Abstract
Plastoglobules (PGs) are oval or tubular lipid-rich structures present in all plastid types, but their specific functions are unclear. PGs contain quinones, α-tocopherol, and lipids and, in chromoplasts, carotenoids as well. It is not known whether PGs contain any enzymes or regulatory proteins. Here, we determined the proteome of PGs from chloroplasts of stressed and unstressed leaves of Arabidopsis (Arabidopsis thaliana) as well as from pepper (Capsicum annuum) fruit chromoplasts using mass spectrometry. Together, this showed that the proteome of chloroplast PGs consists of seven fibrillins, providing a protein coat and preventing coalescence of the PGs, and an additional 25 proteins likely involved in metabolism of isoprenoid-derived molecules (quinines and tocochromanols), lipids, and carotenoid cleavage. Four unknown ABC1 kinases were identified, possibly involved in regulation of quinone monooxygenases. Most proteins have not been observed earlier but have predicted N-terminal chloroplast transit peptides and lack transmembrane domains, consistent with localization in the PG lipid monolayer particles. Quantitative differences in PG composition in response to high light stress and degreening were determined by differential stable-isotope labeling using formaldehyde. More than 20 proteins were identified in the PG proteome of pepper chromoplasts, including four enzymes of carotenoid biosynthesis and several homologs of proteins observed in the chloroplast PGs. Our data strongly suggest that PGs in chloroplasts form a functional metabolic link between the inner envelope and thylakoid membranes and play a role in breakdown of carotenoids and oxidative stress defense, whereas PGs in chromoplasts are also an active site for carotenoid conversions.
Plastoglobules (PGs) are lipid-containing structures present in all types of plant plastids. In chloroplasts, they are primarily attached to thylakoid membranes (Rey et al., 2000), with smaller amounts accumulating in the stroma (Ghosh et al., 1994). The amount of PGs varies during the life cycle of the chloroplast and significantly increases both in number and size in senescing chloroplasts concomitant with thylakoid membrane degradation (Greenwood et al., 1963; Tuquet and Newman, 1980). Mutants blocked in chloroplast development often have increased amounts of electron-dense particles, typically believed to represent PGs (e.g. Kroll et al., 2001).
PGs in chloroplasts and colorless plastids in various plant tissues, such as tapetal cells and roots, are typically oval or round shaped, containing mostly α-tocopherol, plastoquinone, and triacylglycerols, but also sterol esters, mono-, and digalactosyl diacylglycerol, and are virtually devoid of carotenoids and chlorophylls (Bailey and Whyborn, 1963; Greenwood et al., 1963; Steinmuller and Tevini, 1985). PGs in chromoplasts of red bell pepper (Capsicum annuum) fruits, as well as colored flower petals, accumulate high levels of carotenoids (xanthophyll esters), α-tocopherol, and some plastoquinone and often have a fibrillar or tubular shape (Emter et al., 1990; Deruere et al., 1994). Their carotenoids, tocopherols, and quinones are sequestered in the central core, and the hydrophobic tails of the polar galacto- and phospholipids are oriented in a monolayer toward the core, whereas their polar head groups are oriented outward, interacting with a protein coat (Deruere et al., 1994).
PGs in chloroplasts were proposed to serve as reservoirs for α-tocopherol, plastoquinone, and triacylglycerols, particularly in young leaves (Kessler et al., 1999) and during senescence (Tevini and Steinmuller, 1985). PGs were also proposed to play a role in the removal of protein catabolites as part of thylakoid turnover (Ghosh et al., 1994; Smith et al., 2000). In chromoplasts, PGs are the main storage and remodeling site for carotenoids (Deruere et al., 1994). So far, it is not known whether PG proteomes include any enzymes.
The proteome of PGs appears to be composed of more than a dozen proteins, judging from one-dimensional (1-D) SDS-PAGE profiles (Wu et al., 1997; Kessler et al., 1999; Smith et al., 2000). Only one or two proteins belonging to the so-called fibrillin family have been identified in PGs, and these were assigned different names (Pozueta-Romero et al., 1997; Kessler et al., 1999; Vishnevetsky et al., 1999; Langenkamper et al., 2001). The fibrillins were proposed to play a role in stabilizing the globules and preventing their coalescence. In earlier analyses of subfractions of the thylakoid membrane proteome, we identified nine of 13 fibrillins (Peltier et al., 2002, 2004; Friso et al., 2004). Overexpression of a pepper fibrillin (homolog of FIB1a and b) in tobacco (Nicotiana tabacum) resulted in increased accumulation of PGs and more robust growth under higher light intensities (Rey et al., 2000).
In this study, we analyzed the proteome of PGs purified from chloroplasts of Arabidopsis (Arabidopsis thaliana) leaves before and after two different abiotic stress treatments and in the chloroplast protease mutant clpr2-1, which overaccumulates PGs. Stable-isotope labeling was used to quantify the stress response of the PG proteome. Identification of the proteomes of PGs of chromoplasts and chloroplasts showed that they contain unique protein populations. Our findings are conceptually integrated with chloroplast metabolism and function.
RESULTS
Purification, Identification, and Comparison of PG Proteomes from Chloroplasts of Arabidopsis
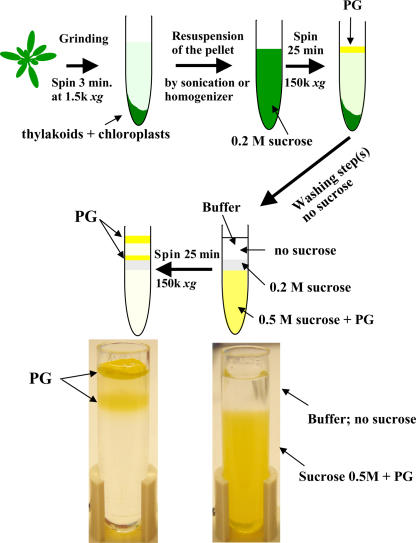
We improved existing PG purification protocols found in the literature with the objective of shortening purification time and improving PG yield. This resulted in highly purified PGs that were light yellow in color (Fig. 1). Using this improved protocol, PGs were purified from wild-type and clpr2-1 plants with reduced expression of the chloroplast ClpR2 protease (A. Rudella, J.M. Alonso, J.R. Ecker, and K.J. van Wijk, unpublished data) grown under optimal conditions. In addition, PGs were prepared from wild-type plants first grown under optimal conditions and then kept for 7 d in complete darkness or exposed to 7 d of high light (HL) flux (1,500 μmol photons m−2 s−1). The protein amount of PGs after HL and dark treatment increased, respectively, 10- to 12- and 3-fold on a total leaf fresh-weight basis.
Figure 1.
Purification scheme and photograph of purified PGs of Arabidopsis chloroplasts (in color).
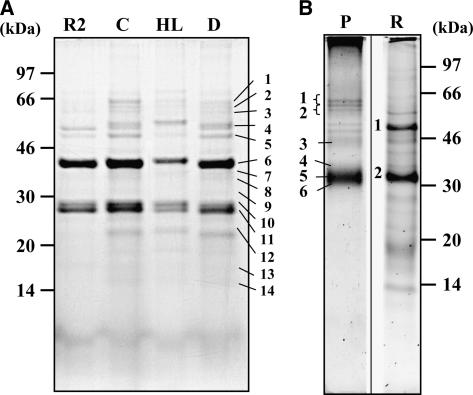
To obtain an overview of the PG proteomes, PG proteins were separated by SDS-PAGE, stained, and the major protein bands were analyzed by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry (MS) peptide mass fingerprinting (Fig. 2A). The chloroplast PG proteomes were dominated by fibrillins in bands 6, 10, and 11 (in particular, FIB1a, b, 2, 4, and 7a) and Fru-bis-P aldolases (FBPA) in band 6 (Table I). (For a complete list and MS scores, see Supplemental Table I.)
Figure 2.
1-D PAGE analysis of PG proteomes from Arabidopsis chloroplasts (A), pepper chromoplasts (B), and low-density lipid structures from rice etioplasts (B). Labeling of the gel lanes is as follows: R2, ClpR2; C, wild-type plant grown under optimal conditions; HL, wild-type plant grown for 7 d at HL (1,500 μE m−2 s−1); D, wild-type plants kept for 7 d in total darkness; P, PGs isolated from red pepper chromoplasts; R, low-density lipid structures from rice etioplasts. Proteins were identified in the numbered bands; accession numbers are listed in Tables I and II (Arabidopsis), and Table III (pepper). Detailed results for MALDI-TOF MS peptide mass fingerprinting on chloroplast PGs are provided in Supplemental Table I.
Table I.
