Abstract
Arabidopsis (Arabidopsis thaliana) contains approximately 100 homeobox genes, many of which have been shown to play critical roles in various developmental processes. Here we characterize the zinc finger-homeodomain (ZF-HD) subfamily of homeobox genes, consisting of 14 members in Arabidopsis. We demonstrate that the HDs of the ZF-HD proteins share some similarities with other known HDs in Arabidopsis, but they contain distinct features that cluster them as a unique class of plant HD-containing proteins. We have carried out mutational analyses to show that the noncanonical residues present in the HDs of this family of proteins are important for function. Yeast (Saccharomyces cerevisiae) two-hybrid matrix analyses of the ZF-HD proteins reveal that these proteins both homo- and heterodimerize, which may contribute to greater selectivity in DNA binding. These assays also show that most of these proteins do not contain an intrinsic activation domain, suggesting that interactions with other factors are required for transcriptional activation. We also show that the family members are all expressed predominantly or exclusively in floral tissue, indicating a likely regulatory role during floral development. Furthermore, we have identified loss-of-function mutations for six of these genes that individually show no obvious phenotype, supporting the idea that the encoded proteins have common roles in floral development. Based on these results, we propose the ZF-HD gene family encodes a group of transcriptional regulators with unique biochemical activities that play overlapping regulatory roles in Arabidopsis floral development.
The homeodomain (HD) is a 60-amino acid DNA-binding domain (BD) found in many transcription factors. HD-containing proteins are found in diverse organisms such as humans, Drosophila, nematode worms, and plants, where they play important roles in development (for example, McGinnis et al., 1984a, 1984b; Scott and Weiner, 1984; Chan et al., 1998; Veraksa et al., 2000). The crystal structures of different HDs bound to DNA have been isolated and the structures have been found to be remarkably similar. The HD consists of three α helices and a flexible N-terminal arm. Helix III, the recognition helix, packs against the DNA major groove and makes specific contacts with the DNA (for review, see Gehring et al., 1994; Wolberger, 1996). HD proteins have been explicitly shown to act as transcription factors because they bind to promoter sequences and act to modulate mRNA synthesis (for example, see Biggin and Tjian, 1989).
In Arabidopsis (Arabidopsis thaliana), about 100 HD-encoding genes have been identified, and they belong to a few specific classes (Chan et al., 1998; Riechmann, 2002). The members of these classes share not only sequence similarity, but in general they play related roles in planta as well. For example, Class I knox genes appear to play similar roles in controlling the balance between meristematic and determinate growth during plant development (Hake et al., 2004). Class I knox genes are typically expressed in the shoot apical meristem and the developing stem, but not in determinate lateral organs such as floral organs and leaves (Smith et al., 1992; Lincoln et al., 1994; Jackson et al., 1994; Long et al., 1996). Mutations in some Class I knox genes (SHOOT MERISTEMLESS in Arabidopsis, KNOTTED1 in maize [Zea mays]) result in the arrest of shoot apical meristem development (Barton and Poethig, 1993; Long et al., 1996; Vollbrecht et al., 2000), whereas mutations in other genes (BREVIPEDICELLUS in Arabidopsis, OSH15 in rice [Oryza sativa]) result in disrupted shoot architecture (Sato et al., 1999; Douglas et al., 2002; Venglat et al., 2002; Mele et al., 2003). Ectopic expression of Class I knox genes results in organs adopting abnormal, or undifferentiated cell fates (for example, Smith et al., 1992; Sinha et al., 1993; Muller et al., 1995; Chuck et al., 1996; Williams-Carrier et al., 1997; Foster et al., 1999; Nishimura et al., 2000; and refs. therein). Taken together, Class I knox genes are thought to be required for the maintenance of meristematic cell fate and proper shoot growth.
The WOX class of HD-containing genes appears to be essential for embryonic patterning, specifying different regions in the embryo (Haecker et al., 2004). STIMPY/WOX9 is required for the continued expression of the WOX gene WUSCHEL (WUS) in the shoot apical meristem, implicating these genes in a shared role in maintaining this stem cell population (Laux et al., 1996; Wu et al., 2005). In flowers, the WOX gene PRESSED FLOWER is involved in promoting cell proliferation at the marginal meristems of the sepals (Matsumoto and Okada, 2001), whereas WUS promotes growth at the center of the flower by repressing AGAMOUS (Lenhard et al., 2001; Lohmann et al., 2001).
Class III HD-Leu zipper (ZIP) proteins are characterized by a START domain and a HD-ZIP at the N terminus. The five Class III HD-ZIP members in Arabidopsis all appear to be regulated by mRNAs (Bowman, 2004). Three members of this family of genes, PHAVOLUTA, PHABULOSA, and REVOLUTA, are involved in determining adaxial identity in leaves and embryos (Talbert et al., 1995; McConnell and Barton, 1998; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003; Prigge et al., 2005). Expression and functional data for the other two members of this class, ATHB8 and ATHB15/CNA, suggest that they play roles in vascular development, with ATHB15/CNA possibly having roles in establishing embryonic and vascular polarity (Baima et al., 1995; Baima et al., 2001; Ohashi-Ito and Fukuda, 2003; Prigge et al., 2005). Based on the functional analyses to date, this class of HDs appears to have a common role in establishing and/or maintaining abaxial/adaxial polarity.
The zinc finger (ZF)-HD family of HDs has not been functionally characterized in Arabidopsis. In Flaveria trinervia, four ZF-HD proteins were identified and found to bind to the regulatory regions of the C4 phosphoenolpyruvate carboxylase gene, using a yeast one-hybrid screen (Windhövel et al., 2001). Here, we characterize the roles of the 14 family members of this class of HDs in Arabidopsis. We show that these HDs have distinct sequence characteristics as compared to other plant HDs studied to date. These amino acid differences have functional consequences, as our mutational analyses show that the noncanonical residues are required for DNA binding. Most of the members of this family heterodimerize with other members of the family, raising the possibility that diverse interaction partners may lead to greater control in the specificity of DNA binding. Intriguingly, these genes are all coordinately expressed in floral tissue; furthermore, our analyses of loss-of-function mutations indicate that at least a subset of these genes have redundant functions. Together these results suggest that the Arabidopsis ZF-HD proteins have unique biochemical properties and play overlapping regulatory roles in floral development.
RESULTS
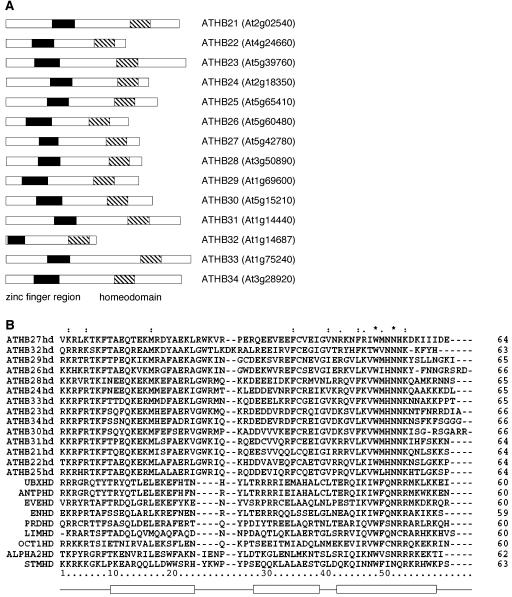
To identify DNA-binding proteins involved in Arabidopsis floral development, we carried out a yeast one-hybrid screen using floral-specific promoter sequences. The DNA-binding protein ATHB33, one of the members of the ZF-HD family, was identified in this screen (Q. K.-G. Tan and V.F. Irish, unpublished data). There are 14 members of this family in Arabidopsis (Fig. 1A) and 11 members had been previously assigned ATHB names (Windhövel et al., 2001).
Figure 1.
