Abstract
The circadian system of Arabidopsis (Arabidopsis thaliana) includes feedback loops of gene regulation that generate 24-h oscillations. Components of these loops remain to be identified; none of the known components is completely understood, including ZEITLUPE (ZTL), a gene implicated in regulated protein degradation. ztl mutations affect both circadian and developmental responses to red light, possibly through ZTL interaction with PHYTOCHROME B (PHYB). We conducted a large-scale genetic screen that identified additional clock-affecting loci. Other mutants recovered include 11 new ztl alleles encompassing mutations in each of the ZTL protein domains. Each mutation lengthened the circadian period, even in dark-grown seedlings entrained to temperature cycles. A mutation of the LIGHT, OXYGEN, VOLTAGE (LOV)/Period-ARNT-Sim (PAS) domain was unique in retaining wild-type responses to red light both for the circadian period and for control of hypocotyl elongation. This uncoupling of ztl phenotypes indicates that interactions of ZTL protein with multiple factors must be disrupted to generate the full ztl mutant phenotype. Protein interaction assays showed that the ztl mutant phenotypes were not fully explained by impaired interactions with previously described partner proteins Arabidopsis S-phase kinase-related protein 1, TIMING OF CAB EXPRESSION 1, and PHYB. Interaction with PHYB was unaffected by mutation of any ZTL domain. Mutation of the kelch repeat domain affected protein binding at both the LOV/PAS and the F-box domains, indicating that interaction among ZTL domains leads to the strong phenotypes of kelch mutations. Forward genetics continues to provide insight regarding both known and newly discovered components of the circadian system, although current approaches have saturated mutations at some loci.
The circadian systems that drive 24-h biological rhythms in many organisms evolved as an adaptation to the Earth's rotation and its attendant changes in light and temperature conditions. The core of a circadian system can be described as an oscillator; oscillator components (clock genes/proteins) rhythmically regulate their own expression/activity (and that of each other) through one or more negative feedback mechanisms at the transcriptional/translational level, generating a self-sustained oscillation (Young and Kay, 2001). Additional clock-associated factors, which do not carry temporal information and which may not be rhythmic, are required to set the period length of this oscillation close to 24 h and to adjust other rhythmic properties. The oscillator regulates the expression of overt rhythms through output pathways. The phase of the oscillator (subjective time) is set to the objective time of the environment by periodic environmental signals (light and temperature) transduced by input pathways. Light signaling to the plant clock is mediated by several photoreceptors of the phytochrome and cryptochrome families, which may themselves be clock regulated (for review, see Fankhauser and Staiger, 2002; Millar, 2003). Increasing light fluence rates shortens the free-running period length in Arabidopsis (Arabidopsis thaliana; Millar et al., 1995b), as in many diurnal animals (Aschoff, 1979).
According to current models (for review, see Hayama and Coupland, 2003; Salome and McClung, 2004, and refs. therein), the central oscillator in Arabidopsis involves the mutual regulation of at least three genes: CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are morning-expressed genes that repress transcription of TIMING OF CAB EXPRESSION 1 (TOC1). TOC1 is proposed to form a second, interlocking loop with GIGANTEA (GI; Locke et al., 2005), which maintains damping, short-period rhythms in the lhy/cca1 double mutant (Alabadi et al., 2002; Mizoguchi et al., 2002; Locke et al., 2005). The evening-expressed genes TOC1, GI, EARLY FLOWERING 3 (ELF3), and EARLY FLOWERING 4 (ELF4) are each required for expression of CCA1/LHY to reach normal levels on the following morning (Hicks et al., 1996; Fowler et al., 1999; Alabadi et al., 2001; Doyle et al., 2002), although the mechanism of this gene activation is unclear.
The majority of known components of the Arabidopsis circadian system have been identified by forward genetics (for review, see Southern and Millar, 2005, and refs. therein), several from limited screens for mutants with altered circadian timing under constant light, and others from flowering time screens. Each gene (including TOC1 and ZEITLUPE [ZTL]) was represented by one or, at most, two alleles. Recently, a large-scale screen was reported for mutants that altered rhythmicity of several clock-regulated luciferase (LUC) reporter genes under constant light (Onai et al., 2004); one of the mutated genes was identified as an allele of a putative DNA-binding protein gene, PHYTOCLOCK1 (Onai and Ishiura, 2005). In contrast, a LUC screen preceded by a screen for long-hypocotyl seedlings and followed by rapid array-based mapping identified several alleles of TOC1, ELF3, ELF4, and the same DNA-binding protein gene, here termed LUX ARRHYTHMO (LUX; Hazen et al., 2005a, 2005b).
ZTL was the first component of the plant circadian system to be implicated in regulated proteolysis. ZTL, LOV KELCH PROTEIN 2 (LKP2), and FLAVIN BINDING, KELCH REPEAT, F-BOX 1 (FKF1) form a protein family with a unique signature of three protein domains: an N-terminal Period-ARNT-Sim (PAS)-like LIGHT, OXYGEN, VOLTAGE (LOV) domain, a central F-box motif, and a C-terminal domain of six kelch repeats that are predicted to form a β-propeller (Kiyosue and Wada, 2000; Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001). F-box proteins are important target-specifier components of the Skp1/Cullin/F-box (SCF) type of E3 ubiquitin ligases (Vierstra, 2003). The F-box motif interacts with the Skp1 protein, while the C terminus is often responsible for binding the target protein. ZTL is assembled into an SCFZTL complex in vivo together with a SKP1-like protein (ASK1), AtCUL1, and AtRBX1, which are core components of characterized SCF complexes in Arabidopsis (Han et al., 2004). Complex formation requires the presence of a functional F-box domain of ZTL. Transient reduction in AtRBX1 levels phenocopies the long-period phenotype of ztl mutants, strongly supporting the role of an SCFZTL complex in the regulation of circadian rhythms (Han et al., 2004). The proposed function of ZTL is to target the TOC1 protein for degradation (Más et al., 2003). Surprisingly, TOC1 can interact with ZTL through the LOV/PAS domain and not the kelch repeats (Más et al., 2003; Yasuhara et al., 2004). The ztl-1 mutation in the kelch domain nonetheless abolished the ZTL-TOC1 interaction, suggesting that the β-propeller supports protein interactions at the LOV/PAS domain.
The unusual LOV/PAS domain of ZTL suggested that ZTL function could be light regulated because the equivalent domain in other proteins (notably FKF1; Imaizumi et al., 2003) can bind a flavin chromophore that is photoactive in blue light. Subsequent results have significantly modified this early proposal, revealing a complex picture. Circadian rhythms in ztl mutants have a long-period phenotype that is light dependent: The circadian period is longer under low rather than high fluence rates of constant red or blue light (Somers et al., 2000). This is broadly consistent with the more rapid degradation of TOC1 protein in darkness than in light (Más et al., 2003). The putative ZTL chromophore was not expected to absorb red light significantly, but a physical interaction was demonstrated between ZTL and the C-terminal fragments of phytochrome B (PHYB) and cryptochrome 1 (CRY1) photoreceptor proteins (Jarillo et al., 2001) such that these photoreceptors might be involved in ZTL function. The known ztl mutants also had short hypocotyls because the red light-controlled inhibition of hypocotyl elongation was hyperresponsive in the mutants; for this phenotype, ztl mutations did not affect blue-light responsiveness (Somers et al., 2000, 2004). Our data discriminate among these overlapping phenotypes, all of which are affected by ZTL function.
