in CAZyme and expansin families in different poplar tissuesa
EXP, Expansins; Tot, total; Cam, cambial region; Dor Cam, dormant cambium; Act Cam, active cambium; Dor Bud, dormant bud; Flw, male and female mature flowers.
| Total | Cam | Dor Camb | Act Camb | TW | WCD | Bark | Root | Leaf Young | Leaf Cold | Leaf Old | Petiole | Apical Bud | SAMb | Dor Bud | Flw Male | Flw Fem | Seed | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ESTs | Total | 2,449 | 269 | 98 | 190 | 394 | 157 | 121 | 112 | 143 | 168 | 156 | 138 | 140 | 138 | 119 | 78 | 170 | 115 |
| GH | 1,115 | 78 | 52 | 67 | 125 | 73 | 59 | 53 | 74 | 65 | 59 | 52 | 77 | 37 | 51 | 32 | 80 | 81 | |
| GT | 970 | 94 | 32 | 64 | 189 | 70 | 34 | 28 | 55 | 58 | 63 | 34 | 37 | 63 | 53 | 23 | 47 | 26 | |
| PL | 66 | 21 | 2 | 1 | 20 | 1 | 1 | 1 | 0 | 5 | 0 | 6 | 0 | 4 | 0 | 1 | 3 | 0 | |
| CE | 117 | 6 | 3 | 10 | 12 | 2 | 8 | 5 | 8 | 15 | 4 | 16 | 3 | 2 | 0 | 10 | 12 | 1 | |
| EXP | 181 | 50 | 0 | 17 | 19 | 4 | 11 | 4 | 1 | 15 | 26 | 11 | 5 | 7 | 0 | 3 | 6 | 2 | |
| Expressed genes | Total | 597 | 133 | 53 | 137 | 139 | 79 | 82 | 68 | 66 | 101 | 71 | 91 | 84 | 93 | 53 | 59 | 101 | 52 |
| GH | 222 | 46 | 20 | 42 | 47 | 34 | 36 | 25 | 25 | 35 | 25 | 33 | 36 | 33 | 28 | 26 | 45 | 27 | |
| GT | 304 | 58 | 25 | 55 | 63 | 37 | 30 | 25 | 30 | 44 | 39 | 30 | 28 | 43 | 21 | 21 | 38 | 18 | |
| PL | 14 | 7 | 2 | 1 | 8 | 1 | 1 | 1 | 0 | 3 | 0 | 2 | 0 | 3 | 0 | 1 | 1 | 0 | |
| CE | 36 | 6 | 3 | 7 | 7 | 1 | 6 | 5 | 6 | 10 | 3 | 10 | 3 | 2 | 0 | 4 | 4 | 1 | |
| EXP | 21 | 9 | 0 | 10 | 6 | 3 | 4 | 4 | 1 | 5 | 1 | 6 | 4 | 4 | 0 | 1 | 4 | 2 |
Numbers of EST clones corresponding to CAZyme-encoding genes and expansins (top half) or the number of genes with at least one corresponding EST (bottom half) in the collection of similar-sized cDNA libraries representing different tissues/developmental stages.
Libraries constructed from amplified cDNA, not directly comparable to other libraries but comparable to each other and internally.
Gene Family Size and Number of Expressed Genes
One of the key questions regarding genes identified within a fully sequenced genome is whether the genes are active or whether they represent degenerate nonfunctional copies. While the presence of an EST indicates that the corresponding gene is transcribed, the absence of an EST implies that it is inactive or expressed at low levels in the tissues sampled or that it is a nonfunctional gene per se. It is likely that a number of genes specifically expressed in particular cell types or under defined circumstances escaped detection in the course of our EST sequencing effort. In Arabidopsis, only 16,125 genes of the total of approximately 25,000 genes were represented by ESTs within a collection of 167,000 ESTs covering over 60 different tissues/organs and various conditions (Rudd, 2003). Thus, even in the large Arabidopsis EST collection gathered over 10 years, only 64.5% of the genes had corresponding ESTs. The poplar EST collection of more than 100,000 allowed us to detect the expression for approximately 600 different CAZyme genes, constituting about 40% of all CAZymes identified in the genome. Small gene families had a higher percentage of genes with ESTs than large gene families (Supplemental Fig. 1), indicating that small families had proportionally more highly and/or ubiquitously expressed genes. The result implies that gene duplication in large CAZyme families was driven by a need for diverse expression and/or biochemical function rather than for high expression. This kind of genetic specialization allows for more rapid evolution of the coding sequence to suit the functional needs of specific organs and tissues. Interestingly, the largest and most diverse gene family, GT1, was proportionally larger in poplar than in Arabidopsis, perhaps due to additional specialized functions and tissues, such as massive wood formation, dormancy, and longevity.
Differential Expression of Major CAZyme Families
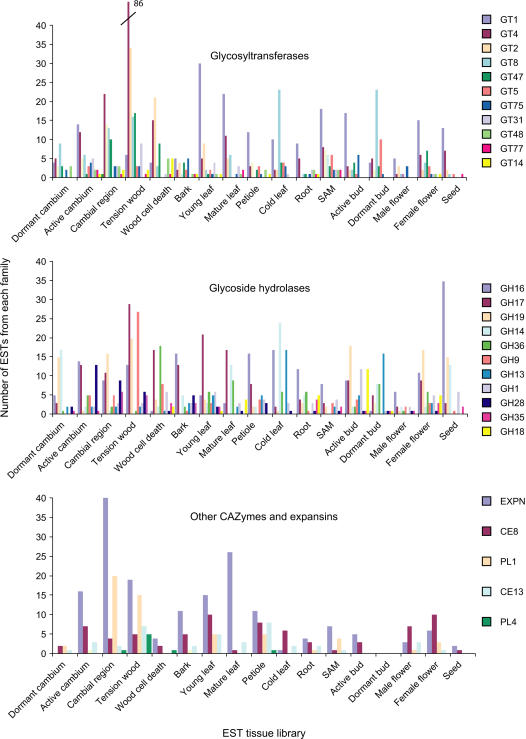
To analyze more specifically the transcript profile of different CAZyme families, the EST number within each cDNA library was compared with the expected number relative to the library size, and the Fisher's exact test statistics were calculated to infer significant overrepresentation of a given family in specific tissues. Of a total of 79 CAZyme families identified, 47 (60%) showed a specific expression pattern in one or more tissues. The families with the highest levels of tissue-specific expression are shown in Figure 3. The enzymes in these families are generally involved in cell wall carbohydrate biosynthesis and modification (GT2, GT8, GT47, expansins, CE8, PL1, GH16, GH19, GH9, GT4) or in starch biosynthesis and turnover (GT5, GH13, GH14). These families will be discussed in detail below. In addition, members of families GT1, GH17, GH36, and GH1 were highly expressed and had specific expression patterns.
Figure 3.
Differential expression of CAZyme families and expansins in poplar. Families significantly overexpressed (Fisher's exact test, P ≤ 5%) in at least one tissue and with more than 20 ESTs for GHs, 15 ESTs for GTs, and eight ESTs for other CAZymes and expansins are shown.
The GT1 family comprises a wide variety of enzymes catalyzing a single sugar addition to diverse compounds such as hormones, including indole-3-acetic acid, zeatin, benzyl adenine, and salicylic acid, monolignols, and secondary metabolites, for example, flavonols and anthocyanins (Ross et al., 2001). This family was most highly expressed in the mature and young leaves and it was also more diverse in poplar than in Arabidopsis (Fig. 2), suggesting more active secondary metabolism of poplar in gene expression.
The GH17 family includes candidate β-1,3-glucanases, many of which carry a putative C-terminal carbohydrate-binding module (P. Coutinho and B. Henrissat, unpublished data), whereas the GH36 family contains enzymes with α-galactosidase activity. These families were overrepresented in TW, the wood cell death (WCD) zone, and mature leaves (Fig. 3), i.e. in tissues containing fully expanded cells. Therefore, GH17 β-1,3-glucanases and GH36 α-galactosidases might be involved in the regulation of mature cell wall functions such as elasticity and/or transport.