The Arabidopsis ZF-HD family. A, The 14 members of the Arabidopsis ZF-HD family are shown schematically, and their corresponding Arabidopsis Genome Initiative locus identifiers listed. The ZF region is shown as a black box, and the HD is represented as a striped box. B, The HD's of the Arabidopsis ZF-HD family members, along with other representative HD-containing proteins are aligned, including Arabidopsis SHOOT MERISTEMLESS (STMHD; At1g62360/Q38874; Long et al., 1996), M. musculus L3 (LIMHD; BAA21649; Matsumoto et al., 1996), as well as the HDs which have been crystallized: D. melanogaster Antennapedia (ANTPHD; CAA43307; Fraenkel and Pabo, 1998), D. melanogaster Even-skipped (EVEHD; AAA28522; Hirsch and Aggarwal, 1995), D. melanogaster Engrailed (ENHD; AAA65478; Kissinger et al., 1990), D. melanogaster Paired (PRD; P06601; Wilson et al., 1995), yeast MATα2 (ALPHA2HD; Q6B2C0; Wolberger et al., 1991), Xenopus laevis Oct-1 (OCT1HD; CAA35051; Klemm et al., 1994), and D. melanogaster Ubx (UBXHD; CAA53803; Passner et al., 1999). Positions of helices 1, 2, and 3 are shown below. Asterisk (*) indicates positions which have a single, fully conserved residue; Colon (:) indicates that one of the following strong groups is fully conserved: STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, and FYW; Period (.) indicates that one of the following weaker groups is fully conserved: CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, and HFY. These are all the positively scoring groups that occur in the Gonnet Pam250 matrix. The strong and weak groups are defined as strong score >0.5 and weak score <0.5, respectively.
Conservation of Domains of ZF-HD Family Members
All the Arabidopsis ZF-HD family members encode small proteins, with the longest sequence containing only 334 amino acids. None of the genes contain introns in their coding sequences. Members of the ZF-HD class contain one conserved region upstream of the HD (Fig. 1A). This upstream region corresponds to the ZF domain and contains conserved His and Cys residues. The ZF domain has been shown to be sufficient in mediating dimerization between different ZF-HD proteins in Flaveria, with the Cys residues being important for interaction (Windhövel et al., 2001). The ZF (CX+NHAX3GX4DGCXEFX8–15CX2CXCHRXFH) is characterized by a CHCC3H2 motif with a variable length spacer between the first CHC residues and the more carboxy terminal C3H2 residues. It forms a distinct group compared with other ZF motifs characterized in Arabidopsis (Takatsuji, 1998; Riechmann, 2002). The length of the spacer may have a crucial role in dimerization, since members with shorter spacers do not appear to effectively dimerize with other members of the ZF-HD family (see below).
The HDs of the Arabidopsis family members are aligned in Figure 1B, together with other representative HDs. The HD in the ZF-HD family is most closely related to the HDs that are associated with LIM domains (Windhövel et al., 2001). LIM-HD transcription factors are involved in tissue patterning and cell type specification in Drosophila, Caenorhabditis elegans, as well as vertebrates (for review, see Curtiss and Heilig, 1998). The LIM domains, via associations with other cofactors, are postulated to modulate the DNA-binding activity of the HD. Similarly, coordinate binding of the ZF-HD family proteins with other transcription factors may serve to regulate transcription. Supporting this idea, an interaction with the MADS domain-containing transcription factor APETALA1 has been demonstrated, using yeast two-hybrid analyses, for ATHB33 (data not shown).
A Noncanonical Residue in the HD Is Required for Function
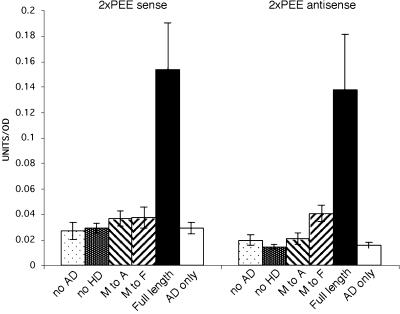
We examined the importance of the HD in DNA binding by generating a variety of mutated versions of ATHB33. While full-length ATHB33 binds to target promoter sequences in yeast one-hybrid assays, removal of the HD from ATHB33 abolished the binding to target promoter sequences (Fig. 2). Most HD-containing transcription factors identified contain a canonical Phe at residue 49 in the HD region (Fig. 1B; Hirsch and Aggarwal, 1995). This residue is not thought to directly contact DNA, but it is important for proper folding of the HD, particularly in stabilizing the hydrophobic core of the HD structure bound to DNA (Kissinger et al., 1990; Wolberger et al., 1991; Hirsch and Aggarwal, 1995; Fraenkel and Pabo, 1998). Intriguingly, the ZF-HD family members do not have a Phe at this position, and instead have a nonpolar residue (Met, Val, Ile, or Leu) in its place (Fig. 1B). When this residue in ATHB33 was mutated to a Phe or Ala, binding to target promoter sequences was abolished (Fig. 2), showing that the substitution of this residue confers important binding characteristics to this family of DNA-binding proteins. ATHB33 also does not contain intrinsic transcriptional activation activity, as demonstrated by the lack of reporter expression in a construct that does not contain the GAL4 activation domain (AD; Fig. 2).
Figure 2.
The noncanonical Met residue is essential for binding to DNA. Various mutations in an ATHB33-GAL4AD fusion protein were assayed for their ability to bind to target DNA sequences using yeast one-hybrid assays; left section, binding to 2XPEE target sequences (−285 to −83) in sense orientation; right panel, binding to 2XPEE target sequences in antisense orientation. No AD, No AD included in construct; no HD, deleted for the HD; M to A, Met 49 to Ala substitution in the HD; M to F, Met 49 to Phe substitution in the HD; Full length, complete coding sequence of ATHB33 fused to GAL4AD; AD only, construct contained only the GAL4AD.
Mapping of the ZF-HD Consensus-Binding Sequence
Yeast one-hybrid assays were used to delimit the domain to which ATHB33 binds to 30 bp of the original 202-bp promoter fragment (data not shown). Within this region, there is a binding sequence for the α2 HD protein (TGTAATT). The α2 HD binds to a consensus sequence motif, TGTANNT (Wolberger et al., 1991). The canonical binding site for most HDs is NNATTA, with the specificity for the first two bases determined by residues as position 50 and 54 (Fraenkel and Pabo, 1998; Connolly et al., 1999). This consensus is also found in the region bound by ATHB33.
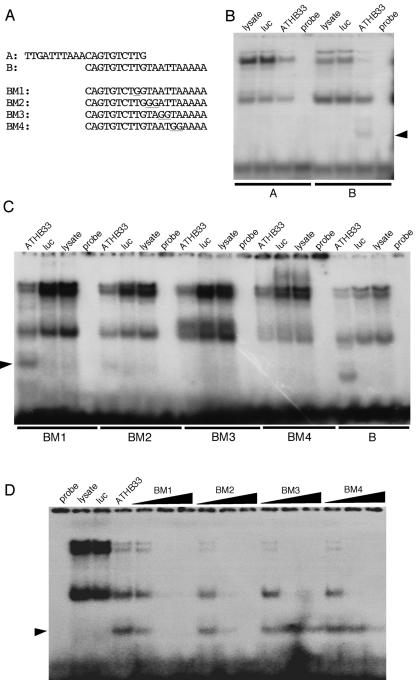
To explicitly map the binding sequence of ATHB33, we carried out electrophoretic mobility shift assays. Two overlapping fragments spanning the 30-bp binding region were initially used to localize the binding of ATHB33 to a 20-bp fragment that contains the consensus NNATTA (Fig. 3A). A series of 20-bp oligonucleotides, in which one or two bases were mutated, were then used to define sequences required for binding (Fig. 3, A and B). Both the BM1 and BM2 mutated oligonucleotides retained the ability to bind ATHB33, while BM3 and BM4 abrogated binding. In addition, competition assays were used to assess the ability of these oligonucleotides to disrupt binding of ATHB33 to the B fragment (Fig. 3C). The relative strength of competition was BM1 > BM2 > BM4 > BM3, indicating that the mutated bases in BM4 and BM3 were the least effective in disrupting binding. Together, these results show that ATHB33 binds to a core consensus sequence of ATTA, similar to that of other HD-containing proteins (Gehring et al., 1994).