We report a large-scale screen to identify circadian clock mutants using a LUC reporter gene under constant darkness. The 104 mutants recovered identify several new clock-affecting genes. In addition, 11 ztl alleles include amino acid substitutions in all three domains of the ZTL protein. Most of the mutants share similar phenotypes. Only mutation of the LOV/PAS domain uncoupled the function of ZTL in red-light responses from its function in the circadian system. Protein interaction assays show that altered interactions with known partner proteins do not explain the phenotype of the ztl mutations, although the strong effects of seemingly disparate mutations are likely due to contributions of multiple domains to each protein interaction.
RESULTS
Isolation of Novel Circadian Mutants
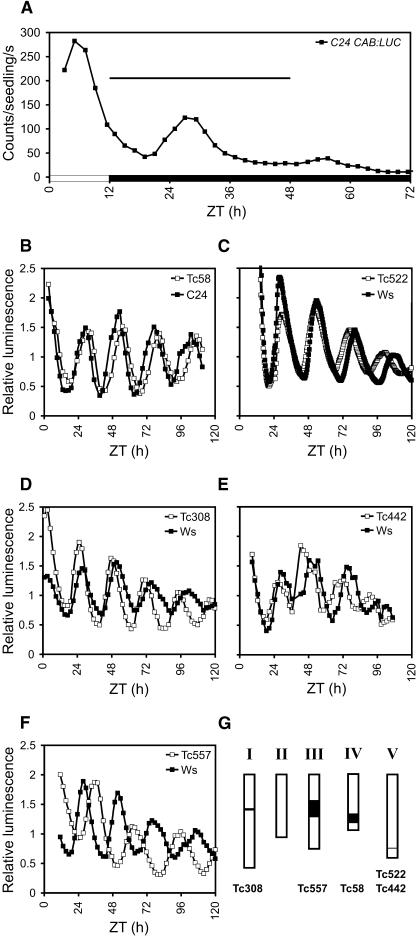
Seeds carrying the CHLOROPHYLL a/b-BINDING PROTEIN (CAB2; also known as LHCB1.1):LUC reporter in the C24 accession (Millar et al., 1995a) or the CAB2:LUC+ reporter in the Wassilewskija (Ws) accession (Hall et al., 2001) were treated with ethyl methane sulfonate (EMS). Seedlings of the M2 generation were tested in two laboratories for LUC activity over 36 h in constant darkness (DD), following entrainment under 12-h-light/12-h-dark photoperiods (LD 12:12). This interval covers the first peak of CAB2:LUC(+) expression in darkness, which occurs at Zeitgeber time (ZT) 26 to 27 in wild-type plants (Fig. 1A). ZT0 is defined as the time of the last dark-light transition; ZT26 is 2 h after the first predicted dawn in darkness. Seedlings with a variety of altered luminescence patterns were selected. Period could not be assessed because the initial screen covered less than two circadian cycles and because the expression of CAB2:LUC damps to a low level in darkness (Fig. 1A). Table I shows that the combined screen identified 104 heritable circadian mutants of several types, with a bias in the Ws screen toward late-phase mutants (64% of the total mutants recovered); few arrhythmic mutants were identified. A higher rate of mutant recovery from the Ws population was due in part to the stronger luminescence of the LUC+ reporter, which allowed more accurate mutant scoring. The Ws population also showed a higher level of mutagenesis as judged by M1 seed mortality, adult plant fertility, and the frequency of visible phenotypes in the M2 generation (A. Hall, unpublished data). Mutant phenotypes were confirmed in the M3 generation in DD and also tested under constant red light to distinguish between period length and phase mutants.
Figure 1.
Identification of additional clock-associated genes and toc1 alleles. A, Luminescence rhythm of transgenic Arabidopsis ecotype C24 carrying the CAB:LUC transgene over one light/dark cycle followed by constant darkness. Luminescence was monitored for the interval ZT12-ZT48 during the genetic screen (horizontal line), capturing only the first peak in darkness. White box, time axis, light interval; black box, darkness. ZT0 is defined as the time of the last light-dark transition. Tc58 (B), Tc308 (D), and Tc557 (F) were identified as mutants displaying late-phase, short- or long-period-length phenotypes, respectively. The short-period mutants Tc442 (E) and Tc522 (C) represent toc1 alleles, named toc1-8 and toc1-9, respectively. The plots show the rhythmic expression of the CAB2:LUC (Tc58, C24 background) or CAB2:LUC+ (the rest of the mutants, Ws background) markers in the mutants and corresponding wild types in constant red light at approximately 5 μmol m−2 s−1 intensity. G, Approximate position and distribution of the mutations among the five chromosomes of the Arabidopsis genome.
Table I.
Summary of the mutant screen
Phenotypes of CAB:LUC expression in constant darkness were classified as: early, Early phase; late, late phase; arrhyt, arrhythmic; low amp, low amplitude; high level, high level of luminescence; and form, altered rhythmic waveform.
| Accession
|
M2 Plants Tested
|
M2 Plants Selected
|
M3 Lines Retained
|
Circadian Phenotype in 36 h DD
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Early | Late | Arrhyt | Low Amp | High Level | Form | ||||
| C24 | 40,000 | 574 | 25 | 11 | 13 | 1 | 0 | 0 | 0 |
| Ws | 5,800 | 545 | 79 | 16 | 54 | 5 | 2 | 1 | 1 |
Mutants were placed on the genetic map. Greater than five mutant lines were identified that did not map close to clock-associated genes, including lines with a long or short period in constant red light and one line with altered phase but no significant period change; three of these lines are shown in Figure 1. To identify mutants that were allelic with known clock-associated genes, the TOC1, ELF3, GI, or ZTL gene was amplified and sequenced from mutant lines that showed both tight genetic linkage to the relevant locus and the phenotype expected for toc1, elf3, gi, or ztl mutants. This identified two new toc1 alleles with predicted mutations in the pseudoresponse regulator domain (Supplemental Fig. 1; Supplemental Table I) and the short-period phenotype characteristic of previously described toc1 alleles (Fig. 1). One elf3 and two gi alleles will be described elsewhere (L. Kozma-Bognár and A. Hall, unpublished data). Here we characterize 11 new alleles of ZTL (Fig. 2), including one with a novel phenotype in red light.
Figure 2.
Predicted mutations in the ZTL protein. ZTL possesses three distinct domains: a LOV/PAS domain, an F-box domain, and a C-terminal domain consisting of six kelch repeats. Amino acid sequences comprising the six kelch repeats are aligned. The newly identified alleles of ztl are named ztl-21 to ztl-31; previously identified ztl-1 and ztl-2 mutations are also indicated. Genetic backgrounds and allele sequences are listed in Supplemental Table I.
