Abstract
Different classes of plant hormones and different wavelengths of light act through specific signal transduction mechanisms to coordinate higher plant development. A specific prephenate dehydratase protein (PD1) was discovered to have a strong interaction with the sole canonical G-protein Gα-subunit (GPA1) in Arabidopsis (Arabidopsis thaliana). PD1 is a protein located in the cytosol, present in etiolated seedlings, with a specific role in blue light-mediated synthesis of phenylpyruvate and subsequently of phenylalanine (Phe). Insertion mutagenesis confirms that GPA1 and the sole canonical G-protein-coupled receptor (GCR1) in Arabidopsis also have a role in this blue light-mediated event. In vitro analyses indicate that the increase in PD1 activity is the direct and specific consequence of its interaction with activated GPA1. Because of their shared role in the light-mediated synthesis of phenylpyruvate and Phe, because they are iteratively interactive, and because activated GPA1 is directly responsible for the activation of PD1; GCR1, GPA1, and PD1 form all of or part of a signal transduction mechanism responsible for the light-mediated synthesis of phenylpyruvate, Phe, and those metabolites that derive from that Phe. Data are also presented to confirm that abscisic acid can act through the same pathway. An additional outcome of the work is the confirmation that phenylpyruvate acts as the intermediate in the synthesis of Phe in etiolated plants, as it commonly does in bacteria and fungi.
The aromatic amino acid Phe is required for protein synthesis and also serves as the precursor to a large number of secondary metabolites critical to normal development and stress responses (Gilchrist and Kosuge, 1980; Chapple et al., 1994; Kliebenstein, 2004; Winkel, 2004), including UV screening and other pigments (Burchard et al., 2000; Ruegger and Chapple, 2001; Casati and Walbot, 2005). Despite the fact that 30% to 40% of the mass of an individual plant can be derived from aromatic amino acids l-Phe and l-Tyr, little is known about their biosynthetic pathway in plants (Lewis and Yamamoto, 1989; Davin et al., 1992; Razal et al., 1994, 1996). In light-grown seedlings, the Shikimate pathway links carbon assimilated via photosynthesis to the biosynthesis of aromatic amino acids, through the formation of chorismate. However, only a small number of studies have addressed the biochemical pathway responsible for the conversion of chorismate to Phe in light-grown plants (Razal et al., 1996; for review, see Schmid and Amrhein, 1995). To our knowledge, there are no such studies in etiolated seedlings. The Shikimate metabolic pathway does appear to function, albeit at low levels, in etiolated seedlings (Razal et al., 1994; Hemm et al., 2004).
Jung et al. (1986) reported the presence of arogenate in the chloroplasts of light-grown plant cells in culture. It was proposed from these data that production of Phe only proceeds via an arogenate intermediate. Subsequent investigations have failed to confirm the presence of arogenate directly from plants in either light- or dark-grown seedlings (Razal et al., 1994).
In bacteria and fungi the pathway is better understood: the product of chorismate mutase, prephenate, is converted to phenylpyruvate, and phenylpyruvate converted to Phe (Haslam, 1993). These processes are catalyzed by the actions of two well-studied enzymatic activities, chorismate mutase (catalyzing the conversion of chorismate to prephenate) and prephenate dehydratase (PD; catalyzing the conversion of prephenate to phenylpyruvate). Depending on the species, the two activities occur in a single, fused protein or as two separate proteins. Synthesis via arogenate can act as an alternative pathway (Patel et al., 1977), and in some organisms, a third enzyme cyclohexadienyl dehydratase, is reported to have both PD and arogenate dehydratase activities (Xia et al., 1991).
Biochemical and genetic studies confirm that Arabidopsis (Arabidopsis thaliana) has three chorismate mutase genes (Romero et al., 1995). Two of the encoded proteins locate to the plastid (CM1 and CM3) and one is cytosolic (CM2; Eberhard et al., 1996; Mobley et al., 1999). The Arabidopsis genome is not known to encode for an arogenate dehydratase or a cyclohexadienyl dehydratase, the enzymatic activities necessary to convert arogenate to Phe. Conversely, the Arabidopsis genome codes for six different PD genes as listed in GenBank and SIGnAL databases, strongly suggesting that Arabidopsis does use PD in the production of Phe.
In higher plants, blue light (BL) is known to affect the phenylpropanoid pathway, which initiates with Phe, and for which Phe can be a limiting component (Bruns et al., 1986; Ohl et al., 1989; Long and Jenkins, 1998; Noh and Spalding, 1998). At least three photoreceptor systems are known to act in BL-mediated phenomena: PHOTOTROPIN, CRYPTOCHROME, and PHYTOCHROME (Ahmad, 1999; Briggs and Huala, 1999; Casal, 2000; Lin, 2000; Chen et al., 2004). None of these are known to have a primary role in the initial rates of Phe synthesis in young (less than 7-d-old) etiolated seedlings. CRYPTOCHROME-effected regulatory pathways appear to regulate the synthesis of several compounds made in the phenylpropanoid pathway, including anthocyanins, when measured in plants grown in continuous BL conditions (Mol et al., 1996; Long and Jenkins, 1998; Noh and Spalding, 1998; Wade et al., 2001).
Separate from the above photosystems, the recently described blue low fluence (BLF) system, consisting of G-protein-coupled receptor 1 (GCR1), G-protein Gα-subunit 1 (GPA1), Pirin 1 (PRN1), and LEAFY COTYLEDON 1 (also known as NF-Y-B9; K.M. Warpeha, S.I. Hawkins, and L.S. Kaufman, unpublished data) utilizes an unknown receptor. In etiolated seedlings, the BLF system has a threshold at 10−1 μmol m−2 of BL and is known to have a role in the induction of transcription of specific members of the Lhcb gene family, alter rates of chlorophyll synthesis, and affect the rate of stem elongation (Warpeha and Kaufman, 1990a, 1990b). The BLF system is activated by a single, short pulse of BL, exhibits reciprocity, has an immediate effect on transcription, and functions in the absence of protein synthesis (Marrs and Kaufman, 1991; Anderson et al., 1999).
