Abstract
The NAR2 protein of Chlamydomonas reinhardtii has no known transport activity yet it is required for high-affinity nitrate uptake. Arabidopsis (Arabidopsis thaliana) possesses two genes, AtNRT3.1 and AtNRT3.2, that are similar to the C. reinhardtii NAR2 gene. AtNRT3.1 accounts for greater than 99% of NRT3 mRNA and is induced 6-fold by nitrate. AtNRT3.2 was expressed constitutively at a very low level and did not compensate for the loss of AtNRT3.1 in two Atnrt3.1 mutants. Nitrate uptake by roots and nitrate induction of gene expression were analyzed in two T-DNA mutants, Atnrt3.1-1 and Atnrt3.1-2, disrupted in the AtNRT3.1 promoter and coding regions, respectively, in 5-week-old plants. Nitrate induction of the nitrate transporter genes AtNRT1.1 and AtNRT2.1 was reduced in Atnrt3.1 mutant plants, and this reduced expression was correlated with reduced nitrate concentrations in the tissues. Constitutive high-affinity influx was reduced by 34% and 89%, respectively, in Atnrt3.1-1 and Atnrt3.1-2 mutant plants, while high-affinity nitrate-inducible influx was reduced by 92% and 96%, respectively, following induction with 1 mm KNO3 after 7 d of nitrogen deprivation. By contrast, low-affinity influx appeared to be unaffected. Thus, the constitutive high-affinity influx and nitrate-inducible high-affinity influx (but not the low-affinity influx) of higher plant roots require a functional AtNRT3 (NAR2) gene.
Nitrate influx into plant roots has been defined kinetically as being composed of at least four additive fluxes; constitutive high-affinity influx (CHATS), nitrate-inducible high-affinity influx (IHATS), constitutive low-affinity influx (CLATS), and inducible low-affinity influx (ILATS; Crawford and Glass, 1998; Forde, 2000). Members of the NRT2 family of transporters are involved in the IHATS in fungi, algae, and plants. The first members of this family were cloned from Aspergillus nidulans and Chlamydomonas reinhardtii (Unkles et al., 1991; Quesada et al., 1994) by use of mutants defective in nitrate transport. Sequence similarities among these genes enabled Trueman et al. (1996) to clone the first members of this family from plants (HvNRT2.1 and HvNRT2.2 from barley [Hordeum vulgare]). Subsequently, AtNRT2.1 and AtNRT2.2 were cloned from Arabidopsis (Arabidopsis thaliana; Filleur and Daniel-Vedele, 1999; Zhuo et al., 1999) along with similar genes from several other plant species (for review, see Crawford and Glass, 1998; Forde, 2000). Evidence that NRT2 genes play an important role in high-affinity nitrate transport came first from strong correlations between AtNRT2.1 and AtNRT2.2 expression and high-affinity influx during induction by nitrate (Zhuo et al., 1999; Okamoto et al., 2003) and during down-regulation by various nitrogen sources (Vidmar et al., 2000; Nazoa et al., 2003). A T-DNA mutant of Arabidopsis disrupted in the AtNRT2.1 and AtNRT2.2 genes exhibited severe and specific impairment of IHATS function, providing further support for this idea (Cerezo et al., 2001; Filleur et al., 2001). However, in C. reinhardtii NRT2 genes do not act alone; two high-affinity nitrate transporter genes (CrNRT2.1 and CrNRT2.2) require a second gene, CrNAR2, to function in nitrate transport (Quesada et al., 1994). A role for CrNAR2 in nitrate uptake was confirmed in Xenopus oocytes. Oocytes injected with CrNAR2 or CrNRT2.1 mRNA separately gave no evidence of nitrate transport activity, whereas coinjection of both mRNAs produced nitrate currents (Zhou et al., 2000). Similar results were reported for barley, where one of three NAR2-like genes, HvNAR2.3, is able to increase nitrate transport compared to water-injected Xenopus oocytes when coexpressed with HvNRT2.1 (Tong et al., 2005). These findings indicate that NAR2 is not required for transcriptional regulation of NRT2.1 but is, instead, facilitating NRT2.1 transport activity perhaps by a direct interaction.
The completion of the Arabidopsis genome-sequencing project enabled us to search for the presence of NAR2 genes in this plant. Two such NAR2-like genes were found. We propose to name them AtNRT3.1 and AtNRT3.2, respectively, since NAR is already reserved for another gene in Arabidopsis. We obtained two T-DNA insertion mutants for AtNRT3.1, one disrupted in the promoter region and the other in the coding region. These mutants were used to investigate whether high-affinity nitrate transport in Arabidopsis requires NRT3 function in planta.
RESULTS
Identification of the AtNRT3 Gene Family
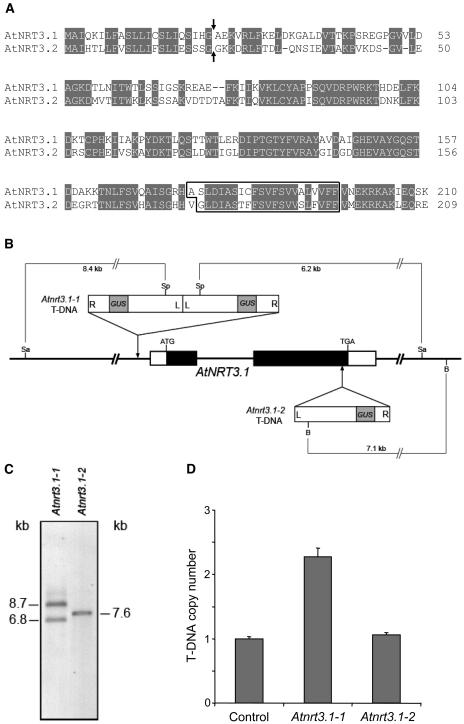
Two genes in Arabidopsis were revealed by a homology search against the Arabidopsis genome database (Arabidopsis Genome Initiative, 2000) using the C. reinhardtii NAR2 sequences, namely, AtNRT3.1 (GenBank ID: AJ310933; Arabidopsis Genome Initiative [AGI] code: At5g50200) and AtNRT3.2 (GenBank ID: BX842351; AGI code: At4g24720). Predicted open reading frames of AtNRT3.1 and AtNRT3.2 encode 210 and 209 amino acids, respectively, sharing 61% identity and 76% similarity with each other (Fig. 1A). SignalP (Nielsen et al., 1999) and PSORT (Nakai and Kanehisa, 1992) predict that both AtNRT3 proteins possess signal peptides, with a predicted cleavage site between amino acids 22 and 23. One transmembrane region was predicted at the C terminus, leaving a long hydrophilic N-terminal end that is predicted to be extracellular.
Figure 1.