New Mutations within the ZTL Gene
The positions of the 11 newly identified and the two previously published ztl mutations within the ZTL protein sequence are shown in Figure 2 and listed in Supplemental Table I. All are caused by G-to-A transitions in the nucleotide sequence, as expected of EMS mutations. In ztl-21, a G-to-D amino acid substitution is predicted in the second half of the LOV/PAS domain in a residue that is conserved within the ZTL/FKF/LKP2 protein family in Arabidopsis. ztl-22 causes an E-to-K change that lies at a conserved position in the F-box domain of ZTL and other F-box proteins from plants, mammals, and fungi (Patton et al., 1998). Nine further mutations were identified within the kelch repeats, six of which cause substitutions of equivalent amino acids within different repeats (ztl-23 and ztl-30; ztl-25, ztl-27, and ztl-28; ztl-26 and the previously identified alleles ztl-1 and ztl-2). Two nonsense mutations (ztl-29 and ztl-31) were identified toward the C-terminal end of the kelch repeat domain.
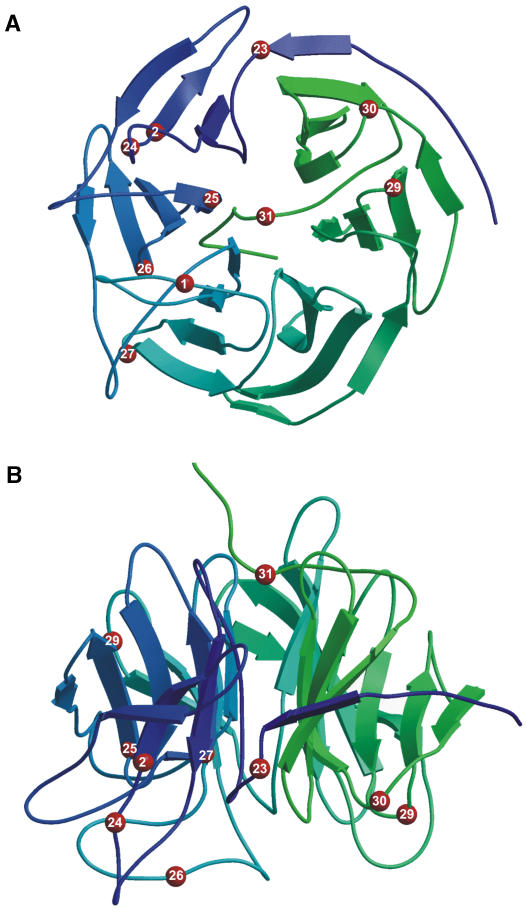
Kelch repeat domains form a β-propeller structure to which each full repeat contributes one propeller blade; a zip-lock blade comprising the partial first and last repeats closes the propeller (Li et al., 2004). The positions of the mutated residues in the ZTL kelch repeats were mapped onto the structure of the six-bladed kelch domain of diisopropylfluorophosphatase (Fig. 3). The mutations are all predicted to fall in loops on the external faces of the propeller, not within the β-strands in the depth of the blades. Six of the seven kelch residues with missense mutations are clustered on a single face of the propeller. In ztl-31, only the extreme C-terminal loop is lost from the predicted product, whereas in ztl-29, the C-terminal blade is lost. Mutations in all three predicted domains of ZTL protein were recovered, which provided an opportunity to obtain more detailed information on the function of the different domains.
Figure 3.
Location of mutated residues in the ZTL kelch repeats. Mutated residues (red spheres) were mapped onto the six-bladed kelch domain structure of diisopropylfluorophosphatase (structure 1e1a). Numbers correspond to ztl alleles where the particular position is mutated (ztl-27 and ztl-28 affect the same position, G452). Ribbon diagrams of the β-propeller are color ramped from blue to green from the N to the C terminus. A, Face view (bottom surface as shown in B). B, Side view (from the top as shown in A). The pictures were drawn in MolScript (Kraulis, 1991; Esnouf, 1997) and rendered in Raster3D (Merritt and Murphy, 1994).
Quantitative Differences in the Circadian Phenotypes of ztl Mutants
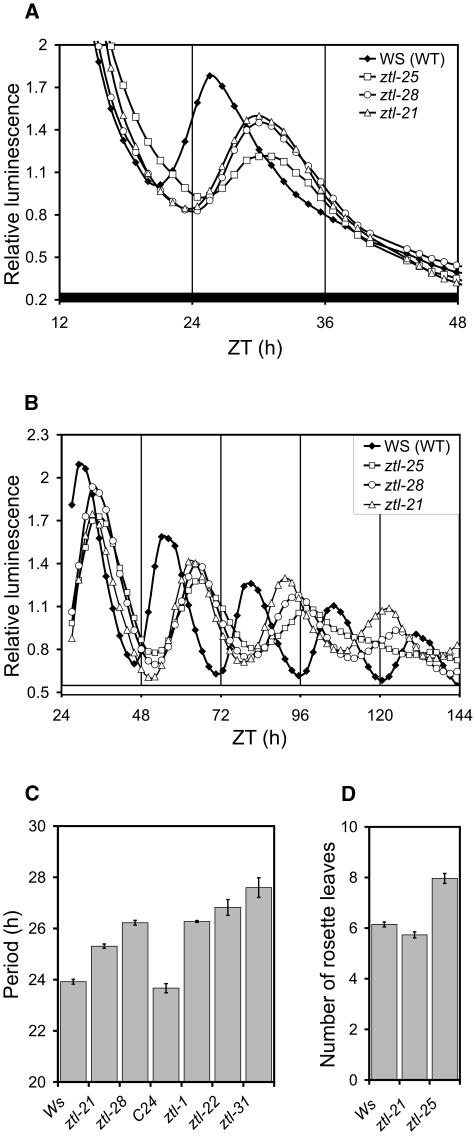
All of the ztl mutants showed a late-phase phenotype of CAB2:LUC(+) expression under the screening conditions (Fig. 4A; data not shown) and all had a long period of CAB2:LUC(+) expression under constant red or blue or red and blue light (Supplemental Table II). We therefore focused on a subset of alleles that affect each ZTL domain and each genetic background. The rhythmic expression of well-characterized promoter:LUC+ fusions (CAB2, CCR2, or CCA1) was examined in the ztl mutants under constant white, red, or blue light and during dark adaptation. All of the ztl alleles displayed long-period phenotypes in all conditions; in general, ztl-21 plants showed a weaker phenotype compared to the other alleles, whereas ztl-31 plants had the longest periods and also lower rhythmic amplitude (Fig. 4B; Supplemental Fig. 2; Supplemental Table II; data not shown). This quantitative difference in phenotype was replicated when we tested a circadian output that is distinct from gene expression, the leaf movement rhythm (Fig. 4C).
Figure 4.
ztl mutations affect several output rhythms under various light conditions. CAB2:LUC+ rhythm of ztl mutants in DD (A) and in constant red light of approximately 5 μmol m−2 s−1 light intensity (B); leaf movement rhythms in ztl mutant and wild-type seedlings under continuous white light (C). Flowering time of the new ztl alleles was determined under long-day conditions (16-h light/8-h dark) and expressed as the number of rosette leaves at the time of bolting (D). Error bars represent se values in C and D.
