Abstract
We have analyzed gene regulation of the Lhc supergene family in poplar (Populus spp.) and Arabidopsis (Arabidopsis thaliana) using digital expression profiling. Multivariate analysis of the tissue-specific, environmental, and developmental Lhc expression patterns in Arabidopsis and poplar was employed to characterize four rarely expressed Lhc genes, Lhca5, Lhca6, Lhcb7, and Lhcb4.3. Those genes have high expression levels under different conditions and in different tissues than the abundantly expressed Lhca1 to 4 and Lhcb1 to 6 genes that code for the 10 major types of higher plant light-harvesting proteins. However, in some of the datasets analyzed, the Lhcb4 and Lhcb6 genes as well as an Arabidopsis gene not present in poplar (Lhcb2.3) exhibited minor differences to the main cooperative Lhc gene expression pattern. The pattern of the rarely expressed Lhc genes was always found to be more similar to that of PsbS and the various light-harvesting-like genes, which might indicate distinct physiological functions for the rarely and abundantly expressed Lhc proteins. The previously undetected Lhcb7 gene encodes a novel plant Lhcb-type protein that possibly contains an additional, fourth, transmembrane N-terminal helix with a highly conserved motif. As the Lhcb4.3 gene seems to be present only in Eurosid species and as its regulation pattern varies significantly from that of Lhcb4.1 and Lhcb4.2, we conclude it to encode a distinct Lhc protein type, Lhcb8.
Light harvesting is one of the most intensively investigated processes in plant biology, and the nuclear genes encoding the chlorophyll a/b-binding proteins (first denoted CAB, later renamed Lhc genes) have been regarded since the birth of plant molecular biology as standard plant genes. The encoded proteins of approximately 20 to 24 kD span the chloroplast thylakoid membrane with three helices and coordinate a number of chlorophylls and carotenoids. Assembling with the two photosystems, they serve to maximize and regulate light harvesting. Together with the rbcS gene, Lhc genes were the first to be cloned (Broglie et al., 1981) and were used in some of the first gene expression studies. Lhc gene expression is primarily regulated by the intensity and quality of light on a transcriptional level (for review, see Brunner and Ruediger, 1995), and Lhc mRNA levels are subject to pronounced circadian and diurnal fluctuations (Kloppstech, 1985; Pichulla, 1988). Lhc gene promoters have subsequently also been used in mutant screening experiments to elucidate the different light-sensing pathways in plants (Chory et al., 1995). In general, Lhc genes are most strongly expressed when light harvesting is limiting for plant growth, i.e. in low light but otherwise optimal conditions. Lhc gene expression levels are very high in leaves but low in nongreen tissues, or even absent in many such tissues. Since the need for efficient light harvesting is lower under high-light conditions, Lhc gene expression is down regulated in strong light.
During efforts to standardize plant gene nomenclature, the Lhc genes were again “guinea pigs.” At this time, individual genes encoding 10 different types of Lhc proteins were recognized, and the Lhc protein types were denoted as Lhca1 to 4 and Lhcb1 to 6 (Jansson et al., 1992; Price, 1994). These Lhc proteins (referred to as abundant Lhc proteins in the following) have been the subject of many studies elucidating their structure, chromophore-binding properties, energy transfer contribution, and both assembly to multimeric complexes (LHCI/LHCII) and with the photosystems. Thus, the abundant three-helix Lhc proteins and their corresponding genes are exceptionally well characterized. When expressed sequence tag (EST) data from Arabidopsis (Arabidopsis thaliana) started to accumulate, the Lhc genes of Arabidopsis were subjected to detailed characterization, and three additional types of genes (denoted as Lhca5, Lhca6, and Lhcb4.3) that appeared to be expressed at very low levels or not at all were identified (Jansson, 1999). These genes (referred to as rarely expressed in the following) have started to gain some attention, but the corresponding proteins have not yet been well characterized, except Lhca5 (Ganeteg et al., 2004; Storf et al., 2004, 2005). The Lhc supergene family of higher plants also contains genes coding for proteins that exhibit sequence similarity to the Lhc proteins, namely, the four-helix protein PsbS and the light-harvesting-like (LIL) proteins. So far, six different classes of Lil genes have been identified (Jansson, 1999; Heddad and Adamska, 2000; Andersson et al., 2003b). The Lil1 genes code for the three-helix early light-inducible proteins (ELIPs), two types of one-helix proteins (OHPs) are encoded by Lil2 (OHP1) and Lil6 (OHP2), and two types of stress-enhanced proteins (SEPs) are encoded by Lil4 (SEP1) and Lil5 (SEP2). PsbS and the LIL proteins do not seem to be constitutively associated with reaction center complexes. Unlike the three-helix Lhc proteins, they have not yet been assigned within the crystal structures of higher plant PSI and PSII holocomplexes (Ben-Shem et al., 2003; Ferreira et al., 2004). However, biochemical characterizations have strongly indicated interactions for some of them, e.g. PsbS with PSII (Funk et al., 1995) or OHP2 with PSI (Andersson et al., 2003b). Therefore, their absence in the structures might be due to the fact that their interactions with the photosynthetic reaction centers are less strong or temporarily limited, but they also might have a function more independently from a close association with the reaction centers.
The three-helix Lhc proteins, however, have all been shown to be integral parts of reaction center antenna (super)complexes that consist of both nuclear and chloroplast-encoded protein subunits (for review, see Dekker and Boekema, 2005). This requires concerted expression of the different protein components and maintenance of stoichiometric balance of the proteins, but how this is regulated is under debate. Promising models propose the chloroplast redox state (for review, see Fey et al., 2005) to give rise to retrograde signals from the chloroplast to the nucleus, altering Lhc gene expression. Lhc gene transcript levels have been shown to correlate with protein levels detected (Anandan et al., 1993; Durnford et al., 2003; Ganeteg et al., 2004), but contrasting data have also been reported (Flachmann and Kühlbrandt, 1995; Flachmann, 1997). Therefore, rising Lhc gene transcript levels alone might not necessarily lead to elevated protein abundance. After translation in the cytosol, protein precursors have to be imported into the chloroplast (for review, see Soll and Schleiff, 2004). Insertion in the thylakoid membrane involves a number of posttranscriptional steps capable of altering functional protein abundance. The acquirement of the cofactors is an excellent example how Lhc protein abundance is regulated by other means than gene expression. The lack of chlorophyll b has been shown to affect the assembly of some Lhcb-type proteins in chlorina mutants of barley (Hordeum vulgare; Preiss and Thornber, 1995), and overexpression of the chlorophyllide a oxygenase enzyme, catalyzing the formation of chlorophyll b, causes an increased Lhc antenna size (Tanaka et al., 2001). Nevertheless, tissue development during ontogenesis as well as the response to environmental conditions clearly includes time- and tissue-specific gene expression. Multivariate analysis of expression patterns has successfully been used to identify regulation associations of genes that encode proteins contributing to the same biochemical pathways (Biehl et al., 2005; Vanderauwera et al., 2005).