Composition and comparative accumulation of the chloroplast PG proteome of stressed (D and HL) and unstressed (C) leaves and clpr2-1 in Arabidopsis
C, control; D, darkness; R2, ClpR2; Env, chloroplast envelope; Thy, thylakoid; Chl, chloroplast; Str, stroma.
| Accession No.a
|
Name
|
Possible Functional Category
|
Stress (D/C or HL/C)b
|
HL/Dc
|
In-Solution Digest (Mascot Scores)d
|
1-D Electrophoresise Gel Band
|
HL/CfAverage ± sd
|
D/CgAverage ± sd
|
HL/DhAverage ± sd
|
PredictioncTPi
|
Experimental Identification
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | R2 | HL | D | Envj | Thyk | Chll | Strm | ||||||||||
| At4g04020.1 | Fibrillin (FIB1a) | PG protein coat | No change | Up | 914 | 1,097 | 548 | 764 | 10 | 1.46 ± 0.29 | 0.74 ± 0.15 | 1.73 ± 0.23 | C | Thy | C | Str | |
| At4g22240.1 | Fibrillin (FIB1b) | PG protein coat | Down in D | No change | 680 | 1,036 | 631 | 801 | 10 | 1.48 ± 0.21 | 0.56 ± 0.01 | 2.16 ± 0.95 | C | Thy | C | ||
| At3g23400.1 | Fibrillin (FIB4) | PG protein coat | Down in HL | No change | 662 | 773 | 512 | 670 | 11 | 0.70 ± 0.01 | 0.97 ± 0.13 | 0.61 ± 0.22 | C | Thy | C | ||
| At2g35490.1 | Fibrillin (FIB2) | PG protein coat | Down in HL | Down | 287 | 620 | 212 | 417 | 6 | 0.86 ± 0.06 | 1.13 ± 0.18 | 0.68 ± 0.01 | C | Thy | C | ||
| At3g58010.1 | Fibrillin FIB7a) | PG protein coat | No change | Down | 126 | 185 | 450 | 10 | 0.92 ± 0.39 | 1.42 ± 0.45 | 0.50 ± 0.11 | C | Thy | ||||
| At2g46910.1 | Fibrillin (FIB8) | PG protein coat | No change | No change | 221 | 57 | 219 | 1.35 ± 0.21 | 0.57 ± 0.44 | C | |||||||
| At2g42130.3 | Fibrillin (FIB7b) | PG protein coat | Up in D | Down | 380 | 1.40 ± 0.12 | 3.05 ± 0.86 | 0.38 ± 0.04 | – | Thy | |||||||
| At4g38970.1 | FBPA-2 | Calvin; glycolysis | No change | No change | 653 | 762 | 467 | 700 | 6 | 0.97 ± 0.31 | 1.24 ± 0.29 | 0.72 ± 0.04 | C | Env | C | Str | |
| At2g21330.1 | FBPA-1 | Calvin; glycolysis | No change | No change | 605 | 612 | 406 | 772 | 6 | 0.85 ± 0.19 | 0.98 ± 0.13 | 0.79 ± 0.01 | C | Env | Thy | C | Str |
| At2g01140.1 | FBPA | Calvin; glycolysis | n.d. | n.d. | 78 | 192 | C | C | |||||||||
| At2g39730.1 | Rubisco activase | Calvin cycle | n.d. | n.d. | 75 | 176 | C | Env | Thy | C | Str | ||||||
| AtCg00490 | Rubisco large subunit | Calvin cycle | n.d. | n.d. | 100 | c-enc | Env | Thy | C | Str | |||||||
| At4g19170.1 | CCD4 | Carotenoid cleavage | No change | Down | 476 | 592 | 370 | 3 | 2.05 ± 1.55 | 3.31 ± 2.90 | 0.46 ± 0.12 | C | C | ||||
| At3g26840.1 | Esterase/lipase/thioesterase | Lipid metabolism | n.d. | n.d. | 149 | 89 | 138 | C | Thy | ||||||||
| At1g54570.1 | Esterase/lipase/thioesterase | Lipid metabolism | Up in D and HL | No change | 175 | 169 | 6.26 ± 4.28 | 1.50 ± 0.05 | 5.50 ± 4.69 | C | |||||||
| At5g42650.1 | AOS | JA synthesis | No change | No change | 343 | 534 | 50 | 679 | 4 | 0.98 ± 0.67 | 1.35 ± 0.78 | 0.64 ± 0.34 | C | Env | C | ||
| At1g78140.1 | UbiE-methyltransferase-related | Quinone synthesis | n.d. | n.d. | 130 | 41 | 146 | 0.90 | 0.44 | 1.17 | M | ||||||
| At2g41040.1 | UbiE-methyltransferase-related | Quinone synthesis | No change | No change | 36 | 131 | 182 | 295 | 8 | 1.83 ± 0.82 | 1.12 ± 0.31 | 1.25 ± 0.13 | C | ||||
| At5g05200.1 | ABC1 kinase | Quinone synthesis | No change | n.d. | 351 | 156 | 124 | 111 | 1.27 ± 0.02 | 0.24 | 3.04 | M | |||||
| At4g31390.1 | ABC1 kinase | Quinone synthesis | Down in D | up | 79 | 123 | 2 | 1.44 ± 0.20 | 0.61 ± 0.10 | 1.88 ± 0.11 | C | ||||||
| At1g79600.1 | ABC1 kinase | Quinone synthesis | No change | No change | 68 | 172 | 107 | 224 | 1 | 1.42 ± 0.13 | 0.73 ± 0.11 | 1.39 ± 0.28 | C | ||||
| At1g71810.1 | ABC1 kinase | Quinone synthesis | n.d. | n.d. | 63 | 52 | 44 | 166 | 0.83 | 1.13 | 0.68 | C | |||||
| At4g32770.1 | VTE1, tocopherol cyclase | Tocopherol synthesis | No change | n.d. | 124 | 86 | 89 | 183 | 5 | 0.68 ± 0.22 | 0.86 ± 0.36 | 0.87 | C | C | |||
| At5g08740.1 | Pyridine nucleotide-disulfide oxidoreductase (DhnA-like) | Unknown | Down in HL | Down | 379 | 332 | 175 | 430 | 5 | 0.56 ± 0.10 | 1.10 ± 0.52 | 0.56 ± 0.06 | C | ||||
| At3g10130.1 | SOUL heme binding | Unknown | No change | No change | 199 | 107 | 86 | 97 | 9 | 2.34 | 0.87 ± 0.33 | 1.71 ± 0.66 | C | Thy | C | ||
| At4g13200.1 | Expressed protein | Unknown | No change | No change | 309 | 175 | 267 | 399 | 13 | 1.26 ± 0.49 | 0.83 ± 0.28 | 0.97 ± 0.23 | C | Thy | C | ||
| At2g34460.1 | Epimerase/dehydratase family | Unknown | No change | Down | 184 | 100 | 76 | 231 | 0.80 ± 0.05 | 1.07 ± 0.11 | 0.60 ± 0.01 | C | Env | Thy | C | ||
| At1g32220.1 | Epimerase/dehydratase family | Unknown | No change | n.d. | 122 | 319 | 240 | 1.40 ± 0.74 | 1.23 ± 0.38 | 1.01 | C | ||||||
| At1g09340.1 | Rap38 | Unknown | Down in HL | n.d. | 332 | 70 | 7 | 0.15 ± 0.09 | 0.63 ± 0.62 | 0.08 | – | Env | C | Str | |||
| At3g63140.1 | Rap41 | Unknown | n.d. | n.d. | 65 | C | Env | C | Str | ||||||||
| At1g26090.1 | Anion-transporting ATPase | Transport | n.d. | n.d. | 58 | 146 | C | ||||||||||
| At1g28150.1 | Expressed protein | Unknown | n.d. | n.d. | 41 | 52 | C | ||||||||||
| At5g01730.1 | Expressed protein | Unknown | n.d. | n.d. | 42 | 39 | 42 | – | |||||||||
| At4g01150.1 | Expressed protein | Unknown | Up in HL | n.d. | 44 | 49 | 3.24 ± 0.01 | 1.69 | 2.41 | C | Env | Thy | C | ||||
Accession numbers in bold indicate that a homolog was also identified in PG from red pepper chromoplasts (listed in Table III).
Response in relative accumulation levels of the protein within the PG proteome after HL stress or dark treatment (n.d., not determined because not enough peptides could be found for quantification).
Response in relative accumulation levels of the protein within the PG proteome comparing HL stress to dark treatment (n.d., not determined because not enough peptides could be found for quantification).
Mascot scores from in-solution digest (higher numbers indicate higher confidence; all scores are significant with P < 0.05).
Identification of the protein by peptide mass fingerprinting from 1-D PAGE (Fig. 2A) is indicated with the number of the gel band (from gel in Fig. 2A).
Relative protein ratio between PGs from HL stress and dark treatment determined from isotope-labeling experiments. The sds calculated from the averages of the independent experiments are indicated. Values in bold indicate significant decrease in the HL-to-D ratio; values in italic indicate significant increase in the HL-to-D ratio. Other numbers show no significant change. Significant is defined as average value of HL/D < 0.75 sd or >1.33 + 1 sd. For average values within each independent replicate and number of peptides used for quantification, see Supplemental Table II.