To assess the impact of the ztl mutations under light/dark cycles, the diurnal expression pattern of CCA1:LUC+ and CCR2:LUC+ was tested under short-day conditions (Supplemental Fig. 2, A and C). CCA1:LUC+ activity in wild-type seedlings started to increase in anticipation of lights on, when acute light induction of CCA1 expression sharply increased activity to a peak 1 to 2 h after lights on. The ztl mutants showed evidence of a late circadian phase, with little or no increase in CCA1 expression before dawn. Together with a reduced acute response to light, this led to peak expression at ZT4-6 compared to ZT1-2 in the wild type (Supplemental Fig. 2A). The reduced light response is consistent with CCA1 RNA data for other ztl alleles (Somers et al., 2004). CCR2:LUC+ activity also peaked 2 to 4 h later in the night in ztl mutants compared to wild type (Supplemental Fig. 2C).
ZTL Protein Is Expressed in ztl Mutants
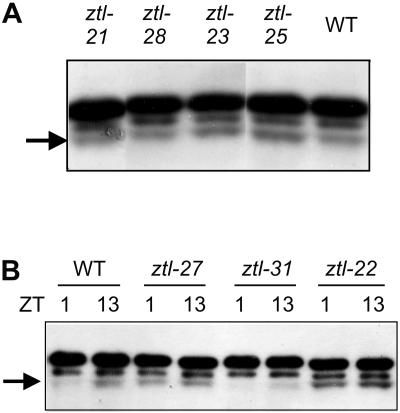
ZTL protein levels were measured in the mutant plants by western analysis. Data obtained from representative mutant lines carrying single point mutations within each functionally important domain are shown in Figure 5. The results show that all mutants tested expressed at least the wild-type level of the protein, except ztl-31 (Fig. 5, A and B). ZTL mRNA levels also showed little or no effect of the mutations (Supplemental Fig. 4). ztl-31 was expected to express a protein truncated by 2.6% (16 amino acids; Fig. 2) and, consistent with this, the ZTL antibody recognized a protein with slightly reduced molecular mass in extracts from ztl-31 (Fig. 5B; data not shown). ZTL levels overall were apparently reduced in this line, although ZTL mRNA levels were unaffected or slightly increased (Supplemental Fig. 3), suggesting that the truncated protein was unstable, although an effect on the polyclonal epitope cannot be ruled out.
Figure 5.
ZTL protein expression levels in the ztl mutant plants. Immunoblots showing ZTL protein levels in extracts from wild-type (WT) and ztl mutant plants growing under LD 12:12. Samples were harvested at 13 h after dawn (ZT13; A) or at ZT1 and ZT13 (B), as indicated. Minimal and maximal ZTL accumulation normally occurs at ZT1 and ZT13, respectively, in these conditions. Arrows show ZTL position.
ZTL protein levels normally oscillate diurnally, with a maximum at ZT13 and a minimum at ZT1 under LD 12:12 cycles. This diurnal oscillation of the protein level is controlled posttranslationally through circadian phase-specific degradation (Kim et al., 2003). Although most of the ztl mutant lines showed wild-type oscillations (data not shown), three lines were aberrant. ztl-22, which carries a mutation in the F-box domain, showed strongly elevated levels of ZTL at ZT1 and moderately elevated levels at ZT13 relative to wild type (Fig. 5B). A G-to-D substitution in the fourth kelch repeat (ztl-27) also enhanced ZTL protein accumulation at ZT1 (Fig. 5B). ztl-31 resulted in much lower levels of mutant protein at both times (Fig. 5B). In these three mutants, particularly in the stronger allele ztl-31, phenotypic differences might relate to variation in ZTL protein levels; in the other lines, any differences in the mutant phenotypes are likely to reflect altered function of the mutant proteins rather than differences in their accumulation.
Novel Light Dependence of the LOV/PAS Mutant ztl-21
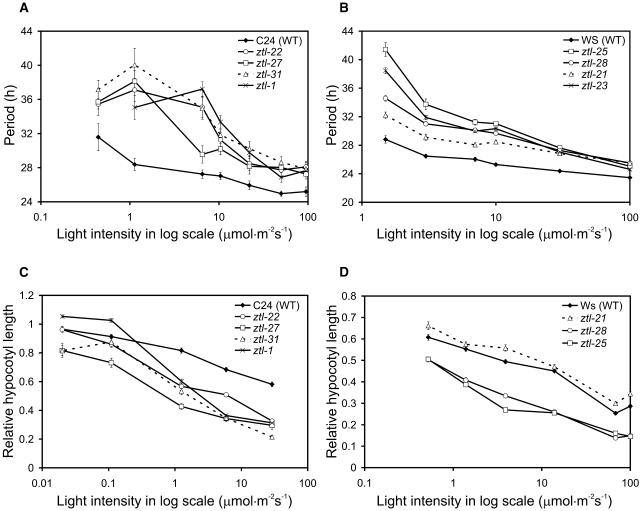
A major aspect of ZTL function is in mediating light responsiveness. The free-running period of circadian rhythms in wild-type Arabidopsis decreases with increasing fluence rates of light. Consequently, fluence rate response curves (FRCs; constructed by plotting period values as a function of the log of fluence rate) for Arabidopsis rhythms show a negative slope. The slope is more negative for ztl-1, ztl-2, and the null ztl-3 mutants than for the wild type because their long-period phenotype is exaggerated at low fluence rates of either red or blue light (Somers et al., 2000, 2004). In contrast, the inhibition of hypocotyl elongation by red light was specifically enhanced in ztl mutants, with little effect on the response to blue light (Somers et al., 2000, 2004).
To characterize the effect of the mutant ZTL proteins on light signaling, FRCs were measured for CAB2:LUC(+) rhythms and hypocotyl elongation under red and blue light (Fig. 6; Supplemental Fig. 4). Hypocotyl length in dark-grown plants was the same in all genotypes, indicating that the differences observed were light dependent (data not shown). The new alleles were similar to ztl-1 for both circadian and hypocotyl phenotypes under blue light (Supplemental Fig. 4). ztl-31 again showed the greatest lengthening of the circadian period, with slightly weaker effects in ztl-21. Under red light, however, ztl-21 showed qualitatively different behavior from the other alleles (Fig. 6).
Figure 6.
Effect of ztl mutations on the free-running period length and hypocotyl elongation under different fluence rates of red light. FRCs were determined from the period of CAB2:LUC (in C24 background) or CAB2:LUC+ (in Ws background) reporter gene activity under continuous red light (A and B). Plants were entrained in LD 12:12 for 7 d prior to free run in constant light. Hypocotyl length of 4-d-old seedlings grown in continuous red light (C and D) of indicated fluence rates. Data were normalized to the hypocotyl length of the corresponding dark-grown controls. Error bars represent se values.