β-Glycosidases of the GH1 family were mostly expressed in the active bud and leaves. In Arabidopsis, this family includes defense-related enzymes, called myrosinases, but no such enzyme was detected in poplar, as expected, because the occurrence of myrosinases is limited to Brassicaceae and a few other species (Rodman, 1991). Poplar probably uses other types of GH1 β-glycosidases and possibly uses them in a different way for defense in developing young leaves.
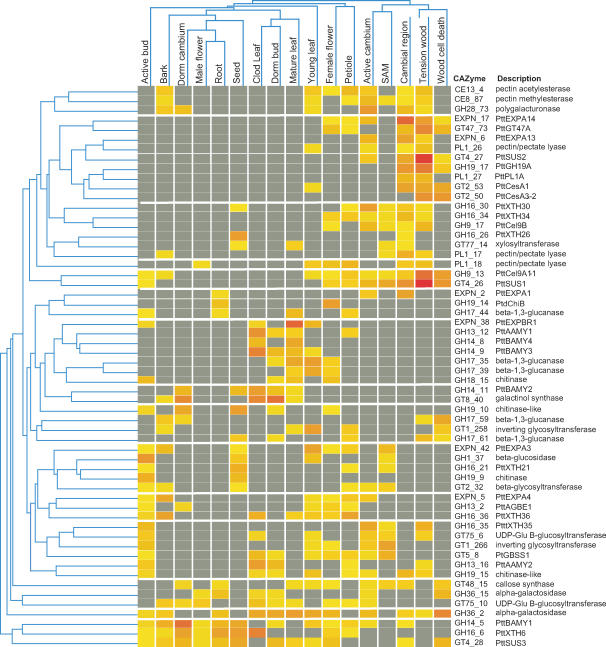
Most Expressed CAZyme-Encoding Genes
A similar analysis of expression patterns was carried out at an individual gene level. Of 680 CAZyme genes with ESTs, 58 genes (3.6% of total poplar CAZyme genes) had a specific expression pattern detectable in at least one tissue/organ by EST frequency analysis after Fisher's exact test statistics (Fig. 4). These genes were clustered according to their frequencies in various cDNA libraries to define genes coexpressed in the same tissues. Figure 4 shows groups of genes with coinciding, tissue-specific expression patterns. Many genes were identified that were similarly expressed in the cambial region, TW, and WCD. Similarly, the active cambium and the shoot apical meristem (SAM) shared many CAZymes, as did cold-stressed leaf, dormant bud, and mature leaf collected in September. However, it was surprising to see that the expression profile for some genes was similar in petiole and female flower, and even more so that gene expression of male flower shared similarities with root and dormant cambium. In addition, active bud was very different from SAM and not very similar to young leaf.
Figure 4.
Tissue-specific expression patterns of most expressed CAZyme-encoding genes. EST clones representing all genes with significant expression pattern (Fisher's exact test, P ≤ 5%) are presented. The clones were grouped according to clustering obtained with Genespring version 6.1, using standard correlation. Shading intensity corresponds to the EST frequency in each cDNA library for easy visualization of most highly expressed genes. Red shading indicates clone frequency ≥8 × 10−3 and yellow shading the frequency ≥0.10 × 10−3.
Comparison of EST Profiles versus cDNA Microarray Data
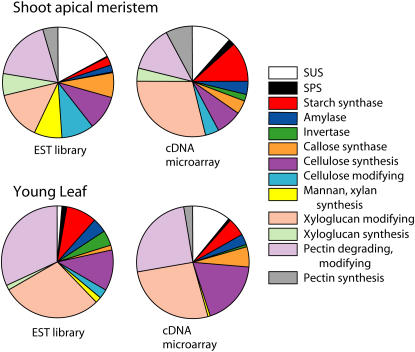
To compare the two approaches in gene expression, EST profiling and cDNA microarray analysis, the expression data previously collected for the CAZymes related to the cell wall carbohydrates and sugar/starch metabolism were compared in identical poplar tissues. Both methods gave similar expression profiles in young leaf and SAM, shown in Figure 5, and also in the other tissues examined (data not shown). Obviously, the datasets differed in cases where the respective genes were absent from the cDNA microarrays (e.g. mannan synthesis-related genes), but also when closely related family members apparently cross-hybridized among themselves, leading to an overestimation of the total expression signals for that family on the microarrays. In such cases, EST profiling should be considered the more reliable method for global transcriptomic analysis. The above-mentioned problems become even more significant when the analysis is carried out for individual genes rather than for the functional groups of genes. Similar problems were evident in the data published by Moreau et al. (2005), who relied heavily on EST data to fish out putative genes related to WCD, and the expression level of a sample of these genes could be confirmed by reverse transcription (RT)-PCR. The problems with cDNA microarray profiling can be avoided by using the oligonucleotide-based arrays with gene-specific probes covering the entire genome. Data from such arrays are presently available for Arabidopsis. Therefore, in subsequent sections, we relied mostly on the expression data obtained by EST profiling for poplar and on the oligoarray data for Arabidopsis to compare the expression of corresponding genes.
Figure 5.
Comparison of CAZyme-encoding gene expression in poplar SAM and young leaf by EST frequency profiling and cDNA microarrays. Genes were grouped by functions, and their combined expression was presented as either the total number of ESTs (left pies) or the combined signal strength on a cDNA microarray (right pies). While some specific differences were observed, the general pattern of expression of CAZyme-encoding genes grouped by functions was similar by two methods. Shown here are data for dissected SAM and leaf, but comparisons of root and bark tissues showed similar results (data not shown).
CAZyme Gene Expression in Suc and Starch Metabolism: Carbon Mobilization and Storage
Suc represents the main form of carbon transported in plants, while starch is an important carbon reserve. In photosynthetic leaves, Suc is formed from UDP-Glc by two enzymes of the GT4 family, Suc synthase (SUS) and Suc phosphate synthase (SPS). Both SUS and SPS catalyze reversible reactions, but SPS cannot break down Suc once it is processed by Suc-P phosphatase (Huber and Huber, 1996). Of the six SPS genes in poplar, only three had EST transcripts, primarily in leaves at relatively low abundance (Table III). SUS forms a larger gene family of 11 members, including SUS1 and SUS2, which were the most abundant CAZyme transcripts in poplar. The SUS transcripts had clear nonoverlapping patterns of differential regulation. The SUS1 and the SUS2 transcripts were predominantly associated with cells forming secondary cell walls found in the cambial region tissues, developing TW, and, during later stages of wood differentiation, in the WCD zone (Fig. 6A). SUS3 transcripts were not abundant in wood-forming tissue and not found at all in TW. In contrast, they were found in abundance in dormant tissues and seeds and had broad expression in virtually all other tissues (Fig. 6A). SUS4 and SUS5 were expressed at low levels in the active cambium, whereas transcripts for the other six genes were not found in the EST collection. Poplar SUS8 to SUS10 are homologous to Arabidopsis SUS2, a distinctive branch of the family, which may have a role under anaerobic conditions (Déjardin et al., 1999). The most important role of SUS1 and SUS2 in poplar seems to be preparatory in the delivery of carbon to cell wall synthesis in wood-forming tissue, whereas SUS3 is the primary metabolic gene for all tissues.
Table III.