Arabidopsis is in most respects an excellent model system for plant genetic and genomic studies, but databases allowing rapid digital northern analysis or comparative analysis of expression data in Arabidopsis have become publicly available only recently (Koo and Ohlrogge, 2002; Mahalingam et al., 2003; Zimmermann et al., 2004). In cases where multiple EST libraries, including sequences originating from the RNA of different tissues or the same tissue subjected to different treatments, have been sequenced, the presence or absence of an EST from a given gene in the different libraries may give valuable indications as to whether or not it is expressed in the respective tissues/treatments. In addition, if the libraries are carefully prepared in a standardized way, the frequency of clones in a library could be taken as an approximation of the transcript level in the tissue. This approach cannot, of course, be used to obtain quantitative data, but it could provide quick indications about the expression patterns of a given gene. Recently, the draft Populus genome sequence (genome.jgi-psf.org/Poptr1/Poptr1.home.html) and an extensive EST database, PopulusDB (www.populus.db.umu.se; Sterky et al., 2004), became publicly accessible. Since poplar and Arabidopsis are quite closely related, most Arabidopsis genes have a clear ortholog in poplar, and the 19 nonnormalized cDNA libraries that have been subjected to EST sequencing in PopulusDB not only serve as a source of gene sequences but also enable digital northern analyses to be performed (Sterky et al., 2004). Here, we have investigated regulation of the Lhc supergene family of poplar using the public poplar and Arabidopsis resources. Including an analysis of the mature Lhc protein sequences of poplar, Arabidopsis, and rice (Oryza sativa), we assigned the Lhc genes to the main Lhc protein classes present in higher plants.
RESULTS
From the poplar genome and EST databases, we extracted all sequences homologous to the Lhc supergene family of Arabidopsis. This family was originally assumed, based only on EST sequences, to consist of 30 members, plus one of the isoforms of ferrochelatase that has a domain with similarity to one of the Lhc protein helices (Jansson, 1999). When the Arabidopsis genome sequence appeared, it was apparent that two of the proposed Lhcb2 genes (Jansson, 1999) were actually two alleles of the same gene, resulting in renaming the Lhcb2.4 (At3g27690) gene to Lhcb2.3 (Legen et al., 2001; Andersson et al., 2003a). On the other hand, a gene coding for an additional OHP that initially escaped detection was subsequently characterized (Andersson et al., 2003b), keeping the number of known genes at 30 in Arabidopsis. In poplar the Lhc supergene family is slightly larger with 39 Lhc genes identified (Table I). Several proteins that are encoded by singletons in Arabidopsis (Lhca1, Lhca2, Lhcb3, Lhcb6, SEP1, SEP2, and OHP2) are each encoded by two highly similar genes in poplar, while others (ELIP and LIL3) that are encoded by two genes in Arabidopsis are encoded by three genes in poplar. However, four genes in poplar encode Lhcb1 (compared to five in Arabidopsis) genes, while the three Arabidopsis genes coding for Lhcb2 only have two poplar counterparts. Two sequences highly similar to Arabidopsis Lhca5 (At1g45474) were found in poplar, but the predicted coding region of Lhca5.2 covers only a fraction of the Lhca5 sequence. There is no EST associated with this gene, which could be either a pseudogene or a gene whose coding region has been wrongly predicted. During the course of this work, we also recognized an additional locus in Arabidopsis (At5g28450) similar to Lhca2, which could either be a pseudogene or a gene that has not been correctly predicted due to the lack of a corresponding EST. The Arabidopsis Lhca6 gene (At1g19150) previously had no corresponding ESTs in public databases, but one EST from an Arabidopsis silique library was deposited in 2004. Seven Lhca6 ESTs were found in young leaves, female catkins, and imbibed seeds from poplar, indicating that this is a true gene rather than a pseudogene. Thus, the evidence gathered to date collectively indicates that ESTs originating from all types of Lhc genes identified in Arabidopsis can be found in poplar.
Table I.
The Lhc supergene family in poplar
Number of Lhc ESTs in EST libraries of poplar (for details, see Supplemental Table I) assigned to the 39 gene models (www.populus.db.umu.se) for the 20 identified Lhc protein classes. The numbers of amino acids refers to the complete coding region encoding the precursor protein.
| Protein | Gene | ESTs | Gene Model | Amino Acids |
|---|---|---|---|---|
| Lhca1 | Lhca1.1 | 31 | estExt_Genewise1_v1.C_LG_X2607 | 243 |
| Lhca1.2 | 2 | estExt_fgenesh4_pg.C_LG_VIII0315 | 244 | |
| Lhca2 | Lhca2.1 | 11 | eugene3.00010471 | 272 |
| Lhca2.2 | 4 | estExt_fgenesh4_pg.C_LG_III1424 | 269 | |
| Lhca3 | Lhca3 | 108 | grail3.0012036701 | 274 |
| Lhca4 | Lhca4 | 24 | grail3.0129002701 | 251 |
| Lhca5 | Lhca5.1 | 1 | estExt_fgenesh4_pg.C_400276 | 267 |
| Lhca5.2 | n.d.ab | eugene3.00021173 | 56 | |
| Lhca6 | Lhca6 | 7 | eugene3.00280046 | 261 |
| Lhcb1 | Lhcb1.1 | 431 | grail3.0002067901 | 264 |
| Lhcb1.2 | 61 | estExt_Genewise1_v1.C_LG_V1774 | 264 | |
| Lhcb1.3 | 37 | eugene3.00110470 | 264 | |
| Lhcb1.4 | 11 | estExt_Genewise1_v1.C_LG_II0002 | 266 | |
| Lhcb2 | Lhcb2.1 | 192 | estExt_fgenesh4_pm.C_LG_II0962 | 265 |
| Lhcb2.2 | 14 | grail3.0012031401 | 265 | |
| Lhcb3 | Lhcb3.1 | 23 | estExt_Genewise1_v1.C_1340185 | 263 |
| Lhcb3.2 | 22 | estExt_fgenesh4_kg.C_LG_XI0033 | 263 | |
| Lhcb4 | Lhcb4.1 | 68 | grail3.0025015101 | 285 |
| Lhcb4.2 | 47 | grail3.0023029001 | 283 | |
| Lhcb4.3 | 13 | estExt_fgenesh4_pm.C_LG_VIII0258 | 275 | |
| Lhcb5 | Lhcb5 | 85 | estExt_Genewise1_v1.C_1250189 | 290 |
| Lhcb6 | Lhcb6.1 | 39 | estExt_Genewise1_v1.C_LG_I9864 | 257 |
| Lhcb6.2 | 39 | estExt_fgenesh4_kg.C_LG_III0002 | 257 | |
| Lhcb7 | Lhcb7 | 3 | estExt_Genewise1_v1.C_LG_V2296 | 338 |
| PsbS | PsbS | 32 | estExt_fgenesh4_pg.C_LG_II0752 | 272 |
| ELIP | Lil1.1 | 351 | estExt_Genewise1_v1.C_LG_VIII0523 | 182 |
| Lil1.2 | 57 | eugene3.00081476 | 182 | |
| Lil1.3 | 26 | estExt_fgenesh4_pg.C_LG_VIII1396 | 182 | |
| OHP1 | Lil2 | 6 | grail3.0023020401 | 119 |
| LIL3 | Lil3.1 | 8 | estExt_Genewise1_v1.C_290436 | 249 |
| Lil3.2 | 4 | eugene3.01180096 | 258 | |
| Lil3.3 | 1 | eugene3.00150905 | 257 | |
| Lil3.4 | n.d. | fgenesh1_pg.C_scaffold_9489000001 | – | |
| SEP1 | Lil4.1 | 11 | estExt_fgenesh4_pg.C_LG_I2581 | 145 |
| Lil4.2 | 3 | estExt_fgenesh4_pg.C_LG_IX0658 | 145 | |
| SEP2 | Lil5.1 | n.d. | estExt_fgenesh4_pg.C_LG_V0240 | 202 |
| Lil5.2 | n.d. | eugene3.00070639 | 202 | |
| OHP2 | Lil6.1 | 15 | estExt_fgenesh4_pg.C_LG_II0599 | 188 |
| Lil6.2 | 2 | estExt_fgenesh4_pg.C_LG_V1127 | 188 |
n.d., No EST detected but predicted gene-model from genome sequence available.