Figure 3.
The ZF-HD ATHB33 protein binds to a core consensus sequence of ATTA. A, Sequences of oligonucleotides used for electrophoretic mobility shift assays. BM1 to 4 represent mutated versions of the B oligonucleotide; mutated residues are underlined. B, Electrophoretic mobility shift assays of radiolabeled A and B oligonucleotides. Specific band shift is indicated by an arrowhead. C, Electrophoretic mobility shift assay using the BM1, BM2, BM3, and BM4 radiolabeled oligonucleotides. D, Competition assay. Increasing concentrations of unlabeled BM1, BM2, BM3, and BM4 oligonucleotides were used to compete for binding of the labeled B oligonucleotide to ATHB33 protein. For B to D, lanes are labeled as follows: probe (labeled oligonucleotide, no protein added); lysate (labeled oligonucleotide plus unprogrammed lysate); luc (labeled oligonucleotide plus luciferase protein control); and ATHB33 (labeled oligonucleotide plus ATHB33 protein).
The ZF-HD Family Comprises a Distinct Family of HDs
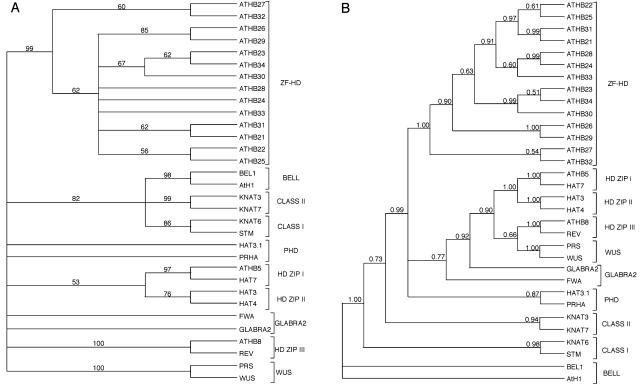
To compare the HD in the 14 Arabidopsis ZF-HD proteins with the HDs found in the other major Arabidopsis HD families, phylogenetic analyses were carried out (Swofford, 2003). Both maximum parsimony and Bayesean analyses show that the ZF-HD proteins form a monophyletic group distinct from the other major classes of plant HDs (Fig. 4). This confirms the preliminary finding of Windhövel et al. (2001) and definitively places this group of factors in a class of their own (Riechmann, 2002). Furthermore, the phylogenetic analyses suggest that certain ZF-HD family members are more closely related to each other than to other members (Fig. 4). Analyses of the Arabidopsis genome indicate that it has undergone a number of ancient and more recent gene duplication events, resulting in multiple regions of synteny across the genome (Blanc et al., 2003). To assess the ancestry of these sequences, we examined whether the ZF-HD family members resided on duplicated segments of the Arabidopsis genome, indicating that they were the result of segmental genome duplication events. We found that the ATHB22 and ATHB25 genes reside in syntenous blocks, as do ATHB21 and ATHB31. ATHB23, ATHB30, and ATHB34 also are in syntenous regions. For two ZF-HD genes, ATHB26 and ATHB27, we could not identify paralogous sequences residing in syntenous blocks, despite the fact that these genes reside in genomic locations that have been duplicated (Blanc et al., 2003). Thus, in general, the duplication history of the ZF-HD genes is consistent with the phylogenetic groupings we identified.
Figure 4.
The ZF-HD family forms a distinct class of HDs. A, Maximum parsimony was used to generate a 50% majority rule consensus using HD sequences of two members of each class of HD proteins in Arabidopsis and the ZF-HD family HD sequences. Bootstrap support of various nodes is shown. B, A Bayesean analysis was carried out using the same dataset. Posterior probabilities are shown for each branch.
BLAST searches revealed that no homologs of the ZF-HD proteins are present in Homo sapiens, C. elegans, Mus musculus, Drosophila melanogaster, and Danio rerio, and no homologs were found when the ZF-HD sequence was used in BLAST searches against 16 fungal and 303 bacterial genomes. However, a hypothetical protein with a region similar to the ZF was found in the fungus Magnaporthe grisea and a Ser protease in the Bdellovibrio bacteriovorus bacterial genome. The HD was most similar to that of LHX8 found in humans and mouse.
The ZF-HD sequence was searched against plant genomic databases (www.plantgdb.org), using tBLASTn against expressed sequence tags, cDNAs, or genomic sequences.
Genes containing both the ZF and HD were found in a variety of plant genomes, including gymnosperms (Pinus taeda), as well as a number of angiosperm species, including monocots (e.g. Triticum aestivum, maize, and rice) and eudicots (e.g. Citrus sinensis, Glycine max, Gossypium arboreum, Medicago truncatula, and Phaseolus vulgaris). Sequences similar to the ZF were found in the fern Ceratopteris richardii, while a noncanonical similar HD sequence was found in the moss Physcomitrella patens. These data suggest that the ZF-HD gene family is specific to the plant kingdom, and implies that such genes have plant-specific functions. Furthermore, the apparent absence of the combination of the ZF and HD motifs in clades other than gymnosperms and angiosperms suggests that the ZF-HD family may have arisen during the evolution of seed plants.
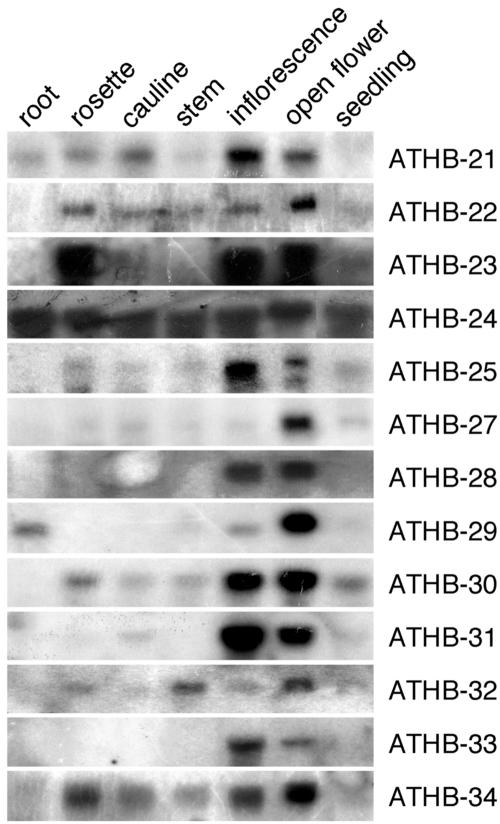
The Arabidopsis ZF-HD Family Members Are All Expressed in Floral Tissues
To analyze the expression patterns of the ZF-HD family members, northern-hybridization analyses were carried out (Fig. 5). Strikingly, RNA of 13 of the family members was detected predominantly or exclusively in floral tissue. The expression of the ATHB26 gene could not be detected by northerns, nor could it be detected by Massively Parallel Signature Sequencing with confidence (Meyers et al., 2004). Digital northern analyses were carried out for ATHB26 using Genevestigator (Zimmermann et al., 2004), which consolidates the data from microarray data performed on Arabidopsis. Using this strategy, no expression data for ATHB26 was recovered. Thus, ATHB26 may be a pseudogene. Some ZF-HD family members are expressed in a floral-specific manner, for example, ATHB33 and ATHB28. The remaining members are expressed most strongly in the flowers and more weakly elsewhere. Although the different members may vary in their expression in vegetative tissue, it is notable that the expression in floral tissue is always strong. Even within the flowers, some members are more highly expressed in younger flowers (ATHB21, ATHB25, and ATHB31) and others are more highly expressed in older flowers (ATHB22, ATHB27, and ATHB29), whereas others are more equally expressed.
Figure 5.
The Arabidopsis ZF-HD genes are all expressed in flowers, with some members being more ubiquitously expressed. Northern-blot analyses using various tissue types were carried out for all ZF-HD family members, with 13 shown; ATHB26 RNA could not be detected in any of the tested tissues. Twenty-five micrograms of total RNA was used for each tissue type: root, rosette (rosette leaves), cauline (cauline leaves), stem, inflorescence (stages 1–12), open flower (stage 13 on), and seedling.