At high fluence rates of red light, ztl-21 and the other ztl mutants had a circadian period about 3.5 h longer than wild-type plants. Only ztl-21 maintained this 3.5-h period lengthening across the whole range of red light intensity (Fig. 6B; see also Fig. 4B), giving a FRC with the same gradient as the wild type. The period of the other alleles increased to 8 to 12 h longer than wild type at the lowest fluence rates, as previously reported. ztl-21 seedlings also showed little or no defect in hypocotyl elongation at any fluence rate of red light (Fig. 6D). Under long photoperiods of white light, ztl-21 plants flowered with the wild type in contrast to the modest delay in flowering time observed for kelch mutations (Fig. 4D; Somers et al., 2000; Kim et al., 2005). Thus, the LOV/PAS mutant ztl-21 affected the circadian period, but did not affect responses to red light either by shortening the circadian period or by inhibiting hypocotyl elongation or photoperiod sensing.
We and others have demonstrated that ZTL affects the circadian period of light-grown plants transferred to constant darkness (dark-adapting plants; Supplemental Fig. 2B). These plants initially have a light-adapted complement of photoreceptors, some of which are thought to remain active for hours to days in darkness. To test the dependence of ZTL function on photoreceptor signaling more stringently, circadian rhythms of CCR2:LUC+ activity were tested in dark-grown seedlings compared to dark-adapting seedlings (Table II; Supplemental Fig. 5). ztl mutant and wild-type seedlings were grown for 3 d in darkness under entrainment from 12 h 26°C/12 h 22°C temperature cycles, then transferred to constant 22°C for period assays also in darkness. ztl mutants showed an identical, long mean period under these conditions as in dark adaptation, indicating that ZTL regulates the circadian system without ongoing photoreceptor signaling (Table II).
Table II.
Free-running period estimates for dark- or light-grown ztl and wild-type seedlings
Seedlings were grown in constant darkness under temperature cycles (12-h 26°C/12-h 22°C) for 3 d or at constant temperature (22°C) under 12-h white light/12-h dark cycles for 6 d. All plants were transferred to constant temperature (22°C) and darkness at ZT12 and rhythmic CCR2:LUC+ activity was recorded during the following 6 d.
| Genotype
|
Temperature-Entrained Etiolated Plants
|
Light-/Dark-Entrained Plants
|
||
|---|---|---|---|---|
| Period (h) | ±sem | Period (h) | ±sem | |
| C24 (wild type) | 27.75 | 0.20 | 27.05 | 0.07 |
| ztl-22 | 33.20 | 0.51 | 33.56 | 0.10 |
| ztl-27 | 36.49 | 0.80 | 36.43 | 0.36 |
Protein Interactions in ztl Mutants
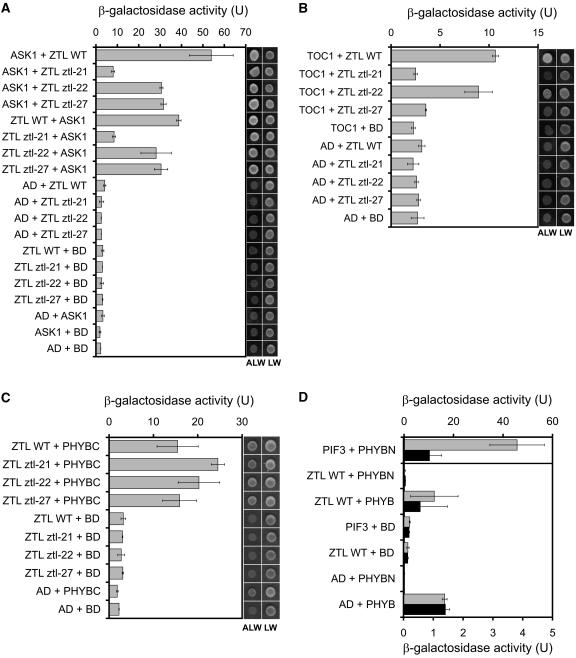
The domain structure of the ZTL protein family, together with published data, suggests that ZTL function might be understood at the molecular level in terms of its physical interactions with degradation targets (including TOC1), the SCF complex (through ASK1), and the phytochrome and cryptochrome photoreceptors. To test this notion, representative mutations located in the three different domains of ZTL (LOV/PAS, ztl-21; F-box, ztl-22; kelch repeats, ztl-27) were introduced into the full-length ZTL cDNA. The corresponding mutant proteins were expressed in yeast (Saccharomyces cerevisiae) and tested for interaction with ASK1, TOC1, and the N-terminal domain (PHYBN; amino acids 1–621), the C-terminal domain (PHYBC; amino acids 645–1,272), or full-length phytochrome B. Protein gel-blot analysis confirmed that the mutated protein fusions were expressed in yeast at similar levels to wild-type protein fusions (data not shown). The ZTL-ASK1 interaction was most severely affected by the mutation in the PAS domain and also reduced by the F-box and kelch mutations (Fig. 7A). The TOC1-ZTL interaction was abolished by the PAS and kelch mutations, as shown by the absence of yeast growth under selective conditions, whereas the F-box mutation had no significant effect (Fig. 7B).
Figure 7.
Mutations in ZTL affect protein-protein interactions. Interaction of full-length wild-type and mutant ZTL proteins with ASK1 (A), with TOC1 (B), with the C-terminal fragment of the Arabidopsis PHYB (PHYBC; C), and with the N-terminal fragment and the full-length PHYB (PHYBN and PHYB, respectively; D). The relative efficiency of the interactions was quantified by measuring β-galactosidase activity. Error bars represent se values. D, Gray bars represent enzyme activity values measured from red light-pulsed (35 μmol m−2 s−1 for 5 min) cultures and black bars represent values from cultures treated with a red light pulse followed by far-red irradiation (25 μmol m−2 s−1 for 5 min). Also, in D, values from the PIF3 + PHYBN interaction are plotted on the axis shown at the top, but values from all the other cultures are plotted on the axis at the bottom. AD and BD correspond to controls transformed with empty vectors expressing the activation and DNA-binding domains of GAL4, respectively. Growth on selective (ALW) and nonselective (LW) media was recorded. Growth on selective and nonselective media for D is shown in Supplemental Figure 6.
The interaction between PHYBC and full-length ZTL protein was not affected by the mutations (Fig. 7C). However, reporter activity was significantly increased when PHYBC, fused to the DNA-binding domain of GAL4 (bait), was coexpressed with ZTL fused to the activation domain of GAL4 (prey). Similar results were reported by Jarillo et al. (2001), who also verified the ZTL-PHYBC interaction with in vitro binding studies. We observed no interaction between PHYBC and ZTL when these proteins were expressed in the inverted configuration (PHYBC as prey, ZTL as bait; data not shown). ZTL failed to interact with PHYBN in the presence or absence of the phycocyanobilin chromophore, regardless of whether reconstituted PHYBN was in the Pr or Pfr conformation (Fig. 7D; Supplemental Fig. 6). In contrast, PHYBN interacted strongly with PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) in a Pfr-dependent manner, as reported previously (Shimizu-Sato et al., 2002). Full-length PHYB showed some transactivation of the β-galactosidase reporter gene when expressed alone as bait (Fig. 7D), but expression of ZTL as prey did not enhance this expression, regardless of the chromophore state (Fig. 7D; Supplemental Fig. 6). In the absence of interaction with full-length PHYB and of any effect of ztl mutations, the significance of the ZTL-PHYBC interaction is unclear.