Amino acid sequences of AtNRT3 proteins and characterization of Atnrt3.1 T-DNA lines. A, Amino acid sequence comparison of AtNRT3.1 and AtNRT3.2. Identical amino acids are shaded. Arrows indicate predicted cleavage sites. The transmembrane region predicted by TMHMM (Krogh et al., 2001) is boxed. B, Schematic diagram of AtNRT3.1 with the T-DNA insertions. In AtNRT3.1 white boxes indicate 5′ or 3′ untranslated region, and black boxes indicate open reading frame. The T-DNAs in the Atnrt3.1-1 and Atnrt3.1-2 mutants are inserted 186 bp upstream of the first putative start codon, and 63 bp before the stop codon, respectively. The diagram is not drawn in scale. R, Right border; L, left border; Sa, SalI; Sp, SphI; B, BamHI. C, The Southern blot of the Atnrt3.1 mutants. Genomic DNAs of Atnrt3.1-1 and Atnrt3.1-2 were digested with SalI/SphI and BamHI, respectively, and probed with a 1.2-kb fragment of GUS gene (Kaiser et al., 2002). D, The T-DNA copy number in the Atnrt3 mutants measured by relative quantitative real-time PCR. Each sample was normalized by nitrite reductase (a single copy gene, AGI code: At2g15620), and expressed relative to the control T-DNA line (M. Galli, unpublished data).
Isolation of Atnrt3 Mutants
To find a T-DNA insertion in AtNRT3.1, a PCR-based screen was carried out with a population of 60,480 T-DNA insertion lines (ecotype Wassilewskija [Ws] background) from the Arabidopsis knockout facility at the University of Wisconsin (see “Materials and Methods” for details). A T-DNA insertion line was isolated, and sequence analysis of the T-DNA-genomic DNA junction regions revealed that the insertion was 184 bp upstream from the predicted start codon of AtNRT3.1; there was a deletion of 51 bp of genomic DNA (−185 to approximately −235) and both 5′ and 3′ ends of the T-DNA were right borders, predicting two copies of T-DNA in a tandem inverted orientation. This T-DNA line was designated as Atnrt3.1-1 (Fig. 1B).
Another putative Atnrt3.1 mutant was identified in the FLAGdb T-DNA lines (Samson et al., 2002). PCR analysis with respective primers of AtNRT3.1 genomic DNA and the T-DNA confirmed a T-DNA insertion near the end of the gene. The T-DNA insertion started at 858 bp from the start codon, deleting 66 bp of exon 2 and 5 bp of the 3′ untranslated region. This second mutant was designated as Atnrt3.1-2 (Fig. 1B).
The T-DNA mutants were backcrossed twice to wild type (ecotype Ws) and the resulting homozygous lines were used for further investigation. To determine T-DNA copy number, Southern-blot and relative quantitative real-time PCR analyses were carried out (Fig. 1, C and D). For Southern-blot analysis, genomic DNAs of Atnrt3.1-1 and Atnrt3.1-2 were digested with SalI/SphI and BamHI, respectively, and hybridized with the T-DNA-specific probe (1.2 kb of β-glucuronidase (GUS) gene; Kaiser et al., 2002). The fragment sizes in the blot are similar to the predicted sizes (Fig. 1, B and C). GUS gene copy numbers were also determined by quantitative real-time PCR assay (Ingham et al., 2001). The results from both methods indicated that Atnrt3.1-1 and Atnrt3.1-2 carry two copies and one copy of T-DNA, respectively.
Expression of AtNRT3.1 and AtNRT3.2 in Wild-Type Plants and Atnrt3.1 Mutants
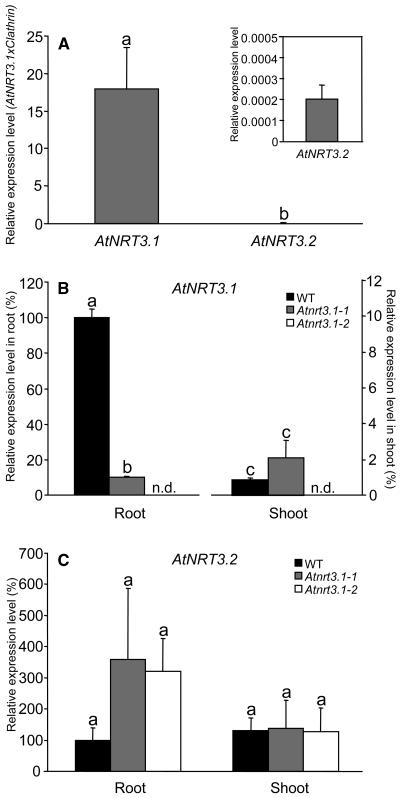
Several expressed sequence tags (ESTs) and full-length cDNA sequence data of AtNRT3.1 are published in The Arabidopsis Information Resource database (http://arabidopsis.org/), indicating that AtNRT3.1 is indeed expressed in Arabidopsis plants, whereas no EST is available for AtNRT3.2. We analyzed the expression level of the two AtNRT3 genes in wild-type roots that had been grown in 1 mm NH4NO3 for 4 weeks, followed by 1 week in 50 μm KNO3 by real-time relative reverse transcription (RT)-PCR. Figure 2A shows relative expression levels of AtNRT3.1 and AtNRT3.2 genes against clathrin, a housekeeping gene (AGI code: At4g24550). Relative to clathrin, AtNRT3.1 showed a much higher level of expression than AtNRT3.2, which was barely detectable. Several more gene-specific primer sets were employed to confirm these results (data not shown). This may explain the absence of an EST for AtNRT3.2, yet we were able to detect expression of this gene due to the sensitivity of the assay.
Figure 2.
Expression analysis of AtNRT3.1 and AtNRT3.2 by relative quantitative real-time RT-PCR. Plants were grown for 4 weeks in 1 mm NH4NO3 and then transferred to 50 μm KNO3 for 1 week. A, Relative expression level of AtNRT3.1 and AtNRT3.2 against clathrin, a housekeeping gene, in wild-type roots. The inset is an expanded-scale plot of AtNRT3.2. B and C, Relative expression profile of AtNRT3.1 (B) and AtNRT3.2 (C) in wild-type plants and Atnrt3.1 mutants. Each sample was normalized by clathrin and relatively expressed to wild-type root. The values are means of four replicates (two biological replicates × two independent reactions). Error bars = se. Different letters above the bars indicate significant difference at P < 0.05 (t test). n.d. indicates the following levels: root (<0.1%) and shoot (0%).
The expression profile of the AtNRT3 genes was also examined in Atnrt3 mutant plants. AtNRT3.1 expression levels in roots of Atnrt3.1-1 and Atnrt3.1-2 mutants were reduced by 90% and 100%, respectively, compared to those of wild-type plants under the same conditions (i.e. 1 mm NH4NO3 for 4 weeks, followed by 1 week in 50 μm KNO3). In Atnrt3.1-1 shoots, levels of AtNRT3.1 were roughly 1% to approximately 2% of those of wild-type roots, whereas in Atnrt3.1-2 there was a complete absence of AtNRT3.1 (Fig. 2B). These results demonstrate that Atnrt3.1-1 is a knockdown mutant and Atnrt3.1-2 is a knockout mutant. They also show that AtNRT3.1 expression is much higher in roots than shoots (about 100-fold higher).