A comparison was carried out between the expression data we obtained and the expression profiles of the ZF-HD family members as assessed by microarray analyses. Using the Gene Atlas feature of the Genevestigator Web site (Zimmermann et al., 2004), which collates data from different microarray experiments (Edgar et al., 2002; Brazma et al., 2003; Craigon et al., 2004), the expression profile of these genes is largely similar to our northern data. One exception is ATHB27, for which we only see high levels of expression in open flowers, but is reported to be fairly ubiquitously expressed by Gene Atlas. High levels of seed expression are observed in Gene Atlas for ATHB22 and ATHB29, which may explain the particularly high expression level we obtained for RNA extracted from open flowers corresponding to these two genes. Strong floral expression is seen for both data sets for ATHB28 and ATHB31, although there are some minor variations in expression levels seen in other tissues.
Redundancy of ZF-HD Family Members
The similarity in expression domains of the ZF-HD family members suggests that they may have similar roles in regulating floral development. To explicitly test this possibility, we identified T-DNA insertional mutations in ATHB22, ATHB23, ATHB25, ATHB29, ATHB31, ATHB32, ATHB33 (two insertions), and ATHB34 (data not shown). Homozygous mutant lines were identified for each of these mutations using PCR genotyping (see “Materials and Methods”); however, none of these nine mutant lines showed any detectable developmental or morphological phenotype (data not shown). Reduced levels of transcripts for the mutations in ATHB31 and ATHB33 were confirmed using northern analyses (data not shown).
The Arabidopsis ZF-HD Family Members Dimerize with Each Other
To characterize the dimerization partners of the Arabidopsis ZF-HD family members, we carried out a matrix of yeast two-hybrid assays, with each family member linked to the yeast GAL4AD or GAL4BD. We carried out these interaction studies for each family member in both combinations. As shown in Table I, the strengths of the interactions between these proteins reveal interesting trends. The Arabidopsis proteins appear to form stronger heterodimers compared to homodimers, as assessed by reporter gene expression. The only exception is ATHB22, which forms a strong interaction with itself, as well as with many other members. Some of the other members, namely ATHB25 and ATHB29, are likewise promiscuous in their interactions. ATHB29 possesses its own AD, since the construct with ATHB29 linked to BD could activate reporter gene expression when coexpressed with just the AD alone. On the other hand, when linked to the AD, ATHB29 interacts with virtually all family members. This is an interesting finding, since ATHB29 could potentially serve as the partner that confers activation of transcription, whereas the other partner could dictate DNA-binding specificity. According to the northern data, the ATHB29 gene is expressed at high levels in flowers only, with low expression in roots (Fig. 5). It is likely that ATHB29 confers floral specificity on the transcriptional complex containing ZF-HD family members.
Table I.
Yeast two-hybrid matrix analysis of interactions between Arabidopsis ZF-HD proteins
Strength of interaction indicated by ++++ (very strong), +++ (strong), ++ (moderate), + (weak), and +−(very weak). AD, GAL4AD; BD, GAL4BD.
| ATHB21 | ATHB22 | ATHB23 | ATHB24 | ATHB25 | ATHB26 | ATHB27 | ATHB28 | ATHB29 | ATHB30 | ATHB31 | ATHB32 | ATHB33 | ATHB34 | AD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATHB21 | – | +++ | +− | – | +++ | – | – | – | + | – | – | – | +− | – | – |
| ATHB22 | ++++ | ++++ | ++++ | ++++ | +++ | – | – | ++++ | ++++ | ++++ | +++ | – | ++++ | ++ | – |
| ATHB23 | ++ | ++++ | ++ | +++ | ++++ | – | – | ++ | + | ++ | +++ | – | +++ | +− | – |
| ATHB24 | – | +++ | ++ | – | ++++ | – | – | – | +++ | – | +− | – | – | – | – |
| ATHB25 | ++ | +++ | ++++ | ++++ | + | – | +− | +++ | +++ | +++ | +++ | – | +++ | + | – |
| ATHB26 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| ATHB27 | – | – | +− | – | +− | – | – | – | + | – | – | – | – | – | – |
| ATHB28 | – | ++++ | + | – | +++ | – | – | – | ++ | – | + | – | – | – | – |
| ATHB29 | +++ | +++ | ++++ | ++++ | ++++ | +++ | +++ | +++ | ++ | +++ | ++++ | +++ | ++++ | +++ | ++ |
| ATHB30 | – | +++ | ++ | +− | +++ | – | – | +− | +++ | +− | + | – | + | – | – |
| ATHB31 | +− | +++ | + | +− | ++++ | – | – | ++ | +++ | + | + | – | ++ | – | – |
| ATHB32 | – | – | – | – | +− | – | – | – | +− | – | – | – | – | – | – |
| ATHB33 | +− | ++++ | ++++ | – | ++++ | – | – | ++ | ++++ | ++ | +++ | – | +− | – | – |
| ATHB34 | – | ++++ | ++ | +− | ++++ | – | – | + | +++ | +− | +− | – | ++ | – | – |
| BD | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
In general, the dimerization characteristics of the proteins are consistent in both combinations tested. The few exceptions are ATHB23/ATHB29, ATHB23/ATHB31, ATHB25/ATHB34, ATHB28/ATHB33, ATHB28/ATHB34, ATHB33/ATHB34, ATHB22/ATHB34, and ATHB23/ATHB34, which differ to some degree in strength.
We only identified one ZF-HD, ATHB26, which does not dimerize at all with any of the other family members. As previously described, ATHB26 does not appear to be expressed. ATHB27 and ATHB32 are two members that form weak dimers with some other members of the family. It may be possible that these weak interactions indicate that ATHB27 and ATHB32 require the presence of cofactors to stabilize the resulting complex. Another possibility is that ATHB27 and ATHB32 contain repressor domains, which overcome the activity of the yeast AD in this assay.
DISCUSSION
A Plant-Specific ZF-HD Family
We have characterized the ZF-HD family of homeobox genes in Arabidopsis. To date, ZF-HD proteins have only been identified in plants, and likely play plant-specific roles. Our phylogenetic analyses support the idea that these genes arose during land plant evolution, and further show that some of the ZF-HD genes likely arose by gene duplication events. We have also shown that members of this family possess a noncanonical residue within the conserved HD, and that this variant residue is required for DNA binding to a core consensus sequence of ATTA. These data, in combination with our findings that all Arabidopsis members of this family are expressed in flowers, and that at least six of these genes have redundant roles, suggest the intriguing possibility that such genes coordinately function in regulating plant reproductive development.
The ZF-HD Proteins Possess a Variant HD
We have shown that the ZF-HD family members contain a noncanonical residue at position 49 in the HD that is required for DNA binding. Mutation of the Met in this position in ATHB33 completely abrogates DNA binding (Fig. 2), demonstrating the importance of this noncanonical residue for binding in the context of these variant HD-containing proteins. Thus, the binding properties of the ZF-HD family members may differ from other HDs with a Phe at position 49. Various HD-DNA structures have been crystallized (Kissinger et al., 1990; Wolberger et al., 1991; Klemm et al., 1994; Hirsch and Aggarwal, 1995; Li et al., 1995; Wilson et al., 1995; Passner et al., 1999; Piper et al., 1999) and the critical residues for contacting DNA have been identified. Based on these studies, the N-terminal arm of the HD forms contacts with the DNA minor groove. Helix III of the HD lies in the major groove where it makes contacts with the sugar-phosphate backbone as well as the bases. In helix III, residues 51 (invariant Asn), 50 (which is thought to determine binding specificity to the DNA), and 47 (usually Val or Ile) contact the DNA bases, and are sometimes aided by residue 54 (which is variant and may help in sequence selection; Wolberger et al., 1991; Klemm et al., 1994; Hirsch and Aggarwal, 1995; Fraenkel and Pabo, 1998). At the same time, the HD makes several contacts with the protein-DNA backbone, usually involving residues Tyr 25, Trp 48, Arg 53, residues in the N-terminal arm, sometimes Lys/Arg 55, as well as others (Hirsch and Aggarwal, 1995; Fraenkel and Pabo, 1998; Piper et al., 1999). From these studies, Phe 49 is not thought to directly contact DNA, however, it is important for proper folding of the HD, particularly in stabilizing the hydrophobic core of the HD structure bound to DNA (Kissinger et al., 1990; Wolberger et al., 1991; Hirsch and Aggarwal, 1995). In the α2 protein, residue 49 is a Val instead of a Phe, but a compensatory change in an interacting residue located in helix I leads to favorable interactions and a similar binding conformation compared to other HDs with a Phe in position 49 (Wolberger et al., 1991).