Pseudogene.
A Gene Coding for a Novel Lhc Protein, Lhcb7, Is Present in Higher Plants
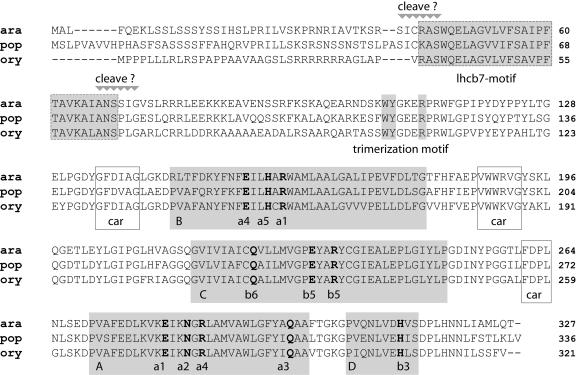
Surprisingly, one poplar gene (with three corresponding ESTs) contained a number of conserved Lhc motifs but did not match any of the previously recognized Lhc genes of Arabidopsis. The corresponding protein sequence is highly homologous to that of a predicted Arabidopsis gene (At1g76570) that has not been assigned as an Lhc protein in the first annotations of the Arabidopsis genome, presumably since the only corresponding EST in public Arabidopsis databases (to date) was deposited in late 2003. The encoded proteins in Arabidopsis, poplar, and rice (AK066997) are most similar to Lhcb5 (CP26) but do not appear to be similar to the short peptide sequence from a CP26 (Lhcb5)-like protein in barley reported by Morishige and Thornber (1994). We tentatively denoted the proteins encoded by the poplar gene and At1g76570 as Lhcb7, although there is no evidence other than sequence homology indicating that it is associated with PSII. By searching databases with the Arabidopsis and poplar Lhcb7 sequences, we identified homologous EST sequences in many other plants, including dicots (Solanum, Ipomoea, Gossypium, Glycine, Prunus) and monocots (Hordeum, Saccharum, Triticum, Zea) as well as conifers (Pinus). A gene model corresponding to this gene has also been reported in Chlamydomonas (Minagawa and Takahashi, 2004). Thus, Lhcb7 appears to be a true Lhc gene that is present in all photosynthetic eukaryotes but has previously escaped detection (or has been incorrectly annotated). An alignment of the deduced Lhcb7 protein sequences of Arabidopsis, poplar, and rice (Fig. 1) reveals full conservation of the well-known Lhc motifs, including the three transmembrane helices A to C, the C-terminal helix D, as well as the carotenoid-binding sites and chlorophyll ligands. The sequence also contains the LHCII trimerization motif present in Lhcb1 to 3, but no putative phosphorylation sites as found in Lhcb1 and Lhcb2. Unequivocal prediction of the position of the cleavage site between the putative transit peptide and mature protein is not (as yet) possible. However, the N-terminal part of the Lhcb7 protein is much larger than that of any other Lhc-protein. In fact, it contains a strongly conserved 27-amino acid motif that is exclusively found in Lhcb7 homologs and is predicted to form a transmembrane helix. As chloroplast transit peptides typically do not exhibit strongly conserved motifs (Bruce, 2000), this helix might be part of a mature Lhcb7 protein with four transmembrane helices.
Figure 1.
Sequences of the Lhcb7 proteins of Arabidopsis (ara), poplar (pop), and rice (ory), indicating helices (A–D, shaded boxes), carotenoid-binding sites (car, white boxes), chlorophyll ligands (bold; according to Caffarri et al., 2004), and two possible positions for the cleavage site (cleave ?) of the transit peptide. The Lhcb7-specific motif that is predicted to be a transmembrane might be part of the mature Lhcb7 protein, which also contains the trimerization motif present in Lhcb1 to 3.
Sequence Comparisons Reveal the Structure of the Lhc Gene Family in Higher Plants
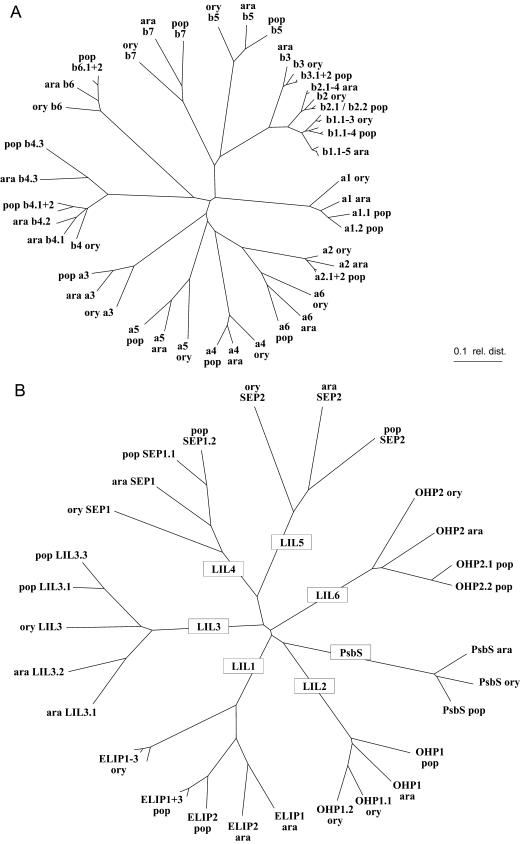
To get an overview of the structure of the whole Lhc supergene family, we analyzed sequences from three species (poplar, Arabidopsis, and rice) of the mature Lhca1 to 6, Lhcb1 to 7, PsbS, and the different LIL proteins using ClustalW. As shown by the data in Figure 2, A and B, each different protein type forms a distinct branch in the unrooted cladograms. These cladograms are not necessarily phylogenetic trees of the Lhc protein family but illustrate their relative sequence similarities. The most similar types within the group of three-helix Lhc proteins (Fig. 2A) are Lhcb1 and Lhcb2, and their divergence has been dated to over 300 million years ago (Jansson and Gustafsson, 1991). It is important to realize that this clustering of the different protein subtypes reflects their conserved evolutionary speciation. For example, the Lhca1 to 4 proteins have been shown to assemble at specific positions within the PSI-LHCI holocomplex. This restricts possible protein alterations as the correct binding highly affects the proper antenna function. In comparison to the Lhc proteins, the distances between the PsbS and the six groups of LIL proteins are substantially larger (Fig. 2B) as this group consists of one-, two-, and four-helix proteins that cluster well apart from the three-helix Lhc proteins. Nevertheless, the relative interspecies similarities between the proteins from the three species analyzed are generally lower for a LIL protein subtype than for a three-helix Lhc protein subtype. For the group of the LIL1 proteins (ELIPs), this is illustrated by the fact that the different ELIP subtypes cluster separately for each species. From this it can be concluded that the different ELIP subtypes in poplar, Arabidopsis, and rice do not share subtype-specific evolutionary conserved functions.