DISCUSSION
To identify components of the Arabidopsis circadian system, we screened approximately 46,000 M2 seedlings from two EMS-mutagenized populations, recovering 104 mutants with altered temporal patterns of CAB:LUC reporter gene expression (Table I). The short duration of the screen (36 h) allowed a relatively high throughput of up to 4,000 seedlings per week per luminescence counter. Forward genetic screens of this type might seem to be approaching saturation, given that we identified alleles of TOC1, ELF3, GI, and ZTL, and a similar approach using prescreening for mutants with long hypocotyls identified alleles of ELF3, ELF4, and TOC1 (Hazen et al., 2005a). The success of this direct screen for altered patterns of dynamic gene expression in Arabidopsis is a testament to the power of LUC reporter methods. The screens are not strictly saturated because both screens also identified mutations in genes not previously described as clock affecting. Characterization of these genes is likely to give new insights into the mechanisms of the Arabidopsis circadian system. The screens have not identified new cca1 or lhy loss-of-function alleles (T. Schultz, personal communication), for example, which had clear phenotypes in our conditions (data not shown). However, the proportion of mutants in previously identified genes is high, suggesting that future screens might retain the LUC reporter approach, but monitor different components of the circadian system (as initiated in Onai et al., 2004), perhaps under different environmental conditions.
Characterization of the New ztl Alleles
The LOV/PAS, F-box, and kelch repeat domains of ZTL and their associated biochemical functions were first predicted on the basis of protein sequence homology. These predictions have recently been tested by analysis of the physical interactions between ZTL and various protein partners (Más et al., 2003; Han et al., 2004; Yasuhara et al., 2004). The set of new ztl alleles with mutations in each domain provided the first opportunity to determine the phenotypic effects of impaired function of the native LOV/PAS and F-box domains in planta in comparison to the previously isolated kelch mutant (ztl-1 and ztl-2) and null (ztl-3) alleles. Each of the alleles tested showed a long-period phenotype (Fig. 4; Supplemental Fig. 2; Table II; Supplemental Table II), regardless of the lighting conditions (constant red, blue, or white light; darkness) and the particular overt rhythm (CAB2, CCR2, or CCA1 expression; leaf movement). The amplitude of the rhythms was slightly reduced by the mutations, particularly in DD (Supplemental Fig. 2B). ztl-31 (expressing a truncated ZTL protein at very low levels; Fig. 5B) displayed the strongest circadian phenotype and ztl-21 (the LOV/PAS mutant), the weakest. However, these differences were quantitative, indicating that ZTL function was significantly reduced in each of the mutants. Mutant ZTL protein accumulated to significant levels in each line, except ztl-31 (Fig. 5); thus all three protein domains contribute to ZTL function in the Arabidopsis circadian system.
The F-Box Domain and SCF Interaction Mutated in ztl-22
The F-box mutant (ztl-22) alone had constitutively higher ZTL levels relative to wild type, although the period was similar to the strongest allele (ztl-31), which had very low levels of ZTL (Fig. 5). This suggests that ZTLztl-22 protein is functionally inactive and that inactivity stabilized the protein. The ztl-22 mutation had no effect on the TOC1-ZTL interaction (Fig. 7B), indicating that reduced interaction with this degradation target did not contribute to the phenotype. A double amino acid substitution in the F-box domain has previously been shown to eliminate interaction with ASK1 in yeast and abrogate SCFZTL complex formation in vivo; the mutant ZTL protein was also stabilized in vivo (Han et al., 2004). This is consistent with reduced participation in the SCF complex, leading to reduced degradation of ZTLztl-22 protein both in the light and in the dark (Fig. 5). However, the ztl-22 mutation reduced, but did not eliminate, the interaction of full-length ZTL with full-length ASK1 (Fig. 7A). Given its strong phenotype in vivo, it is likely that the ztl-22 mutation disrupts the interaction with Skp1-like proteins other than ASK1 more severely, indicating that other ASK proteins are important in the SCFZTL complex (see also Han et al., 2004).
The Kelch Domain and ZTL Interactions
The predicted clustering of the new ztl mutations on one face of the β-propeller formed by the kelch repeats, including several mutations in equivalent residues of different repeats, strongly suggests that the mutated residues normally contribute to an interaction surface that is important for ZTL function (Fig. 3). One of the C-terminal truncations (ztl-29) eliminates the closing blade of the propeller, also potentially disrupting an interaction surface. No partner protein that interacts specifically with the ZTL kelch domain has been identified. We replicated the PHYB-ZTL interaction (Jarillo et al., 2001), but only in one bait/prey configuration and not with full-length PHYB (Fig. 7, C and D; Supplemental Fig. 6; data not shown). Because the mutations tested had no effect on this interaction, we have no evidence that any ztl phenotype is due to impaired interaction with PHYB.
Our data suggest that the kelch domain is important to support intermolecular interactions at the LOV/PAS domain (as previously proposed by Más et al., 2003) and also at the F-box domain. The kelch mutation ztl-27 eliminated the TOC1-ZTL interaction (Fig. 7B) despite the fact that the LOV domain alone was sufficient to interact with TOC1 in yeast, but the kelch domain alone was not (data not shown). The ztl-27 mutation also reduced the interaction of ZTL with ASK1, although this interaction is mediated primarily by the F-box domain (Fig. 7A). ztl-27 was the only new kelch mutation to affect the accumulation of ZTL protein in LD cycles (Fig. 5B; data not shown). ZTLztl-27 protein levels were increased during the photoperiod (ZT1), with little effect on skotoperiod (ZT13) levels. This effect has not been previously observed in the ztl-1 and ztl-2 kelch mutations (Somers et al., 2004) and is consistent with a stabilization of ZTL protein through an altered interaction with SCFZTL. Thus, some determinants of ZTL stability lie outside the F-box domain: The kelch domain is probably required to maintain the proper structure of the entire ZTL protein, allowing normal interactions with both TOC1 and Skp1-like proteins. This structural role might depend on intramolecular interaction of the other ZTL domains with the surface of the β-propeller.
The LOV/PAS Mutant ztl-21 Uncouples Red Light Signaling from Circadian Function
ZTL controls the circadian period under all lighting conditions, including in seedlings grown in complete darkness (Table II), resulting in a long-period phenotype in ztl mutants. However, the period phenotype of most ztl alleles is light dependent, being more pronounced at low fluence rates of constant red or blue light (Figs. 4 and 6; Supplemental Fig. 4; Somers et al., 2000, 2004). ZTL presumably enhances target degradation under all conditions, although a proportion of its activity is modulated by light (Más et al., 2003). In contrast, the inhibition of hypocotyl elongation in ztl mutants is hyperresponsive to red light, but little different from wild type in blue light (Fig. 6; Supplemental Fig. 4; Somers et al., 2004). Taken together, these results provide circumstantial evidence that the short hypocotyl under red light is not secondary to altered clock function because hypocotyl elongation was unaffected under constant blue light despite the clear long-period phenotype.