In terms of residues that contact the major groove of DNA, the ZF-HD proteins contain a conserved Asp at position 51, as in all HDs. Position 47 in the ZF-HD family is occupied by Val or Ile in most of the family members in Arabidopsis, which is similar to HD proteins that contact DNA bases using this residue. For most of the ZF-HD family members, residue 54 is a polar group (Gln, Asp), suggesting that this residue may confer some binding similarity to the family, since this residue may mediate some DNA-binding specificity (Wolberger et al., 1991). Residue 50, which determines binding sequence determination in HD proteins (for review, see Gehring et al., 1994), is either a His or an Asp in the Arabidopsis ZF-HD class of proteins. His 50 is also found in the Cut class of HD proteins (Gehring et al., 1994). This class of proteins has one or more Cut DNA-BDs in addition to a Cut HD. The Drosophila Cut gene is important in cell type specification in various tissues, and there is evidence that Cut factors can function either as repressors or activators of trancription, most likely due to their interaction with different factors (for review, see Nepveu, 2001). There is some evidence that a Cut HD, in conjunction with one or more Cut DNA-BDs, can bind to the consensus ATNNAT (Nepveu, 2001).
As for the other residues in the HD involved in contacting DNA, the ZF-HD family possesses the conserved Trp at position 48. The ZF-HD proteins also contains a Lys at position 53, which has a positive side chain and thus is similar to the Arg at position 53 in other HD proteins, and likely makes the same kind of contacts to the DNA backbone. The residue at position 25 for the family is not a Tyr, as in many HDs crystallized to date, but a positive residue, in 11 of the 14 family members. It is interesting to note that a Lys residue also resides in the equivalent position in the Oct-1 HD, and this residue also makes phosphate contacts similar to the others with a Tyr in this position (Klemm et al., 1994). Instead of a Lys/Arg residue at position 55, most of the ZF-HD family members have a small, uncharged polar group (Ser, Thr) at this position, which may also mediate some form of electrostatic interaction with the phosphate backbone. The N-terminal arm of the ZF-HD proteins is also made up of many basic residues, which likely facilitates contacts with the minor groove of DNA, as in the other HD proteins.
As also noted by Windhövel et al. (2001), the ZF-HD family is also characterized by a four-amino acid insertion between helices I and II of the sequence GW(K/R)(I/M). In the three-amino acid loop extension (TALE) superclass of HD factors, the insertion of the three extra amino acids is also located between helix I and II (for review, see Burglin, 1997). The KNOX and BEL class genes in plants, as well as the yeast α2 gene, the vertebrate Pbx genes, and Drosophila Extradenticle (Exd) gene all belong to this superclass. The crystal structure of α2 reveals that the three-residue insertion does not change the fold of the HD (Wolberger et al., 1991). The binding specificity of the ZF-HD family may differ from that of the TALE class, since residue 50 in the TALE HDs is usually a small, nonpolar residue (Burglin, 1997), and residue 47's contacts with the DNA are also modified due to the presence of Asp at this position (for the TALE HDs), instead of the usual Val/Ile for other HDs, as well as HDs in the ZF-HD family (Passner et al., 1999).
All in all, the HDs crystallized to date, although varying in the identity of some residues that contact DNA, appear to bind DNA in a very similar spatial manner. Supporting this idea, we have found that the ZF-HD family member ATHB33 binds to a core consensus sequence of ATTA, similar to that of other HD-containing proteins (Gehring et al., 1994). Together, these data suggest that the ZF-HD class of proteins may have a distinct DNA-binding protein architecture that nonetheless, can recognize target sequences in vitro similar to that of other HDs. These differences may play important roles in vivo in modulating binding to target sites through interactions with cofactors or in terms of target site context.
The ZF-HD Family Members Are All Florally Expressed
For all but one of the ZF-HD family members, we could detect strong expression in flowers (Fig. 5). We could not detect any expression for ATHB26 using northern hybridization (data not shown), and since the expression of ATHB26 is not identified by microarray experiments as well (Zimmermann et al., 2004), ATHB26 may be a pseudogene.
The northern data raises the interesting possibility that the ZF-HD family may share important roles in floral development. This idea is consistent with the observation that members of some classes of transcription factors perform similar functions in plants. For example, the YABBY family of transcription factors in Arabidopsis specifies abaxial identity in various organs (Bowman, 2000). However, some members may also have distinct roles; for example, within the YABBY family, CRABS CLAW has a unique function in specifying nectary development (Baum et al., 2001). In a similar manner, the ZF-HD family members may have both redundant and unique functions in the Arabidopsis flower. This possibility is consistent with the observed expression of the family members in floral tissue, combined with their varied expression elsewhere in the plant. One possibility is that the ZF-HD proteins act in a combinatorial manner to modulate transcription, with different members contributing either specificity or transcriptional activation to the complex.
It is likely that the Arabidopsis ZF-HD members have largely overlapping or redundant roles. For instance, ATHB23, ATHB34, and ATHB30 are closely related to each other (Fig. 4) and have similar patterns of expression (Fig. 5). As for the factors that are largely floral specific, ATHB21 and ATHB31 are close relatives (Fig. 4). This suggests that members that are closely related to each other may play similar roles in plant development. Because it is likely that the ZF-HD gene products play redundant roles, loss-of-function mutations in multiple family members will likely have to be generated to identify their functions. Individual loss-of-function mutations have been obtained in ATHB23, ATHB25, ATHB31, ATHB32, ATHB33, and ATHB34, but none of them display mutant phenotypes (data not shown), supporting the idea that there is a high degree of redundancy in this family.
ZF-HD Dimerization: A Form of Transcriptional Regulation?
Using yeast two-hybrid analyses, we found that the ZF-HD family members in general form strong heterodimeric interactions. This raises the intriguing possibility that heterodimerization between different ZF-HD members serves to regulate the transcription of floral genes, since all the members are expressed in flowers. Heterodimerization of HD proteins is one mechanism that can modulate the specificity of target gene activation. For instance, the HD proteins Aristaless-like4 (Alx4) and Goosecoid are capable of self dimerization, as well as forming heterodimers with each other. The homodimers and heterodimer preferentially bind to different DNA sequences (Tucker and Wisdom, 1999). Although Alx4 homodimers activate transcription, Alx4/Goosecoid heterodimers repress transcription (Tucker and Wisdom, 1999). Hence, dimerization affects the DNA binding, as well as activity of Alx4. In another example, Exd/Pbx is a class of HDs that interacts with other HDs to modulate their DNA-binding activity (Mann, 1995). Exd/Pbx selectively interact with some HDs and not others, and interaction depends on the slight differences in DNA-binding site sequence (Mann, 1995). It has been suggested that Exd/Pbx-Hox complex functions as a transcriptional activator, whereas HD protein or Exd/Pbx binding to DNA as homodimers may confer repressive abilities (Pinsonneault et al., 1997).