EST tissue distributiona of CAZyme-encoding genes involved in major pathways of carbohydrate synthesis/metabolism
Cam Reg, Cambial region; Dor Cam, dormant cambium; Act Cam, active cambium; Dor Bud, dormant bud; Flw, male and female mature flowers.
| Genes | Dor Camb | Act Camb | Cam Reg | TW | WCD | Bark | Root | Young Leaf | Mature Leaf | Leaf Cold | Petiole | SAMb | Act Bud | Dor Bud | Male Flw | Fem Flw | Seed |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SUS, GT4 | 3 | 9 | 21 | 84 | 14 | 2 | 3 | 1 | 2 | 0 | 2 | 6 | 2 | 4 | 1 | 5 | 7 |
| SPS, GT4 | 2 | 2 | 0 | 2 | 1 | 0 | 1 | 1 | 3 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Invertase, GH32 and GH100 | 1 | 2 | 0 | 4 | 1 | 3 | 4 | 3 | 3 | 1 | 0 | 1 | 1 | 5 | 7 | 3 | 1 |
| Starch synthesis | 17 | 2 | 0 | 3 | 0 | 1 | 0 | 6 | 0 | 4 | 3 | 6 | 2 | 10 | 0 | 5 | 0 |
| Amylases | 4 | 0 | 1 | 1 | 3 | 6 | 5 | 3 | 15 | 39 | 6 | 2 | 6 | 22 | 2 | 3 | 13 |
| Callose synthases, CAS, GT48 | 3 | 2 | 3 | 0 | 5 | 1 | 2 | 1 | 1 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | 0 |
| Secondary wall CesAs, GT2 | 0 | 0 | 9 | 24 | 17 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other CesAs, GT2 | 1 | 2 | 3 | 4 | 0 | 2 | 0 | 1 | 2 | 2 | 0 | 3 | 0 | 1 | 2 | 2 | 0 |
| KOR-like cellulases, GH9 | 0 | 1 | 3 | 22 | 8 | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 0 | 1 | 1 |
| ELP1-like, GH19 | 0 | 1 | 16 | 20 | 2 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 0 |
| CSLD, GT2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Other cellulases, GH9 | 0 | 4 | 2 | 5 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 3 | 0 | 2 | 2 | 0 |
| CSLA, GT2 | 0 | 1 | 1 | 1 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 2 |
| Galactomannan α-1,6-galactosyl transferases, GAT, GT34 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Xylanase PttXyn10A, GH10 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Xylan glucuronate transferase, Fra8-like, GT47 | 0 | 0 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mannanases, GH5 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | 0 |
| Xyloglucan α-1,6-xylosyltrasnferases XT, GT34 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| RGP1, GT75 | 1 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 2 | 1 | 0 |
| Xyloglucan galactosyltransferase, GAT, subfamily A, GT47 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| XTH subfamily 3, GH16 | 0 | 0 | 0 | 4 | 1 | 1 | 0 | 2 | 1 | 0 | 2 | 0 | 0 | 2 | 2 | 1 | 0 |
| XTH subfamilies 1 and 2, GH16 | 6 | 13 | 8 | 8 | 0 | 10 | 9 | 3 | 1 | 17 | 15 | 6 | 9 | 0 | 3 | 7 | 33 |
| EXPA | 0 | 15 | 48 | 18 | 1 | 10 | 2 | 11 | 0 | 0 | 6 | 6 | 3 | 0 | 3 | 6 | 2 |
| EXPB | 0 | 1 | 1 | 1 | 3 | 1 | 1 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| QUA1-like, GT8 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| β-Glucuronyltransferase, GUT, GT47 | 0 | 0 | 4 | 14 | 4 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 4 | 0 |
| RG II xylosyltransferase, XT, GT77 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 |
| Pectate lyases, PL1 | 2 | 1 | 20 | 15 | 0 | 1 | 1 | 5 | 0 | 0 | 5 | 4 | 0 | 0 | 1 | 3 | 0 |
| RG II lyases, PL4 | 0 | 0 | 1 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Polygalacturonases, GH28 | 2 | 13 | 8 | 6 | 1 | 5 | 1 | 2 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 0 |
| Pectin acetylesterases, CE13 | 1 | 3 | 2 | 7 | 0 | 2 | 2 | 5 | 3 | 2 | 8 | 1 | 0 | 0 | 3 | 1 | 0 |
| Pectin methylesterase, CE8 | 2 | 8 | 4 | 5 | 2 | 5 | 3 | 10 | 1 | 6 | 8 | 1 | 3 | 0 | 7 | 10 | 1 |
Numbers of clones in each EST library.
Libraries constructed from amplified cDNA, not directly comparable to other libraries but comparable to each other and internally.
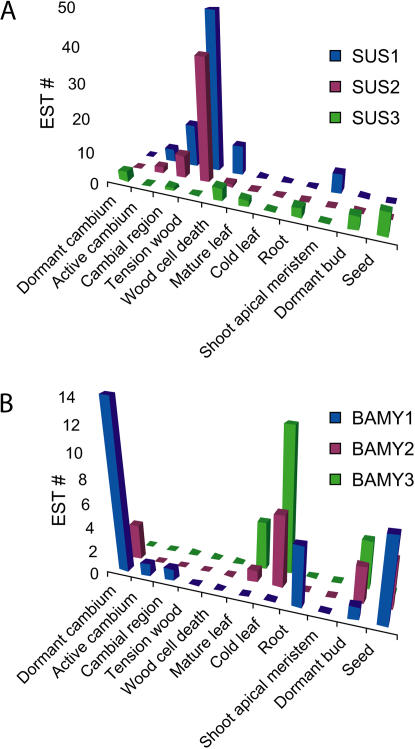
Figure 6.
Differential regulation of SUS and β-amylase genes. The three most abundantly expressed SUS and β-amylase genes show strong tissue-specific expression patterns. These patterns are not observable in cDNA microarray data due to the high degree of cross-hybridization of these highly homologous sequences (data not shown). SUS1 and SUS2 are predominantly expressed during wood formation, while SUS3 has a broad expression and is excluded from wood-forming cells. BAMY1 is expressed in dormant tissues, such as seasonally dormant cambium and seeds, and also in roots. Other β-amylases are found in mature leaves and induced strongly upon cold stress.
Beside SUS, the breakdown of Suc can be catalyzed by invertases, which fall into two distinct groups based on preferred pH and are classified into two distinct GH families. Basic/neutral (GH100) and acidic (GH32) invertases represent multigene families in poplar with 19 and nine members, respectively, and have rather broad expression in every library (Table III). Slightly more invertase ESTs were found in male flowers.
CAZymes involved in starch synthesis include starch synthases (GT5) and branching enzymes (GH13), whereas those degrading starch are starch phosphorylases (GT35), disproportionating enzymes (DPE, GH77; Chia et al., 2004), amylases (GH13 and 14), and debranching enzymes (GH13). Starch synthases (GT5) were expressed in many tissues, but most strongly in dormant cambium and buds (Table III). Generally, genes involved in starch breakdown were expressed predominantly in seasonally dormant tissues, in storage tissues and seeds, and during cold shock (Table III). The three most highly expressed CAZymes in starch metabolism are β-amylases (GH14). These also showed strong differential regulation, especially in sink tissue and storage organs, such as root, seed, and dormant cambium, where BAMY1 was the predominant transcript, and during cold response in cold-shocked leaf, mature leaf collected in September, and dormant apical buds, which were the domains of BAMY2 and BAMY3 expression (Fig. 6B).
CAZyme Gene Expression in Cell Wall Biosynthesis and Modification
Callose
Callose is a β-1,3-glucan synthesized by a plasmalemma-spanning callose synthase (GT48) and broken down by β-1,3-glucanase (GH17). We identified 19 callose synthase genes in poplar, eight of which had corresponding ESTs. Collectively, the genes had a broad expression pattern in all active meristematic tissues (Table III), consistent with the expected function of callose deposition to the nascent cell walls after cytokinesis (Hong et al., 2001; Verma and Hong, 2001). In addition, callose synthase transcripts were detected in dormant cambium, probably associated with the deposition of dormancy callose in the adjacent overwintering sieve tubes. An unexpected observation was the expression of callose synthase (Table III) and β-1,3-glucanase (Fig. 3) in the maturing wood cells (i.e. in the WCD region), suggesting a role for callose turnover in the sapwood. A β-1,3-glucanase of GH17 was shown to regulate the plasmodesmatal transport in cotton (Gossypium hirsutum) fibers (Ruan et al., 2004). Callose synthases and callose hydrolases expressed in mature wood perhaps also regulate the symplastic transport taking place in the rays (Kozlowski, 1992).