Figure 2.
The Lhc protein family in Arabidopsis (ara), poplar (pop), and rice (ory). A, Lhca (a1–a6) and Lhcb (b1–b7) proteins; B, PsbS and LIL proteins; unrooted cladograms were generated using ClustalW alignments of mature protein sequences; ELIP (LIL1), OHP (OHP1, LIL2; OHP2, LIL6), SEP (SEP1, LIL4; SEP2, LIL5).
Lhc Genes with Low EST Coverage in Arabidopsis Are Not Primarily Expressed in Green Leaves in Poplar
The number of ESTs representing a specific gene in a cDNA library is likely to be proportional to the occurrence of the homologous RNA in the mRNA pool. Although the low number of observations typically makes it necessary to find rather large differences in EST numbers in order to get statistical significance in these comparisons (Audic and Claverie, 1997), the generally high expression level of the Lhc genes facilitates such an analysis. Since PopulusDB enables digital northern analyses to be performed (Sterky et al., 2004), we determined the frequencies of ESTs assigned to the 13 types of Lhc genes, PsbS, and the Lil genes in the poplar EST libraries prepared from different tissues. Each library contains about 5,000 EST clones (except for the library prepared from virus/fungus-infected leaves, Y, which contains about 1,000 ESTs). In Supplemental Table I, the number of ESTs in each library for each type of gene is presented. Of about 100,000 ESTs sequenced, 1,789 ESTs were assigned to 35 of the 39 gene models encoding 20 different protein types, and the most highly represented was Lhcb1 with 540 ESTs assigned to four genes (Lhcb1.1 to 4). If one protein type was coded by more than one gene in all cases (except Lhcb3 and Lhcb6), one of the genes was highly overrepresented both in the total dataset as well as in the different libraries. For example, almost 80% (431) of the 540 Lhcb1 ESTs were assigned to the Lhcb1.1 gene model. Twenty-nine percent of the ESTs (528) were found in the library prepared from young leaves (C), which was by far the most enriched in ESTs coding for proteins of the Lhc family. No ESTs in any library corresponded to Lil5.1, Lil5.2, Lil3.4, and Lhca5.2. ESTs for the Lhca1 to 4 and Lhcb1 to 6 genes were significantly enriched in the C library according to calculations using the equation of Audic and Claverie (1997). However, Lhca5, Lhca6, Lhcb7, and Lhcb4.3 ESTs were not found most frequently in the C library. Lhca5 and Lhcb7 ESTs were entirely absent in library C, and the highest numbers of ESTs for Lhca6 and Lhcb4.3 were found in libraries prepared from imbibed seeds (S) and bark (N), respectively. Lil1 ESTs coding for ELIPs were also absent in the young leaf library, but highly abundant in the senescing leaf (I) and cold-stressed leaf (L) libraries.
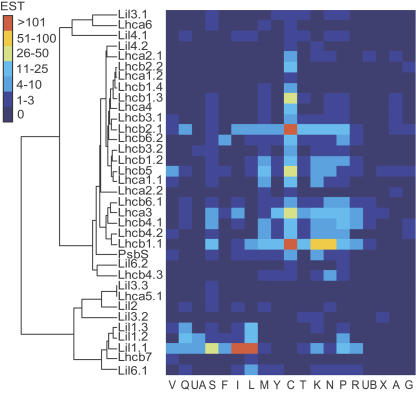
The digital expression data were clustered according to the methods of Ewing et al. (1999) to identify genes with similar expression patterns, and this analysis revealed two main groups (Fig. 3). The first group contained two clusters, one containing all abundantly expressed Lhc genes as well as Lil4.2, PsbS, Lhcb4.3, and Lil6.2. The last two form a subcluster distinct from the main cluster where PsbS clusters separately from all the other abundantly expressed Lhc genes. There is only one Lil gene, Lil4.2, present in this main cluster. The second cluster contains Lil3.1, Lil4.1, and Lhca6 well apart from the first cluster, almost closer to the second group than to the main cluster within the first group. This second group, containing Lhca5, Lhcb7, and the remaining Lil genes (Lil1–Lil3 and Lil6), had a very different expression pattern. Lhcb7 and Lil1 clustered together with Lil6; Lhca5 clustered close to Lil2 and Lil3. It is important to note that one of the main characteristics of this group is the almost complete absence of expression in young leaves (library C). From this analysis it can be concluded that the genes Lhca5, Lhca6, and Lhcb7, coding for the rarely expressed three-helix Lhc proteins, all had an expression pattern that was very different from that of the abundant Lhc genes, and that the Lhcb4.3 expression pattern may be somewhat divergent. It also shows that none of the LIL proteins had an expression pattern as strongly biased as the ELIPs. Quite surprisingly, Lhc ESTs were found in all poplar cDNA libraries, including those prepared from wood tissue (libraries G and X). It is, at this stage, not possible to discriminate between two possible explanations for this finding: that the tissues sampled included a few chloroplast-containing cells or that some Lhc genes are also expressed at very low levels in cells lacking chloroplasts. Such leaky expression is perhaps not unexpected since the expression levels of the Lhc genes are generally very high. The 35 Lhc gene models represent less than 0.1% of the 40,000 genes predicted in poplar but account for almost 1.8% of the identified ESTs in the our dataset.
Figure 3.
Digital expression profiling in PopulusDB. Clustered correlation map according to Ewing et al. (1999) showing the distribution of ESTs from the genes in the Lhc supergene family in 19 poplar cDNA libraries; Lil1 genes code for ELIP; Lil2 (OHP1) and Lil6 (OHP2) genes code for OHP; and Lil4 (SEP1) and Lil5 (SEP2) genes code for SEP. Library codes: A, cambial zone (A + B); C, young leaves; F, flower buds; G, tension wood; I, senescing leaves; UA, active cambium; UB, dormant cambium; K, apical shoot; L, cold-stressed leaves; M, female catkins; N, bark; P, petioles; Q, dormant buds; R, roots; S, imbibed seeds; T, shoot meristem; V, male catkins; X, wood cell death; Y, virus/fungus-infected leaves; for a detailed library description, see Sterky et al. (2004) or www.populus.db.umu.se.