Heterotrimeric G-protein-mediated cell signaling is one of the most highly conserved signaling mechanisms in eukaryotes. In contrast to animals, where they may number in the hundreds, plant G-protein subunits are coded by only one or sometimes two genes (Ma et al., 1990; Marsh and Kaufman, 1999; Assmann, 2002). Analysis of Arabidopsis plants mutant in GPA1 gene reveal that G proteins are likely to be involved in a number of processes critical for proper plant development, including hypocotyls and leaf formation (Ullah et al., 2001), stomatal opening (Wang et al., 2001), abscisic acid (ABA) responsiveness/germination (Colucci et al., 2002; Ullah et al., 2002; Lapik and Kaufman, 2003), and both ABA and BL regulation of gene expression (K.M. Warpeha, unpublished data). The importance of G proteins in cell signaling and early development in Arabidopsis suggests the occurrence of many effector proteins.
As part of a study designed to identify the effect of GPA1-interacting proteins on G proteins and their BL regulation (Lapik and Kaufman, 2003), we identified physical interactions between GPA1 and three potential effectors, PRN1, a cupin-fold superfamily member, and NF-Y (CCAAT-box-binding protein) interactive protein (Dunwell et al., 2004; Pang et al., 2004) with a role in the BLF system-mediated transcription (K.M. Warpeha, unpublished data), PRK1, a CTR1-like raf kinase (K.M. Warpeha, Y.R. Lapik, R. Muppavarapu, and L.S. Kaufman, unpublished data), and PD1, a member of the PD family.
The data presented herein demonstrate that, in etiolated Arabidopsis seedlings, a single short pulse of low-fluence BL, acting through a signal transduction mechanism involving GCR1, GPA1, and PD1, results in the specific activation of PD1 and the subsequent accumulation of Phe. This BLF system induction of PD1 activity occurs through the specific interaction of PD1 with activated GPA1. The data also confirm that etiolated plants can synthesize Phe via a phenylpyruvate intermediate as is commonly observed for other organisms. Data are also presented to confirm that ABA can potentially act through the same pathway.
RESULTS
PD1 Has a Specific Interaction with GPA1
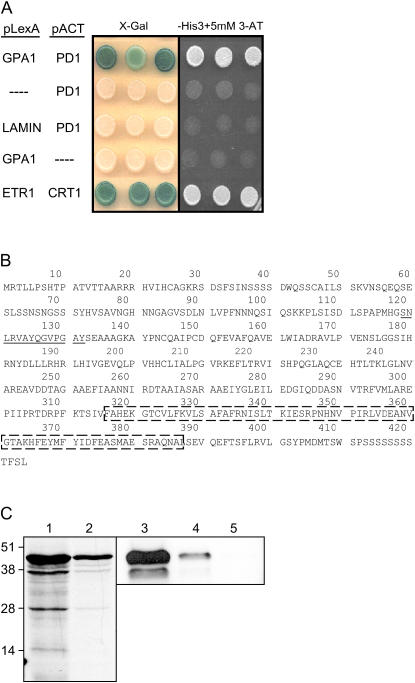
A yeast (Saccharomyces cerevisiae) two-hybrid screen was used to identify transcripts derived from etiolated Arabidopsis seedlings that encode for proteins with the potential to interact with GPA1. The screen identified several different transcripts encoding potential partners (Lapik and Kaufman, 2003) including PD1, a putative cytosolic PD approximately 46 kD, encoded by At2g27820 (Fig. 1, A and B). The potential for physical interaction between GPA1 and PD1 was confirmed using an in vitro pull-down procedure (Fig. 1C). The Arabidopsis genome is reported as having five additional genes coding for PD (GenBank; http://www.ncbi.nlm.nih.gov; http://signal.salk.edu).
Figure 1.
Protein-protein interactions between GPA1 and PD1. A, PD1 binds to GPA1 in the yeast two-hybrid assay. PD1 was identified in a yeast two-hybrid screen as a potential GPA1-interacting protein. The figure represents the results of a directed two-hybrid assay to confirm the interaction. Bait and prey plasmids used were pLexA-NLS-AGA (fusion to the LexA DNA-binding domain) and pACT (fusion to the GAL4 activation domain), respectively. The left section shows growth of yeast colonies on synthetic media containing X-gal; blue colonies indicate a positive interaction resulting in the expression a LacZ reporter gene. The right section shows yeast growth on the SD-His 3-AT-containing media, colony growth indicates a positive interaction resulting in the expression of a HIS3 reporter gene. Proteins expressed in either vector are depicted on the figure. Use of the pACT vector with no insert and the pLexA-NLS vector with a lamin insert represent negative controls. pLexA-NLS-ETR1 and CTR1pACT coexpression is used as a positive control. B, Predicted amino acid sequence of PD1 in Arabidopsis. The predicted amino acid sequence of PD1 (Gene: At2g27820; AAC73018.1). The sequences corresponding to the PD domain (underlined sections) and the Phe-binding domain (boxed-in sections) of E. coli P protein (AAC75648.1) are indicated on the figure. C, In vitro interaction of PD1 and GPA1. Lanes 1 and 2 represent an SDS-PAGE analysis of the initial and partially purified PD1 derived from a coupled in vitro transcription/translation reaction, respectively. Lanes 3 to 5 represent the autoradiograph of an SDS-PAGE analysis showing in vitro translated 35S-labeled PD1 protein alone (lane 3), the column-purified products of the interaction between in vitro translated 35S-labeled PD1 protein and a glutathione S-transferase-GPA1 fusion protein (lane 4), and the column-purified products of the interaction between in vitro translated 35S-labeled PD1 protein and glutathione S-transferase alone (lane 5).
BL Stimulates the Production of Phe
Because GPA1 has a role in BL signal transduction (Warpeha et al., 1991; K.M. Warpeha, unpublished data), and because of the well-characterized and committed role of PD in the production of Phe in bacteria and fungi (Haslam, 1993), the levels of several amino acids were measured in mock-irradiated in darkness (DK) and BL-treated (BL, single pulse of 104 μmol m−2) 6-d-old dark-grown wild-type Arabidopsis seedlings.