Our results demonstrate directly that ZTL function in the control of hypocotyl elongation by red light is distinct from its role in the circadian system because the LOV/PAS mutant (ztl-21) uncouples these phenotypes. ztl-21 plants had a long circadian period, but showed no defect in hypocotyl elongation under red light (Fig. 6D). The circadian clock in ztl-21 showed no defect in sensitivity to red light: The change in period length across the range of red-light fluence rates was the same as in the wild type (Fig. 6B). Flowering time in long days was also unaffected in ztl-21, although the kelch mutation ztl-25 delayed flowering (Fig. 4D), as previously reported for the null allele ztl-3 (Kim et al., 2005) and the kelch mutation ztl-1 in the C24 genetic background (Somers et al., 2000). In blue light, ztl-21 phenotypes were qualitatively similar to the other alleles, but with a slightly weaker effect (Supplemental Fig. 4). Thus ztl-21 links the red light-signaling functions of ZTL in control of the circadian period and hypocotyl elongation, leaving them intact and separating them from a red light-independent (but blue light-modulated) portion of ZTL circadian function. It follows that the role of ZTL in red light signaling does not require an intact LOV/PAS domain, whereas the latter, red light-independent function of ZTL does.
The LOV/PAS domain alone is sufficient to mediate ZTL interaction with its degradation target TOC1 (data not shown; Más et al., 2003) and, as expected, the LOV/PAS mutation in ztl-21 eliminated this interaction (Fig. 7B). The ztl-21 mutation also reduced the ASK1-ZTL interaction (Fig. 7A), suggesting that the LOV/PAS domain can enhance ZTL interaction with the SCF complex. Although the activity of ZTL in targeting TOC1 for degradation is modulated by light (Más et al., 2003), our results indicate that this quantitative change alone does not account for the various phenotypes of ztl mutants under different light conditions. At high fluences of red light, for example, ztl-21 displays a period length identical to the other alleles (Fig. 6B), which could be taken to indicate similar, elevated levels of TOC1 (Más et al., 2003). This level of TOC1 is insufficient to alter hypocotyl elongation because ztl-21 shows no hypocotyl phenotype under these conditions (Fig. 6D). The hypocotyl phenotype of the other alleles is more likely caused by altered degradation of a target protein other than TOC1, interacting with a ZTL domain other than LOV/PAS. Most generally, the complexity of ztl mutant phenotypes is due to the functions of several target proteins that have differing requirements for interaction with ZTL, all of which are affected by strong ztl mutations. Analysis of allelic series in other clock-related genes has also separated circadian function from the other roles of TOC1 and GI genes (Más et al., 2003; Mizoguchi et al., 2005).
MATERIALS AND METHODS
Plant Material and Growth Conditions
ztl alleles were isolated from EMS-mutagenized populations of the C24 accession carrying the CAB2:LUC reporter gene (Millar et al., 1995a) and of the Ws ecotype carrying the CAB2:LUC+ reporter gene (Hall et al., 2001). The mutant lines were backcrossed three times to the corresponding parental lines; the mutations segregated as single, partially dominant loci (data not shown). The CCR2:LUC+ and CCA1:LUC+ (Doyle et al., 2002) reporter gene constructs were transformed into the mutant and wild-type lines via Agrobacterium-mediated transformation (Clough and Bent, 1998). Expression of the reporter genes was analyzed in at least five independent transgenic lines in each background, with similar results in all transgenic lines of the same genotype.
Unless otherwise indicated, seedlings were grown under LD 12:12 at 22°C prior to analysis and all measurements were carried out at constant 22°C. Illumination was provided by cool-white fluorescent tubes or, where indicated, by monochromatic LED light sources (red, λmax = 667 nm; blue, λmax = 470 nm). FRCs for hypocotyl elongation were obtained as described (Bauer et al., 2004). Hypocotyl lengths at different fluences of light were normalized to the corresponding dark-grown hypocotyl length to reflect solely the light-dependent regulation.
Measurement of Flowering Time
Seeds were sown on soil and incubated for 2 d in darkness at 4°C. They were subsequently transferred to long-day (16-h white light/8-h dark) conditions. Light sources were fluorescent (cool-white) tubes producing a fluence rate of approximately 60 μmol m−2 s−1. Flowering time was recorded as the number of rosette leaves at the time when inflorescences reached 1-cm height. The experiment was repeated twice using 30 to 40 plants per genotype.
Analysis of Luminescence and Leaf Movement Rhythms
LUC activity was measured either by low-light video imaging (groups of 5–10 seedlings) or with an automated luminometer (single seedlings), essentially as described (Hall et al., 2003). For FRCs, circadian periods were measured in seedlings transferred to constant illumination at the fluence rates indicated. For Table II, seedlings were grown in darkness under 12-h 22°C/12-h 26°C for three cycles and transferred to constant 22°C at the time of the predicted warm-cold transition for luminescence imaging. Leaf movement rhythms were measured as described (Dowson-Day and Millar, 1999). All rhythm data were analyzed with the Biological Rhythms Analysis Software System (BRASS; Southern et al., 2006; available at http://www.amillar.org), running fast Fourier transform nonlinear least-squares estimation (Plautz et al., 1997). Variance-weighted mean periods within the circadian range (15–40 h) and ses were estimated as described (Hall et al., 2003), from 10 to 36 traces per genotype. Measurements for producing FRCs were repeated three times; all the other results are representative of two independent experiments.
RNA Analysis
Total RNA extraction, northern blotting, and quantification of ZTL and 18S rRNA specific signals were performed as described (Viczian et al., 2005). A portion of the 18S rRNA gene was amplified using the 18S rRNA-F (5′-gggcctcgtcctttctctctttccattgc-3′) and 18S rRNA-R (5′-ttggagctcgctttcgcagttgttcgtctttcataa-3′) primers and used as a template for random primed labeling to produce the 18S rRNA-specific probe. The full-length ZTL cDNA was used as a template to generate the ZTL-specific probe.
Immunoblotting
Extracts were prepared from 10-d-old Arabidopsis (Arabidopsis thaliana) seedlings grown in LD 12:12. Ground tissue was resuspended in extraction buffer (50 mm Tris-Cl, pH 7.5, 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 5 μg/mL leupeptin, 1 μg/mL aprotinin, 1 μg/mL pepstatin, 5 μg/mL antipain, and 5 μg/mL chymostatin) by vortexing and clarified by centrifugation at 14,000g for 10 min at 4°C. Supernatant protein concentration was determined (Bio-Rad), concentrated by TCA precipitation, and resuspended in urea/SDS loading buffer (40 mM Tris-Cl, pH 6.8, 8 m urea, 5% SDS, 1 mm EDTA, and 2% 2-mercaptoethanol) to a final concentration of 5 μg/μL. Forty micrograms of protein were separated by SDS-PAGE (8%) and subjected to immunoblot analysis as described previously (Kim et al., 2003).