Expression Profiling in Arabidopsis Reveals a Distinct Expression Pattern for Lhca5, Lhca6, Lhcb7, and Lhcb4.3
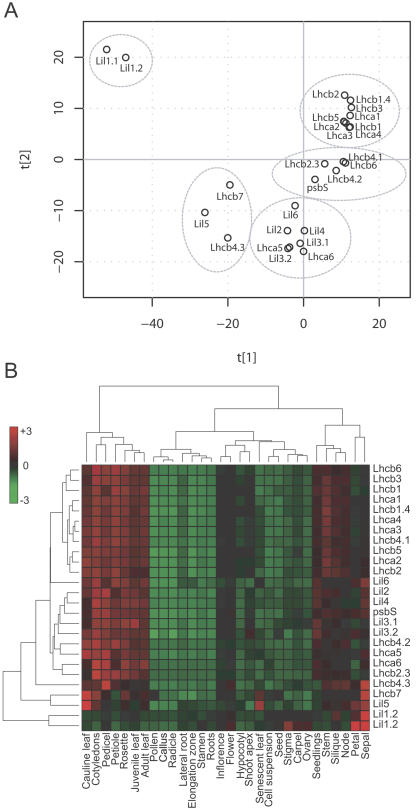
We used publicly available expression data from Arabidopsis to extract expression profiles for all genes of the Lhc gene family (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004). Multivariate statistical analysis of the 2,119 arrays (see “Materials and Methods”) was used to separate the expression profiles into five groups (Fig. 4A). To avoid clustering effects due to the absolute expression differences between indvidiual Lhc genes, which can be higher than an order of magnitude, we transformed the data and used the relative expression levels. All abundantly expressed Lhc genes cluster close to each other, with Lhcb4.1, Lhcb4.2, Lhcb6, Lhcb2.3, and PsbS forming their own cluster very close to this group. None of the Lil genes clusters close to the abundantly expressed Lhc genes. Instead, they form three distinct clusters, one containing Lil2 to Lil4, and Lil6 together with Lhca5 and Lhca6 and a second containing Lil5, Lhcb7, and Lhcb4.3. The profiles for the two ELIP genes, Lil1.1 and Lil1.2, form a third cluster completely distinct from all other Lhc genes. We then analyzed the expression pattern of the Lhc genes with tissue-specific resolution (Fig. 4B). Again, the main cluster contains none of the Lil genes but all of the abundantly expressed Lhc genes. The expression profiles of PsbS, Lil3.1, Lil3.2, Lil4, and Lhca5 are closely related. The patterns of Lhca6, Lhcb2.3, Lil6, Lhcb4.3, Lhcb7, and Lil5 are different from the main cluster, and again the ELIP genes Lil1.1 and Lil1.2 form their own group completely separated from all other genes analyzed. The tissue-resolved expression analysis also shows that the four rarely expressed Lhc protein genes have high relative expression levels in petals and sepals of flowers. These are also the organs where the Lil1 genes are highest expressed.
Figure 4.
Global (A) and tissue-specific (B) clustering of Lhc gene expressions patterns in Arabidopsis based on publicly available data from 2,119 arrays (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004). Lhcb1: Lhcb1.1, Lhcb1.2 and Lhcb1.3; Lhcb2: Lhcb2.1 and Lhcb2.2; Lil1 genes code for ELIP, Lil2 (OHP1) and Lil6 (OHP2) genes code for OHP, and Lil4 (SEP1) and Lil5 (SEP2) genes code for SEP.
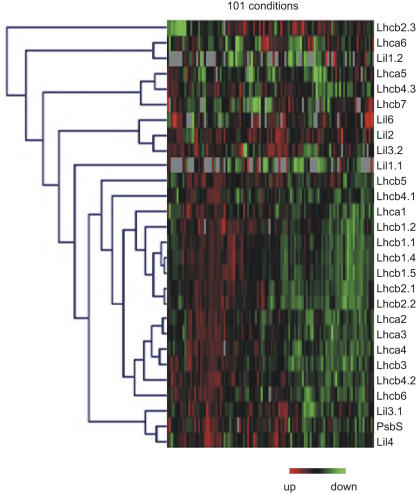
To control for reliability of this pattern, we also reinvestigated DNA macroarray data for chloroplast-related genes that we acquired from samples representing 101 different sets of genetic and environmental conditions in Arabidopsis (Biehl et al., 2005). This dataset is not part of the publicly available dataset analyzed above. The expression data for all genes from the Lhc supergene family in the 101 different conditions were clustered to visualize the similarities and differences in their expression profiles (Fig. 5). In this dataset the Lhcb2.3 gene exhibits the most distinct pattern in comparison to the main cluster of abundantly expressed Lhc genes. The patterns for the other genes were very consistent with the Arabidopsis data presented above. Lhca6 clustered closest together with Lil1.2, Lhca5, Lhcb7, and Lhcb4.3, also clustered closely together, as did Lil2, Lil3, and Lil6, whereas Lil3 and Lil4 clustered closest to PsbS. Based on these two datasets, we concluded that the Lhc genes coding for the four rarely expressed three-helix light-harvesting proteins (Lhca5, Lhca6, Lhcb4.3, Lhcb7) have a regulation pattern in Arabidopsis that clearly differs from the pattern of the genes coding for the abundantly expressed Lhc proteins.
Figure 5.
Expression of the genes of the Arabidopsis Lhc supergene family under 101 different genetic and experimental conditions as investigated by Biehl et al. (2005); Lil1 genes code for ELIP, Lil2 (OHP1) and Lil6 (OHP2) genes code for OHP, and Lil4 (SEP1) and Lil5 (SEP2) genes code for SEP.
DISCUSSION
The public plant genomic resources that have been created, most prominently for Arabidopsis, rice, and poplar, greatly facilitate gene identification and annotation but can also give rapid information about gene regulation. Our study provides the first comprehensive analysis of the expression characteristics of all the genes in the Lhc supergene family, and we report data obtained from two species and from different tissues, developmental stages, and under environmental conditions. We could identify a previously unknown Lhc gene in higher plants, Lhcb7, and show that four Lhc genes (Lhca5, Lhca6, Lhcb4.3, Lhcb7) exhibit a different expression pattern than the genes coding for the 10 abundant Lhc protein types. The abundantly expressed Lhca1 to 4 and the Lhcb1 to 6 genes are to a large extent coregulated in poplar and Arabidopsis, which is consistent with Lhc gene expression regulation data reported for tomato (Lycopersicon esculentum; Kellmann et al., 1993). However, minor differences occur within this group. Lhcb6, Lhcb4.1, and Lhcb4.2, as well as the Lhcb2.3 gene, are obviously expressed with a slightly different pattern than the bulk of the Lhc genes. As there is no Lhcb2.3 ortholog with distinct expression in poplar, we can only speculate if this pattern reflects a function for the Lhcb2.3 protein. The main difference in Arabidopsis is that Lhcb2.3 (At3g27690) is less expressed in caulin leaves, siliques, stem, and bud tissue than Lhcb2.1 (At2g05100) and Lhcb2.2 (At2g05070).
The clustering of the PsbS, Lhcb4, and Lhcb6 gene expression patterns may include additional information about the regulation of nonphotochemical quenching in plants. Nonphotochemical quenching decreases the excitation pressure on PSII under conditions where the absorbed light exceeds the electron transfer capacities of the thylakoid complexes contributing to primary photochemistry (for review, see Szabo et al., 2005). The PsbS protein has clearly been shown to be involved in this process (Li et al., 2000; for review, see Horton and Ruban, 2005), but there is also biochemical evidence for a contribution of Lhcb4 (CP29), Lhcb5 (CP26), and Lhcb6 (CP24; Färber et al., 1997; Dall'Osto et al., 2005). The similarity in the expression profiles of the genes coding for Lhcb4 and PsbS might be due to a cooperative function of the proteins under conditions where elevated capacity for nonradiative energy dissipation is of physiological relevance. Recently, it has been found that a phosphorylated form of Lhcb4 (CP29) may associate with PSI-LHCI in Chlamydomonas under state transitions (Kargul et al., 2005). Balancing of excitation energy between the photosystems also contributes to the plant response to unfavorable light conditions. Though the degree of state transitions is much higher in Chlamydomonas than in higher plants, it is possible that the Lhcb4 gene product is involved in energy balancing or energy dissipating processes even in higher plants.