Cellulose
Cellulose microfibrils are synthesized by the plasma membrane-located rosette complexes built of possibly three different CesA proteins (GT2) and associated with the KORRIGAN1 (KOR1)-type cellulase (GH9), SUS (GT4, see above), and other yet unidentified proteins (Doblin et al., 2002). Eighteen genes encoding different CesA proteins were recently identified in the P. trichocarpa genome (Djerbi et al., 2005). In this study, we annotated the same 18 gene models as CesAs (Supplemental Table I) and found four additional fragmented models that may correspond to additional copies of CesA genes. However, as these gene models do not cover the highly conserved areas of true CesA genes, they may turn out to be nonfunctional gene copies. Six gene models, of which four had ESTs, bear close resemblance with Arabidopsis CesA genes involved in secondary cell wall formation, denoted irx1, irx3, and irx5 (Taylor et al., 2003). Consistent with a role in secondary wall biosynthesis, ESTs were detected for all four genes, CesA1, CesA3-2, CesA3-1, and CesA9-1, in the wood-forming tissues (Table III; Fig. 7A). Real-time RT-PCR experiments demonstrated a high expression of these genes in developing wood and TW (Djerbi et al., 2004), confirming that our approach of using EST profiling to decipher transcription profiles of CAZymes was correct. The EST distribution for the remaining CesA genes suggests a broad pattern of expression in all tissues, including dormant bud and cambium, where CesA8-3 transcripts were detected.
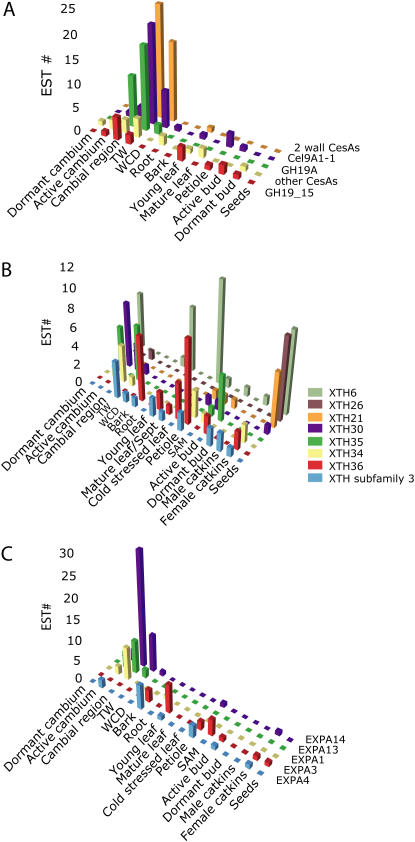
Figure 7.
Differential expression of the highly expressed cell wall-related CAZyme-encoding genes. A, Differential expression of secondary wall-related CesA-encoding genes (PttCesA1, PttCesA3-1, PttCesA3-2, and PttCesA9-1) and other CesA-encoding genes. Note that PttCel9A1-1 and PttGH19A follow the expression of secondary wall CesAs, whereas the other related chitinase, PttGH19_15, follows the expression of other CesA-encoding genes. B, Differential expression of XTH genes. Specific XTH genes are expressed in the cambium, cambial region, TW, dormant cambium, seeds, bark, and petioles. PttXTH34 has been previously published as PttXET16A (Bourquin et al., 2002). C, Wood, leaves, and bark and petiole-specific expansins, PttEXPA1 and PttEXPA3, were previously published as PttEXP1 and PttEXP3, respectively (Gray-Mitsumune et al., 2004).
In addition to CesA genes, we have identified cellulose synthase-like (CSL) genes in subgroup D resembling Arabidopsis KOJAK (Favery et al., 2001). KOJAK is thought to be involved in cellulose deposition during tip growth. However, none of the poplar CSLD homologs had ESTs present in wood-forming tissues, making them unlikely candidates for glycan synthases of the tip-growing wood fibers.
Thirty-three putative cellulases in the GH9 family were identified (J. Takahashi, E. Master, U. Rudsander, T.T. Teeri, B. Sundberg, and E.J. Mellerowicz, unpublished data). Seven cellulases closely resembled the membrane-bound Arabidopsis KOR1, but only two, Cel9A1-1 and Cel9A1-2, had corresponding ESTs. Cel9A1-1, formerly known as Cel9A (Rudsander et al., 2003; Master et al., 2004), was highly expressed in wood-forming tissues, especially in TW, and exhibited low expression levels in other tissues (Fig. 7A). Enzymatic characterization of recombinant protein revealed specificity toward unsubstituted, noncrystalline cellulose and a low specific activity on cello-oligosaccharides that could be correlated with specific features of the enzyme active site (Master et al., 2004). It is thus possible that this class of plant cellulases has an editing function during cellulose biosynthesis needed for the formation of crystalline cellulose or, as suggested earlier, a role in removing putative, membrane-bound precursors from the end of the growing cellulose chain (Peng et al., 2002). Other cellulases in GH9, lacking the membrane-spanning domain, had ESTs that were broadly distributed among poplar tissues (Table III). These cellulases may act on amorphous cellulose in the cell wall and, by doing so, increase cell wall plasticity (Ohmiya et al., 2000, 2003; Park et al., 2003).
ELP1, which belongs to GH19 family chitinases but is lacking a catalytic acid moiety, is thought to be involved in cellulose biosynthesis in Arabidopsis (Zhong et al., 2002). Two ELP1-like genes, GH19_15 and PttGH19A, were found in poplar. Similar genes were expressed in secondary wall-forming cotton fibers (Zhang et al., 2004). GH19_15 was broadly expressed, but PttGH19A was specifically and highly expressed in secondary wall-forming wood tissues, particularly TW and the cambial region (Fig. 7A), consistent with our earlier microarray study (Aspeborg et al., 2005). In sum, the genes related to cellulose biosynthesis were expressed predominantly in wood-forming tissues containing secondary wall-depositing cells.
Hemicelluloses
Hemicelluloses are cell wall polysaccharides with a backbone composed of 1,4-linked β-d-pyranosyl residues, similar to cellulose, but unlike cellulose containing diverse side chains (O'Neill and York, 2003). The backbone structures are probably synthesized by CSL enzymes, distantly related to CesAs, but to date an enzymatic activity has only been demonstrated biochemically in subgroup A of CSLs. These enzymes can produce the backbone of mannan, glucomannan, and galactomannan (Dhugga et al., 2004; Liepman et al., 2005). Poplar CSLA-like genes had ESTs that were broadly distributed in active tissues but absent in dormant meristems (Table III). The side chains of galactomannan are synthesized by an α-1,6-galactosyl transferase belonging to the GT34 family (Edwards et al., 2002; Reid et al., 2003). Two homologous genes were found in poplar, with only one EST detected in the TW tissues (Table III).
4-O-Methylglucuronoxylan is a component in both primary and secondary walls in poplar (Mellerowicz et al., 2001). Xylan synthesis genes are still unknown except for a glucuronyl transferase FRA8, recently identified in Arabidopsis (Zhong et al., 2005). The poplar genome contains one highly similar gene, Ptt47C, that was observed to be expressed in the secondary wall-forming zone (Aspeborg et al., 2005) and down-regulated during TW formation (Andersson-Gunnerås et al., 2006), in agreement with its putative role in xylan biosynthesis. Xylan-degrading xylanases form a small multigene family in Arabidopsis (GH10; Suzuki et al., 2002). The poplar genome encodes seven GH10 xylanases, but only PttXyn10A had detectable ESTs in the wood-forming tissues (Table III). This gene was found specifically up-regulated during secondary wall biosynthesis (Aspeborg et al., 2005), suggesting a role in remodeling of secondary cell walls.