Relative protein ratio between PGs from dark treatment and control plants determined from isotope-labeling experiments. The sds calculated from the averages of the independent experiments are indicated. Average values and the number of peptides used for each independent experiment are listed in Supplemental Table II. For bold and italic text, see footnote f.
Relative protein ratio between PGs from HL and dark treatment and control plants determined from isotope-labeling experiments. The sds calculated from the averages of the independent experiments are indicated. Average values and the number of peptides used for each independent experiment are listed in Supplemental Table II. For bold and italic text, see footnote f.
Predicted cTP using TargetP or chloroplast encoded (c-enc). C, Chloroplast; M, mitochondria.
Identification in proteome analysis of chloroplast envelopes. PubMed numbers for cross-reference papers are 12938931 and 12177442.
Identification in proteome analysis of thylakoids (and associated PGs). PubMed numbers for cross-reference papers are 11719511, 11826309, and 15322131.
Identification in proteome analysis of total chloroplasts. PubMed number for cross-reference paper is 15028209.
Identification in proteome analysis of purified stroma. PubMed number for cross-reference paper is 16207701.
To more fully identify the PG proteomes, we used a protocol compatible with the hydrophobic nature of PGs (Peltier et al., 2004). In short, proteomes were delipidized by SDS solubilization and acetone precipitation, followed by resolubilization of the proteins in dimethyl sulfoxide (Me2SO), trypsin digestion, and nano liquid chromatography (nanoLC)-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) analysis of the peptides (Tables I and II). Thirty-two proteins, not part of the thylakoid photosynthetic complexes, were identified in the chloroplast PGs from unstressed and stressed conditions and clpr2-1 (Table I). PGs purified from the chloroplast protease mutant clpr2 were very similar in composition to those from dark-treated wild-type plants.
Table II.
Composition and comparative accumulation of the chloroplast PG proteome of stressed (D and HL) and unstressed (C) leaves and clpr2-1 in Arabidopsis, the photosynthetic apparatus
See Table I legend for definitions of abbreviations.
| Accession No.
|
Name
|
Functional Category
|
In-Solution Digest (Mascot Scores)a
|
1-D Electrophoresisb Gel Band
|
HL/Dc Average ± sd
|
|||
|---|---|---|---|---|---|---|---|---|
| C | R2 | HL | D | |||||
| At2g34420.1 | LHCII-1.5 | Light-harvesting PSII | 113 | 106 | 171 | 156 | 1.31 ± 0.28 (i) | |
| At1g29910.1d | LHCII-1.1–3 | Light-harvesting PSII | 113 | 106 | 151 | 162 | 1.31 ± 0.28 (i) | |
| At2g34430.1 | LHCII-1.4 | Light-harvesting PSII | 50 | 49 | 108 | 105 | 1.31 ± 0.28 (i) | |
| At2g05100.1e | LHCII-2.1–4 | Light-harvesting PSII | 50 | 98 | 89 | 63 | ||
| At1g15820.1 | LHCII-6 CP24 | Light-harvesting PSII | 36 | 47 | 0.39 ± 0.11 | |||
| At4g10340.1 | LHCII-5 CP26 | Light-harvesting PSII | 71 | 12 | ||||
| At3g50820.1f | psbO OEC33-like | PSII | 62 | 105 | 50 | |||
| AtCg00280 | psbC CP43 | PSII | 36 | 133 | ||||
| AtCg00680 | psbB CP47 | PSII | 128 | 1.10 ± 0.09 | ||||
| AtCg00270 | psbD D2 | PSII | 121 | |||||
| At1g31330.1 | psaF subunit III | PSI | 156 | 130 | 1.00 | |||
| AtCg00020 | psbA D1 | PSI | 102 | 81 | ||||
| At2g20260.1g | psaE-2 subunit IV | PSI | 69 | 14 | 1.58 ± 0.54 | |||
| AtCg00340 | psaB subunit Ib | PSI | 67 | 56 | 1.06 ± 0.11 | |||
| AtCg00350 | psaA subunit Ia | PSI | 48 | 46 | ||||
| At1g52230.1h | psaH-2 subunit VI | PSI | 38 | 48 | 1.80 ± 1.11 | |||
| At3g47470.1 | LHCI-4 LHCI-730 | Light-harvesting PSI | 73 | 51 | ||||
| AtCg00480 | CF1b atpB | ATP synthase | 522 | 444 | 4 | 2.95 ± 1.04 | ||
| AtCg00120 | CF1a atpA | ATP synthase | 383 | 216 | 4 | 2.89 ± 0.81 | ||
| At4g09650.1 | CF1d atpD | ATP synthase | 75 | |||||
Mascot scores from in-solution digest (higher numbers indicate higher confidence; all scores are significant with P < 0.05).
Identification of the protein by peptide mass fingerprinting from 1-D PAGE is indicated with the number of the gel band (from the gel in Fig. 2A).
Response in relative accumulation levels of the protein within the PG proteome comparing HL stress to dark treatment. Value in bold indicates significant decrease in HL-to-D ratio; values in italics indicate significant increase in HL-to-D ratio. Other numbers show no significant change. Significant is defined as average value of HL/D < 0.75 sd or >1.33 + 1 sd. For average values within each independent replicate and number of peptides used for quantification, see Supplemental Table II.
Cannot distinguish between At1g29910.1, At1g29920.1, and At1g29930.1.
Cannot distinguish between At2g05070.1, Atg05100.1, and At3g27690.1.
Cannot distinguish between At5g66570.1 and At3g50820.1.
Cannot distinguish between At4g28750.1 and At2g20260.1.
Cannot distinguish between At3g16140.1 and At1g52230.1.
iPeptides shared between all lhcb1 were used for the quantification.
These identified proteins were cross-correlated to large-scale Arabidopsis proteomics data from envelope (Ferro et al., 2002, 2003; Froehlich et al., 2003) and other Arabidopsis subcellular proteomes (Borderies et al., 2003; Nuhse et al., 2003; Carter et al., 2004; Heazlewood et al., 2004; Marmagne et al., 2004) and to other literature. Most of the proteins were never identified in the stromal or envelope proteome, clearly indicating that PGs contain a specific protein population (Table I). A number of proteins were identified during our previous in-depth analyses of the thylakoid membranes (Friso et al., 2004; Peltier et al., 2004), which is not surprising because PGs are associated with the thylakoid membrane (Table I). Only one (At4g01150) of the 33 PG proteins has predicted transmembrane domains. The absence of transmembrane domains is consistent with the absence of a lipid bilayer in the PGs. The documented lipid monolayer cannot accommodate transmembrane domains. This is another strong indication that the proteome identified here is not a random collection of hydrophilic thylakoid or envelope (bilayer) proteins.
The PG proteins were tentatively assigned to four functional classes, namely (1) fibrillins (seven proteins) forming the protein coat of the PGs; (2) lipid metabolism or mobilization of fatty acids (two proteins); (3) quinone synthesis and regulation (six proteins); and (4) no obvious function (11 proteins). In addition, we identified three proteins involved in synthesis of the hormone jasmonic acid (JA), tocopherol, and a protein likely involved carotenoid cleavage. Finally, we identified two FBPAs with very high scores; it is highly unlikely that they represent stromal contamination because we hardly observed other abundant stromal proteins. We will comment on the unique localization and functional assignments of most of the identified proteins later in this article.
We also identified a number of proteins of the thylakoid photosynthetic apparatus, in particular, after 7 d of continued HL stress and, to a lesser extent, also after 7 d of continued darkness (Table II). Only some of the very abundant light-harvesting proteins were found in the control and clpr2-1 PG preparations with relatively low scores. It is most likely that they represent dismantled thylakoid membrane fragments.
We were interested in determining whether and how the PG proteome changes after prolonged light stress (HL) or after degreening during prolonged darkness. Under these conditions, significant breakdown of the thylakoid proteome and possibly the lipid bilayer occurs. To determine differential protein accumulation due to these dark and HL stress treatments, peptides for each of the PG samples were labeled with either formaldehyde (HCHO) or its deuterated form (DCDO). The principles of this comparative proteomics technique were initially described by Hsu et al. (2003). We optimized the technique for peptide recovery and labeling efficiency and also adapted it for membrane proteins. Three pairwise comparisons between the three PG samples were carried out and included a label switch within each pair, as outlined in Figure 3. The experiment was carried out with two independent biological replicates, and the data are summarized in Tables I and II and discussed further below. Detailed data can be found in Supplemental Table II.
Figure 3.
Outline of differential accumulation analysis using stable-isotope labeling with DCDO and HCHO. For a pairwise comparison of dark and HL samples, 2.5 μg of each digested proteome were labeled with DCDO or HCHO. Each quantification was repeated with a switch of isotope labels so as to minimize possible isotope biases on the quantification (A). Comparison between the different stress treatments (dark and HL) and the unstressed condition were always pairwise (B). C, Detailed m/z isotope envelopes for peptide GDGGLFVLAR labeled with HCHO (mass shift of 28 D per free amine) or DCDO (mass shift of 32 D per free amine).