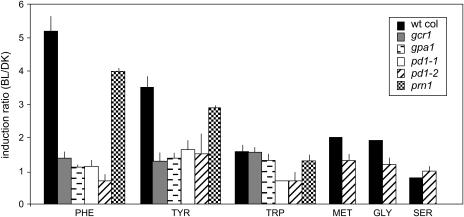
The data presented in Figure 2 indicate that a single pulse of low-fluence BL enhances the accumulation of Phe and Tyr in etiolated wild-type Arabidopsis by approximately 6- and 4-fold, respectively, 2 h after irradiation. In wild-type seedlings, the levels of Trp, whose synthetic pathway branches at chorismate, the immediate precursor to prephenate (the presumed substrate for PD1) and other amino acids (e.g. Ser, Gly, and Met) are generally unaffected by the BL treatment. The data indicate that BL enhances the synthesis of the aromatic amino acids and may be a key regulator downstream of the chorismate branch point.
Figure 2.
BL-induced amino acid accumulation in etiolated wild-type seedlings and pd1-1, pd1-2, gcr1, gpa1, and prn1 mutant seedlings. Six-day-old dark-grown seedlings were irradiated with no light (DK) or 104 μmol m−2 of BL. Two hours after irradiation period, the aerial portions of the seedlings were frozen and ground in liquid nitrogen, and hydrolyzed in preparation for HPLC analysis of amino acid content. Data are shown for Phe, Tyr, Trp, Met, Gly, and Ser. Levels are indicated for T-DNA insertion mutants in these genes (GCR1, GPA1, PD1, and PRN1) and wild-type (Col) seedlings. Data for wild-type Col, and pd1-1 and pd1-2 mutants represent three independent replicates, and error bars represent SEM. Data for gcr1, gpa1, and prn1 mutants represent the average of two independent replicates, and error bars represent the sd. Data for MET, SER, and GLY are only shown for pd1-2. wt, Wild type.
BL-Induced Phe and Tyr Accumulation Occurs via a PD1-Requiring Pathway
To determine if PD1 is in fact responsible for the demonstrated BL-induced increase in Phe and Tyr accumulation, we tested T-DNA insertion mutants of PD1 for their ability to accumulate Phe, Tyr, and Trp in response to BL treatment. The data (Fig. 2) indicate that neither Phe nor Tyr accumulates in the pd1-1 or pd1-2 mutants 2 h after BL irradiation. The levels of Trp and other amino acids also remain unaffected by the BL. These data confirm that PD1 is responsible for BL-induced accumulation of the aromatic amino acids in etiolated Arabidopsis and strongly supports the idea that phenylpyruvate, the immediate product of PD, can act as the immediate precursor to Phe.
Cytosolic PD Activity Is Increased by BL
The derived amino acid sequence for PD1 suggests that it codes for a cytosolic PD. To test for PD activity in a plastid-free cytosolic fraction and to determine if that activity is BL regulated, we adapted a standard cell extract assay based on Euverink et al. (1995), for use with Arabidopsis. Wild type and insertion mutants of PD1 Arabidopsis seedlings were grown for 6 d in complete DK, given a mock BL treatment (DK), or irradiated with a single brief pulse of BL (total fluence 104 μmol m−2) and returned to the dark for 2 h. Cytosolic extracts were tested for their ability to convert exogenously added prephenate (the precursor) to phenylpyruvate (the product) by measuring the change in A320 over time (Euverink et al., 1995).
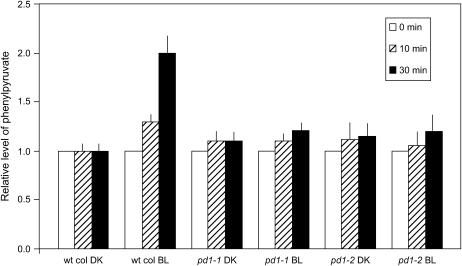
The data presented in Figure 3 confirm the presence of a PD activity in the cytosolic fraction and indicate that the activity is enhanced only in those extracts derived from the BL-treated wild-type seedlings. Extracts obtained from pd1-1 and pd1-2 mutant seedlings do not indicate BL-induced phenylpyruvate synthesis, suggesting that PD1 is the sole cytosolically located, BL-induced PD activity in etiolated seedlings.
Figure 3.
BL treatment leads to an increase in the rate of conversion of prephenate to phenylpyruvate in the cytoplasm. Six-day-old dark-grown wild-type Col and pd1 mutant seedlings were irradiated with no light (DK) or a single pulse of low-fluence BL (104 μmol m−2). Cell-free extracts (at 5 mg/mL protein), were made 2 h after irradiation, prephenate was added to 1.0 mm, and the resulting levels of phenylpyruvate were measured spectrophotometrically at the time points indicated on the figure. Data represent at least three independent replicates, and error bars represent the SEM. wt, Wild type.
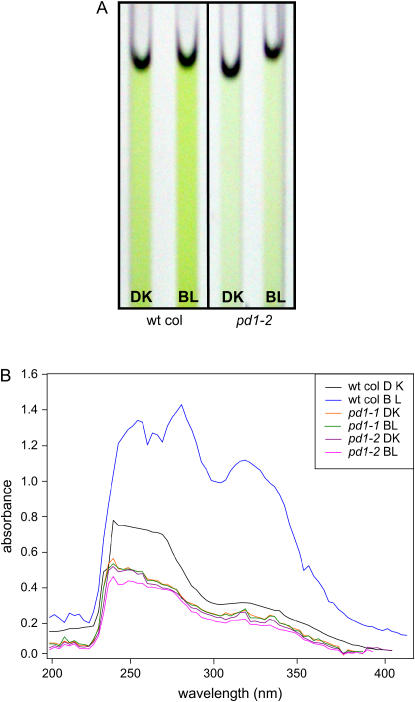
Phenylpyruvate, the immediate product of PD1, is a committed precursor to Phe. Phenyl-alanine itself is a precursor to those compounds in phenylpropanoid pathway, many of which absorb in the UV/BL range (e.g. anthocyanins). The resultant absorption spectra obtained from the cytosolic extract assay of BL-irradiated wild-type seedlings show a profound increase in, and difference in, compounds that absorb light in the UV/BL region. Changes in pigment composition and quantity of absorbing molecules can be seen in the cytosolic extracts themselves (Fig. 4A) and in the absorption spectra of those extracts (Fig. 4B). Absorption spectra of extracts from the pd1-1 and pd1-2 mutants with and without BL irradiation are highly similar to extracts from dark-grown wild-type seedlings, suggesting that PD1 is responsible for the Phe that in turn leads to the production of those BL-induced compounds responsible for the spectral changes in etiolated wild-type seedlings.