Xyloglucan (XG) has a β-1,4-glucan backbone substituted with α-1,6-Xyl, which is substituted at specific positions with β-1,2-Gal, which may be further substituted with α-1,2-Fuc (O'Neill and York, 2003). In Arabidopsis, the Xyl is added by an α-1,6-xylosyltransferase AtXT1 of the GT34 family (Faik et al., 2002). Poplar had three genes with high similarity to the AtXT1, expressed in the wood-forming tissues, including the WCD zone (Table III), confirming the idea that XG is synthesized during secondary wall deposition (Bourquin et al., 2002). A galactose moiety is added at specific Xyl residues by different members of subfamily A of the GT47 family (Madson et al., 2003; Li et al., 2004). Eight genes were identified within this subfamily in poplar, with only one EST found in the dormant cambium (Table III). Fuc residue is added by the α-1,2-fucosyltransferase of the GT37 family, such as FUT1 in Arabidopsis and pea (Pisum sativum; Perrin et al., 1999; Faik et al., 2000). Phylogenetic analysis of the poplar GT37 family (data not shown) indicates that poplar has one FUT1-like gene, PttGT37_7, and more distantly related genes similar to Arabidopsis AtFUT3 (Sarria et al., 2001); however, none of them had corresponding ESTs. A protein denoted the reversibly glycosylated peptide 1 (RGP1) in the GT75 family has been proposed to participate in XG biosynthesis based on its ability to reversibly self-glycosylate using UDP-Glc, UDP-Xyl, and UDP-Gal and on its similarity with nonprocessive GTs (Dhugga et al., 1997). Two of three RGP1-like genes found in poplar had ESTs in the wood-forming tissues, bark, and dormant/cold-stressed organs (Table III).
XG is incorporated and modified in the cell wall network by xyloglucan endotransglycosylases (XETs)/hydrolases (XEHs), jointly renamed as XTHs, of the GH16 family (Rose et al., 2002). Forty-one gene models of the GH16 family were found in the poplar genome. Eleven putative genes were classified in subfamily 3 of XTH proteins characterized by having both XEH and XET activity. The corresponding ESTs were detected in the leaves, TW, and the WCD zone, dormant bud, and flowers (Table III; Fig. 7B). Remaining XTH genes corresponded to subfamilies 1 and 2 of genuine XETs. They were among the most highly expressed cell wall-related CAZymes, and several genes exhibited specific expression patterns (Fig. 7B). XTH30, XTH34 (previously published as XET16A; Bourquin et al., 2002; Kallas et al., 2005), and XTH35 were highly expressed in wood-forming tissues, including TW. XTH36 was expressed in organs containing collenchyma, such as petioles and bark. XTH6, XTH26, and XTH21 were especially active in seeds. In addition, XTH6 seemed to be specifically up-regulated in dormant cambium and cold-stressed organs.
Interaction of XG, xylan, and other hemicelluloses with cellulose is affected by expansins that presumably disrupt hydrogen bonds between these components (McQueen-Mason and Cosgrove, 1994, 1995). Expansins form a large family, including α- and β-expansins (EXPA and EXPB, respectively; Kende et al., 2004), and more distantly related expansin-like proteins of unknown function. Poplar had 31 EXPAs, many of which were highly and specifically expressed in poplar tissues (Fig. 7C). EXPA1, formerly known as EXP1 (Gray-Mitsumune et al., 2004), EXPA13, and EXPA14 were highly expressed in the cambial region and TW. Interestingly, they all belonged to a specific subgroup A of the EXPA family (Gray-Mitsumune et al., 2004). Other EXPAs were expressed in the bark and petioles (EXPA4) or in developing leaves and bud (EXPA3, formerly reported as EXP3; Gray-Mitsumune et al., 2004). The subfamily of EXPBs contained six genes, with few ESTs found in the wood-forming tissues, including the WCD zone, and in most actively growing tissues (Table III). Expansins were not found in the cDNA libraries from dormant meristems.
Pectins
Pectins constitute major carbohydrates of the primary walls in poplar (Mellerowicz et al., 2001) and include chemically diverse polymers such as homogalacturonan (HG), RG I and II, arabinan, and galactan. Only a few genes responsible for pectin biosynthesis have been identified so far. Arabidopsis QUASIMODO 1 (QUA1) glycosyltransferase of the GT8 family was proposed to be involved in HG biosynthesis (Bouton et al., 2002). We found two poplar homologous genes, one with few ESTs in the wood-forming tissues and flowers (Table III); however, considering the importance of HG in all primary walled tissues of poplar, the low EST coverage of the putative major pectin synthetic gene is surprising. A glucuronyl transferase involved in RG II biosynthesis has been described in tobacco (Nicotiana tabacum; Iwai et al., 2002), and corresponding genes were identified in Arabidopsis in subgroup E of the GT47 family (Li et al., 2004). Poplar has four members in this subgroup; three were broadly expressed, whereas the fourth member, GT47A, was highly expressed in TW and was among the most expressed CAZyme genes (Fig. 4). It was also found up-regulated in TW compared to normal wood (Andersson-Gunnerås et al., 2006), making it a likely candidate for the glycosylating enzyme of the arabinogalactan proteins known to be up-regulated in TW (Lafarguette et al., 2004). α-Xylosyl transferases involved in the addition of RG II side chains have recently been identified in Arabidopsis (Egelund et al., 2004) and classified in the GT77 family (http://afmb.cnrs-mrs.fr/CAZY). Sixteen GT77 gene models were identified in P. trichocarpa, with broadly distributed ESTs (Table III). Overall, the putative pectin biosynthetic genes identified in poplar were found broadly expressed at low levels in all tissues except dormant meristems.
Pectins undergo substantial remodeling in the cell wall (Micheli, 2001). In Arabidopsis, pectin remodeling/degrading enzymes form some of the most numerous CAZyme families (Henrissat et al., 2001). Poplar also has abundant gene families of pectin remodeling/degrading enzymes (e.g. GH28, and CE8; Table II), although relatively fewer than in Arabidopsis (Fig. 2). Pectin methyl esterases (CE8 family) and pectin acetyl esterases (CE13 family) catalyze HG de-esterification and de-acetylation in the wall. Their transcripts were found in all tissues, including the dormant cambium. Pectin degradation in the walls is mediated by pectin/pectate lyases and hydrolases. The lyases that degrade HG (PL1) and RG II (PL4) were highly expressed in the wood-forming tissues, including TW but excluding the WCD zone (Table III). This suggests a unique role of these lyases in pectin remodeling at the early stages of secondary wall biosynthesis. Poplar had 89 genes encoding polygalacturonases (GH28), but only 11 of them were found expressed, most highly in the active cambium (Table III), implying a role in cell expansion. Interestingly, polygalacturonases and pectin lyases had contrasting expression patterns in wood-forming tissues. The polygalacturonase transcripts were found in the cambium, whereas the pectate lyase transcripts were more prominent in tissues containing secondary wall-depositing cells such as the cambial region and TW. Another interesting observation was the fact that the pectin modifying and degrading enzymes were found expressed at the mRNA level in all tissues (Table III), which, along with a presence of corresponding enzymatic activities (Micheli et al., 2000), points to a constant active remodeling of cell wall pectins in cells at all developmental stages and even during dormancy.
CAZyme Transcriptome Reveals Differences in Regulation of Carbon Fluxes to Major Carbohydrates between a Tree and an Herb
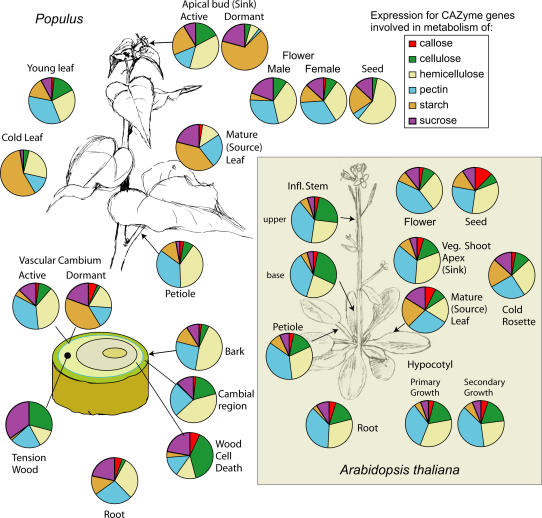
The activities of the sugar- and starch-metabolizing CAZymes and the cell wall carbohydrate-acting CAZymes discussed above regulate fluxes of carbon to major plant carbohydrates. Transcriptome analysis of the corresponding genes revealed their tissue-specific regulation. We were interested in seeing how this regulation of gene expression differs between a tree and an annual herbaceous species. For this purpose, the EST profile data for poplar tissues/organs and the data from the oligoarrays for corresponding Arabidopsis tissues/organs (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl; M. Bennett, personal communication) were utilized and compared. CAZyme genes were grouped according to function into Suc, starch, cellulose, pectin, callose, and hemicellulose-related genes, and their relative transcriptional activity was compared (Fig. 8).