It is possible that AtNRT3.2, a homolog of AtNRT3.1, might have compensated for the loss of AtNRT3.1 function in the Atnrt3.1 mutants. However, expression levels of AtNRT3.2 in roots and shoots of Atnrt3.1 mutant plants remained at low levels, though somewhat higher than in wild-type roots (Fig. 2C).
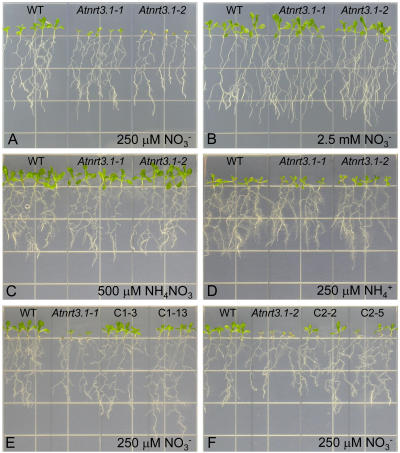
Phenotypes of the Atnrt3.1 Mutants and Restoration of Wild-Type Phenotype with an AtNRT3.1 cDNA
There was no obvious phenotype when the Atnrt3.1 mutants were grown in peat-based soil, but phenotypic differences became apparent when external nitrogen sources were controlled. When grown on plates containing 250 μm NO3− as the sole source of available nitrogen for 10 d, mutant plants grew poorly, shoot growth being particularly affected (Fig. 3A). By contrast to these phenotypic differences, mutant growth was normalized when plants were grown on 2.5 mm NO3−, 500 μm NH4NO3, or 250 μm NH4+ as sole nitrogen sources (Fig. 3, B–D).
Figure 3.
Plant growth on various nitrogen sources. A to D, Wild-type plants and Atnrt3.1 mutants were grown on vertical plates containing 250 μm KNO3 (A), 2.5 mm KNO3 (B), 500 μm NH4NO3 (C), or 125 μm (NH4)2 succinate (D) with other nutrients for 10 d. E and F, Complemented lines (C1-3, C1-13, C2-2, and C2-5) with 35S-NRT3.1 in the Atnrt3.1-1 (E) and Atnrt3.1-2 (F) mutant background were grown in 250 μm KNO3 for 10 d.
When mutants (both Atnrt3.1-1 and Atnrt3.1-2) and wild-type plants were grown in a common tank containing 1 mm NH4NO3 for 4 weeks, followed by 1 week without a source of nitrogen, to deinduce the IHATS in preparation for influx measurements, shoot-to-root ratios were consistently and significantly lower (P < 0.05) in the mutant plants than in wild-type plants. Absolute values for shoot-to-root ratios varied from one experiment to another. Table I provides representative values for wild-type plants and Atnrt3.1 mutants as well as shoot and root fresh weights (FWs). Shoot:root ratios were 3.5 (wild type), 1.4 (Atnrt3.1-1), and 1.5 (Atnrt3.1-2), respectively, differences that were highly significant (P < 0.05), as were shoot biomasses, but root weights were not different. However, in some experiments using older plants FWs of mutant roots were greater than those of wild-type plants (data not shown).
Table I.
Shoot and root FWs and shoot:root ratios in wild-type and Atnrt3.1 mutants
Plants were grown for 4 weeks in 1 mm NH4NO3 and then nitrogen deprived for 1 week. The values are the means of five (wild type) or 12 (Atnrt3.1 mutants) plants ± se. Differences between wild-type and mutant shoot weights and shoot:root ratios were highly significant (P > 0.05, t test).
| Genotype
|
FW
|
Shoot:Root Ratio
|
|
|---|---|---|---|
| Shoot | Root | ||
| g plant−1 | |||
| Wild type | 0.46 ± 0.03 | 0.13 ± 0.02 | 3.5 |
| Atnrt3.1-1 | 0.23 ± 0.02 | 0.17 ± 0.05 | 1.4 |
| Atnrt3.1-2 | 0.15 ± 0.03 | 0.10 ± 0.03 | 1.5 |
Genetic complementation of the two mutant lines by introducing a AtNRT3.1 cDNA driven by the cauliflower mosaic virus (CaMV) 35S promoter (lines C1-3, C1-13, C2-2, and C2-5) restored their growth on 250 μm NO3− to that of wild-type plants (Fig. 3, E and F).
Nitrate Responses of AtNRT3 and Nitrate Transporter Genes
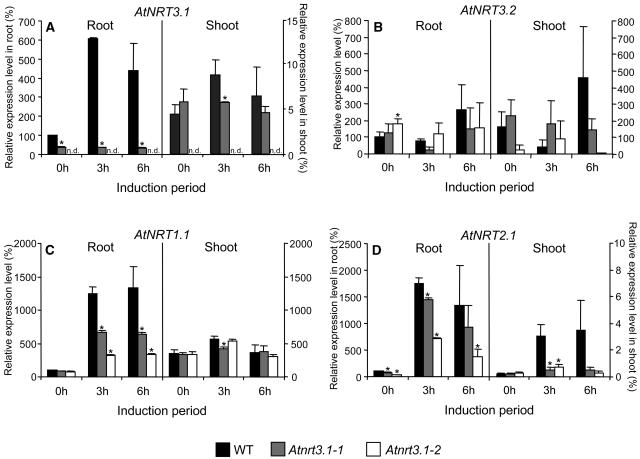
The induction by nitrate of AtNRT3 and the major nitrate transporter genes was examined in plants previously deprived of nitrogen and subsequently exposed to 1 mm KNO3 for 3 or 6 h. Growth and nitrogen deprivation conditions were the same as used for the nitrate uptake experiments described below. Following this treatment, RNA was extracted and analyzed by relative real-time RT-PCR. In wild-type roots, AtNRT3.1 was strongly induced by 1 mm KNO3 (6-fold by 3 h); in shoots there is much less AtNRT3.1 mRNA and little response to the same treatment (Fig. 4A). The Atnrt3.1-1 mutant showed significantly less AtNRT3.1 mRNA in roots prior to reexposure to KNO3 (<40% of wild type), and no response to inducing conditions (1 mm KNO3); shoots showed expression levels similar to those of the wild type. Compared to wild type (100%), the level of AtNRT3.1 mRNA in the Atnrt3.1-2 mutant was essentially 0% in both roots and shoots as expected for a null mutant. AtNRT3.2 showed no consistent pattern of expression in wild-type and mutant plants except for a slight induction after 6 h in wild-type plants (Fig. 4B).