Purification and Identification of the PG Proteome from Red Pepper Chromoplasts
We also purified and analyzed PGs from chromoplasts of red pepper fruits using similar procedures as for the chloroplasts. 1-D electrophoresis gel analysis showed that the chromoplast PG proteome was dominated by several fibrillins (bands 1 and 6), FBPAs (band 4), and others (Fig. 2B; Table III). The proteomes of PG pepper chromoplasts were also analyzed by in-solution digestion and nanoLC-ESI-MS/MS, and MS data searched against the National Center for Biotechnology Information (NCBI) database and the Solanaceae database (downloaded from http://www.sgn.cornell.edu). Twenty-eight proteins were identified in the PGs of pepper chromoplasts (Table III). To allow better cross-correlation to the chloroplast PG data, protein accessions identified in the pepper PG were BLAST searched against the predicted Arabidopsis proteome (Table III). The two PG types have 12 proteins in common, as indicated in bold in Tables I and III (for discussion, see below).
Table III.
Composition of the PG proteome of red pepper chromoplasts
*, Cannot distinguish between U197257 and 82130.
| Accession No. | Species | Protein Name | Possible Function | Mascot Score | 1-D Gela | Homologb | E Value |
|---|---|---|---|---|---|---|---|
| gi|1076575 | Capsicum annuum | Fibrillin (FIB1a) | Protein coat | 1,399 | 6 (976) | At4g04020 | 3.00E-96 |
| Fibrillin (FIB1b) | Protein coat | 1,399 | At4g22240 | 1.20E-92 | |||
| sgn|U247199 | Solanum tuberosum | Fibrillin (FIB4) | Protein coat | 340 | 1 (149) | At3g23400 | 2.90E-84 |
| sgn|U197362 | Capsicum annuum | Fibrillin (FIB2) | Protein coat | 62 | At2g35490 | 4.90E-25 | |
| gi|12643508 | Capsicum annuum | LYC-β | Carotenoids | 1,307 | 2 (394) | At3g10230 | 5.10E-142 |
| gi|1583601 | Capsicum annuum | ZDS | Carotenoids | 436 | 1 (67) | At3g04870 | 1.30E-244 |
| sgn|U265491 | Solanum tuberosum | VTE1, tocopherol cyclase | Carotenoids | 66 | At4g32770 | 5.40E-67 | |
| gi|2956671 | Capsicum annuum | CrtR-β | Carotenoids | 66 | At4g25700 | 7.60E-100 | |
| sgn|U209549 | Petunia hybrida | CrtR-β | Carotenoids | 55 | At5g52570 | 8.50E-37 | |
| sgn|U196115 | Capsicum annuum | FBPA-2 | Glycolysis | 688 | 3 (276) | At4g38970 | 4.40E-175 |
| sgn|U196550 | Capsicum annuum | FBPA | Glycolysis | 528 | 3 (362) | At2g01140 | 2.60E-177 |
| gi|7436610 | Solanum tuberosum | FBPA-1 | Glycolysis | 318 | 3 (204) | At2g21330 | 5.80E-173 |
| sgn|U204835 | Capsicum annuum | Expressed protein | Unknown | 247 | At1g32220 | 1.10E-57 | |
| sgn|U213348 | Lycopersicon esculentum | Aldolase | Unknown | 142 | At3g52930 | 4.30E-161 | |
| sgn|U198272 | Capsicum annuum | Expressed protein | Unknown | 137 | At4g13200 | 3.90E-32 | |
| sgn|U212944 | Lycopersicon esculentum | Aldo/keto reductase family | Unknown | 110 | At1g06690 | 1.10E-82 | |
| sgn|U251738 | Solanum tuberosum | ABC1 family protein, kinase domain | Quinone | 99 | 2 (45) | At5g05200 | 2.10E-97 |
| sgn|U220570 | Lycopersicon esculentum | Esterase/lipase/thioesterase | Lipid metabolism | 59 | At1g54570 | 6.70E-53 | |
| sgn|U201138 | Capsicum annuum | Thioredoxin m4 | Redox | 115 | At3g15360 | 8.00E-41 | |
| sgn|U197257 | Capsicum annuum | Glucan endo-1,3-β-glucosidase | Cell wall | 286 | 5 (173)* | At3g57270 | 4.30E-97 |
| gi|82130 | Nicotiana plumbaginifolia | Glucan endo-1,3-β-glucosidase | Cell wall | 218 | 5 (173)* | At3g57260 | 2.40E-96 |
| gi|42564093 | Capsicum annuum | Glucan endo-1,3-β-glucosidase | Cell wall | x | 4 (246) | At4g16260 | 7.00E-47 |
| sgn|U212883 | Lycopersicon esculentum | Basic endochitinase | Other | 128 | At3g12500 | 5.10E-133 | |
| gi|12004153 | Primula palinuri | CF1b, atpB | Photosynthesis | 84 | 5 (42) | ATCG00480 | 5.20E-220 |
| sgn|U245166 | Solanum tuberosum | Rubber elongation factor family | Other | 63 | At1g67360 | 2.60E-50 | |
| gi|10638269 | Thuja plicata | Phytochrome C | Other | 62 | At5g35840 | 7.80E-69 | |
| gi|6721571 | Citrus unshiu | ATPase 70 kD | Unknown | 60 | At1g78900 | 5.90E-164 | |
| sgn|U196125 | Capsicum annuum | ADP, ATP carrier protein 1 | Transport | 60 | At3g08580 | 5.40E-170 |
Purification and Identification of the Proteome of Low-Density Lipids of Rice Etioplasts
To compare the PG proteomes with prolamellar bodies (Staehelin, 2003) and possibly PGs in etioplasts, we grew rice (Oryza sativa) seedlings in complete darkness and purified the etioplasts. We then used the PG purification procedure to collect low-density lipid particles and membranes from these etioplasts. SDS-PAGE and MS analysis of the low-density fractions showed that they were dominated by protochlorophyllide reductase a/b (band 2), as expected in prolamellar bodies (Sperling et al., 1998), and a mixture of proteins unrelated to PGs in band 1 (Fig. 2B). Further analysis by in-solution digestion and nanoLC-ESI-MS/MS again identified protochlorophyllide reductase a/b with very high MOWSE scores and also identified one fibrillin (PAP10/FIB10) and an aldolase (homolog to At4g38970/At2g21330). This fibrillin was identified so far only in thylakoids (Peltier et al., 2004). No further overlap was observed with the PG proteomes from either chloroplasts or chromoplasts (data not shown). These low-density membranes thus mostly represented prolamellar bodies with some associated or contaminating proteins (data not shown) and not much plastoglubular protein.
DISCUSSION
Many of the PG proteins in chloroplasts and chromoplasts have not been experimentally identified in plants, whereas others have been observed but their precise subplastid localization was so far unknown. The identified PG proteins are clearly involved in different aspects of plastid metabolism, and the challenge is to understand their individual and collective functions. In the sections below, we first discuss the possible function of the identified proteins based on published literature and predicted functional domains, and, if determined, we comment on their relative accumulation in response to stress (dark or HL). Subsequently, we will integrate this collective information into a summarizing functional model of PGs and their place in plastid metabolism, biogenesis, and stress response.
Fibrillins
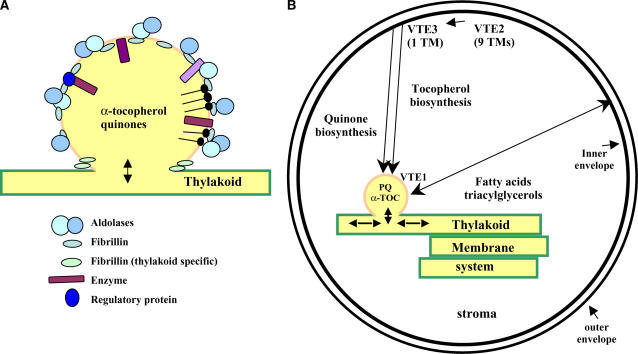
The most prominent proteins in PGs are fibrillins, as suggested previously in the analysis of PGs from several plant species. The Arabidopsis genome has 13 fibrillin genes that are all predicted to encode plastid-localized proteins (Laizet et al., 2004). In chloroplast PGs, we identified six earlier observed fibrillins (FIB1a, 1b, 2, 4, 7a, and 7b) but also FIB8, not previously observed before. In chromoplast PGs, we identified homologs of FIB1a, 1b, 2, and 4. Fibrillins have one or more hydrophobic regions, which have been proposed to play a role in stabilizing the globules and preventing coalescence (Deruere et al., 1994). Importantly, Camara, Kuntz, and colleagues (Deruere et al., 1994) reported that FIBs themselves have no enzymatic activity but should be viewed as structural (Fig. 4A). The relative quantification by stable-isotope labeling showed that most identified members of the fibrillin family responded differentially to the HL and dark treatment, with FIB1b down and FIB7b up after the dark treatment and FIB2 and FIB4 down after the HL treatment, as compared to control plants (Table I). However, considering that the amount of PG protein after HL and dark treatment increased severalfold on a total leaf basis, it is clear that expression of all observed fibrillins increased after stress treatment on a total cellular basis.