Figure 8.
Global expression analysis of CAZyme-encoding genes involved in synthesis/metabolism of major carbohydrates in poplar and Arabidopsis. CAZyme-encoding genes were grouped by metabolic function for the major carbohydrates found in plants. Pies represent relative transcript abundance (poplar) or total signal intensity (Arabidopsis) for all genes of each group from each tissue and treatment in the poplar EST library and Arabidopsis microarrays (see “Materials and Methods”). Note that some poplar tissues have two pies for seasonally active (spring/summer) and dormant (fall/winter) tissues. Plants were subjected to 5°C for 3 to 4 d (poplar) or 4°C for 24 h (Arabidopsis) for cold leaf pie charts. Arabidopsis hypocotyls were collected after 7 d (from AtGenExpress) or after 3 months when a large amount of wood with fibers was produced (courtesy of Dr. Malcolm Bennett).
Poplar tissues showed much more diversified CAZyme transcript profiles than Arabidopsis tissues (Fig. 8). Cold treatment and dormancy in poplar resulted in the most profound changes in CAZyme gene expression (compare active to dormant meristems, cold to young leaves). The CAZyme transcriptome in dormant or cold-treated tissues was dominated by starch, mostly due to the activation of α-amylase and starch synthase genes (Fig. 6), in agreement with the proposed role of starch breakdown in cryoprotection (Kozlowski, 1992; Kaplan and Guy, 2004). Transcripts related to Suc metabolism were also abundant in dormant meristems, where a specific isoform of SUS, SUS3, was induced (Fig. 6A). In contrast, the transcripts related to cellulose, hemicellulose, and pectin metabolism were reduced. However, some cellulose biosynthetic activity might be possible in dormant meristems as evidenced by the presence of transcripts of cellulose synthase (CesA8-3) and GH19_15 in dormant cambium and buds (Fig. 7A). In addition, both dormant cambium and cold-stressed leaves exhibited intense hemicellulose metabolic activity in their transcript profile, mostly related to the expression of XTH6 (Fig. 7B). XG galactosyl transferase and RGP1-like transferase were also found expressed in the dormant cambium (Table III), implying a possibility of XG biosynthesis. These observations corroborate the ultrastructural study showing secondary wall-like layers containing cellulose microfibrils in the dormant cambium (Chaffey et al., 1998) and may explain the well-established phenomenon of cell wall thickening during cambial dormancy (Mellerowicz et al., 2001).
In contrast to poplar, the Arabidopsis cold response does not involve a comparable induction of a CAZyme transcriptome related to starch metabolism (Fig. 8, compare cold-stressed rosette to mature leaf) but a slight increase of gene expression in hemicellulose metabolism, including XTHs of subfamily 3 (AtXTH28 and AtXTH30) and subfamily 2 (TCH4). Interestingly, no induction of the closest homolog (At4g25810) of the cold-induced poplar XTH gene (PttXTH6) was observed in Arabidopsis. These observations may imply a different mechanism to cope with cold between poplar and Arabidopsis.
Another distinct feature of transcript profiling in poplar was the difference between tissues with primary wall-forming cells, such as the active vascular cambium, SAM, leaves, root, and tissues with secondary wall-forming cells, including the cambial region, TW, and WCD. In tissues with secondary wall-forming cells, there was very little or no expression of starch metabolic genes (Fig. 8). This suggests that the carbon from Suc arriving to these tissues is entirely diverted into the cell wall biosynthetic machinery or immediately consumed for energy. Differences in the cell wall composition between primary and secondary wall-forming tissues were also reflected in the CAZyme transcriptomic profile. There was a larger proportion of Suc and cellulose metabolism-related transcripts found in secondary wall-forming tissues, related to expression of SUS1 and SUS2 genes, which may metabolically couple to specific cellulose synthase gene products. In addition, the secondary wall-forming tissues had less XET-, expansin-, and pectin-related transcripts than primary wall-forming tissues. Indeed, at a transition from the primary to the secondary wall-forming stage in poplar, the cells stop expanding and shut down their pectin biosynthetic machinery while increasing their cellulose biosynthesis (Mellerowicz et al., 2001), and these changes are reflected in transcription levels (Fig. 8; Djerbi et al., 2004; Aspeborg et al., 2005).
In Arabidopsis, the secondary wall-forming cells are abundant in hypocotyls of about 3-month-old plants, which produce xylary fibers and vessels (Chaffey et al., 2002). When the CAZyme transcriptome was compared between such hypocotyls and a 7-d-old hypocotyl containing mostly primary walled cells, surprisingly only a slight up-regulation of pectin metabolism transcripts in the former could be detected (Fig. 8). This up-regulation involved some pectin methyl esterase and polygalacturonase genes. The inflorescence stem that has also been used as a model for secondary wall formation (Oh et al., 2003) had a similar expression profile to the hypocotyl and did not exhibit a pattern typical for secondary wall-forming tissues of poplar either. It is therefore possible that woody species reprogram their CAZyme transcriptome in wood-forming tissues to a much larger degree than herbaceous species.
Poplar storage organs, such as the bark and the roots, have a relatively large proportion of starch synthetic transcripts (Fig. 8). In contrast, Arabidopsis roots had more SUS and CesA transcripts (Table III), an indication of rapidly growing sink tissue. This might imply real differences in metabolic strategy between a perennial plant and an annual plant: A perennial plant stores carbon reserves in the roots; in contrast, an annual plant extends its roots to grow as quickly as possible before setting seed.
Female flowers in poplar showed higher proportional expression of starch genes than male flowers or the bisexual flowers in Arabidopsis (Fig. 8). Presumably, this is due to greater demand for starch accumulation in developing seeds. However, expression of starch synthase in mature poplar seeds was similar to that in Arabidopsis. Callose genes were not expressed in poplar seeds; in contrast, callose genes were the fourth largest group of CAZyme transcripts in Arabidopsis seeds.
CAZyme transcript profiles of all tissues in both species revealed the unexpected importance of hemicellulose and pectin metabolism genes. The observation of corresponding enzymatic activities in the fully grown organs (Micheli et al., 2000) suggests that the cell wall remodeling involving hemicelluloses and pectins may largely contribute to internal carbon recycling.
Conclusions and Perspectives
This work identified the CAZyme genome of a woody plant using poplar, a model tree species. Poplar had more CAZyme genes than any other species characterized to date, and, moreover, the wood-forming tissues were the richest source of various CAZyme transcripts. These findings underscore the importance of CAZymes for the woody habit. Some interesting differences between poplar and Arabidopsis CAZyme genomes were revealed and would be useful in addressing a variety of questions concerning the physiology and evolution of the different life strategies. For example, poplar had many more genes encoding enzymes that glycosylate secondary metabolites, which could point to more diversified defense mechanisms in woody perennial species. A lower relative abundance of pectin- and xylan-related genes in poplar points to some distinct differences in cell wall metabolism and the role of these carbohydrates between the two developmental habits.
The second important aspect of this work was the global transcript profile analysis of all CAZyme genes using a set of over 100,000 ESTs from 17 different tissues/developmental stages. The profile revealed striking and sometimes unexpected differences in CAZyme transcriptomes analyzed by enzyme function between tissues and developmental stages of poplar. Most remarkably, the analysis showed that the transcriptome of dormant, cold-treated, and storage organs was dominated by starch metabolism, whereas the transcriptome of secondary wall-forming tissues was characterized by a high representation of transcripts related to cellulose and Suc metabolism and almost a complete lack of transcripts related to starch metabolism. Transcripts of some cell wall biosynthetic genes were detected in the dormant meristems, especially genes for the synthesis of cellulose and XG. It will be of interest to find out whether this indeed corresponds to the active cellulose and XG biosynthesis during dormancy.