Figure 4.
Expression analysis of AtNRT3, AtNRT1.1, and AtNRT2.1 genes in response to nitrate by relative quantitative real-time RT-PCR. A to D, Plants were grown for 4 weeks in 1 mm NH4NO3 and then nitrogen deprived for 1 week before being reexposed to 1 mm KNO3 for 0, 3, and 6 h. Each sample was normalized by clathrin and relatively expressed to wild-type root at 0 h. Relative expression values of AtNRT3.1 (A), AtNRT3.2 (B), AtNRT1.1 (C), and AtNRT2.1 (D) are means of four replicates (two biological replicates × two independent reactions). Error bars = se. Asterisk (*) indicates significant difference of the mutants from wild type (t test, P < 0.05). n.d. indicates the following levels: root (<0.3%) and shoot (0%).
Two NRT nitrate transporters, a dual-affinity nitrate transporter, AtNRT1.1 (CHL1) (Tsay et al., 1993; Wang et al., 1998; Liu et al., 1999; Liu and Tsay, 2003), and a high-affinity nitrate transporter, AtNRT2.1, were also analyzed under the same conditions as described above. In wild-type roots, both AtNRT1.1 and AtNRT2.1 were induced approximately 13- and 18-fold, respectively, by 1 mm KNO3, peaking at 6 and 3 h, respectively (Fig. 4, C and D). AtNRT1.1 mRNA levels were estimated to be 20- to 35-fold lower than those for AtNRT2.1 in induced roots based on real-time RT-PCR cycle number comparisons (data not shown). AtNRT1.1 expression level in wild-type shoots was higher than roots at 0 h, and showed a slight induction by KNO3, whereas AtNRT2.1 expression levels were <1% of roots but were induced significantly by 3 h. Similarly both NRT genes were strongly induced by KNO3 in the Atnrt3.1 mutant roots, indicating that the defect in AtNRT3.1 did not prevent induction of these nitrate transporter genes at the transcriptional level. Nevertheless, the expression levels of both AtNRT1.1 and AtNRT2.1 in the mutants were significantly lower than those in wild type. AtNRT1.1 levels in Atnrt3.1-1 and Atnrt3.1-2 peaked with 8- and 4-fold induction, respectively, giving expression levels that were 50% and 75% lower than wild type, respectively. AtNRT2.1 was induced about 20-fold in both mutants. However, the expression levels were also lower than wild type in the same order (wild type > Atnrt3.1-1 > Atnrt3.1-2) at all time points. In shoots of the mutants, AtNRT1.1 levels were similar to wild type, whereas AtNRT2.1 levels remained low and showed a small induction (Fig. 4, C and D).
Tissue Nitrate Analysis
To determine tissue NO3− accumulation during the standard induction period (i.e. 6 h pretreatment in 1 mm KNO3, as used for the gene expression analyses described above), root and shoot samples of wild-type and Atnrt3.1-2 mutant plants were weighed, boiled in distilled water for 5 min, and tissue NO3− concentrations determined by the Cataldo method (Cataldo et al., 1975). As shown in Table II, wild-type roots and shoots accumulated significantly more NO3− than mutant during 6 h, specifically 5.4-fold higher in wild-type roots (7.5 μmol g−1 FW versus 1.4 μmol g−1 FW increases in wild type and mutant, respectively), and 52-fold more in wild-type shoots (5.2 μmol g−1 FW versus 0.1 μmol g−1 FW increases in wild type and mutant, respectively).
Table II.
Tissue nitrate concentration in wild type and the Atnrt3.1-2 mutant
Plants were grown for 4 weeks in 1 mm NH4NO3 and then nitrogen deprived for 1 week. Nitrate contents (μmol g−1 FW) were then measured at time 0 (0 h) and after 6 h of incubation with 1 mm KNO3. The values are the means of five plants ± se. Differences between wild-type and mutant root or shoot were significant at 6 h (P > 0.05, t test).
| Genotype | Treatment (1 mm KNO3) | Root | Shoot |
|---|---|---|---|
| Wild type | 0 h | 0.38 ± 0.1 | 5.0 ± 0.1 |
| 6 h | 7.9 ± 0.2 | 10.2 ± 0.2 | |
| Atnrt3.1-2 | 0 h | 0.3 ± 0.1 | 6.3 ± 0.15 |
| 6 h | 1.7 ± 0.1 | 6.4 ± 0.1 |
High-Affinity Nitrate Influx
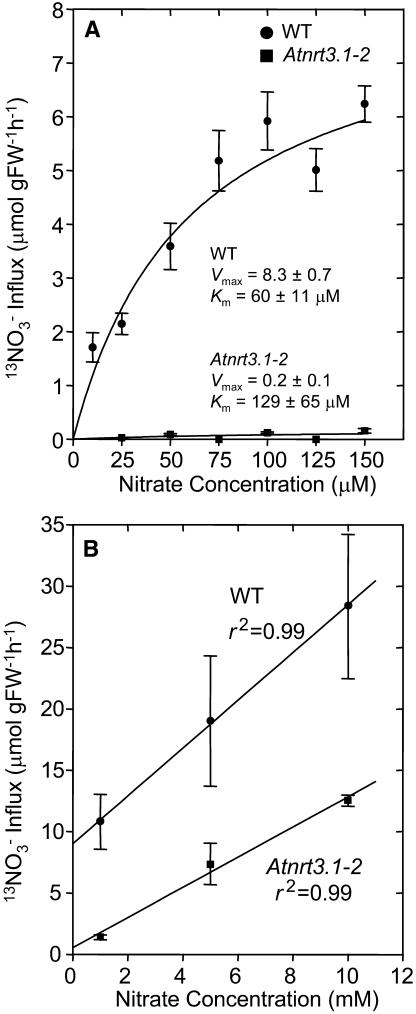
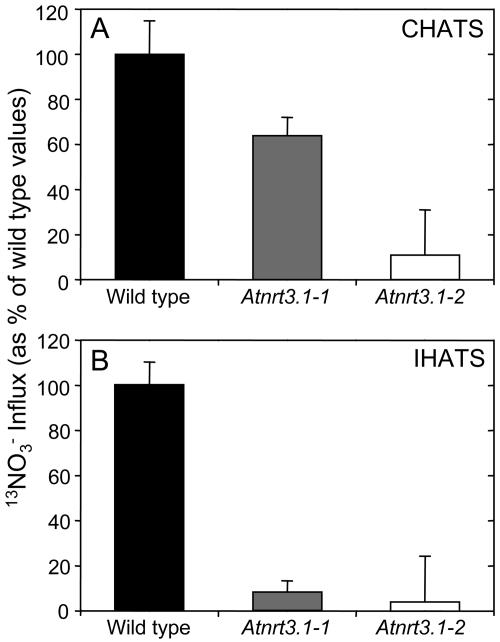
High-affinity nitrate influx into intact roots of wild-type and mutant plants was examined using 13NO3− to determine the effect of the Atnrt3.1 mutations. Figure 5A shows high-affinity nitrate influx into roots of wild-type and Atnrt3.1-2 mutant plants, measured at external NO3− concentrations from 10 to 150 μm. In this experiment, plants were grown for 4 weeks in 1 mm NH4NO3 and then transferred to solutions without nitrogen for 7 d before being reexposed to 1 mm KNO3. This treatment has previously been shown to deinduce IHATS and ensure that root NO3− stores are consumed (Okamoto et al., 2003). Subsequently, the provision of 1 mm KNO3 for 6 h to plants pretreated in this fashion, caused AtNRT2.1 transcript and high-affinity nitrate influx to reach peak values within 3 and 6 h, respectively. Thereafter, transcript abundance and influx gradually decline (Okamoto et al., 2003). Compared to wild-type plants, nitrate influx into roots of mutant plants was reduced across the whole range of NO3− concentration examined. Vmax values for wild-type and mutant plants were 8.3 ± 0.7 and 0.2 ± 0.1 μmol g FW−1 h−1, respectively, while corresponding Km values were 60 ± 11 and 129 ± 65 μm. The Vmax for influx was reduced by 98%. Similar results were obtained with Atnrt3.1-1 mutant plants (data not shown), however the high-affinity influx was reduced by only 80% in this mutant. To