Figure 4.
Schematic overview of proposed organization (A) and functional role of the PG and its proteome (B). PGs consist of a monolayer of lipids and sequester different hydrophilic small molecules, such as quinones and tocopherols. Structural proteins (fibrillins) and enzymes are attached to or embedded in the monolayer, but proteins lack transmembrane domains (A). Integration of PG functions in plastid metabolism (B).
Carotenoid Biosynthesis and Apocarotenoids
In chloroplast PGs, we identified At4g19170 (assigned CCD4 for carotenoid cleavage dioxygenase; also named NCED4 for 9-cis-epoxy-carotenoid dioxygenase); its actual substrate and cleavage product are not known. It has not been identified in the envelope or thylakoid proteome. The stable-isotope experiments suggested twice as much accumulation in PGs after the dark treatment than after the HL treatment, suggesting an active role in dark-induced breakdown of carotenoids (Table I). CCD4 is part of a family of nine monooxygenases; five of these nine genes were shown to encode for chloroplast-localized proteins that differ in subplastid location (Tan et al., 2003). Just recently, the zinc-finger protein VAR3 (At5g17790) was shown to interact with CCD4 and localizes to chloroplasts in punctated spots, as viewed by green fluorescent protein fusion and microscopy (Naested et al., 2004). Based on our results, we believe that these spots represent PGs.
In red bell pepper, more than one-half of the carotenoids accumulate as esterified capsanthin (Deruere et al., 1994), but so far no carotenoid biosynthetic enzymes have been identified in PGs of chromoplasts. Fibril formation is dependent on a minimal concentration of bicyclic carotenoids (carotene and xanthophylls). We identified ζ-carotene desaturase (ZDS), lycopene β-cyclase (LYC-β or CYC-β), and two β-carotene β-hydroxylases (CrtR-β), operating in series in bicyclic carotenoid biosynthesis, with ζ-carotene as the first substrate and zeaxanthin and lutein as potential end products (Hirschberg, 2001). In particular, LYC-β was identified with very high Mascot scores (1,307 from in-solution; 394 from 1-D gels); this suggests that PGs in chromoplasts have a specific function in carotenoid synthesis, in addition to the well-known carotenoid storage/sequestering.
JA and Triacylglycerol Metabolism
We identified four enzymes possibly involved in different aspects of lipid and hormone metabolism, but the functions of most of them are unknown and functional domain predictions do not suggest clear-cut function. The exception was allene oxide synthase (AOS; At5g42650), the first and abundant enzyme in the lipoxygenase pathway leading to the formation of JA (Laudert et al., 1996; von Malek et al., 2002). Biosynthesis of JA starts with the oxygenation of linolenic acid released from membrane lipids through the action of an unknown lipase. Lipoxygenase then converts linolenic acid to 13-hydroperoxylinolenic acid, which subsequently serves as a substrate for AOS, converting it into an unstable epoxide. We speculate that the high quinone and α-tocopherol in PGs will help to stabilize the epoxide against oxidation. The function of PG proteins At3g26840 and At1g54570 is unknown, but they have predicted acyltransferase, diacylglycerol transferase and thioesterase domains. Relative accumulation of At5g54570 was increased severalfold after prolonged HL treatment and increased 50% after degreening, suggesting involvement in disassembly of the thylakoid bilayer (Fig. 4B). A homolog for At1g54570 was also identified in pepper PGs.
Tocopherol Cyclase
Tocochromanols are a group of four tocopherols and four tocotrienols that constitute vitamin E and are only synthesized by oxygenic photosynthetic organisms. Chloroplasts accumulate predominantly α-tocopherols (DellaPenna, 2005). We identified tocopherol cyclase (also named VTE1) both in chromoplast and chloroplast PGs. VTE1 converts 2-methyl-6-phytyl-quinol (MPBQ) and 2,3-dimethyl-5-phytyl-5-phytylquinol (DMBQ) to δ- and γ-tocopherol, respectively (Porfirova et al., 2002; Sattler et al., 2003). VTE1 (without any predicted transmembrane domains) had not been identified in chloroplast proteomics studies. VTE3, a MPBQ and DMBQ methyltransferase (also named APG1 or E37; with one predicted transmembrane domain) immediately upstream of VTE1 in the tocopherol and plastoquinone pathway (Van Eenennaam et al., 2003), was identified as a relatively abundant inner envelope membrane protein (Froehlich et al., 2003; Motohashi et al., 2003; Fig. 4B). VTE4, γ-tocopherol-methyltransferase immediately downstream of VTE1, was characterized, but its precise subchloroplast location is unknown (Bergmuller et al., 2003). We also anticipate that VTE4 is localized in PGs and that PGs not only store significant amounts of tocopherols but also are actively involved in their synthesis.
Quinone Biosynthesis and ABC1 Kinases
We identified two proteins, At2g41040 and At1g78140, which have predicted ubiquinone methyltransferase domains and are part of the so-called UbiE family. The Arabidopsis genome has at least nine proteins in this family; one of them is VTE3, mentioned above. Small-molecule analysis of PGs from chloroplasts has shown that PGs have very significant amounts of plastoquinones and smaller amounts of phylloquinone (vitamin K1) and α-tocopherol quinone (oxidized α-tocopherol; Bailey and Whyborn, 1963; Steinmuller and Tevini, 1985). Plastoquinone is the lipid-soluble electron carrier between PSII and the cytochrome b6f complex and the most abundant quinone in the membrane, whereas phylloquinone is the A1 electron acceptor in PSI. Plastoquinone has nine isoprene units and phylloquinone has only four isoprenes. The only reported commonality in their biosynthetic pathway is their requirement for isopentenyl diphosphate and dimethylallyl diphosphate, both products of the nonmevalonate pathway. Interestingly, an Arabidopsis deletion mutant in 1,4-dihydroxy-2-napathoic acid phytyltransferase (abc4) lacked phylloquinone but also accumulated only 3% of plastoquinone (Shimada et al., 2005). A possible explanation is that accumulation of phytol diphosphate leads to feedback regulation of the nonmevalonate pathway. It appears that the biosynthetic pathway of plastoquinone is rather simple and seems resolved (Cheng et al., 2003; Sattler et al., 2004). In contrast, several enzymes in the phylloquinone pathway have not yet been identified. It would be worth testing whether At2g41040 and At1g78140 function in phylloquinone biosynthesis.
We also identified four proteins of the ABC1 kinase family in chloroplast PGs (At5g05200, At1g79600, At1g71810, and At4g31390); a homolog of one of them (At5g05200) was also identified in pepper PGs. Sequence analysis and homology modeling revealed that all four have the typical ABC1 kinase domain (D.R. Ripoll, unpublished data). Relative accumulation levels of one of these ABC1 proteins (At4g31390) within PGs increased after prolonged HL treatment, but decreased after prolonged darkness (Table I). The unifying theme of ABC1 proteins in diverse species (Providencia stuarti, Escherichia coli, Saccharomyces cerevisiae, etc.) is that inactivation leads to deficiency in quinone synthesis (Cardazzo et al., 1998; Leonard et al., 1998; Poon et al., 2000; Iiizumi et al., 2002). The published data on E. coli suggest that ABC1 proteins (UbiB) regulate the first monooxygenation of the quinone precursor (Poon et al., 2000). The perspective of identifying regulators of quinone synthesis in plants is exciting and warrants functional studies on the four identified ABC1 proteins.
Finally, we identified At5g08740 with a pyridine nucleotide-disulfide oxidoreductase domain and related to mitochondrial rotenone-insensitive NADH-ubiquinone oxidoreductase. The function of At5g08740 is unclear, but is likely to involve electron transfer to and from PG-localized quinones. Relative accumulation was reduced 2-fold after the HL stress as compared to control and degreening treatment (Table I).
Aldolases
Surprisingly, two known and abundant stromal FBPAs (At2g21330 and At4g38970; class I) involved in the Calvin cycle and/or glycolysis were found as major components in PGs (Fig. 2A; Table I). We identified an additional class I FBPA (At2g01140) in the PGs, which was not earlier identified in the stroma of chloroplasts. Homologs for all three were identified in the (nonphotosynthetic) chromoplast PGs (Table III). Relative accumulation levels of FBPA1 and 2 within PGs did not significantly change after the HL or dark treatments as compared with normal conditions (Table I). The high abundance of aldolase within chloroplast PGs and the absence of significant accumulation of other abundant Calvin cycle/glycolytic enzymes strongly suggest that the aldolases are truly part of the PG proteome in chloroplasts and chromoplasts and make an unknown functional contribution to the (metabolism/structure) of PGs. We speculate that aldolases are somehow involved in carbon flux to and from the PGs, respectively, during synthesis or degradation of hydrophilic small molecules.