Compared to poplar, the transcriptome variability among similar tissues in Arabidopsis was not nearly as diversified, and sometimes substantial differences in transcriptomes were revealed between species for specific tissues/developmental stages, as, for example, in the case of root or cold response. These differences between species suggest that profound transcriptome remodeling separates the perennial from the annual growth strategy.
In both poplar and Arabidopsis, a large portion of a transcriptome constituted pectin and hemicellulose-related genes, suggesting constant cell wall matrix remodeling/recycling. Although the idea of pectin and XG recycling is not new, the large scale of this phenomenon as revealed in the transcriptome was not anticipated. Pectin and XG remodeling genes form some of the largest gene families, suggesting their diverse, but presently not well known, roles in plant development and an elucidation of their function should be an important direction for further study.
MATERIALS AND METHODS
Identification of CAZyme Gene Models
Approximately 55,000 Populus trichocarpa gene models (filtered set released September 2004; DOE Joint Genome Institute Web address, http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) were subjected to pairwise comparisons with each of the approximately 33,000 entries of the CAZyme database (CAZy at http://afmb.cnrs-mrs.fr/CAZy/) using BLAST (Altschul et al., 1997). After elimination of false-positive hits due to the presence of noncatalytic modules, inspection of the best hits with other plant sequences revealed that P. trichocarpa gene models sometimes corresponded to fragments (e.g. different exons of the same gene). To eliminate some of these supernumerary gene fragments, exon-intron junctions were identified and putative coding sequences were aligned with corresponding Arabidopsis (Arabidopsis thaliana) orthologs. The linkage group and the length of each gene model were identified, and gene models that shared linkage groups and were obviously exons of the same gene based on Arabidopsis orthologs or poplar (Populus spp.) EST sequence were merged with the longest gene model name chosen as representative.
Phylogenetic Analysis of CAZyme Families
The protein sequences corresponding to the catalytic modules of CAZymes from plant origin were first aligned by the MUSCLE program (Edgar, 2004), and subfamilies were then identified by Secator (Wicker et al., 2001). The poplar CAZyme-encoding protein models were then aligned against plant CAZymes in the profile mode of ClustalW (Thompson et al., 1994). The resulting MUSCLE and ClustalW combination alignment was used for tree construction by ClustalW using 1,000 bootstrap steps. Finally, the trees were visualized by the Treedyn graphics editor for phylogenetic or classification trees (www.treedyn.org).
Comparisons of Genetic Divergence among Poplar Species
All full-length Populus alba, Populus alba × Populus tremula, Populus tremuloides, and P. tremula × P. tremuloides CAZyme-encoding cDNAs were identified from GenBank (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi), as well as among the full-length sequences provided by the University of British Columbia to the poplar genome project (http://genome.jgi-psf.org) and the Populus database (http://www.Populus.db.umu.se). The nucleotide sequences of other poplar species were aligned using BLAST to their most similar P. trichocarpa gene model available at DOE Joint Genome Institute (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html). All the mismatched percentages from one poplar species (ranging from 2–26 samples, depending on the availability of cDNA sequences; see Table II) were averaged with a se.
Mapping of ESTs to the Poplar Gene Models
The poplar ESTs available in GenBank were mapped to the transcript models predicted within the poplar genome project (http://genome.jgi-psf.org/Poptr1) with a BLAST-based protocol. An EST was considered mapped to the BLASTN high-scoring transcript model if the best high-scoring segment pairs showed an identity of at least 95%. Due to the high frequency of closely related paralogs in poplar, ESTs that showed less than 95% identity in the best high-scoring segment pairs were not considered unambiguously mapped and were therefore excluded.
Expression Analysis by Comparing EST Frequencies
Expression analysis was performed only on ESTs coming from the 17 non-normalized cDNA libraries sequenced in Sweden (Sterky et al., 2004). This dataset constitutes a large, similarly processed collection of sequences covering various tissues and developmental stages whose EST frequencies can be directly compared to each other. A table of frequencies of mapped ESTs for each model and each library was made. Since some cDNA clones had been sequenced more than once, all analyses were made on the clone level. Gene models with a specific enrichment of ESTs from one or a few of the libraries were identified with a set of Fisher's exact tests (Audic and Claverie, 1997) using the R-package (http://www.r-project.org). We compared the proportion of clones in each library with the proportion of clones in all libraries for each model.
Microarray Expression Analysis
Poplar cDNA microarray experiments for genome expression in young leaves, SAM, roots, TW, WCD, cambium region, bark, and flowers were meta-analyzed from published or personally communicated sources from the in-house available database (www.upscbase.db.umu.se). RNA extraction, microarray preprocessing, normalization, and identification of gene expression levels were all preformed by the original authors, and that information is available within the database. These results were used for validation and comparison to EST analysis and were not otherwise used to interpret CAZyme expression patterns. Unique poplar CAZyme-encoding genes were assigned to each microarray spot, although considerable cross-hybridization was suspected. Genes were grouped by function, and the sum expression (signal strength) of each group was presented in relative terms (100% = sum expression of all groups for a tissue) and compared between tissues.
Arabidopsis oligo microarray data were downloaded from the Nottingham Arabidopsis Stock Center (NASC; http://affymetrix.Arabidopsis.info/narrays/experimentbrowse.pl). Experiments on tissue-specific genomic expression were chosen from the expression atlas of Arabidopsis development provided by the AtGenExpress project (http://www.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenex.htm) and used with permission. The spots in these arrays were based on unique oligonucleotides, which did not suffer from cross-hybridization, and represented the majority of Arabidopsis CAZyme-encoding genes. We totaled the Microarray Suite scaled and normalized signal values for each group of CAZyme-encoding genes (grouped by gene function) and thus determined relative expression in each Arabidopsis tissue (all methods for microarray handling and normalization were done by AFGN and Affymetrix and are published online within the database).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed in Supplemental Table I online.
Supplementary Material
Acknowledgments
We thank Dr. Malcolm Bennett and Dr. Göran Sandberg for the release of microarray data prior to publication.