determine the component fluxes (CHATS and IHATS) of the measured high-affinity influx it is necessary to measure influx in uninduced plants. Thus, influx was also measured in nitrate-deprived plants during the first 10 min of exposure to 100 μm KNO3 after 7 d of nitrogen deprivation. The results of this experiment are shown in Figure 6A as percentages of the wild-type fluxes. Absolute influx values before induction were 1.9 ± 0.1 μmol g FW−1 h−1 (wild type) and 1.26 ± 0.1 (Atnrt3.1-1 mutant), respectively, a reduction of 34%. In separate experiments corresponding values for wild type and the Atnrt3.1-2 mutant were 2.7 ± 0.4 and 0.3 ± 0.1 μmol g FW−1 h−1, respectively, a reduction of 89%. Using the same batch of plants, 13NO3− was measured after 6 h of exposure to 1 mm KNO3. The measured fluxes (CHATS plus IHATS) had increased to 9.84 ± 1.0 μmol g FW−1 h−1 in wild-type roots, 1.9 ± 0.32 in the Atnrt3.1-1 mutant, and 0.32 ± 0.2 μmol g FW−1 h−1 in the Atnrt3.1-2 mutant. Thus, IHATS influx (measured flux in induced plants minus that of uninduced plants) was reduced by 92% and 96% in the Atnrt3.1-1 and Atnrt3.1-2 mutants, respectively (Fig. 6B).
Figure 5.
13NO3− influx into wild-type and Atnrt3.1-2 mutant roots. A, High-affinity 13NO3− influx. Plants were grown for 4 weeks in 1 mm NH4NO3 and then nitrogen deprived for 1 week before being reexposed to 1 mm KNO3 for 6 h. Nitrate influx was then measured at 10, 25, 50, 75, 100, 125, and 150 μm NO3− as described in “Materials and Methods.” Data values are means ± se of five replicates. The fitted curve was obtained by direct fit to the Michaelis-Menten equation. Estimated Km and Vmax values are indicated in the figure. B, Low-affinity 13NO3− influx. Plants were grown for 4 weeks in 1 mm NH4NO3 and then nitrogen deprived for 1 week before being reexposed to 1 mm KNO3 for 6 h. Nitrate influx was then measured at 1, 5, and 10 mm NO3−. Data values are means ± se of four replicates.
Figure 6.
13NO3− influx by CHATS and IHATS. A, 13NO3− influx due to CHATS in wild-type and Atnrt3.1 mutants as percentages of wild-type fluxes. Plants were grown for 4 weeks in 1 mm NH4NO3 and then nitrogen deprived for 1 week. Nitrate influx was then measured at 100 μm NO3−. B, 13NO3− influx due to IHATS in wild-type and Atnrt3.1 mutants as percentages of wild-type fluxes. Plants were grown for 4 weeks in 1 mm NH4NO3 and then nitrogen deprived for 1 week before being reexposed to 1 mm KNO3 for 6 h. Nitrate influx was then measured at 100 μm NO3−. IHATS influxes were calculated by subtracting the measured CHATS flux from the flux measured after induction. Data values are means ± se of four replicates.
Low-Affinity Nitrate Influx
To determine if the low value of influx in the mutant plants was due in part to disruption of the LATS influx, 13NO3− influx into roots of both lines of mutant plants, deprived of nitrate for 7 d, and then exposed to 1 mm KNO3 for 6 h, was determined from solutions containing 1, 5, and 10 mm K13NO3 (Fig. 5B). In both lines 13NO3− influx in the LATS concentration range was found to be unaffected by the AtNRT3.1 mutation, except that measured fluxes at all three concentrations were reduced by the extent to which the IHATS flux had been reduced. Data for the Atnrt3.1-2 mutant line is shown in Figure 5B. Results for the Atnrt3.1-1 mutant (data not shown) were essentially identical to those for Atnrt3.1-2.
DISCUSSION
The goal of this study was to determine if high-affinity nitrate transport into roots of Arabidopsis requires the presence of a functional NAR2-like gene (i.e. AtNRT3) in addition to members of the NRT2 family. A requirement for the presence of functional AtNRT2.1 and AtNRT2.2 genes was established previously by demonstrating that inducible high-affinity nitrate influx in a T-DNA mutant disrupted in AtNRT2.1 and AtNRT2.2 was strongly impaired compared to control plants (Cerezo et al., 2001; Filleur et al., 2001). The same study revealed that low-affinity influx was unaffected by this disruption.
Expression Patterns of AtNRT3 and AtNRT2.1 Genes
The absence of ESTs for AtNRT3.2 and the present expression analyses of AtNRT3.1 and AtNRT3.2 suggest that only AtNRT3.1 would be detected under the conditions examined. The pattern of expression of AtNRT3.1 is quite similar, with respect to induction by nitrate, to AtNRT2.1, which is thought to encode an inducible high-affinity nitrate transporter (Zhuo et al., 1999; Cerezo et al., 2001; Filleur et al., 2001; Okamoto et al., 2003). In the Atnrt3.1 T-DNA mutant plants, AtNRT3.1 transcript levels were low or virtually undetectable compared to wild-type plants deprived of nitrate (Fig. 4). After exposure to 1 mm KNO3 to induce expression of nitrate-inducible genes, AtNRT3.1 transcript abundance actually declined in the Atnrt3.1-1 mutant. By contrast AtNRT3.1 transcript abundance increased 6-fold in wild-type plants during the first 3 h after resupply of 1 mm KNO3, and tended to decline by 6 h. This decline was observed to continue until 24 h had elapsed, when the expression level was down to preinduced level (data not shown). This pattern of AtNRT3.1 expression in wild-type plants following reexposure to KNO3 is similar to that of AtNRT2.1, which typically peaks in abundance after 3 h then declines (Fig. 4; see Zhuo et al., 1999; Okamoto et al., 2003).