Heterodimerization likely occurs through sequences in the ZF region, although dimer stability in vivo may involve sequences in the HD. Heterodimerization through the ZF domain has been found for the ZF-HD family members in Flaveria (Windhövel et al., 2001). Although the Flaveria ZF-HD proteins dimerized in a two-hybrid assay in the absence of the HD, and the HD alone does not seem to confer dimerization capabilities in yeast two-hybrid assays (Windhövel et al., 2001), the HD sequence may confer specificity and dimer stability, as well as interact with cofactors, in vivo. Dimerization and interaction with cofactors through the HD has been characterized for a variety of HD proteins. For example, the mouse ZFs and homeoboxes 2 protein heterodimerizes with ZFs and homeoboxes 3, using the most N-terminal HD of both proteins (Kawata et al., 2003). In another example, the Arabidopsis ovate family of proteins has been identified as regulators of BELL and KNOX function by binding to their HDs and modulating their subcellular localization (Hackbusch et al., 2005).
Some of the interactions between HDs and other proteins occur through the three-amino acid extension (between helix I and II) in the TALE class of HD proteins (for review, see Burglin, 1997). For instance, these three residues in Exd form part of the pocket that binds the Ultrabithorax YPWM motif, and this interaction results in cooperative binding of these two factors, in that they bind more specifically and with greater affinity than either protein alone (Passner et al., 1999). Similarly, the three-amino acid insertion of Pbx1 is involved in binding the TFDWMK motif (equivalent to the motif in Ubx mentioned above) in HoxB1 as well (Piper et al., 1999). In fact, when the three amino acids in Pbx1 are deleted, the interaction with Hox proteins is abrogated (Peltenburg and Murre, 1997). This raises the intriguing possibility that members of the ZF-HD class may also use the four-amino acid insertion to interact with other proteins that may confer transcriptional activation properties. This would not be unexpected, since most of the family members do not contain intrinsic ADs, as assayed by yeast two-hybrid analysis (Table I).
In addition to the 14 family members, it is very intriguing that there are two expressed sequences in Arabidopsis that only contain the upstream ZF regions (At1g74660 and AAM63930, data not shown) and not the HD. Because these factors do not contain the HD, they presumably cannot bind DNA (this study; Windhövel et al., 2001). One possibility is that these sequences play a regulatory role in titrating the function of the ZF-HD class. It could be envisioned that these non-HD-containing factors may heterodimerize with the ZF-HD proteins, and modify their DNA-binding activity through modulating target site specificity or affinity, or through affecting interactions with cofactors.
ATHB29 is expressed predominantly in floral tissues and possesses its own AD. The insertional mutant of ATHB29 does not have obvious floral phenotypes (data not shown), so it is likely that ATHB29 is not the only protein that confers transcriptional activation to the rest of the family. Other ZF-HD members may have transcriptional activation capabilities that we could not detect in the two-hybrid assay. Moreover, HD-containing proteins are known to interact with other activators/repressors to regulate transcriptional specificity or activity. For example, the yeast α2 HD binds to different operators with either MCM1 or a1. α2 interacts with MCM1 using a region N terminus to the HD (Mead et al., 1996; Tan and Richmond, 1998) and to a1 using a region C terminus to the HD (Li et al., 1995). Both a1 and MCM1 increase the binding affinity and specificity of α2 (Smith and Johnson, 1992; Li et al., 1995, and refs. therein). Whereas the α2/a1 heterodimer represses transcription of haploid-specific genes in diploid cells, the α2/MCM1 heterotetramer serves to repress transcription of a-specific genes in haploid α cells (Jin et al., 1999, and refs. therein). When the cooperative binding between MCM1 and α2 is disrupted, transcriptional repression is severely compromised in vivo (Smith and Johnson, 1992). HD-containing factors may also associate with coactivators such as CBP/p300, indicating that associations with such histone acetylases (HATs) may lead to transcriptional activation by the HD proteins (Chariot et al., 1999). There has also been evidence of the Hox proteins and NF-kB/IkB-α proteins working synergistically together to activate transcription, and they may regulate the nuclear localization of each other (Chariot et al., 1999). As such, identifying other cofactors that interact with this florally expressed family of ZF-HD proteins should shed light on the mechanisms whereby such factors regulate floral gene transcription. Additionally, functional analyses of the ZF-HD family members will be useful in defining the degree of redundancy between different members and their specific roles in floral development.
MATERIALS AND METHODS
Phylogenetic Analyses
The HD sequences were downloaded from GenBank. HD sequences from two members from each homeobox class in Arabidopsis (Arabidopsis thaliana) were used for the alignment (classes were selected based on Chan et al., 1998; Sessa et al., 1998; Haecker et al., 2004). The Arabidopsis Genome Initiative and GenBank accession numbers used for the phylogenetic analyses are as follows: BEL1, At5g41410/A57632; AtH1, At4g32980, NP_195024; KNAT3, At5g25220, CAA63130; KNAT7, At1g62990, AAG40858; KNAT6, At1g23380, BAB69679; STM, At1g62360, Q38874; HAT3.1, At3g19510, NP_188582; PRHA, At4g29940, AAA32843; ATHB5, At5g65310, AAG40406; HAT7, At5g15150, AAA56906; HAT3, At3g60390, NP_191598; HAT4, At4g16780, AAA32815; FWA, At4g25530, AAK28350; GLABRA2, At1g79840, AAG52245; ATHB8, At4g32880, AAM20482; REV, At5g60690, AAF42938; PRS, At2g28610, BAB79446; and WUS, At2g17950, CAA09986.
The HD sequences were aligned with ClustalX (1.81), and alignments refined by hand using MacClade 4.03.68K (Maddison and Maddison, 1989).
Phylogenetic analyses to generate maximum parsimony trees were performed using PAUP 4.0b10 (Swofford, 2003). Maximum parsimony trees were generated using a heuristic search method of 1,000 random stepwise additions employing a tree-bisection reconnection branch swapping algorithm. Gaps in sequences were treated as missing data, and all characters were equally weighted. Multiple equal length parsimony trees were collapsed into a 50% majority rule consensus tree. Bootstrap support was estimated by carrying out 1,000 heuristic searches using the same criteria as above.
Bayesean analyses were carried out using MrBayes 3.1 (Ronquist and Huelsenbeck, 2003) utilizing amino acid alignments. Posterior probability distribution was approximated by Markov Chain Monte Carlo (or MCMC) of 1 million generations with a fixed rate model estimated for our amino acid data. Trees were sampled at every 100th generation and a 50% majority rule consensus tree was generated after trees were removed as “burn-in.” Posterior probabilities are presented under each branch.
Identification and Analysis of Insertion Lines
SLAT lines: The SLAT collection of Arabidopsis insertion lines was created by Dr. Jonathan Jones, Sainsbury Laboratory, John Innes Center, Norwich, UK. The lines are of the Columbia ecotype containing dSpm (a nonautonomous Spm derivative) insertions in the genome. DNA from the iPCR products of the SLAT plants (approximately 48,000 transposants) was arranged into superpools and subpools and spotted onto a filter. The filter was probed with the gene of interest, and plants from the subpool with a hit were grown, genomic DNA was isolated from these plants, and PCR was used to isolate the plant with the insert. dSpm primers and PCR protocols were obtained from the SLAT line Web site (http://arabidopsis.info/CollectionInfo?id=51).
SAIL lines: The SAIL T-DNA lines were generated by the Torrey Mesa Research Institute/Syngenta Biotechnology, San Diego, CA (Sessions et al. 2002). Protocols for analyses are available on their Web site (http://www.tmri.org/en/partnership/sail_collection.aspx).
SALK Lines: The SALK SiGNAL T-DNA lines (Alonso et al., 2003) were analyzed according to protocols obtained from the Web site (http://signal.salk.edu/tabout.html).
For all mutant lines, plant tissues were ground in liquid nitrogen using an eppendorf tube pestle. Three-hundred microliters of extraction buffer (200 mm Tris, pH 7.5; 250 mm NaCl; 25 mm EDTA; and 0.5% SDS) was added to the ground tissue and the tube was incubated at 50°C for 20 to 30 min. An equal volume of phenol:chloroform was then added to the mixture, mixed, and then centrifuged for 5 min. Three-hundred microliters of isopropanol was added to the supernatant, mixed, and frozen at −20°C for 30 min. The DNA was obtained by a 10-min centrifugation, and it was washed with 70% ethanol, dried, dissolved in Tris-EDTA, and used in PCR reactions. Taq polymerase was obtained from Promega or NEB and Pwo polymerase was obtained from Roche. PCR reactions were performed as recommended by the respective Web sites.