This work was supported by grants from the European Commission (to B.H.), Formas (to E.J.M. and B.S.), the Swedish Research Council (L.A.K.), the Wallenberg Foundation, European project Eden (QLK5–CT–2001–00443), the Wood Ultrastructure Research Centre, and the Umeå Plant Science Center Excellence Centre.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ewa J. Mellerowicz (ewa.mellerowicz@genfys.slu.se).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.072652.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson-Gunnerås S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B (2006) Biosynthesis of cellulose enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45: 144–165 [DOI] [PubMed]
- Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, Nilsson P, Denman S, Amini B, Sterky F, et al (2005) Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol 137: 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie J-M (1997) The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14: 3073–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker MT, Talbotec J, Granier F, Lahaye M, Hofte H, Truong HN (2002) Quasimodo1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14: 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffey N, Cholewa E, Regan S, Sundberg B (2002) Secondary xylem development in Arabidopsis: a model for wood formation. Physiol Plant 114: 594–600 [DOI] [PubMed] [Google Scholar]
- Chaffey NJ, Barlow PW, Barnett JR (1998) A seasonal cycle of cell wall structure is accompanied by a cyclical rearrangement of cortical microtubules in fusiform cambial cells within taproots of Aesculus hippocastanum (Hippocastanaceae). New Phytol 139: 623–635 [Google Scholar]
- Chia T, Thorneycroft D, Chapple A, Messerli G, Chen J, Zeeman SC, Smith SM, Smith AM (2004) A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J 37: 853–863 [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003. a) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Stam M, Blanc E, Henrissat B (2003. b) Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci 8: 563–565 [DOI] [PubMed] [Google Scholar]
- Déjardin A, Sokolov LN, Kleczkowski LA (1999) Sugar/osmoticum levels modulate differential abscisic acid-independent expression of two stress-responsive sucrose synthase genes in Arabidopsis. Biochem J 344: 503–509 [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomas D, Nichols S, Anderson P (2004) Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303: 363–366 [DOI] [PubMed] [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM (1997) A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA 94: 7679–7684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djerbi S, Aspeborg H, Nilsson P, Sundberg B, Mellerowicz E, Blomqvist K, Teeri TT (2004) Identification and expression analysis of genes encoding putative cellulose synthases (CesA) in the hybrid aspen, Populus tremula (L.) × P. tremuloides (Michx.). Cellulose 11: 301–312 [Google Scholar]
- Djerbi S, Lindskog M, Arvestad L, Sterky F, Teeri TT (2005) The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta 221: 739–746 [DOI] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43: 1407–1420 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards ME, Marshall E, Gidley MJ, Reid JSG (2002) Transfer specificity of detergent-solubilized fenugreek galactomannan galactosyltransferase. Plant Physiol 129: 1391–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Skjot M, Geshi N, Ulvskov P, Petersen BL (2004) A complementary bioinformatics approach to identify potential plant cell wall glycosyltransferase-encoding genes. Plant Physiol 136: 2609–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Bar-Peled M, DeRocher AE, Zeng W, Perrin RM, Wilkerson C, Raikhel NV, Keegstra K (2000) Biochemical characterization and molecular cloning of an α-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in pea. J Biol Chem 275: 15082–15089 [DOI] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K (2002) An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci USA 99: 7797–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Mitsumune M, Mellerowicz EJ, Abe H, McQueen-Mason S, Winzéll A, Sterky F, Blomqvist K, Schrader J, Teeri TT, Sundberg B (2004) Expansins abundant in secondary xylem belong to subgroup A of the α-expansin gene family. Plant Physiol 135: 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Coutinho PM, Davies GJ (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47: 55–72 [PubMed] [Google Scholar]
- Henrissat B, Davies GJ (2000) Glycoside hydrolases and glycosyltransferases: families, modules and implications for genomics. Plant Physiol 124: 1515–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Delauney AJ, Verma DP (2001) A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Huber JL (1996) Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 431–444 [DOI] [PubMed] [Google Scholar]
- Iwai H, Masaoka N, Ishii T, Satoh S (2002) A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc Natl Acad Sci USA 99: 16319–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallas AM, Piens K, Denman SE, Henriksson H, Faldt J, Johansson P, Brumer H, Teeri TT (2005) Enzymatic properties of native and deglycosylated hybrid aspen xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem J 390: 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2004) β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135: 1674–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, Bradford KJ, Brummell DA, Cho H-T, Cosgrove DJ, Fleming A, Gehring C, Lee Y, McQueen-Mason S, Rose JKC, et al (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55: 311–314 [DOI] [PubMed] [Google Scholar]
- Konishi T, Ohmiya Y, Hayashi T (2004) Evidence that sucrose loaded into the phloem of a poplar leaf is used directly by sucrose synthase associated with various β-glucan synthases in the stem. Plant Physiol 134: 1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski TT (1992) Carbohydrate sources and sinks in woody plants. Bot Rev 58: 107–222 [Google Scholar]
- Lafarguette F, Leplé JC, Déjardin A, Laurans F, Costa G, Lesage-Descauses MC, Pilate G (2004) Poplar genes encoding fasciclin-like arabinogalactan proteins are highly expressed in tension wood. New Phytol 164: 107–121 [DOI] [PubMed] [Google Scholar]
- Li X, Cordero I, Caplan J, Mølhøj M, Reiter W-D (2004) Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferases. Plant Physiol 134: 940–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Master ER, Rudsander UR, Zhou W, Henriksson H, Divne C, Denman S, Wilson DB, Teeri TT (2004) Recombinant expression and enzymatic characterization of PttCel9A, a KOR homologue from Populus tremula x tremuloides. Biochemistry 43: 10080–10089 [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91: 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ (1995) Expansin mode of action on cell-walls—analysis of wall hydrolysis, stress-relaxation, and binding. Plant Physiol 107: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Bojeran W (2001) Unraveling cell wall formation in the woody dicot stem. Plant Mol Biol 47: 239–274 [PubMed] [Google Scholar]
- Micheli F (2001) Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Micheli F, Sundberg B, Goldberg R, Richard L (2000) Radial distribution pattern of pectin methylesterases across the cambial region of hybrid aspen at activity and dormancy. Plant Physiol 124: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau C, Aksenov N, Lorenzo MG, Segerman B, Funk C, Nilsson P, Jansson S, Tuominen H (2005) A genomic approach to investigate developmental cell death in woody tissues of Populus trees. Genome Biol 6: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Park S, Han K-H (2003) Transcriptional regulation of secondary growth in Arabidopsis thaliana. J Exp Bot 54: 1–14 [DOI] [PubMed] [Google Scholar]
- Ohmiya Y, Nakai T, Park YW, Aoyama T, Oka A, Sakai F, Hayashi T (2003) The role of PopCel1 and PopCel2 in poplar leaf growth and cellulose biosynthesis. Plant J 33: 1087–1097 [DOI] [PubMed] [Google Scholar]
- Ohmiya Y, Samejima M, Shiroishi M, Amano Y, Kanda T, Sakai F, Hayashi T (2000) Evidence that endo-1,4-beta-glucanases act on cellulose in suspension-cultured poplar cells. Plant J 24: 147–158 [DOI] [PubMed] [Google Scholar]
- O'Neill MA, York WS (2003) The composition and structures of primary cell walls. In JKC Rose, ed, The Plant Cell Wall, Annual Plant Reviews, Vol 8. CRC Press, Boca Raton, FL, pp 1–54
- Park YW, Tominaga R, Sugiyama J, Furuta Y, Tanimoto E, Samejima M, Sakai F, Hayashi T (2003) Enhancement of growth by expression of poplar cellulase in Arabidopsis thaliana. Plant J 33: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Peng L, Kawagoe Y, Hogan P, Delmer D (2002) Sitosterol-beta-glucoside as primer for cellulose synthesis in plants. Science 295: 147–150 [DOI] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Reid JS, Edwards ME, Dickson CA, Scott C, Gidley MJ (2003) Tobacco transgenic lines that express fenugreek galactomannan galactosyltransferase constitutively have structurally altered galactomannans in their seed endosperm cell walls. Plant Physiol 131: 1487–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodman JE (1991) A taxonomic analysis of glucosinolate-producing plants. Syst Bot 16: 598–618 [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Ross J, Li Y, Lin EK, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2: 3004.1–3004.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Xu SM, White R, Furbank RT (2004) Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol 136: 4104–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd S (2003) Expressed sequence tags: alternative or complement to whole genome sequences? Trends Plant Sci 8: 321–329 [DOI] [PubMed] [Google Scholar]
- Rudsander U, Denman S, Raza S, Teeri TT (2003) Molecular features of family GH9 cellulases in hybrid aspen and the filamentous fungus Phanerochaete chrysosporium. J Appl Glycoscience 50: 253–256 [Google Scholar]
- Sarria R, Wagner TA, O'Neill MA, Faik A, Wilkerson CG, Keegstra K, Raikhel NV (2001) Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferase1. Plant Physiol 127: 1595–1606 [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Jonsson-Lindvall J, Tandre K, Strauss SH, et al (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101: 13951–13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kato A, Nagata N, Komeda Y (2002) A xylanase, AtXyn1, is predominantly expressed in vascular bundles, and four putative xylanase genes were identified in the Arabidopsis thaliana genome. Plant Cell Physiol 43: 759–767 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibbon TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DP, Hong Z (2001) Plant callose synthase complexes. Plant Mol Biol 47: 693–701 [DOI] [PubMed] [Google Scholar]
- Wicker N, Perrin GR, Thierry JC, Poch O (2001) Secator: a program for inferring protein subfamilies from phylogenetic trees. Mol Biol Evol 18: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2004) Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol 45: 1111–1121 [DOI] [PubMed] [Google Scholar]
- Zhang DS, Hrmova M, Wan CH, Wu CF, Balzen J, Cai W, Wang J, Densmore LD, Fincher GB, Zhang H, et al (2004) Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol Biol 54: 353–372 [DOI] [PubMed] [Google Scholar]
- Zhong R, Peña MJ, Zhou GK, Nairn CJ, Wood-Jones A, Richardson EA, Morrison WH III, Darvill AG, York WS, Ye ZH (2005) Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17: 3390–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong RQ, Kays SJ, Schroeder BP, Ye ZH (2002) Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14: 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.