Within the Lhc gene family, the Lil expression pattern is by far the most extreme: The Lil1 genes coding for ELIPs are simply not expressed in unstressed green leaves, but have very high expression levels under some conditions or in flower tissues. ESTs coding for ELIPS are, for example, the most abundant ESTs of all in poplar leaves in autumn (Bhalerao et al., 2003). Changes in PsbS and other Lil gene expression levels are much less pronounced; they seem to be affected by the same environmental factors as the genes encoding ELIPs, but the amplitudes are much smaller. The differences found among the different Lil genes indicate that they may have specific, not yet defined, functions that can probably be best addressed using reverse genetics. It has already been demonstrated that the ELIPs and the different SEP and OHP proteins are more likely involved in other pigment-related processes than light harvesting (Krol et al., 1995; Adamska, 1997; Jansson et al., 2000; Hutin et al., 2003). This might even be illustrated by the lower degree of sequence similarity within the LIL subtypes compared to the Lhc protein subtypes (Fig. 2, A and B). This also implies that the functions for the LIL proteins are not primarily determined through a fastidious positional specificity within multiprotein complexes as is the case for Lhc proteins. In plant reaction center antenna systems, the Lhc protein function is highly determined by protein interactions that ensure a correct association of the Lhc protein to the holocomplex. Consequently, as the Lhc proteins with the lowest interspecies similarities are also coded by the rarely expressed genes Lhca5, Lhca6, Lhcb7, and Lhcb4.3, it can be deduced that those proteins might not exhibit equivalent close associations to the reaction centers as the abundant Lhc proteins.
Lhcb4.3, a Dicotyledon-Specific Lhc Gene: Lhcb8?
The gene (At2g40100) encoding a protein quite similar to, but shorter than, Lhcb4 identified in Arabidopsis (Jansson, 1999) was denoted Lhcb4.3 to indicate its close relationship to Lhcb4.1 and Lhcb4.2. However, the Lhcb4.3 protein sequence differs more from that of the other two Lhcb4 genes (Lhcb4.1 and Lhcb4.2) than Lhcb1 differs from Lhcb2 (Fig. 2A). Their nearest neighbor is Lhcb3, which is known to have a slightly different function from Lhcb1 and Lhcb2, which are found in the mobile LHCII fraction that attaches to PSI while Lhcb3 is not. Therefore, it is reasonable to propose that all the protein subtypes branching at distances comparable to the distance between the Lhcb1/2 and Lhcb3 groups have different functions. Lhca5, Lhca6, Lhcb7, and Lhcb4.3 clearly branch more deeply from their nearest neighbors than Lhcb3 branches from Lhcb1/2, supporting the assumption that they are unique Lhc protein subtypes with a distinct function(s).
A homologous Lhcb4.3 gene was also found in poplar, and database searches identified a large number of homologous ESTs in several other plant species (e.g. Brassica napus, Poncirus trifoliata, Descurainia sophia, and Citrus sinensis). Interestingly, no ESTs from monocots were found, nor was a homolog found in the genome sequences of indica (Yu et al., 2002) or japonica (Goff et al., 2002) rice, showing that this gene seems to be confined to dicotyledonous plants. Moreover, the plant species (including poplar) in which homologous sequences were found all belong to the classes Eurosids I and II, suggesting that this gene may be found only among Eurosids. Whether or not this hypothesis is true will become clear as more sequences accumulate in the databases. If so, the most likely evolutionary scenario is that the Lhcb4.3 gene arose from duplication of an Lhcb4 gene in the lineage leading to Eurosids I and II after the split from the other lineages, i.e. 100 million years ago or later (Wikström et al., 2001). However, the Lhcb4.3 sequences compared in this study differ more from each other than Lhcb1 differs from Lhcb2, although Lhcb1 and Lhcb2 appear to have diverged more that 300 million years ago. Apparently, therefore, there has been less evolutionary pressure to conserve Lhcb4.3 protein sequences than Lhcb1 and Lhcb2. Alternatively, Lhcb4.3 may have been present in the common ancestors of all higher plants, but orthologs have not yet been found or have been lost in plants outside Eurosids I and II. Although we cannot rule out the latter possibility, we believe it to be less likely. This is probably also true for the Lhca5, Lhca6, and Lhcb7 proteins, the sequences of which differ more from each other in Arabidopsis, poplar, and rice than those of the abundant Lhc proteins (Fig. 2A). The high degree of conservation among dicots and its regulation pattern suggest that the Lhcb4.3 gene product has a distinct function from Lhcb4.1 and Lhcb4.2, and that the gene should be given a unique name, Lhcb8. Nevertheless, based on its sequence similarity to Lhcb4, we hypothesize that the Lhcb8 protein might associate with PSII, but this has to be verified with biochemical methods.
Why Are There Rarely and Abundantly Expressed Lhc Proteins?
Clearly, the light-harvesting antenna of poplar contains the same types of proteins as the Arabidopsis antenna. But as not all Lhc genes are expressed with the same pattern, it is appropriate to discuss how distinct regulation patterns might reflect the flexibility in the light-harvesting antenna of higher plants. In fact, our data indicate that in tissues other than green, unstressed leaves, the materials typically used in biochemical studies of the photosynthetic apparatus, it seems as if the light-harvesting apparatus may have a different polypeptide composition. Although association with PSI or PSII has not been unequivocally demonstrated for Lhca6, Lhcb7, and Lhcb4.3/Lhcb8, it is quite unlikely that proteins found in the middle of the Lhc clade have evolved completely new functions and associations, so we assume that these proteins interact in some way with the photosynthetic light-harvesting antenna of PSI and/or PSII. It is not possible, using the expression data generated so far, to draw any firm conclusions about the possible functions of the encoded proteins, but they may have a function during some stages of plastid development (although that would probably have shown up more clearly in our expression studies); they may accumulate under conditions when light harvesting does not have to be optimized or in cell types where the photosystems, for unknown reasons, require a light-harvesting apparatus different from that in typical mesophyll cells.
For one of these rare Lhc proteins, Lhca5, we have performed a more detailed analysis (Ganeteg et al., 2004). It seems that the Lhca5 protein is present in substoichiometric amounts and that it interacts with the PSI/LHCI complex but that this interaction is weaker than the corresponding interaction of the Lhca1 to 4 proteins. Presumably, under certain conditions Lhca5 accumulates in the thylakoids relative to the other Lhca proteins and can, under those conditions, either partially replace some of the Lhca proteins that are typically bound quite tightly to the PSI core or alternatively bind to novel binding sites, perhaps next to some of the Lhca proteins since the amount of Lhca5 is changed when Lhca proteins are depleted by genetic manipulations (Ganeteg et al., 2004; Klimmek et al., 2005). Lhca5 has also been detected at the protein level in tomato (Storf et al., 2004), and reconstituted Lhca5 has been found to bind photosynthetic pigments in amounts similar, although not identical, to the other Lhca proteins (Storf et al., 2005). Plants lacking Lhca5 have no obvious phenotype, showing that the Lhca5 protein is not essential for the plant under standard laboratory conditions. For the Lhca6, Lhcb7, and Lhcb4.3/Lhcb8 proteins, no such data are available, but it is possible that a detailed analysis would yield similar results. Reverse genetic investigations and a more careful analysis of the expression patterns, preferably on the level of single cells, will be needed to understand their functions.