Figure 4.
Pigment accumulation and spectral absorption characteristics of pd1 insertion mutants as compared to wild-type Col. A, Pigments accumulating in cytosolic extracts. Six-day-old dark-grown wild-type Col and insertion mutants (pd1) of PD1 gene were irradiated with no light (DK) or a single pulse of low-fluence BL (104 μmol m−2). Cell-free extracts (approximately 5 mg/mL protein) were made 2 h after irradiation, prephenate was added to 1.0 mm, and the solutions were incubated for an additional 30 min in the dark, then photographed in quartz cuvettes. Sample data for pd1-2 is shown. B, Absorption spectra of cytosolic extracts. Absorption spectra between 200 and 400 nm of the cytosolic extracts described in part A, for both pd1-1 and pd1-2 mutants compared to wild type. wt, Wild type.
GCR1, GPA1, and PD1 Form a Signaling Chain That Effects BL-Induced Phe Accumulation
The data presented in Figures 1 to 4 demonstrate that GPA1 can interact with PD1 and that PD1 is responsible for the BL induction accumulation of phenylpyruvate, Phe, and a range of UV- and BL-absorbing compounds. Pandey and Assmann (2004) recently demonstrated that GCR1 can interact with GPA1, and we recently demonstrated that both GCR1 and GPA1 participate in the BL induction of Lhcb RNA accumulation (K.M. Warpeha, unpublished data). Together, these data suggest that GCR1, GPA1, and PD1 form all or part of a signal transduction mechanism responsible for BL-induced Phe accumulation in etiolated Arabidopsis.
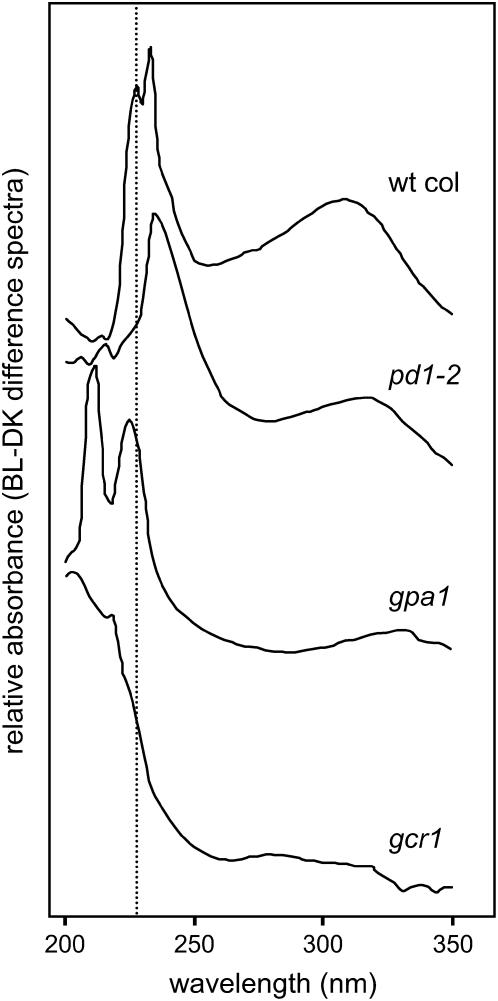
To test this hypothesis, we measured the ability of etiolated T-DNA single-insertion mutants in GCR1 and GPA1 genes to accumulate the amino acids Phe, Tyr, and Trp, as well as UV- and BL-absorbing compounds in response to irradiation with BL. The data shown in Figure 2 indicate that neither mutant accumulates Phe or Tyr, confirming a role for GCR1 and GPA1 in this BL response. The difference spectra shown in Figure 5 indicate that the cytoplasmic constituents of BL-irradiated gcr1 and gpa1 mutants differ considerably from wild type, each other, and the pd1 mutants. The general trend appears to be one in which the pd1 mutant has lost the widest range of UV-absorbing material (actually resembling unirradiated wild-type seedlings), the gcr1 mutant has lost the least, and the gpa1 mutant is somewhat intermediate.
Figure 5.
BL-DK difference spectra of cytosolic extracts of insertion mutants pd1, gpa1, gcr1, and wild-type Col. Six-day-old dark-grown wild-type Col, and pd1, gpa1, and gcr1 mutant seedlings were irradiated with no light (DK) or a single pulse of low-fluence BL (104 μmol m−2). Extracts were made 2 h after irradiation by the same methods as for amino acid studies. The difference spectra between 200 and 350 nm are shown for BL-DK absorption. The dotted line at 231 nm corresponds to the absorption peak for Phe. The example spectrum for pd1 mutants is pd1-2. wt, Wild type.
Activated GPA1 Can Increase PD1 Activity
The data presented in Figure 1 confirm that GPA1 and PD1 can interact, but do not address the biochemical consequences of that interaction. The fact that a single pulse of BL can activate both Gα (Warpeha et al., 1991) and PD1, and that BL activation of PD1 activity is lost in the gpa1 mutant suggests that the interaction between activated (GTP-bound) GPA1 and PD1 is responsible for the BL-induced increase in PD1 activity.
PD1 and GPA1 were synthesized and purified as previously described (Lapik and Kaufman, 2003). GPA1 is generally thought to be synthesized in an inactive state (i.e. resembling the structure when GDP is bound, even though no GDP is present). This structure can be changed to an active one by incubating GPA1 with the nonhydrolyzable GTP analog GTPγS (Lapik and Kaufman, 2003).
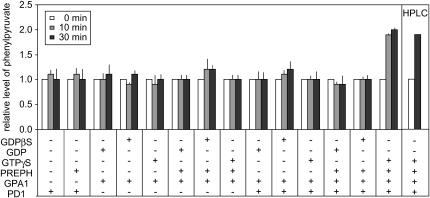
GPA1 was preincubated with GTPγS (producing activated GPA1), GDP, or GDPβS (a nonhydrolyzable analog of GDP-producing inactive GPA1), prior to incubation with PD1 and prephenate, and subsequent monitoring of PD1 activity. The data presented in Figure 6 indicate that incubation of PD1 with activated GPA1 results in a 2-fold increase in PD1 activity when compared with inactive (incubation with GDP or GDPβS) or no GPA1. The increase in activity occurs quickly as approximately 90% of the measured increase in activity occurs within the first 10 min of incubation. These data are confirmed by HPLC analysis of the prephenate-to-phenylpyruvate conversion rate (Fig. 6).