Northern Blotting
Seedling tissue from Arabidopsis Landsberg erecta was collected at 6 d postgermination, grown on Murashige and Skoog plates (all plant media from Sigma) or soil. Root tissue was collected from plants grown on Murashige and Skoog plates. The other tissues were collected from soil-grown plants. All plants were grown in standard long-day conditions (16-h light/8-h dark; 22°C). RNAs from tissues were prepared using Trizol (Invitrogen) following the manufacturer's protocol.
For RNA blots, 25 μg of RNA was loaded per lane; the gels were then treated with 50 mm NaOH/10 mm NaCl, and subsequently with 100 mm Tris, pH 7.5. They were blotted onto a Hybond-N membrane (GE Healthcare Amersham). The probes used for hybridization were unique regions for each gene. Hybridization was carried out at 42°C overnight in a solution containing 50% formamide, 3× SSC, 0.1 mg/mL ssDNA, 5× Denhardt's, 5% Dextran Sulphate, and 25 mm EDTA, pH 8. The filters were then washed in SSC. All chemicals were obtained from Sigma.
Generation of Constructs Used in the Yeast One-Hybrid Assay
The −285 to −83 region of the promoter of APETALA3 (At3g54340; Hill et al., 1998) was dimerized and ligated into the β-galactosidase reporter gene-containing vector pLacZi (BD Biosciences Clontech). The promoter fragments were cloned in the sense or antisense orientation with respect to the reporter genes. These reporter constructs were then linearized with the appropriate enzymes and transformed into yeast (Saccharomyces cerevisiae) strain YM4271. The resultant yeast strain had the promoter-LacZ reporter integrated into its genome.
For the no-HD construct, the HD of ATHB33 was removed using BglII and ligated to the pGAD424 vector, which contains the yeast GAL4AD (BD Biosciences Clontech). This construct lacks the HD sequence and 77 bp 5′ to the HD. As for the FL construct, the full-length ATHB33 fragment (coding region and 33 bp of 3′ untranslated region) was first cloned into pCR2.1 using the TOPO TA cloning kit (Invitrogen). The insert was then digested with EcoRI, and cloned into the EcoRI site in pGAD424 to give FL. For the no-AD construct, the AD sequence in FL (in pGAD424) was cut out using KpnI and a partial digest of EcoRI, and the plasmid was then blunt ended with Klenow polymerase (NEB) and religated. The MtoA (Met 49 to Ala) and MtoF (Met 49 to Phe) mutations were engineered using PCR, as described in Innis et al. (1990). The primers used for the MtoA construct are: 5′CAAAGTTTGGGCGCATAACAACAAG (forward) and 5′CTTGTTGTTATGCGCCCAAACTTTG (reverse). Primers for MtoF are: 5′CAAAGTTTGGTTTCATAACAACAAG (forward) and 5′CTTGTTGTTATGAAACCAAACTTTG (reverse). The bases that have been altered from the ATHB33 sequence are underlined. The polymerase and buffer for PCR were obtained from GE Healthcare Amersham. All restriction enzymes were purchased from New England Biolabs.
LacZ liquid assays provide a quantitative measure of the amount of reporter expression. For the liquid assay, the yeast colonies were grown on synthetic dextrose (SD)/Leu liquid media for 2 d at room temperature. They were then transferred to 2 mL of yeast peptone dextrose and grown for 3 to 4 h at room temperature. One milliliter of the culture was then centrifuged and resuspended in 800 μL of Z buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, 1 mm MgSO4, and 50 mm β-mercaptoethanol, pH 7). The rest of the culture was used for the OD600 reading. One drop of 0.1% SDS and two drops of chloroform were added to the resuspended cells and the cells were vortexed for 15 s. This mixture was incubated at 30°C for 15 min. One-hundred sixty microliters of 4 mg/mL ONPG (Sigma) was then added, and the mixture was vortexed for 10 s and placed at 30°C for the reaction to take place. After a few hours, the reaction was quenched with 400 μL of 1 m sodium carbonate. The OD420 and OD550 of the mixture were noted. To calculate units, the following formula was used:
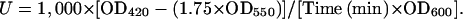 |
Each construct was assayed using five independent transformants and the assays were performed at least twice for consistency.
Yeast Two-Hybrid Assay
The open reading frame sequences of all ZF-HD family members were obtained from Ler wild-type tissue using reverse transcription-PCR. Five micrograms of the total RNA was reverse transcribed in a 20 μL reaction, using 0.5 μg of oligo dT primer and Superscript II (Invitrogen), according to the manufacturer's recommendations. The cDNA was diluted 20×, and 5 μL was used in PCR reactions using the Expand HiFidelity PCR system (Roche). PCR products were cloned into pCR2.1 (Invitrogen) and sequenced. All the constructs were ligated into either the pGAD424 vector or the pGBT9 vector (containing the GAL4BD sequence). The plasmids containing the AD sequence were transformed into Y187 yeast strain (MATα)), and colonies were selected on SD medium lacking Leu. The plasmids with the BD sequence were transformed into the Y190 yeast strain (MATa), and colonies were selected on SD medium lacking Trp. Colonies to be mated were placed in yeast peptone dextrose medium, and incubated at 30°C overnight, with shaking. An aliquot was then plated on SD dropout medium lacking both Leu and Trp to select for yeast with two plasmids. Colonies were then restreaked on fresh SD/Leu/Trp plates, and replica plated onto Whatman 3MM filter paper, which was then placed on an SD/Leu/Trp plate. Alternatively, the colonies were streaked directly on filter paper placed on an SD/Leu/Trp plate. The colonies were grown for 2 d at 30°C. The filters were then dipped in liquid nitrogen for 10 s to lyse the cells. Filters were then left to thaw, and then placed on top of a piece of filter paper that had been soaked with Z buffer/X-gal solution. The time it took for colonies to turn blue was recorded. The assay was performed for a total of six times. All yeast protocols, vectors, and strains were obtained from BD Biosciences Clontech.
Electrophoretic Mobility Shift Assays
The ATHB33 coding region was cloned into pSP72A (Promega) and a transcription reaction was performed with SP6 RNA polymerase (Roche) at 37°C for 7 h. The mixture was then treated with DNAse I (Roche). Five micrograms of RNA was used for in vitro translation using the Rabbit Reticulocyte Lysate system (Promega). 35S Met (Amersham) was used to label the protein, and the reaction was carried out following the manufacturer's protocol. As a control, the Luc control RNA was also translated, and a control reaction containing no RNA was also set up. The integrity of the proteins was checked on a polyacrylamide gel.
Oligonucleotides were generated by the Keck Facility at Yale University. To anneal, complementary pairs of oligonucleotides were dissolved in 0.05 m NaCl, boiled, then slowly cooled to room temperature. A total of 1.5 units of shrimp alkaline phosphatase (USB) were then added to dephosphorylate the ends of the DNA. After the reaction, the phosphatase was inactivated and removed, and the DNA was precipitated. The DNA was labeled with gamma32P ATP (Amersham) using 10 units of T4 polynucleotide kinase (NEB). Labeled DNA was spotted onto DE81 filters to measure the specific activity of the labeling reaction. The excess probe was then removed using sephadex G50 microspin columns (Amersham). Electrophoretic mobility shift reactions were run on an 8% polyacrylamide gel. For binding reactions, 6 fmol of each probe was used in a mixture containing 0.8 μg poly dIdC, 20 mm HEPES, pH 7.8, 50 mm NaCl, 10 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, and 10% glycerol (Windhövel et al., 2001). Two microliters of the translated protein mix was added, and the reaction was incubated at room temperature for 1 h. For the competition assays, cold probe (BM1-4) was added at 1,000×, 10,000×, and 50,000× excess. Control reactions were carried out using lysate alone with no added protein (lysate), Luc RNA added to lysate (Luc), or water added in place of lysate and protein (probe).