Other Proteins
We identified three proteins (At2g34460, At1g32220, and At1g09340) with predicted NAD-dependent epimerase/dehydratase domains (E−05 to E−08) and several other predicted domains with less significant E values. This suggests that these three proteins play a role in conversion of different carbohydrates somewhat similar to the three identified aldolases. At1g09340 is a homolog of RAP38 isolated in Chlamydomonas reinhardtii chloroplasts. The stable-isotope labeling showed that this protein had a 5- to 6-fold reduced accumulation after prolonged HL (Table I). It likely partners with RAP41 (At3g63140; Peltier et al., 2006), also identified here in the PGs. C. reinhardtii homologs of RAP41 (At3g63140) and RAP38 (At1g09340) were copurified with 70S chloroplast ribosomes (Yamaguchi and Subramanian, 2003), and curiously a homolog in spinach (Spinacia oleracea) was reported to be an RNA nuclease (Yang et al., 1996; Bollenbach and Stern, 2003).
We identified five additional proteins with unknown functions in the PGs from chloroplasts (At4g13200, At4g01150, At3g10130, At1g28150, and At5g01730; Table I). Pepper homologs were identified for two of those (At1g32220 and At4g13200; Table III). Three of them (At4g13200, At4g01150, and At3g10130) were also identified in our earlier studies on the hydrophobic thylakoid proteome (Friso et al., 2004; Peltier et al., 2004). At4g01150 showed a 3-fold increased relative accumulation in PGs after HL. At3g10130 has a predicted SOUL motif. The Arabidopsis genome has six proteins with a SOUL motif; two of them (At5g20140 and At3g10130) are predicted to be plastid localized and both were identified earlier as thylakoid associated (Peltier et al., 2004). SOUL proteins are heme-binding proteins identified in mammalian cells (Taketani et al., 1998; Zylka and Reppert, 1999; Sato et al., 2004) and may be involved in heme transfer or binding of free heme to prevent damage by reactive oxygen species.
Changes in PG Proteome Composition after Prolonged HL Stress and Degreening
We were able to determine quantitative changes (with independent biological replicates and isotope label switch within each replicate) for more than 20 proteins for stress/control comparisons (Tables I and II). Plotting the averages for each of the three comparisons showed that there was not a bias in up- or down-regulation (data not shown). The fibrillin family responded most differentially to the stress treatments, with decreased HL-to-dark ratios for FIB3, 7a, and 7b, and an increased HL-to-dark ratio for FIB1a. We speculate that the differential expression of the fibrillins might be to (1) accommodate small-molecule content of the PGs; (2) accommodate a suitable environment for metabolic enzymes; and (3) regulate thylakoid membrane interaction and shape and size of the PGs. FIB10 was not identified in PGs from chloroplasts or chromoplasts, but found in thylakoids and rice etioplasts, suggesting that they might have a role in the formation of the PGs from the thylakoid bilayer. FIB8 seems to have a specific role important in the protease mutant and during prolonged darkness; this possibly indicates a role in proteome homeostasis or imbalance between lipid and protein accumulation. The increased accumulation after HL and dark of esterase/lipase/thioesterase is very interesting, and we speculate that the enzyme is involved in lipid breakdown during the extensive stress periods. It is relevant to note that very little is known about thylakoid lipases. The direct experimental comparisons between the two stress treatments (HL/dark) were consistent with the stress/control comparisons.
In particular, after prolonged HL and dark treatments, multiple transmembrane proteins of the abundant thylakoid photosynthetic machinery were found in chloroplast PGs (Table II). The stable isotope-based quantification showed that the HL treatment resulted in more accumulation of photosynthetic products than after degreening (Table II). This suggests either contamination by degraded thylakoid membranes or a role of PG in thylakoid protein recycling (not proteolysis, but rather sequestering similar to inclusion bodies) under extreme stress. PGs from chloroplasts have been proposed to play a role in the removal of protein catabolites as part of thylakoid turnover (Ghosh et al., 1994; Smith et al., 2000). We do note that all PG preparations were light yellow but never green (Fig. 1).
Integration of PG Function with Chloroplast Metabolism and the Unique Nature of the PG Proteome
Most of the proteins identified here in PGs from chloroplasts were not found in the envelope membrane or in the stroma, clearly indicating that the PGs contain a specific protein population. Most of these proteins have no known function but seem to be involved with metabolism of isoprenoid-derived molecules (quinones and tocopherol) and lipids. It is firmly established in several independent studies (cited in the introduction) that PGs in chloroplasts contain tocopherol, at least two different quinones, as well as triacylglycerols, but also mono- and digalactosyl diacylglycerol. The identification in this study of proteins (tentatively) involved with lipids/fatty acids, tocopherols, and quinones is therefore consistent. Importantly, many of the proteins newly identified have not been assigned to any other location and nearly all of them have a predicted N-terminal chloroplast transit peptide, supporting their chloroplast location. It is also very striking that only one (At4g01150) of the 33 proteins identified in the PGs has predicted transmembrane domains. The absence of transmembrane domains is completely consistent with the absence of a lipid bilayer in the PGs (they have a documented monolayer); this is a strong indication that the proteome identified here is not just a random collection of hydrophilic thylakoid or envelope bilayer proteins. Figure 4B shows a schematic overview of how PGs and their proteomes can be integrated in plastid functions.
Furthermore, plastoquinone and phylloquinone are critical for photosynthetic function as electron acceptors and donors, whereas tocopherol is an important protector against oxidative damage to the thylakoid membrane (Havaux et al., 2005; Kanwischer et al., 2005). The presence of proteins somehow involved with lipid metabolism and/or breakdown is also consistent with the proposition that PGs form a lipid/fatty acid storage space (Fig. 4B).
It is often cited that quinones and tocopherol are synthesized in the inner envelope with typical citation of papers from Lichtenthaler et al. (1981) and Soll et al. (1985). However, there is very little experimental evidence for the localization of the known enzymes in tocopherol and plastoquinone synthesis (HPPD, HPT1, or VTE2, VTE3, VTE1, and VTE4; see DellaPenna, 2005). In fact, only VTE3 (also named APG and E37) has clearly been shown to be in the inner envelope (Teyssier et al., 1996) and was also found in large-scale proteome studies on the chloroplast envelope (Ferro et al., 2003; Froehlich et al., 2003). There is little, if any, experimental information on the precise experimental localization of VTE1 and VTE4 functioning downstream of VTE3. Thus, the consistent identification of VTE1 in all four chloroplast PG preparations (control, HL, dark, and from clpr2) and in PGs from chromoplasts makes a good case for its localization in PGs. Finally, envelope-localized VTE2 and VTE3 have one or more predicted transmembrane domains, whereas VTE1 and VTE4 have none. We expect that VTE4 is also localized in PGs. We also identified CCD4 in the PGs, which is indicative of carotenoid breakdown in PGs.
It will be important to experimentally integrate the observed PG proteome information with chloroplast function and metabolism. This will require time-consuming and specialized enzyme and small-molecule analysis. In the absence of such measurements, we propose that PGs form a functional bridge between the thylakoid membrane and the inner envelope membrane in the metabolism of hydrophobic small molecules that are critical in thylakoid function and protection (summarized in Fig. 4B).
Integration of PG Function with Chromoplast Metabolism
The chromoplasts of ripe red peppers do not contain thylakoid membranes and hence lack the need for plasto/phylloquinone production. In contrast, chromoplasts accumulate very large amounts of carotenoids that are mostly sequestered in the fibrillous PGs (Deruere et al., 1994), but no carotenoid biosynthetic enzymes had been identified in PGs of chromoplasts (or any other plastid type). The identification of ZDS, LYC-β, and two CrtR-βs operating in series in bicyclic carotenoid biosynthesis thus makes perfect sense. This suggests that PGs in chromoplasts have a specific enzymatic function in carotenoid biosynthesis in addition to the well-known carotenoid storage/sequestering.
CONCLUSION
This proteome analysis of PGs in chloroplasts and chromoplasts clearly shows that these low-density particles contain enzymes in various pathways. This strongly suggests that PGs are not only a storage compartment for lipophilic thylakoid membrane components, but additionally serve as an active site of synthesis and recycling. The identification of several newly identified enzymes, in particular the four ABC1 kinases with a possible regulatory function in quinone/tocopherol synthesis, warrants future functional studies. The differential labeling with HCHO and DCDO clearly provides a useful tool for non-gel-based comparative proteomics.
MATERIALS AND METHODS
Plant Material and Stress Treatments
Arabidopsis (Arabidopsis thaliana Col-0) was grown for 57 d under conditions optimized for vegetative growth (10 h at 250 μE m−2 s−1 per 14-h dark cycle at 25°C/17°C) or 50 d under standard conditions followed by 1 week in complete darkness or 1 week under continuous light stress (1,500 μE m−2 s−1). Plants were in their vegetative stage prior to bolting. The clpr2-1 mutant with reduced ClpR2 expression (A. Rudella, J.M. Alonso, J.R. Ecker, and K.J. van Wijk, unpublished data) was grown on soil at 100 μE m−2 s−1 under otherwise similar conditions as wild type. Ripe (red) pepper (Capsicum annuum) fruits were purchased from the local store; 30-d-old etiolated rice (Oryza sativa var. Nipponbare) seedlings were grown in complete darkness.