Figure 6.
Preactivated GPA1 can increase PD1 activity by 2-fold. The figure depicts an in vitro assay where in vitro-translated GPA1 and PD1 were combined with a number of nonhydrolyzable analogs to mimic either inactive (GDP or GDPβS) GPA1 or activated (GTPγS) GPA1 to test if active or inactive GPA1 was capable of interacting with PD1 to metabolize added prephenate. Purified prephenate was used and added to final concentration of 1.0 mm where indicated (PREPH). Progress of reaction was determined at 0, 10, and 30 min postinitiation by methods as indicated in Figure 3, then quantifying phenylpyruvate formation by absorption characteristics also as described for Figure 3. Results were confirmed by HPLC. SEM is indicated.
pd1, gcr1, and gpa1 Mutants Do Not Synthesize Specifically Located UV- and BL-Absorbing Compounds in Response to Treatment with BL
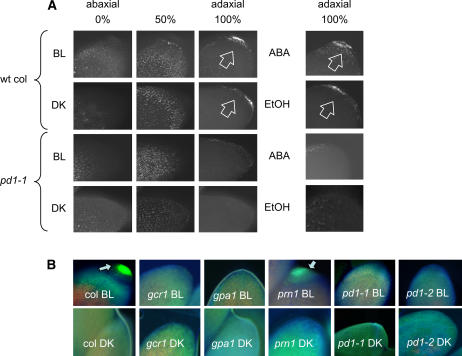
The absorption spectra and visual data presented in Figures 4 and 5 indicate that BL treatment of wild-type Arabidopsis results in a significant increase in the amount and range of UV- and BL-absorbing compounds, and that PD1 activity is required for the synthesis of these compounds. We used deconvolution microscopy and optical sectioning to help define the location(s) of the absorbing compounds synthesized in wild-type seedlings in response to BL treatment, and as a consequence of PD1 activity. The results shown in Figure 7A indicate the presence of a set of cells in the upper layers of the cotyledon tip containing UV- and BL-absorbing fluorescent material, which increases in accumulation in response to the BL treatment. This fluorescent material is absent from unirradiated (DK) and irradiated (BL) pd1 mutants.
Figure 7.
Natural fluorescence in the cotyledons of 7-d-old wild-type Col and mutant seedlings. A, Seedlings of wild-type Col and pd1 mutant seedlings were grown for 6 d in complete DK. Seedlings were mock irradiated with either no light (DK) or irradiated with 104 μmol m−2 of BL or ABA (in ethanol) or just ethanol (ethanol), returned to the dark for 24 h after which the cotyledons were photographed on a deconvoluting microscope. The images shown are natural fluorescence and represent optical sections at 0%, 50%, and/or 100% thickness from abaxial to adaxial surfaces of the cotyledon using 4′,6-diamino-phenylindole and fluorescein isothiocyanate excitation-blocking filter sets. The arrows indicate fluorescent material at the tip of the cotyledon present in wild-type seedlings that is absent in pd1 mutant seedlings (example for pd1 shown is pd1-1). B, Seedlings of wild-type Col, and gcr1, gpa1, prn1, pd1-1, and pd1-2 mutant seedlings were grown, irradiated, and photographed as described in A. Photographs shown represent the whole cotyledon. The arrows indicate fluorescent materials at the tip of the cotyledon. wt, Wild type.
Because ABA is able to elicit Lhcb transcription in etiolated cotyledons using a signal transduction pathway identical to that of BL and requiring both GCR1 and GPA1 (K.M. Warpeha, unpublished data) we tested application of ABA to etiolated cotyledons to determine the effect on accumulation of UV- and BL-absorbing fluorescent material. Figure 7A demonstrates that the ABA treatment results in a phenotype identical to that observed in wild-type seedlings for BL irradiation, and, like BL, the response to ABA is dependent upon PD1.
Figure 7B shows the natural fluorescence of whole cotyledons and confirms that gcr1 and gpa1 mutants also lack the BL-induced UV-absorbing fluorescent material at the tip of the cotyledon observed for pd1-1 and pd1-2 mutants. The prn1 mutant appears to have the same fluorescence pattern as wild-type seedlings.
Dark-Grown pd1 Mutants Do Not Accumulate Anthocyanins
The data presented in Figure 4 suggest that similar UV- and BL-absorbing compounds are synthesized in etiolated wild-type (nonirradiated) and pd1-1 and pd1-2 mutant seedlings, although a greater quantity is synthesized in wild-type seedlings. The data also suggest that an additional set of UV- and BL-absorbing compounds are synthesized in wild-type seedlings via a BL-activated PD1-dependent pathway. We do not yet know the specific compounds made. Others have demonstrated that continuous BL will stimulate anthocyanin (a UV/BL-absorbing product of the PD1-dependent phenylpropanoid pathway) synthesis in Arabidopsis (Mol et al., 1996; Noh and Spalding, 1998).
The data in Table I show that etiolated pd1-1, pd1-2, and wild-type seedlings accumulate anthocyanin and that the level of accumulation in wild type is approximately 30% greater then in the pd1 mutant seedlings, and this difference is significantly different according to the Student's t test. This is consistent with the overall difference in UV- and BL-absorbing compounds seen in Figure 4. Table I also shows that measurable anthocyanins in wild-type seedlings exposed to continuous BL for 48 h does not differ appreciably from that measured for pd1-1 or pd 1-2 seedlings similarly treated.
Table I.
Anthocyanins accumulation (absorbance/g fresh weight)
Anthocyanins production in dark-grown seedlings. Seedlings were grown for 7 d in complete DK or for 5 d in complete DK followed by 48 h of continuous BL at 100 μmol m−2 s−1. Anthocyanins extraction and quantification were performed on fresh tissue according to Noh and Spalding (1998). Data derive from three independent replicates. Errors represent the SEM. Replicates (three) and means were analyzed with the Student's t test.
| Col Wild Type | pd1-1 | pd1-2 | |
|---|---|---|---|
| DK | 0.03a ± 0.004 | 0.02a ± 0.002 | 0.02a ± 0.002 |
| Continuous BL | 1.54 ± 0.056 | 1.31 ± 0.013 | 1.25 ± 0.056 |
The means of the mutant anthocyanins levels are significantly different from wild-type levels (>95% confidence level).