In conclusion, the data presented here strengthen the concept of adaptive flexibility in the light-harvesting antennas of higher plant PSI and PSII. Hence, the antenna that is typically studied by various biochemical and spectroscopic methods is not necessarily “the” light-harvesting antenna of higher plants but rather a version of it, the one that is most abundant in green leaves of plants grown under nonstressed, low-light conditions in the laboratory. Under natural conditions, which tend to be much more stressful for plants, in which the abundant Lhc genes are relatively lower expressed, it is likely that the rarely expressed Lhc proteins, Lhca5, Lhca6, Lhcb7, and Lhcb4.3/Lhcb8, have a stronger influence on antenna characteristics.
MATERIALS AND METHODS
Sequence Comparisons
The Populus genome browser (genome.jgi-psf.org/Poptr1/Poptr1.home.html) and PopulusDB (www.populus.db.umu.se) were BLAST searched with plant Lhc protein sequences to identify all Lhc homologs. Partial protein sequences for Lhcb7 homologs were identified in Gossypium raimondii (GenBank accession no. CO072671), barley (Hordeum vulgare; CD662315), tomato (Lycopersicon esculentum; AW034099, BE433158, AI483199, AW649809), Prunus dulcis (BU574046), Saccharum officinarum (CA203631), Solanum tuberosum (CK269614), Triticum aestivum (CK217073), and Zea mays (AI600320, BG267854). Full-length protein sequences for Lhcb7 homologs were identified in Arabidopsis (Arabidopsis thaliana; At1g76570) and rice (Oryza sativa; AK066997) or constructed from EST data for Glycine max (BI468979 and BU927047) and Ipomoea nil (BJ558234 and BJ573799). Lhc protein sequences from poplar (Populus spp.), Arabidopsis, and rice were compared using mature protein sequences. Cleavage sites were predicted using ChloroP (version 1.1; Emanuelsson et al., 1999) with manual adjustments where biochemical data were available. Unrooted cladograms were generated using TreeView (version 1.6; Page, 1996).
Digital EST Expression Profiling in Poplar
Dendrograms and clustered correlation maps were prepared following procedures described by Ewing et al. (1999) and Sterky et al. (2004) with some modifications. First, the number of ESTs in each poplar gene model was counted and the ESTs were classified according to their libraries. All gene models corresponding to genes of interest were used in the calculations. The similarity between clusters and between libraries was estimated by calculating Pearson's correlation coefficients, and the Manhattan distance between each pair of objects was calculated. Dendrograms were constructed from the pair-wise distances using the average agglomeration method. The original data set was reordered and plotted according to the ordering in the dendrograms. All steps were performed with the in-house scripts in the R software package (www.r-project.org).
Arabidopsis Gene Expression Profiling
For the first dataset, digital northern information about Lhc genes from 2,119 ATH1 Affymetrix slides was extracted from Genevestigator (Zimmermann et al., 2004; https://www.genevestigator.ethz.ch). The data were gene-wise scaled to unit variance to get information about expression pattern and not expression level. The complete scaled dataset was analyzed with principal component analysis in SIMPCA-P 10 (Umetrics AB). Data were also extracted from the meta-analyzer section of Genevestigator for information about plant organ specificity of Lhc gene expression. Hierarchical clustering of UV-scaled data was performed in the software R (www.r-project.org) with Euclidean distance and complete measurement method. The second dataset contained Lhc gene expression data generated from 3,292 glutathione S-transferase nylon arrays, enriched for nuclear chloroplast genes (Richly et al., 2003; Biehl et al., 2005). At least three experiments with cDNA probes from independent plant pools were carried out for each set of conditions (n = 101). cDNA synthesis was primed by an oligonucleotide mixture matching the 3,292 genes in antisense orientation, and hybridized to the array (Kurth et al., 2002; Richly et al., 2003; Biehl et al., 2005). Hybridization images were read by a phosphorimager (Storm 860; Molecular Dynamics), imported into ArrayVision (version 6.0; Imaging Research), and statistically evaluated using ArrayStat (version 1.0 Rev. 2.0; Imaging Research) as described (Pesaresi et al., 2003; Richly et al., 2003). Data were normalized with reference to all spots on the array (Kurth et al., 2002), and average expression ratios derived from at least three independent experiments were analyzed by hierarchical clustering using Genesis (version 1.5.0 b1; Sturn et al., 2002).
Supplementary Material
Acknowledgments
The authors acknowledge Edouard Pesquet for important discussions.
This work was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning, Swedish Research Council; and the European Community's Human Potential Program (contract no. HPRN–CT–2002–00248 [PSICO]).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stefan Jansson (stefan.jansson@plantphys.umu.se).
The online version of this article contains Web-only data.
References
- Adamska I (1997) ELIPs: light induced stress proteins. Physiol Plant 100: 798–805 [Google Scholar]
- Anandan S, Morishige DT, Thornber JP (1993) Light-induced biogenesis of light-harvesting complex I (LHCI) during chloroplast development in barley (Hordeum vulgare). Plant Physiol 101: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J, Wentworth M, Walters RG, Howard CA, Ruban AV, Horton P, Jansson S (2003. a) Absence of the Lhcb1 and Lhcb2 proteins from the light-harvesting complex of photosystem II: effects on photosynthesis, grana stacking and fitness. Plant J 35: 350–361 [DOI] [PubMed] [Google Scholar]
- Andersson U, Heddad M, Adamska I (2003. b) Light stress-induced one-helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol 132: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM (1997) The significance of digital expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426: 630–635 [DOI] [PubMed] [Google Scholar]
- Bhalerao R, Keskitalo J, Sterky F, Erlandsson R, Björkbacka H, Johnsson Birve S, Karlsson J, Gustaffson P, Lundeberg J, Jansson S (2003) Gene expression in autumn leaves. Plant Physiol 131: 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehl A, Richley E, Noutsos C, Salamini F, Leister D (2005) Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and co-ordination of nuclear and plastid gene expression. Gene 344: 33–41 [DOI] [PubMed] [Google Scholar]
- Broglie R, Bellemare G, Bartlett SG, Chua NH, Cashmore AR (1981) Cloned DNA sequences complementary to mRNAs encoding precursors to the small subunit of ribulose-1,5-bisphosphate carboxylase and a chlorophyll a/b binding polypeptide. Proc Natl Acad Sci USA 78: 7304–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce BD (2000) Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol 10: 440–447 [DOI] [PubMed] [Google Scholar]
- Brunner H, Ruediger W (1995) The greening process in cress seedlings IV. Light regulated expression of single Lhc genes. J Photochem Photobiol B 27: 257–263 [Google Scholar]
- Caffarri S, Croce R, Cattivelli L, Bassi R (2004) A look within LHCII: differential analysis of the Lhcb1-3 complexes building the major trimeric antenna complex of higher-plant photosynthesis. Biochemistry 43: 9467–9476 [DOI] [PubMed] [Google Scholar]
- Chory J, Li HN, Mochizuki N (1995) Molecular methods for isolation of signal transduction pathway mutants. Methods Cell Biol 49: 441–454 [DOI] [PubMed] [Google Scholar]