Purification and Yield of PGs
Crude Arabidopsis chloroplast pellets and intact chloroplasts purified through Percoll gradients were obtained (as described in Peltier et al., 2002). These chloroplasts were resuspended in medium R (50 mm HEPES-KOH, pH 8, 5 mm MgCl2, and a cocktail of protease inhibitors, as in Peltier et al., 2002) with 0.5 m Suc and subsequently sonicated (two times for 5 s) or homogenized (Dounce homogenizer), followed by 25-min centrifugation at 150,000g. The resulting floating pad of crude PGs (based on the low density of PGs) was harvested and resuspended in 1 mL of medium R with 0.5 m Suc, overlaid first with 0.3 mL of medium R with 0.2 m Suc and then with 0.3 mL of medium R. After a 25-min centrifugation at 380,000g, pure PGs were collected and stored at −80°C for further analysis. PGs from chromoplasts of ripe pepper fruit or low-density membrane/particles from etioplasts from rice were purified through the same procedure. The PG protein yield per fresh weight of Arabidopsis leaves was about 3 times higher from dark-treated plants and 10 to 12 times higher from HL-treated plants, as compared to wild-type plants kept under optimal conditions.
SDS-PAGE, Protein, and Chlorophyll Determinations
Chlorophyll concentrations and protein determinations were determined (as described in Smith et al., 1985; Porra et al., 1989). Proteins were separated on tricine-SDS-PAGE gels (12% acrylamide) and stained with the fluorescent dye SYPRO ruby.
Protein Identification by MS
For in-solution digestion, proteins were precipitated for 12 to 15 h at −20°C (in batches of 5 or 10 μg) with 100% acetone. The precipitates were collected by centrifugation and washed with 80% acetone, 10% methanol, and 0.2% acetic acid and incubated at −20°C for 30 min. The pellets were centrifuged again, supernatants were removed, and residual acetone was removed by evaporation. The pellets were then dissolved with 20 μL DMSO and diluted to 50 mm NH4HCO3 and 30% DMSO. Trypsin was added to a final protease:protein ratio of 1:20, and proteins were digested overnight at 37°C. The peptide mixtures were dried down and resuspended in 5% formic acid (FA). For identification of proteins from SDS-PAGE gels, stained protein spots were manually excised, washed, and digested with trypsin (as described in Shevchenko et al., 1996). Proteins were analyzed by peptide mass fingerprinting (Arabidopsis 1-D gel samples only) using a MALDI-TOF mass spectrometer (Voyager DE-STR; Applied Biosystems) and/or by MS/MS using a CapLC-ESI-MS/MS (Q-TOF1; Waters). Peptides were loaded on a guard column (LC Packings; MGU-30-C18PM), followed by separation on a PepMap C18 reverse-phase nano column (LC Packings nan75-15-03-C18PM), using 90-min gradients with 95% water, 5% acetonitrile (ACN), 0.1% FA (solvent A), and 95% ACN, 5% water, 0.1% FA (solvent B), and a flow rate of 0.2 μL/min. MS/MS spectra were processed using Mascot distiller (version 1.1.2.0), and the proteins were identified by searching different databases using in-house Mascot (Matrix Science). MS/MS data from pepper proteins were searched against NCBI (all green plants) and the Solanaceae database (downloaded from http://www.sgn.cornell.edu). For Arabidopsis protein data, the Arabidopsis genome was searched (version 5.0 of ATH1.pep). For rice proteome data, we searched the OsGi database downloaded from The Institute for Genomic Research (TIGR; http://www.tigr.org). Criteria for positive identification from peptide mass fingerprinting are described by Friso et al. (2004) and for MS/MS data by Peltier et al. (2004). Identified proteins without unique peptides (i.e. peptides that are not shared with any other protein entries) were kept in the dataset, but denoted ambiguous.
Stable-Isotope Labeling and Quantification
Stable-isotope labeling with HCHO and DCDO was done according to Hsu et al. (2003), with some changes. Peptides (2.25 μg) from each sample were dried down and resuspended with 100 μL labeling buffer (75% ACN, 25 mm MES-KOH, pH 5.5). The peptides were labeled by adding 2 μL of 20% HCHO or DCDO and 1 μL of 260 mm sodium cyanoborohydride, followed by incubation at room temperature for 45 min. The reactions were stopped by adding 1 μL of 4% NH4OH. The HCHO- or DCDO-labeled peptides from different treatments were mixed, dried down, and desalted using C18 microcolumns (Gobom et al., 1999). Peptides were eluted with 95% ACN and 5% FA, dried down, and resuspended in 5% FA for analysis. These mixtures of HCHO- and DCDO-labeled proteomes were analyzed for quantification in MS mode in the Q-TOF1. The area of the peptides was calculated using MassLynx 4.0, followed by normalizing the ratio of the total area of heavy and light isotope-labeled peptides to 1. The area of peptides originating from the same sequence (i.e. different charge states, oxidized Met, and, in a few cases, partially labeled peptides) were added together before calculating ratios between the different treatments to avoid biasing the quantification to a particular peptide. Peptide ratios that were outliers were removed and protein averages calculated. The average and sd of the two replicates were calculated (listed in Tables I and II; for more detail, see Supplemental Table II). Significant change in accumulation levels required that the average ratio (HL/dark, dark/control, or HL/control) was <0.75 sd or >1.33 + 1 sd.
Plastid Proteome Database
The construction of the Plastid Proteome Database (PPDB; http://ppdb.tc.cornell.edu) was originally described by Friso et al. (2004). The PPDB interface was improved, curated information was added, and search functions expanded since its inception in 2004. Mascot scores, number of matching peptides, and highest peptide score for each identification as well as functional classification are listed. Ambiguous identifications of members of multigene families can be directly viewed in PPDB.
Supplementary Material
Acknowledgments
We thank Dr. Daniel Ripoll for theoretical analysis and homology modeling of the ABC1 kinases and Dr. Susan McCouch for providing rice seeds. We also thank Andrea Rudella for providing material of the clpr2-1 mutant, both Andrea Rudella and Dr. Giulia Friso for help with repair and optimization of the CapLC-Q-TOF, and Dr. Wojciech Majeran and Heidi Rutschow for assistance with gels and helpful discussions.
This work was supported by grants from the U.S. Department of Agriculture (USDA-Biochemistry; grant no. 2003–35100–13579) and New York Science and Technology and Research to K.J.v.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Klaas J. van Wijk (kv35@cornell.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.076083.