The BL-Induced Accumulation of Phe Is Not Part of the Post-GPA1-Signaling Mechanism That Leads to the BL Regulation of Lhcb
The iterative interaction between GCR1, GPA1, and PD1, the common phenotype of the T-DNA insertion mutants, and the ability of activated GPA1 to activate PD1 defines PD1 as a GPA1 effector and GCR1, GPA1, and PD1 as a signal transduction chain for the BLF system activation of PD1 activity.
A single pulse of low-fluence BL also induces the immediate expression of Lhcb in etiolated pea (Pisum sativum) and Arabidopsis (Marrs and Kaufman, 1991; Gao and Kaufman, 1994). The signal transduction mechanism responsible for this gene expression is comprised in part by a GCR1, GPA1, PRN1, and NF-Y-signaling chain, wherein PRN1 acts as the GPA1 effector (K.M. Warpeha, unpublished data). To confirm the divergence of these two signaling mechanisms at the level of the effectors, we tested a T-DNA insertion mutant in the PRN1 gene for ability to accumulate the amino acids Phe, Tyr, and Trp in response to BL treatment (Fig. 2), and ability to produce fluorescent material at the tip of the cotyledon in response to BL treatment (Fig. 7B). In both instances, the prn1 mutant appears to respond like wild type: The prn1 mutant accumulates both Phe and Tyr, and the tip of the cotyledon has natural fluorescence similar to that observed for wild-type seedlings.
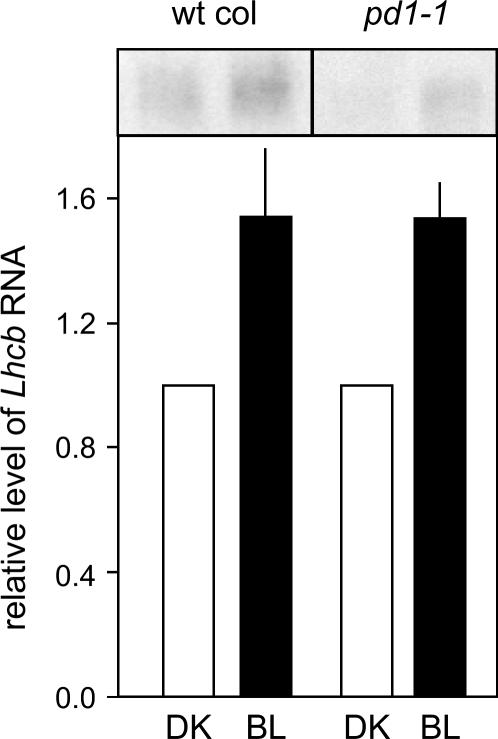
We also tested the ability of pd1 insertion mutants to accumulate Lhcb RNA in response to BL treatment (Fig. 8). The data indicate that BL-induced accumulation of Lhcb RNA for pd1 mutants is similar to the level of accumulation observed for wild-type seedlings.
Figure 8.
PD1 is not involved in the BL induction of Lhcb. Six-day-old dark-grown wild-type Col and pd1 mutant seedlings were irradiated with a single pulse of low-fluence BL (102 μmol m−2) or no light (DK), and were placed back in the dark for 2 h after which total RNA was extracted and used for northern analysis (Lapik and Kaufman, 2003). Dark levels are set to 1.0. Example mutant shown is pd1-1. Northern blots shown are representative. Plotted data derive from at least three independent replicates. Error bars represent the SEM. wt, Wild type.
DISCUSSION
Phe-derived compounds can account for as much as 30% to 40% of the mass of a plant (Lewis and Yamamoto, 1989; Davin et al., 1992). As far as we are aware, the actual pathway by which Phe is produced or the various ways in which that pathway is regulated have not been explored by modern genetic and molecular-genetic means.
The two main synthetic pathways by which prokaryotes and fungi produce Phe rely on the committed synthesis of prephenate from chorismate-via-chorismate mutase. The major synthetic pathway proceeds via the conversion of prephenate to phenylpyruvate by PD and the subsequent conversion phenylpyruvate to Phe by phenylpyruvate amino transferase (Haslam, 1993). Chorismate mutase and PD activities can occur as two separate proteins or, in the case of P protein in Escherichia coli, as a fused protein with both activities (Zurawski et al., 1977; Hudson and Davidson, 1984; Romero et al., 1995). An alternative pathway proceeds via the conversion of chorismate to arogenate via the action of a prephenate amino transferase and the subsequent conversion of arogenate to Phe via the action of arogenate dehydratase (Patel et al., 1977). Cyclohexadienyl dehydratase is reported to have both PD and arogenate dehydratase activities (Xia et al., 1991).
It has been the assumption that in plants, unlike fungi wherein synthesis takes place in the cytosol, that the postchorismate steps occur in the chloroplast via an arogenate intermediate (for review, see Schmid and Amrhein, 1995). Examination of the Arabidopsis genome reveals no copies of arogenate dehydratase or cyclohexadienyl dehydratase. Further, Razal et al. (1994) were unable to isolate or detect arogenate from any plant grown under any light condition.
In contrast, the Arabidopsis genome contains three genes coding for chorismate mutase, two chloroplastic and one cytosolic (Eberhard et al., 1996; Mobley et al., 1999). Six PD genes are described for Arabidopsis, where sequence analysis predicts four have signal peptides, suggesting they are chloroplastic, and two less likely to have signal peptides (including PD1), suggesting they are cytosolic (K.M. Warpeha, unpublished data). Given the importance of Phe to plants and the large investment of genetic material in phenylpyruvate-based pathways in both the chloroplast and cytoplasm, it would seem likely that the process of Phe production can occur via phenylpyruvate both in the cytosol and in the mature plant, the chloroplast. Our findings herein confirm that in etiolated seedlings, this pathway can occur in the cytoplasm and can occur via a phenylpyruvate intermediate.