- Dall'Osto L, Caffarri S, Bassi R (2005) A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17: 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Durnford DG, Price JA, McKim SM, Sarchfield ML (2003) Light-harvesting complex gene expression is controlled by both transcriptional and post-transcriptional mechanisms during photoacclimation in Chlamydomonas reinhardtii. Physiol Plant 118: 193–205 [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, Ben Kahla A, Poirot O, Lopez F, Audic S, Claverie JM (1999) Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res 9: 950–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Färber A, Young AJ, Ruban AV, Horton P, Jahns P (1997) Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants. Plant Physiol 115: 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303: 1782–1784 [DOI] [PubMed] [Google Scholar]
- Fey V, Wagner R, Brautigam K, Pfannschmidt T (2005) Photosynthetic redox control of nuclear gene expression. J Exp Bot 56: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Flachmann R (1997) Composition of photosystem II antenna in light-harvesting complex II antisense tobacco plants at varying irradiances. Plant Physiol 113: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachmann R, Kühlbrandt W (1995) Accumulation of plant antenna complexes is regulated by post-transcriptional mechanisms in tobacco. Plant Cell 7: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C, Schroeder WP, Napiwotzki A, Tjus SE, Renger G, Andersson B (1995) The PSII-S protein of higher plants: a new type of pigment-binding protein. Biochemistry 34: 11133–11141 [DOI] [PubMed] [Google Scholar]
- Ganeteg U, Klimmek F, Jansson S (2004) Lhc5: an LHC-type protein associated with photosystem I. Plant Mol Biol 54: 641–651 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Heddad M, Adamska I (2000) Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc Natl Acad Sci USA 97: 3741–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Ruban A (2005) Molecular design of the photosystem II light-harvesting antenna: photosynthesis and photoprotection. J Exp Bot 56: 365–373 [DOI] [PubMed] [Google Scholar]
- Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M (2003) Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc Natl Acad Sci USA 100: 4921–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Jansson S, Andersson J, Kim SJ, Jackowski G (2000) An Arabidopsis thaliana protein homologous to cyanobacterial high-light-inducible proteins. Plant Mol Biol 42: 345–351 [DOI] [PubMed] [Google Scholar]
- Jansson S, Gustafsson P (1991) Evolutionary conservation of the chlorophyll a/b-binding proteins: cDNAs encoding type I, II and III LHC I polypeptides from the gymnosperm scots pine. Mol Gen Genet 229: 67–76 [DOI] [PubMed] [Google Scholar]
- Jansson S, Pichersky E, Bassi R, Green BR, Ikeuchi M, Melis A, Simpson DJ, Spangfort M, Staehelin LA, Thornber JP (1992) A nomenclature for the genes encoding the chlorophyll a/b-binding proteins of higher plants. Plant Mol Biol Rep 10: 242–253 [Google Scholar]
- Kargul J, Turkina MV, Nield J, Benson S, Vener AV, Barber J (2005) Light-harvesting complex II protein CP29 binds to photosystem I of Chlamydomonas reinhardtii under State 2 conditions. FEBS J 272: 4797–4806 [DOI] [PubMed] [Google Scholar]
- Kellmann JW, Merforth N, Wiese M, Pichersky E, Piechulla B (1993) Concerted circadian oscillations in transcript levels of nineteen Lha/b (cab) genes in Lycopersicon esculentum (tomato). Mol Gen Genet 237: 439–448 [DOI] [PubMed] [Google Scholar]
- Klimmek F, Ganeteg U, Ihalainen JA, van Roon H, Jensen PE, Scheller HV, Dekker JP, Jansson S (2005) Structure of the higher plant light harvesting complex I: in vivo characterization and structural interdependence of the Lhca proteins. Biochemistry 44: 3065–3073 [DOI] [PubMed] [Google Scholar]
- Kloppstech K (1985) Diurnal and circadian rhythmicity in the expression of light-induced plant nuclear messenger RNAs. Planta 165: 502–506 [DOI] [PubMed] [Google Scholar]
- Koo AJ, Ohlrogge JB (2002) The predicted candidates of Arabidopsis plastid inner envelope membrane proteins and their expression profiles. Plant Physiol 130: 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol M, Spangfort MD, Huner NP, Öquist G, Gustafsson P, Jansson S (1995) Chlorophyll a/b-binding proteins, pigment conversions, and early light-induced proteins in a chlorophyll b-less barley mutant. Plant Physiol 107: 873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth J, Varotto C, Pesaresi P, Biehl A, Richly E, Salamini F, Leister D (2002) Gene-sequence-tag expression analyses of 1,800 genes related to chloroplast functions. Planta 215: 101–109 [DOI] [PubMed] [Google Scholar]
- Legen J, Misera S, Herrmann RG, Meurer J (2001) Map positions of 69 Arabidopsis thaliana genes of all known nuclear encoded constituent polypeptides and various regulatory factors of the photosynthetic membrane: a case study. DNA Res 8: 53–60 [DOI] [PubMed] [Google Scholar]
- Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Gomez-Buitrago A, Eckardt N, Shah N, Guevara-Garcia A, Day P, Raina R, Fedoroff NV (2003) Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 4: R20 [DOI] [PMC free article] [PubMed]
- Minagawa J, Takahashi Y (2004) Structure, function and assembly of Photosystem II and its light-harvesting proteins. Photosynth Res 82: 241–263 [DOI] [PubMed] [Google Scholar]
- Morishige DT, Thornber JP (1994) Identification of a novel light-harvesting complex II protein (LHC IIc'). Photosynth Res 39: 33–38 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Gardner NA, Masiero S, Dietzmann A, Eichacker L, Wickner R, Salamini F, Leister D (2003) Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. Plant Cell 15: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichulla B (1988) Plastid and nuclear mRNA fluctuations in tomato leaves: diurnal and circadian rhythms during extended dark and light periods. Plant Mol Biol 11: 345–353 [DOI] [PubMed] [Google Scholar]
- Preiss S, Thornber JP (1995) Stability of the apoproteins of light-harvesting complex I and II during biogenesis of thylakoids in the chlorophyll b-less barley mutant chlorina f2. Plant Physiol 107: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CA (1994) A nomenclature for sequenced plant genes: a brief description. Plant Mol Biol Rep 12: S4–S9 [Google Scholar]
- Richly E, Dietzmann A, Biehl A, Kurth J, Laloi C, Apel K, Salamini F, Leister D (2003) Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep 4: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J, Schleiff E (2004) Protein import into chloroplasts. Nat Rev Mol Cell Biol 5: 198–208 [DOI] [PubMed] [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH, et al (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101: 13951–13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storf S, Jansson S, Schmid VHR (2005) Pigment binding, fluorescence properties, and oligomerization behavior of Lhca5, a novel light-harvesting protein. J Biol Chem 280: 5163–5168 [DOI] [PubMed] [Google Scholar]
- Storf S, Stauber EJ, Hippler M, Schmid VHR (2004) Proteomic analysis of the photosystem I light-harvesting antenna in tomato (Lycopersicon esculentum). Biochemistry 43: 9214–9224 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Szabo I, Bergantino E, Giacometti GM (2005) Light and oxygenic photosynthesis: energy dissipation as a protection mechanism against photo-oxidation. EMBO Rep 6: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Koshino Y, Sawa S, Ishiguro S, Okada K, Tanaka A (2001) Overexpression of chlorophyllide a oxygenase (CAO) enlarges the antenna size of photosystem II in Arabidopsis thaliana. Plant J 26: 365–373 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, Van Breusegem F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström N, Savolainen V, Chase MV (2001) Evolution of angiosperms: calibrating the family tree. Proc R Soc Lond B Biol Sci 268: 2211–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.