References
- Bailey JL, Whyborn AG (1963) The osmophilic globules of chloroplasts. II. Globules of the spinach-beet chloroplast. Biochim Biophys Acta 78: 163–174 [Google Scholar]
- Bergmuller E, Porfirova S, Dormann P (2003) Characterization of an Arabidopsis mutant deficient in gamma-tocopherol methyltransferase. Plant Mol Biol 52: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Stern DB (2003) Secondary structures common to chloroplast mRNA 3′-untranslated regions direct cleavage by CSP41, an endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. J Biol Chem 278: 25832–25838 [DOI] [PubMed] [Google Scholar]
- Borderies G, Jamet E, Lafitte C, Rossignol M, Jauneau A, Boudart G, Monsarrat B, Esquerre-Tugaye MT, Boudet A, Pont-Lezica R (2003) Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: a critical analysis. Electrophoresis 24: 3421–3432 [DOI] [PubMed] [Google Scholar]
- Cardazzo B, Hamel P, Sakamoto W, Wintz H, Dujardin G (1998) Isolation of an Arabidopsis thaliana cDNA by complementation of a yeast abc1 deletion mutant deficient in complex III respiratory activity. Gene 221: 117–125 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Sattler S, Maeda H, Sakuragi Y, Bryant DA, DellaPenna D (2003) Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15: 2343–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D (2005) Progress in the dissection and manipulation of vitamin E synthesis. Trends Plant Sci 10: 574–579 [DOI] [PubMed] [Google Scholar]
- Deruere J, Romer S, d'Harlingue A, Backhaus RA, Kuntz M, Camara B (1994) Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell 6: 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emter O, Falk H, Stitte P (1990) Specific carotenoids and proteins as prerequisites for chromoplast tubule formation. Protoplasma 157: 128–135 [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2: 325–345 [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Riviere-Rolland H, Vermat T, Seigneurin-Berny D, Grunwald D, Garin J, Joyard J, Rolland N (2002) Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc Natl Acad Sci USA 99: 11487–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS (2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res 2: 413–425 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hudak KA, Dumbroff EB, Thompson JE (1994) Release of photosynthetic protein catabolites by blebbing from thylakoids. Plant Physiol 106: 1547–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobom J, Nordhoff E, Mirgorodskaya E, Ekman R, Roepstorff P (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom 34: 105–116 [DOI] [PubMed] [Google Scholar]
- Greenwood AD, Leech RM, Williams JP (1963) The osmiophylic globules of chloroplasts. I. Osmiophylic globules as a normal component of chloroplasts and their isolation and composition in Vicia faba L. Biochim Biophys Acta 78: 148–162 [Google Scholar]
- Havaux M, Eymery F, Porfirova S, Rey P, Dormann P (2005) Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17: 3451–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Hsu JL, Huang SY, Chow NH, Chen SH (2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal Chem 75: 6843–6852 [DOI] [PubMed] [Google Scholar]
- Iiizumi M, Arakawa H, Mori T, Ando A, Nakamura Y (2002) Isolation of a novel gene, CABC1, encoding a mitochondrial protein that is highly homologous to Yyast activity of bc1 complex. Cancer Res 62: 1246–1250 [PubMed] [Google Scholar]
- Kanwischer M, Porfirova S, Bergmuller E, Dormann P (2005) Alterations in tocopherol cyclase activity in transgenic and mutant plants of Arabidopsis affect tocopherol content, tocopherol composition, and oxidative stress. Plant Physiol 137: 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Schnell D, Blobel G (1999) Identification of proteins associated with plastoglobules isolated from pea (Pisum sativum L.) chloroplasts. Planta 208: 107–113 [DOI] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P (2001) VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA 98: 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laizet Y, Pontier D, March R, Kuntz M (2004) Subfamily organization and phylogenetic origin of genes encoding plastid-lipid-associated proteins of the fibrillin type. J Genome Sci Technol 3: 19–28 [Google Scholar]
- Langenkamper G, Manac'h N, Broin M, Cuine S, Becuwe N, Kuntz M, Rey P (2001) Accumulation of plastid lipid-associated proteins (fibrillin/CDSP34) upon oxidative stress, ageing and biotic stress in Solanaceae and in response to drought in other species. J Exp Bot 52: 1545–1554 [DOI] [PubMed] [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Hollander-Czytko H, Weiler EW (1996) Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol 31: 323–335 [DOI] [PubMed] [Google Scholar]
- Leonard CJ, Aravind L, Koonin EV (1998) Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res 8: 1038–1047 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Prenzel U, Douce R, Joyard J (1981) Localization of prenylquinones in the envelope of spinach chloroplasts. Biochim Biophys Acta 641: 99–105 [DOI] [PubMed] [Google Scholar]
- Marmagne A, Rouet MA, Ferro M, Rolland N, Alcon C, Joyard J, Garin J, Barbier-Brygoo H, Ephritikhine G (2004) Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol Cell Proteomics 3: 675–691 [DOI] [PubMed] [Google Scholar]
- Motohashi R, Ito T, Kobayashi M, Taji T, Nagata N, Asami T, Yoshida S, Yamaguchi-Shinozaki K, Shinozaki K (2003) Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J 34: 719–731 [DOI] [PubMed] [Google Scholar]
- Naested H, Holm A, Jenkins T, Nielsen HB, Harris CA, Beale MH, Andersen M, Mant A, Scheller H, Camara B, et al (2004) Arabidopsis VARIEGATED 3 encodes a chloroplast-targeted, zinc-finger protein required for chloroplast and palisade cell development. J Cell Sci 117: 4807–4818 [DOI] [PubMed] [Google Scholar]
- Nuhse TS, Stensballe A, Jensen ON, Peck SC (2003) Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics 2: 1234–1243 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Cai Y, Sun Q, Zabrouskov V, Giacomelli L, Rudella A, Ytterberg AJ, Rutschow H, van Wijk KJ (2006) The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol Cell Proteomics 5: 114–133 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, et al (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Ytterberg AJ, Sun Q, van Wijk KJ (2004) New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J Biol Chem 279: 49367–49383 [DOI] [PubMed] [Google Scholar]
- Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, Clarke CF (2000) Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol 182: 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porfirova S, Bergmuller E, Tropf S, Lemke R, Dormann P (2002) Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc Natl Acad Sci USA 99: 12495–12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Pozueta-Romero J, Rafia F, Houlne G, Cheniclet C, Carde JP, Schantz ML, Schantz R (1997) A ubiquitous plant housekeeping gene, PAP, encodes a major protein component of bell pepper chromoplasts. Plant Physiol 115: 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P, Gillet B, Romer S, Eymery F, Massimino J, Peltier G, Kuntz M (2000) Over-expression of a pepper plastid lipid-associated protein in tobacco leads to changes in plastid ultrastructure and plant development upon stress. Plant J 21: 483–494 [DOI] [PubMed] [Google Scholar]
- Sato E, Sagami I, Uchida T, Sato A, Kitagawa T, Igarashi J, Shimizu T (2004) SOUL in mouse eyes is a new hexameric heme-binding protein with characteristic optical absorption, resonance Raman spectral, and heme-binding properties. Biochemistry 43: 14189–14198 [DOI] [PubMed] [Google Scholar]
- Sattler SE, Cahoon EB, Coughlan SJ, DellaPenna D (2003) Characterization of tocopherol cyclases from higher plants and cyanobacteria. Evolutionary implications for tocopherol synthesis and function. Plant Physiol 132: 2184–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Cheng Z, DellaPenna D (2004) From Arabidopsis to agriculture: engineering improved vitamin E content in soybean. Trends Plant Sci 9: 365–367 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Shimada H, Ohno R, Shibata M, Ikegami I, Onai K, Ohto MA, Takamiya K (2005) Inactivation and deficiency of core proteins of photosystems I and II caused by genetical phylloquinone and plastoquinone deficiency but retained lamellar structure in a T-DNA mutant of Arabidopsis. Plant J 41: 627–637 [DOI] [PubMed] [Google Scholar]
- Smith MD, Licatalosi DD, Thompson JE (2000) Co-association of cytochrome f catabolites and plastid-lipid-associated protein with chloroplast lipid particles. Plant Physiol 124: 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Soll J, Schultz G, Joyard J, Douce R, Block MA (1985) Localization and synthesis of prenylquinones in isolated outer and inner envelope membranes from spinach chloroplasts. Arch Biochem Biophys 238: 290–299 [DOI] [PubMed] [Google Scholar]
- Sperling U, Franck F, van Cleve B, Frick G, Apel K, Armstrong GA (1998) Etioplast differentiation in Arabidopsis: both PORA and PORB restore the prolamellar body and photoactive protochlorophyllide-F655 to the cop1 photomorphogenic mutant. Plant Cell 10: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA (2003) Chloroplast structure: from chlorophyll granules to supra-molecular architecture of thylakoid membranes. Photosynth Res 76: 185–196 [DOI] [PubMed] [Google Scholar]
- Steinmuller D, Tevini M (1985) Composition and function of plastoglobuli. I. Isolation and purification from chloroplasts and chromoplasts. Planta 163: 201–207 [DOI] [PubMed] [Google Scholar]
- Taketani S, Adachi Y, Kohno H, Ikehara S, Tokunaga R, Ishii T (1998) Molecular characterization of a newly identified heme-binding protein induced during differentiation of urine erythroleukemia cells. J Biol Chem 273: 31388–31394 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tevini M, Steinmuller D (1985) Composition and function of plastoglobuli. II. Lipid composition of leaves and plastoglobuli during beech senescence. Planta 163: 91–96 [DOI] [PubMed] [Google Scholar]
- Teyssier E, Block MA, Douce R, Joyard J (1996) Is E37, a major polypeptide of the inner membrane from plastid envelope, an S-adenosyl methionine-dependent methyltransferase? Plant J 10: 903–912 [DOI] [PubMed] [Google Scholar]
- Tuquet C, Newman DW (1980) Aging and regreening in soybean cotyledons. 1 Ultrastructural changes in plastids and plastoglobuli. Cytobios 29: 43–59 [PubMed] [Google Scholar]
- Van Eenennaam AL, Lincoln K, Durrett TP, Valentin HE, Shewmaker CK, Thorne GM, Jiang J, Baszis SR, Levering CK, Aasen ED, et al (2003) Engineering vitamin E content: from Arabidopsis mutant to soy oil. Plant Cell 15: 3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnevetsky M, Ovadis M, Zuker A, Vainstein A (1999) Molecular mechanisms underlying carotenogenesis in the chromoplast: multilevel regulation of carotenoid-associated genes. Plant J 20: 423–431 [DOI] [PubMed] [Google Scholar]
- von Malek B, van der Graaff E, Schneitz K, Keller B (2002) The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wu SS, Platt KA, Ratnayake C, Wang TW, Ting JT, Huang AH (1997) Isolation and characterization of neutral-lipid-containing organelles and globuli-filled plastids from Brassica napus tapetum. Proc Natl Acad Sci USA 94: 12711–12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2003) Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur J Biochem 270: 190–205 [DOI] [PubMed] [Google Scholar]
- Yang J, Schuster G, Stern DB (1996) CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell 8: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Reppert SM (1999) Discovery of a putative heme-binding protein family (SOUL/HBP) by two-tissue suppression subtractive hybridization and database searches. Brain Res Mol Brain Res 74: 175–181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.