In this study, AtNRT2.1 abundance in roots of the Atnrt3.1-1 and Atnrt3.1-2 mutants increased approximately 13- and 6-fold, respectively, after reexposure to 1 mm KNO3 (Fig. 4D), demonstrating that AtNRT3.1 is not required for transcription of AtNRT2.1. We also examined expression patterns of AtNRT1.1 (CHL1), another nitrate-inducible gene encoding the inducible dual-affinity nitrate transporter. Transcript abundances increased as a result of this protocol both in the Atnrt3.1 mutants and wild-type plants, although the extent of inductions were less in the mutants as seen in AtNRT2.1 (Fig. 3C). The most likely cause of reduced expression of these nitrate-induced genes is the reduced root NO3− concentration associated with disruption of high-affinity nitrate influx. Indeed, as demonstrated in the “Results” section, 13NO3− influx into roots of mutant plants and tissue NO3− concentration were substantially reduced compared to wild-type plants (Figs. 5 and 6; Table II). Indeed, the opposite result, namely elevated levels of NRT2.1 transcript, have been reported in nitrate reductase (NR) mutants or in plants treated with tungstate to block NR activity, due to elevated tissue NO3− concentration in the absence of downstream products of nitrate assimilation (Filleur and Daniel-Vedele, 1999; Vidmar et al., 2000). If this interpretation is correct, it suggests that induction of nitrate-dependent genes requires the entry of nitrate into the roots rather than external sensing of nitrate, a hypothesis that has been suggested earlier (Unkles et al., 2001), because roots of mutant plants were exposed to the same NO3− concentration as wild-type plants during induction.
There are examples where one gene in a gene family can compensate for the loss of other homologous gene(s). Examples of this functional compensation include the phosphate transporter PHO family in Saccharomyces cerevisiae (Wykoff and O'Shea, 2001) and the ammonium transporter AMT family in Arabidopsis (Kaiser et al., 2002). In this study, however, AtNRT3.2 failed to compensate for the disrupted AtNRT3.1 gene in the mutant, since AtNRT3.2 (normally expressed at substantially lower levels than AtNRT3.1) showed no apparent sign of nitrate induction in either wild-type or the Atnrt3.1 mutants.
Nitrate Influx
The first indication that high-affinity nitrate uptake was disrupted in the Atnrt3.1 mutants was the poor growth on low-nitrate media (Fig. 3A). In particular, shoot growth was impaired. This was confirmed quantitatively in plants used for influx studies that had been grown on 1 mm NH4NO3 under hydroponic conditions. Shoot-to-root ratios were strongly reduced in mutant plants, even though 50% of nitrogen came from NH4+ (Table I). Such low shoot-to-root ratios are typical of nitrogen deprivation. By contrast, mutant growth was restored to wild-type levels when plants were grown on elevated concentrations of KNO3, or on (NH4)2 succinate. Normalization of plant growth on 2.5 mm NO3− in agar suggests that influx via the LATS (which was shown to function normally in the Atnrt3.1 mutants, see below) was sufficient to satisfy plant demand. Likewise, growth was normalized in the mutant lines that had been rescued with an AtNRT3.1 cDNA (Fig. 3, E and F).
To evaluate the role of AtNRT3 in high-affinity nitrate influx directly, plants that had been maintained on 1 mm NH4NO3 for 4 weeks were completely deprived of nitrate for 7 d and then reexposed to 1 mm KNO3 for 6 h to induce expression of nitrate-inducible genes. NH4NO3 was used in the prior growth period to optimize growth, especially that of mutant plants that exhibited impaired growth on KNO3 as the sole source of nitrogen. However, in the standard induction NH4+ was omitted since it is well known to inhibit nitrate influx. 13NO3− influx was then measured using various concentrations of KNO3 before and after the 6 h period of induction. The data presented in Figure 5A show that disruption of AtNRT3.1 caused a virtual elimination of high-affinity influx in nitrate-induced plants across the whole range of NO3− concentration investigated. Influx values before induction (at time 0 h), presumably due to the CHATS alone or in combination with a CLATS, were also reduced but to different extents in the two mutant lines. Expressing wild-type fluxes as 100%, the reductions of CHATS associated with the T-DNA insertion were 34% in the Atnrt3.1-1 mutant and 89% in the Atnrt3.1-2 mutant. These results indicate that the AtNRT3.1 gene is required not only for normal activity of the IHATS but also for the CHATS. The relatively small reduction of CHATS in the Atnrt3.1-1 mutant may be the result of leakiness in this mutation associated with the location of the disruption in the promoter region of AtNRT3.1. The influx values for wild-type plants are similar to those reported in earlier studies for nitrate-deprived Arabidopsis roots (Zhuo et al., 1999; Okamoto et al., 2003). Following induction, 13NO3− influx increased from 1.9 ± 0.1 to 9.84 ± 1.0 μmol g FW−1 h−1 in wild-type plants, from 1.26 ± 0.1 to 1.90 ± 0.1 μmol g FW−1 h−1 in the Atnrt3.1-1 mutant, and from 0.3 ± 0.1 to 0.32 ± 0.2 in the Atnrt3.1-2 mutant. Thus the inducible component of HATS influx (IHATS: measured flux in induced plants minus that of uninduced plants) was reduced by 92% and 96% in the Atnrt3.1-1 and Atnrt3.1-2 mutant lines, respectively (Fig. 6B). Again, we suggest that the small increase of influx in the Atnrt3.1-1 mutant (from 1.26–1.90 μmol g FW−1 h−1) may have been due to a leaky mutation in this line. By contrast, the large increase in wild-type plants after reexposure to NO3− (from 1.9–9.84 μmol g FW−1 h−1) represents the combined contribution of the large IHATS influx together with any contribution from the ILATS.
It is evident from the foregoing discussion that the IHATS is essentially absent in the Atnrt3.1 mutants, but (as stated above) it was possible that the ILATS and/or the CLATS might also have contributed to the calculated IHATS and have a requirement for coexpression of AtNRT3.1. To evaluate this possibility, 13NO3− influx was measured in wild-type and the Atnrt3.1 mutant plants at 1, 5, and 10 mm KNO3, concentrations that are typical of the LATS. Figure 5B indicates an essentially normal concentration response of the Atnrt3.1-2 mutant plants, suggesting that LATS function is not disrupted in these plants, and does not require coexpression of AtNRT3.1. The absolute value of influx was lower in the case of the mutants because IHATS function had been disrupted. Note that the intercept values for influx correspond quite closely to the values obtained for HATS influx measured at 100 μm. Identical results were obtained for Atnrt3.1-1 mutant (data not shown).