Acknowledgments
We thank members of the Irish lab for their constructive comments during the course of this work, and thank Sang-Tae Kim and Irvin Pan for help with the phylogenetic analyses. The SLAT filter was kindly generated and donated by Drs. Stephen Rutherford and Jane Langdale.
This work was supported by a grant from the U.S. Department of Agriculture (grant no. 2001–35304–2226 to V.F.I.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Vivian F. Irish (vivian.irish@yale.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070565.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G (2001) The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126: 643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Poethig RS (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Baum SF, Eshed Y, Bowman JL (2001) The Arabidopsis nectary is an ABC-independent floral structure. Development 128: 4657–4667 [DOI] [PubMed] [Google Scholar]
- Biggin MD, Tjian R (1989) A purified Drosophila homeodomain protein represses transcription in vitro. Cell 58: 433–440 [DOI] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 3: 17–22 [DOI] [PubMed] [Google Scholar]
- Bowman JL (2004) Class III HD-zip gene regulation, the golden fleece of ARGONAUTE activity? Bioessays 26: 938–942 [DOI] [PubMed] [Google Scholar]
- Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushenksy M, Kemmeren P, Lara GG, et al (2003) ArrayExpress–a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 31: 68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR (1997) Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res 25: 4173–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RL, Gago GM, Palena CM, Gonzalez DH (1998) Homeoboxes in plant development. Biochim Biophys Acta 1442: 1–19 [DOI] [PubMed] [Google Scholar]
- Chariot A, Gielen J, Merville MP, Bours V (1999) The homeodomain-containing proteins: an update on their interacting partners. Biochem Pharmacol 58: 1851–1857 [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JP, Augustine JG, Francklyn C (1999) Mutational analysis of the engrailed homeodomain recognition helix by phage display. Nucleic Acids Res 27: 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acid Res 32: D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss J, Heilig JS (1998) DeLIMiting development. Bioessays 20: 58–69 [DOI] [PubMed] [Google Scholar]
- Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD (2002) KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14: 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Foster T, Yamaguchi J, Wong BC, Veit B, Hake S (1999) Gnarley1 is a dominant mutation in the knox4 homeobox gene affecting cell shape and identity. Plant Cell 11: 1239–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel E, Pabo CO (1998) Comparison of x-ray and NMR structures for the Antennapedia homeodomain-DNA complex. Nat Struct Biol 5: 692–697 [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Affolter M, Burglin T (1994) Homeodomain proteins. Annu Rev Biochem 63: 487–526 [DOI] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA 102: 4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hill T, Day CD, Zondlo SC, Thackeray A, Irish VF (1998) Transcriptional regulation of the Arabidopsis floral homeotic gene APETALA3 is mediated by discrete spatial and temporal cis-acting elements. Development 125: 1711–1721 [DOI] [PubMed] [Google Scholar]
- Hirsch JA, Aggarwal AK (1995) Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity. EMBO J 14: 6280–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis MA, Gelfand DH, Sninsky JJ, White TJ (1990) PCR Protocols: A Guide to Methods and Applications. San Diego Academic Press, San Diego
- Jackson D, Veit B, Hake S (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jin Y, Zhong H, Vershon AK (1999) The yeast a1 and alpha2 homeodomain proteins do not contribute equally to heterodimeric DNA binding. Mol Cell Biol 19: 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata H, Yamada K, Shou Z, Mizutani T, Miyamoto K (2003) The mouse zinc-fingers and homeoboxes (ZHX) family; ZHX2 forms a heterodimer with ZHX3. Gene 323: 133–140 [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO (1990) Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell 63: 579–590 [DOI] [PubMed] [Google Scholar]
- Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO (1994) Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77: 21–32 [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Li T, Stark MR, Johnson AD, Wolberger C (1995) Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science 270: 262–269 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR (1989) Interactive analysis of phylogeny and character evolution using the computer program MacClade. Folia Primatol (Basel) 53: 190–202 [DOI] [PubMed] [Google Scholar]
- Mann RS (1995) The specificity of homeotic gene function. Bioessays 17: 855–863 [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Okada K (2001) A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev 15: 3355–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Tanaka T, Furuyama T, Kashihara Y, Mori T, Ishii N, Kitanaka J, Takemura M, Tohyama M, Wanaka A (1996) L3, a novel murine LIM-homeodomain transcription factor expressed in the ventral telencephalon and the mesenchyme surrounding the oral cavity. Neurosci Lett 204: 113–116 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- McGinnis W, Garber RL, Wirz J, Kuroiwa A, Gehring WJ (1984. a) A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37: 403–408 [DOI] [PubMed] [Google Scholar]
- McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ (1984. b) A conserved DNA sequence in homoeotic genes of the Drosophila antennapedia and bithorax complexes. Nature 308: 428–433 [DOI] [PubMed] [Google Scholar]
- Mead J, Zhong H, Acton TB, Vershon AK (1996) The yeast alpha2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins of higher eukaryotes. Mol Cell Biol 16: 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S (2003) The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD (2004) Arabidopsis MPSS: an online resource for quantitative expression analysis. Plant Physiol 135: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller KJ, Romano N, Gerstner O, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W (1995) The barley hooded mutation caused by a duplication in a homeobox gene intron. Nature 374: 727–730 [DOI] [PubMed] [Google Scholar]
- Nepveu A (2001) Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene 270: 1–15 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Tamaoki M, Sakamoto T, Matsuoka M (2000) Over-expression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant Cell Physiol 41: 583–590 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H (2003) HD-zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol 44: 1350–1358 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK (1999) Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397: 714–719 [DOI] [PubMed] [Google Scholar]
- Peltenburg LT, Murre C (1997) Specific residues in the Pbx homeodomain differentially modulate the DNA-binding activity of Hox and Engrailed proteins. Development 124: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Pinsonneault J, Florence B, Vaessin H, McGinnis W (1997) A model for extradenticle function as a switch that changes HOX proteins from repressors to activators. EMBO J 16: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C (1999) Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96: 587–597 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL (2002) Transcriptional regulation: a genomic overview. In CR Somerville, EM Meyerowitz, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0085, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M (1999) Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J 18: 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP, Weiner AJ (1984) Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci USA 81: 4115–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Steindler C, Morelli G, Ruberti I (1998) The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 38: 609–622 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NR, Williams RE, Hake S (1993) Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev 7: 787–795 [DOI] [PubMed] [Google Scholar]
- Smith DL, Johnson AD (1992) A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell 68: 133–142 [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S (1992) A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116: 21–30 [DOI] [PubMed] [Google Scholar]
- Swofford DL (2003) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA
- Takatsuji H (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci 54: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L (1995) The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tan S, Richmond TJ (1998) Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature 391: 660–666 [DOI] [PubMed] [Google Scholar]
- Tucker SC, Wisdom R (1999) Site-specific heterodimerization by paired class homeodomain proteins mediates selective transcriptional responses. J Biol Chem 274: 32325–32332 [DOI] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA 99: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraksa A, Del Campo M, McGinnis W (2000) Developmental patterning genes and their conserved functions: from model organisms to humans. Mol Genet Metab 69: 85–100 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S (2000) Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier RE, Lie YS, Hake S, Lemaux PG (1997) Ectopic expression of the maize kn1 gene phenocopies the hooded mutant of barley. Development 124: 3737–3745 [DOI] [PubMed] [Google Scholar]
- Wilson DS, Guenther B, Desplan C, Kuriyan J (1995) High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell 82: 709–719 [DOI] [PubMed] [Google Scholar]
- Windhövel A, Hein I, Dabrowa R, Stockhaus J (2001) Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol Biol 45: 201–214 [DOI] [PubMed] [Google Scholar]
- Wolberger C (1996) Homeodomain interactions. Curr Opin Struct Biol 6: 62–68 [DOI] [PubMed] [Google Scholar]
- Wolberger C, Vershon AK, Liu B, Johnson AD, Pabo CO (1991) Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell 67: 517–528 [DOI] [PubMed] [Google Scholar]
- Wu X, Dabi T, Weigel D (2005) Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 15: 436–440 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]