Data presented herein deriving from the use of T-DNA insertion mutants and biochemical and protein interaction studies, considered along with interaction data recently published by Pandey and Assmann (2004), confirm that GCR1, GPA1, and PD1 form all or part of a BL-induced signal transduction mechanism resulting in an immediate increase in the rate of phenylpyruvate synthesis and the subsequent increase in the levels of Phe. It remains unclear if GCR1 acts as, or downstream of, the responsible BL receptor. The fact that pd1 insertion mutants lack the BL-induced responses indicates that none of the other five genes coding for PD has the capacity to compensate the PD1 activity in etiolated seedlings. All of our plant material is derived from dark-grown seedlings, where BL irradiations are less than 2 min, so we are examining early fast responses to discrete pulses.
We have recently demonstrated that PRN1, like PD1, can act as a GPA1 effector. In this signaling chain, the BL-induced PRN1-mediated pathway leads to the induction of Lhcb transcription (K.M. Warpeha, unpublished data). GCR1 is also involved in this BL-mediated pathway and, similarly, with an undefined role. The data presented herein support the hypothesis that the two BL-signaling pathways branch immediately downstream of GPA1, via its separate interaction with PD1 or PRN1.
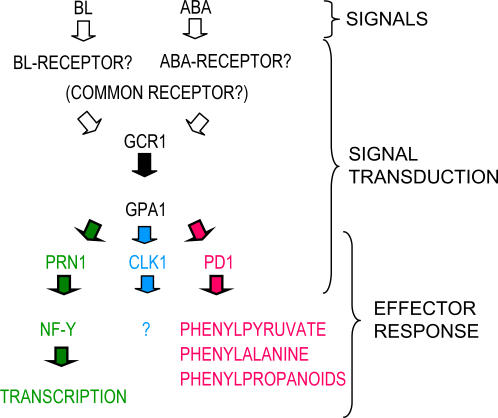
Because GPA1 is the only Gα-subunit in Arabidopsis, and because it is known to be involved in several different signal transduction pathways (for review, see Jones and Assmann, 2004; Perfus-Barbeoch et al., 2004), it is likely to interact with many different effector proteins. Proteins known to both interact with and participate in the same signaling pathways as GPA1 include PRN1 (Lapik and Kaufman, 2003), Phospholipase C (Apone et al., 2003), and Phospholipase Eα1 (Zhao and Wang, 2004). This study adds PD1 to that list. Based on our data and what is known about PDs we have designed a model of Phe formation in etiolated seedlings (Fig. 9).
Figure 9.
A proposed model for the GCR1, GPA1, and PD1 signal transduction mechanism responsible for BL-enhanced Phe synthesis in etiolated seedlings. The figure depicts the potential interface between the BL-activated GCR1, GPA1, and PD1 signal transduction chain and the cytosolic biosynthetic pathway leading from chorismate to Phe. Also depicted is the major signaling branch point that occurs immediately downstream of GPA1, as there now appear to be several different GPA1 effector proteins.
Etiolated Arabidopsis seedlings have relatively small pools of prephenate or Phe. At this point we have no understanding of the specific biological role of this BL-induced pathway, or the exact identities of the BL- and UV-absorbing compounds that seem to result from its activation. Phe is known to be the first committed precursor for the phenylpropanoid pathway, itself responsible for the synthesis of a large number of UV- and BL-absorbing compounds. It would seem likely that many of the BL- and UV-absorbing compounds that occur as a result of the BL activation of the GCR1, GPA1, and PD1 pathway intimately derive from the phenylpropanoid pathway. In the only other study that we are aware of examining early phenylpropanoid production in etiolated tissue, albeit in roots, Hemm et al. (2004) noted that phenylpropanoid production is miniscule. Irradiation resulted in a greater than 100-fold induction in phenylpropanoid synthesis. This is consistent with published studies where BL initiates phenylpropanoid production (Long and Jenkins, 1998; Noh and Spalding, 1998), and preirradiation of parsley (Petroselinum crispum) cells in culture prior to UV treatment reduces a lag period for expression of Phe derivatives (Bruns et al., 1986; Ohl et al., 1989). Lag elimination is often indicative of precursor synthesis.
Phe is the precursor for Phe-derived compounds including the light-induced UV and visible protective pigments (Long and Jenkins, 1998; Noh and Spalding, 1998; Hemm et al., 2004; Agati et al., 2005; Bussotti et al., 2005). The seedling mutants for PD1 fail to accumulate the phenylpropanoid precursor Phe and display either little or no fluorescence (mock-irradiated, i.e. no light), or diffuse reddish fluorescence (BL-irradiated) upon excitation by UV and blue wavelengths. The compounds causing the fluorescence observed in wild-type seedlings, which are absent from the PD1-deficient seedlings, are unknown. Likely candidates include the anthocyanins (Agati et al., 2002, 2005; Bussotti et al., 2005) and sinapolymalate (Chapple et al., 1992). Sinapolymalate, a UV-fluorescent compound, accumulates in the upper leaf epidermis and can cause a blue-green fluorescence under UV light (Chapple et al., 1992) similar to what we have observed in the tip of the cotyledon herein. Anthocyanins are present in etiolated pd1-1 and pd1-2 albeit at levels about 30% less than measured in etiolated wild-type seedlings.
Growth of pd1-1 and pd1-2 for 48 h in continuous BL results in anthocyanins levels approximating those measured for wild-type seedlings. It is uncertain if other members of the PD family, or some other biochemical pathway, is responsible for the Phe used in the synthesis of these anthocyanins.
There is clear evidence that GCR1 is a critical component of the BLF signal transduction mechanism; it is required for the responses mediated through both the PD1 and PRN1 effector systems and is also required for ABA activation of the so-called BLF system. It is not certain if GCR1 acts as either the direct BL and/or ABA receptor or neither. Given that ABA derives from an asymmetric cleavage of a carotenoid and that the cyclic head group of ABA is similar to that of the carotenoids from which it derives, it is possible that one protein could bind both ABA and a member of the carotenoid group from which it derives, and act as a single receptor. There are data to support a role for carotenoids in certain BL-mediated responses (Quinones and Zeiger, 1994; Quinones et al., 1998).
While it is unclear if a single receptor or two independent receptors are responsible for the responses we observe to ABA and BL, it is clear that they feed into a single signal transduction system that relies on GCR1, GPA1, and a set of effector systems, one of which is PD1, as described herein.