In summary, this study establishes that Arabidopsis possesses two NAR2-like genes, AtNRT3.1 and AtNRT3.2. Of these only AtNRT3.1 is expressed at significant levels and AtNRT3.1 proved to be highly responsive to induction by nitrate. In the Atnrt3.1 mutants described, AtNRT3.1 transcript was expressed at very low levels compared to the wild type and was not induced by exposure to nitrate. In these mutants, both high-affinity nitrate transport systems (CHATS and IHATS) are functionally impaired, even though transcript abundance of AtNRT2.1 was strongly expressed after induction. Therefore, in Arabidopsis both the CHATS and the IHATS appear to require coexpression of the AtNRT3.1 gene. By contrast, LATS function was shown to be independent of the AtNRT3.1 expression.
MATERIALS AND METHODS
Plant Growth Conditions and Influx Determinations
Plants used for gene expression and influx studies were maintained in an environment chamber with light/dark periods of 8/16 h, 25°C/20°C, and relative humidity = 70%, with photon flux of 150 to 200 μE m−2 s−1. Plants were grown hydroponically in nutrient solution with 1 mm KH2PO4, 0.5 mm MgSO4, 0.25 mm CaSO4, 20 μm Fe-EDTA, 25 μm H3BO3, 2 μm ZnSO4, 2 μm MnSO4, 0.5 μm CuSO4, and 0.5 μm (NH4)6Mo7O24 in 8-L plastic containers. The pH of the nutrient solution was maintained with CaCO3 around 6.2 for all the experiments. One-centimeter holes were cut in Styrofoam platforms (1.25 cm thickness) and nylon mesh was placed over the holes. Fine sand was placed in the mesh and seeds were planted on top of the sand. The platforms and seeds were then floated on the nutrient solutions. After seeding onto the platform, seeds were imbibed in a cold room at 4°C for 3 to 4 d.
For the nitrate influx analyses through HATS, plants were grown at 1 mm NH4NO3 for 4 weeks and then transferred to media lacking any source of nitrogen for 1 week. To measure CHATS activity roots of plants were transferred to fresh medium containing 100 μm KNO3 for 5 min before being exposed to chemically identical solutions, except that the KNO3 was labeled with 13NO3−. After exposure to tracer for 5 min, roots were transferred back to identical nonlabeled solutions for 3 min to desorb tracer from the cell walls. To measure induced fluxes, plants were grown on 1 mm NH4NO3 for 4 weeks, transferred to −N solution for 1 week, and then reinduced with 1 mm KNO3 for up to 6 h. Roots of plants used for experiments designed to measure concentration-dependent fluxes were induced in the same manner as described above and then roots were pretreated for 5 min at the concentration to be used to measure tracer fluxes. The same standard desorption protocol (described above) was used in these experiments. After desorption, roots and shoots were separated, weighed, and put into vials for counting using a γ-counter (MINAXI Auto-Gamma 5000 series, Packard Instruments). Generation and purification of 13NO3− and other details regarding influx analyses using 13NO3− are described by Siddiqi et al. (1989).
Continuous light was provided for the growth study on plates. The nutrient medium was the same as described above except for nitrogen whose concentration was as indicated in the figures. In addition, the nutrient solution contained 0.5% (w/v) Suc and 0.5 g/L of MES (pH 5.7), and 0.7% (w/v) agarose (Invitrogen).
Isolation of Atnrt3.1 Mutant Lines
The Atnrt3.1-1 mutant was isolated from a population of 60,480 T-DNA inserted lines (Ws background) from the Arabidopsis knockout facility at the University of Wisconsin. The methodology was described by Krysan et al. (1996, 1999). Primers from the right border of the T-DNA (XR-2: 5′-TGGGAAAACCTGGCGTTACCCAACTTAAT), and from AtNRT3.1 (forward: 5′-CTCTTCTCTTCCTCAGCCTTATTTTTCTG, reverse: 5′-GAAGAAGTGTGCAACAAGACAAAAGGAAT) were used for the PCR-based screening and the subsequent genotyping. During the screening, putative PCR products were also verified by Southern-blot analysis using 1.1 kb AtNRT3.1 genomic DNA fragment (a PCR product of the AtNRT3.1 forward and reverse primers above) as a probe (data not shown). PCR products, which also showed positive in the Southern-blot analysis, were cloned into TOPO TA cloning vector (Invitrogen) and sequenced.
The Atnrt3.1-2 mutant was identified in the FLAGdb T-DNA lines (Samson et al., 2002). Primer sets for the T-DNA (left border: 5′-TCCAGGGCGTGTGCCAGGTGC, right border: 5′-CCAGACTGAATGCCCACAGGCCGTC) and for AtNRT3.1 (forward: 5′-AGCCAAGTTGACCGACCATGG, reverse: 5′-GATTTCTCTTTGAAAGTAAGAGGTGAAG) were used for PCR-based genotyping and sequencing of the T-DNA/genome flanking region.
Quantitative Real-Time PCR
To determine T-DNA copy number in the mutant lines, genomic DNA was isolated from mature leaves of homozygous Atnrt3.1 mutants by DNeasy plant mini kit (Qiagen) and analyzed by quantitative real-time PCR assay (Ingham et al., 2001). Primers were designed specific to GUS and nitrite reductase (AGI code: At2g15620) for transgene and endogenous control gene, respectively. As a control line we included a transgenic plant in which the GUS gene copy number had already been determined (M. Galli, unpublished data). Quantitative real-time PCR was performed by using LightCycler (Roche) with the SYBR Green I detection system, under the following conditions: 95°C for 10 min; 45 cycles of 95°C for 5 s, 63°C for 5 s, and 72°C for 10 s; followed by melting curve analysis. PCR mixture of a final volume of 10 μL contained 2 μL of gDNA, 0.5 μm of each primer, 4 mm Mg2+, and 1 μL of LightCycler-FastStart DNA Master SYBR Green I mixture (Roche). The following primer sets were used: GUS (forward: 5′-CGTTTCGATGCGGTCACTC; reverse: 5′-CGTCGGTAATCACCATTCCC) and NR (forward: 5′-CCGGTAGCCAGTTCTGCG; reverse: 5′-CCTATTCGTCCCCCGACGT).