Yeast Two-Hybrid Tests for Protein Interactions
All experimental procedures related to Figure 7, A to C, were performed as described by Chien et al. (1991). Individual point mutations were introduced into the full-length ZTL cDNA clones by site-directed mutagenesis. The phyB N-terminal and C-terminal fragments contain amino acids 1 to 621 and 645 to 1,972, respectively. Yeast (Saccharomyces cerevisiae) cells were cotransformed with different cDNA fragments cloned into pGADT7 and pGBKT7 vectors (CLONTECH) in frame with the GAL4 transcriptional activator domain or with the GAL4 DNA-binding domain, respectively. Transformed cells were plated on Leu/Trp (LW) and Ade/Leu/Trp (ALW) complete supplement mixture (CSM) agar plates and grown at 30°C for 5 d. For β-galactosidase enzyme activity assay, three independent colonies were picked up from LW or ALW plates and inoculated into LW CSM liquid media, and were shaken at 30°C until density reached OD600 of approximately 0.8, when the assay (using O-nitrophenyl-β-d-galactopyranoside as substrate) was carried out (Yasuhara et al., 2004). For documentation of the growth on selective medium, single colonies from LW or ALW plates were diluted in 100 μL sterile water and 5 μL of them were dropped on fresh LW and ALW plates. After 5 d of growth, plates were scanned by a flat-bed scanner. Expression of the mutated ZTL proteins in yeast was confirmed by western blotting using anti-c-Myc and anti-HA antibodies (Sigma).
All manipulations involving reconstituted phytochromes (Fig. 7D; Supplemental Fig. 6) were performed as described (Shimizu-Sato et al., 2002).
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Vilmos Fülöp for protein structure mapping; Tom Schultz, Sam Hazen, and Steve Kay for helpful discussions; Tim Kunkel for providing the purified phycocyanobilin chromophore; and Paul Goode, Nazir Shariff, and Katalin Jószai for expert technical assistance. The empty BD vector, the PHYB-N and PHYB clones in the BD vector, and the PIF3 clone in the AD vector used for yeast two-hybrid assays were kindly provided by Peter H. Quail.
This work was supported in part by a long-term fellowship of the European Molecular Biology Organization (to L.K.B.), by a Marie Curie fellowship of the European Union (to M.E.E.), in part by a Japanese Society for the Promotion of Science postdoctoral fellowship (to S.H.), by a Biotechnology and Biological Science Research Council (BBSRC) postgraduate studentship (to M.M.S.), by a studentship from the Gatsby Charitable Foundation (to R.M.B.), by a Department of Energy Biosciences fellowship of the Life Science Research Foundation (to S.J.D.), by the National Science Foundation (grant no. IBN 0344377) and U.S. Department of Agriculture/Cooperative State Research, Education and Extension Service (grant no.CRIS 2002 35304 12594 to D.E.S.), and by the Alexander von Humboldt Foundation (grant no. IV–UNG/1118446 STP to A.V.); work in Szeged was supported by the Hungarian Scientific Research Fund (grant no. OTKA T046710) and the Howard Hughes Medical Institute (grant no. INTNL 55000325 to F.N.); work in Warwick was supported by the BBSRC (grant nos. G08667, G13967, and G15231) and the imaging facility was supported by grants from the Gatsby Charitable Foundation, the BBSRC, and the Royal Society (to A.J.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andrew J. Millar (andrew.millar@ed.ac.uk).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074864.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12: 757–761 [DOI] [PubMed] [Google Scholar]
- Aschoff J (1979) Circadian rhythms: influences of internal and external factors on the period measured in constant conditions. Z Tierpsychol 49: 225–247 [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Ádám É, Fejes E, Schäfer E, et al (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S (1991) The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA 88: 9578–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17: 63–71 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Esnouf RM (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph 15: 133–138 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Staiger D (2002) Photoreceptors in Arabidopsis thaliana: light perception signal transduction and entrainment of the endogenous clock. Planta 216: 1–16 [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, et al (2003) The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognar L, Toth R, Nagy F, Millar AJ (2001) Conditional circadian regulation of PHYTOCHROME A gene expression. Plant Physiol 127: 1808–1818 [PMC free article] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE (2004) Formation of an SCF complex is required for proper regulation of circadian timing. Plant J 40: 291–301 [DOI] [PubMed] [Google Scholar]
- Hayama R, Coupland G (2003) Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 6: 13–19 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Borevitz JO, Harmon FG, Pruneda-Paz JL, Schultz TF, Yanovsky MJ, Liljegren SJ, Ecker JR, Kay SA (2005. a) Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol 138: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005. b) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner R, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410: 487–490 [DOI] [PubMed] [Google Scholar]
- Kim WY, Geng R, Somers DE (2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc Natl Acad Sci USA 100: 4933–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Hicks KA, Somers DE (2005) Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol 139: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Wada M (2000) LKP1 (LOV kelch protein 1): a factor involved in the regulation of flowering time in Arabidopsis. Plant J 23: 807–815 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MolScript: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Li X, Zhang D, Hannink M, Beamer LJ (2004) Crystal structure of the kelch domain of human Keap1. J Biol Chem 279: 54750–54758 [DOI] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Sys Biol doi/10.1038/msb4100018 [DOI] [PMC free article] [PubMed]
- Más P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Murphy MEP (1994) Raster3D version 20: a program for photorealistic molecular graphics. Acta Crystallogr D 50: 869–873 [DOI] [PubMed] [Google Scholar]
- Millar AJ (2003) A suite of photoreceptors entrains the plant circadian clock. J Biol Rhythms 18: 217–226 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua N-H, Kay SA (1995. a) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua N-H, Kay SA (1995. b) The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163–1166 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340 [DOI] [PubMed] [Google Scholar]
- Onai K, Ishiura M (2005) PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10: 963–972 [DOI] [PubMed] [Google Scholar]
- Onai K, Okamoto K, Nishimoto H, Morioka C, Hirano M, Kami-Ike N, Ishiura M (2004) Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput real-time bioluminescence monitoring system. Plant J 40: 1–11 [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M (1998) Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet 14: 236–243 [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Salome PA, McClung CR (2004) The Arabidopsis thaliana clock. J Biol Rhythms 19: 425–435 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH (2002) A light-switchable gene promoter system. Nat Biotechnol 20: 1041–1044 [DOI] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock photomorphogenesis and flowering time. Plant Cell 16: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Southern MM, Brown PE, Hall A (2006) Luciferases as reporter genes. Methods Enzymol (in press) [DOI] [PubMed]
- Southern MM, Millar AJ (2005) Circadian genetics in the model higher plant, Arabidopsis thaliana. Methods Enzymol 393: 23–35 [DOI] [PubMed] [Google Scholar]
- Viczian A, Kircher S, Fejes E, Millar AJ, Schafer E, Kozma-Bognar L, Nagy F (2005) Functional characterization of phytochrome interacting factor 3 for the Arabidopsis thaliana circadian clockwork. Plant Cell Physiol 46: 1591–1602 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8: 135–142 [DOI] [PubMed] [Google Scholar]
- Yasuhara M, Mitsui S, Hirano H, Takanabe R, Tokioka Y, Ihara N, Komatsu A, Seki M, Shinozaki K, Kiyosue T (2004) Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J Exp Bot 55: 2015–2027 [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA (2001) Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.