For gene expression analysis total RNA was isolated from approximately 100 mg FW with RNeasy plant mini kit (Qiagen). RNase-free DNase treatment was also carried out during the isolation. Gene expression levels were analyzed by two-step real-time RT-PCR. cDNAs were synthesized from 250 ng of total RNA by Transcriptor (Roche), and the reaction mixture was diluted 20 times for subsequent PCR. The conditions for the quantitative real-time PCR were same as above. As a control, no RT (omitting reverse transcriptase in RT step) reaction was included. PCR mixture of a final volume of 10 μL contained 2 μL of cDNA, 0.5 μm of each primer, and 2 μL of LightCycler-FastStart DNA Plus Master SYBR Green I mixture (Roche). The following primer sets were used: AtNRT3.1 (forward: 5′-GACCTGCCCACACAAGATCA; reverse: 5′-TGGAGGCAATATCTAGGGACGC); AtNRT3.2 (forward: 5′-CATGAGATTGTGTCCAAGGCATA; reverse: 5′-TATGTCTAGCCCCACGTGATGA); AtNRT1.1 (forward: 5′-AAAGCTGCCACACACTGAAC; reverse: 5′-ATTGTGCGACTGATAATGTCGT); AtNRT2.1 (forward: 5′-CCACAGATCCAGTGAAAGG; reverse: 5′-CATTGTTGGGTGTGTTCTCA); and Clathrin-At4g24550 (internal control gene; forward: 5′-ATACGCGCTGAGTTCCC; reverse: 5′-CTGACTGGCCCTGCTT). Quantitative data analysis was performed with the LightCycler software 4.0 (Roche).
Rescue of the Atnrt3.1 Mutants
To rescue the Atnrt3.1 mutants the AtNRT3.1 cDNA driven by the CaMV 35S promoter was transformed into the mutant plants. First, the cDNA AtNRT3.1 was amplified by RT-PCR from wild-type (Ws) root total RNA using a pair of primers (forward: 5′-AAGGATCCATGGCGATCCAGAAGA; reverse: 5′-TCCCGGGTAAACGACTCATTTGCTTTGCT), introducing BamHI and SmaI sites at 5′ and 3′ ends of the cDNA, respectively. The PCR product was cloned into pGEM-Teasy (Promega) and sequenced to check its integrity. The NRT3.1 BamHI/SmaI fragment was cloned into the BamHI/SmaI sites of two binary vectors, the pGreen0229 (Hellens et al., 2000) in which the CaMV 35S-cassette was introduced, and the pBIN-JIT (Kwak et al., 2001), designated as pGreen-NRT3.1 and pBIN-JIT-NRT3.1, respectively. The pBIN-JIT-NRT3.1 was then partially digested with KpnI and HindIII to produce an approximately 2.5-kb fragment of the 35S promoter-NRT3.1 terminator cassette. The DNA cassette was cloned into the pCAMBIA1303 binary vector (CAMBIA) at the KpnI/HindIII sites (designated as pCAMBIA-NRT3.1). The pGreen-NRT3.1 and pCAMBIA-NRT3.1 were transformed into Atnrt3.1-1 and Atnrt3.1-2 mutant lines, respectively, by the floral-dip procedure (Clough and Bent, 1998) with Agrobacterium tumefaciens strain C58. Transformants were selected either in pots by spraying with 300 μm glufosinate ammonium for the pGreen-NRT3.1, or on Murashige and Skoog plates containing 27 μg/mL hygromycin (A.G. Scientific) for the pCAMBIA-NRT3.1. More than six transgenic lines that showed 3:1 ratio to the selection markers in the T2 generation were characterized, and two representative lines for each vector were presented in this paper.
Acknowledgments
We thank the Tri-University Meson Facility at the University of British Columbia for the provision of 13N, and Mary Galli for providing T-DNA lines.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (to A.D.M.G) and by the National Institutes of Health (grant no. GM40672 to N.M.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Anthony D.M. Glass (aglass@interchange.ubc.ca).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074385.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Cataldo DA, Haroon M, Schrader LE, Youngs VL (1975) Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal 6: 71–80 [Google Scholar]
- Cerezo M, Tillard P, Filleur S, Munos S, Daniel-Vedele F, Gojon A (2001) Major alterations of the regulation of root NO3− uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3: 389–395 [Google Scholar]
- Filleur S, Daniel-Vedele F (1999) Expression analysis of a high-affinity nitrate transporter isolated from Arabidopsis thaliana by differential display. Planta 207: 461–469 [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F (2001) An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett 489: 220–224 [DOI] [PubMed] [Google Scholar]
- Forde BG (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Ingham DJ, Beer S, Money S, Hansen G (2001) Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques 31: 132–140 [DOI] [PubMed] [Google Scholar]
- Kaiser BN, Rawat SR, Siddiqi MY, Masle J, Glass ADM (2002) Functional analysis of an Arabidopsis T-DNA “knockout” of the high-affinity NH4+ transporter AtAMT1;1. Plant Physiol 130: 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR (1996) Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA 93: 8145–8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Murata Y, Baizabal-Aguirre VM, Merrill J, Wang M, Kemper A, Hawke SD, Tallman G, Schroeder JI (2001) Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in Arabidopsis. Plant Physiol 127: 473–485 [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazoa P, Vidmar JJ, Tranbarger TJ, Mouline K, Damiani I, Tillard P, Zhuo DG, Glass ADM, Touraine B (2003) Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Mol Biol 52: 689–703 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Brunak S, von Heijne G (1999) Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng 12: 3–9 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Vidmar JJ, Glass ADM (2003) Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol 44: 304–317 [DOI] [PubMed] [Google Scholar]
- Quesada A, Galvan A, Fernandez E (1994) Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J 5: 407–419 [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi MY, Glass ADM, Ruth TJ, Fernando M (1989) Studies of the regulation of nitrate influx by barley seedlings using 13NO3−. Plant Physiol 90: 806–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Zhou JJ, Li ZS, Miller AJ (2005) A two-component high-affinity nitrate uptake system in barley. Plant J 41: 442–450 [DOI] [PubMed] [Google Scholar]
- Trueman LJ, Richardson A, Forde BG (1996) Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene 175: 223–231 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Unkles SE, Hawker KL, Grieve C, Campbell EI, Montague P, Kinghorn JR (1991) crnA encodes a nitrate transporter in Aspergillus nidulans. Proc Natl Acad Sci USA 88: 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unkles SE, Zhou D, Siddiqi MY, Kinghorn JR, Glass ADM (2001) Apparent genetic redundancy facilitates ecological plasticity for nitrate transport. EMBO J 20: 6246–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidmar JJ, Zhuo D, Siddiqi MY, Schjoerring JK, Touraine B, Glass ADM (2000) Regulation of high-affinity nitrate transporter genes and high-affinity nitrate influx by nitrogen pools in roots of barley. Plant Physiol 123: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford NM (1998) The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc Natl Acad Sci USA 95: 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, O'Shea EK (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159: 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, Fernandez E, Galvan A, Miller AJ (2000) A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett 466: 225–227 [DOI] [PubMed] [Google Scholar]
- Zhuo DG, Okamoto M, Vidmar JJ, Glass ADM (1999) Regulation of a putative high-affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana. Plant J 17: 563–568 [DOI] [PubMed] [Google Scholar]