MATERIALS AND METHODS
Plant Materials and Accessions
Matched seed lots of wild-type Columbia (Col) Arabidopsis (Arabidopsis thaliana) and siblings carrying T-DNA insertions within coding regions of GCR1 (SALK_027808), GPA1 (SALK_066823), PRN1 (SALK_006939), and PD1 (SALK_013392, called pd1-1; SALK_029949, called pd1-2; both mutants have identical responses in our experiments, although in some cases data is shown for one mutant for brevity) were obtained from the Arabidopsis Biological Resource Center (Alonzo et al., 2003). Plants intended for seed stocks were grown in Scott Metromix 200 (Scotts) in continuous white light as described elsewhere (Lapik and Kaufman, 2003). All lines are homozygous for the reported insertion. Gene sequence accessions were obtained from GenBank (http://www.ncbi.nlm.nih.gov) and SIGnAL (http://signal.salk.edu), and compared by CLUSTALX program.
Plant Growth and Preparation of Tissue
Six-day-old dark-grown seedlings of Arabidopsis wild-type Col or insertion mutants were grown on 0.8% agarose plates containing only 0.5× Murashige and Skoog media as described in Lapik and Kaufman (2003). The growth media contain no additional sugar, hormones, vitamins, or other nutrients. Seedlings were irradiated with a single pulse (100 s or less) of low-fluence BL (total fluence of 104 μmol m−2 or 102 μmol m−2 [RNA only]; Warpeha and Kaufman, 1990) or no light (DK), and placed back in the dark for 2 or 24 h as indicated. Aerial portions were subsequently harvested to measure PD activity, amino acid analysis, and spectral analysis as described. RNA analysis has been described in full in Lapik and Kaufman (2003). ABA treatments (800 nm in ethanol) were conducted as described previously (K.M. Warpeha, unpublished data). Seedlings were harvested 24 h after treatment and treated in a manner to that described for seedlings irradiated with BL.
Chemicals
All chemicals unless otherwise noted were obtained from Sigma.
In Vitro GPA1-PD1 Activation Assays
Full-length GPA1 and PD1 templates were amplified, prepared, and purified by the methods described (Lapik and Kaufman, 2003). GPA1 and PD1 proteins were individually produced by coupled in vitro transcription/translation using TNT T7 coupled wheat germ extract system (Promega) as directed and as modified previously (Lapik and Kaufman, 2003). In vitro association assays were conducted by mixing approximately equimolar concentrations of GPA1 with PD1 in PD assay buffer at 4°C (Euverink et al., 1995) with modifications for plants: 50 mm K2PO4 pH 7.5, 1.0 mm dithiothreitol, 100 mm phenylmethylsulfonyl fluoride, and 0.5% protease inhibitor cocktail for plants. Activated GPA1 was achieved by preincubation with 100 μm GTPγS (a nonhydrolyzable GTP analog). Inactivated GPA1 was achieved by preincubation with GDP, or GDPβS (a nonhydrolyzable GDP analog). At time zero, prephenate at concentration of 10.0 mm (final concentration 1.0 mm) dissolved in 50 mm K2PO4 pH 7.5 was added to the reaction mixture. Reaction mixtures were stopped at time zero and various time points thereafter by adding 0.5 volume of 1n NaOH (Euverink et al., 1995). Absorbance values were read immediately afterward at 320 nm to assess conversion of prephenate to phenylpyruvate as described in Euverink et al. (1995). For controls, activity assays with conditions such as buffer only, no protein, one protein (GPA1 or PD1), or GPA1 + PD1 + no prephenate, were conducted in the same manner as described above.
Amino Acid HPLC Preparation and Spectral Analysis Experiments
Amino acid analysis was conducted as described (Razal et al., 1994) with these modifications: Amino acid content was assessed in the aerial portions of wild-type Col and insertion mutant seedlings (pd1-1 and pd1-2) 2 h after BL irradiation treatment or mock treatment (no BL). Seedlings were grown and irradiated as described above, aerial portions harvested in liquid nitrogen, ground to a fine powder, and stored at −80°C until amino acid analysis could be performed. All procedures hereafter were conducted at 4°C or on ice unless specified. Preweighed ground samples were dissolved in methanol:water:triethylamine (2:2:1 by vol) at approximately 13 mL per 0.5 g of sample by shaking in the dark at 4°C for 24 h. Two mL of the solution was centrifuged for 6 min at 12,000 rpm. The supernatant was transferred to a new 2.0 mL tube and dried using a using a speed vac (Savant). Quantity of 1.5 mL of acetone at −80°C was added to the evaporated samples, and the mixture was ultrasonicated until the sample was fully dissolved. The acetone samples were kept at −80°C for 30 min and centrifuged at 12,000 rpm for 5 min to remove precipitated proteins and other acetone-insoluble materials. The supernatant, containing the amino acids, was dried in a speed vac and used for amino acid or absorption spectra analysis.
Amino Acid Analysis
The acetone-dried samples were resuspended in 50 μL methanol:water:triethylamine (2:2:1 by volume), redried in the speed vac, resuspended in 50 μL of ethanol:triethylamine:water:phenylisothiocyanate (Pierce; 7:1:1:1 by volume), incubated for 20 min, and dried in a speed vac. The dried phenylisothiocyanate-labeled samples were dissolved in 200 μL of disodium hydrogen phosphate (pH 7.4) buffer and passed over a pico tag column (Waters) per the manufacturer's specifications. Amino acid standard H (Pierce) was used as the standard to assign peak position.
Anthocyanins Extraction and Quantification
Seedlings were grown in continuous DK for 7 or 5 d followed by 48 h of continuous BL irradiation at a fluence rate of 100 μmol m−2 s−1. BL irradiation materials have been described in full (Warpeha and Kaufman, 1990a). Anthocyanins were extracted and quantified according to Noh and Spalding (1998). Replicates (three) and means were analyzed with the Student's t test.
Absorbance Spectra
Absorbance spectra were obtained from samples in assay buffer as described above for the in vitro activation assays. Additional spectral analyses were carried out using samples prepared in the same manner as for amino acid analysis. Acetone dried samples were mixed with disodium hydrogen phosphate (pH 7.4) buffer and filtered once using a 0.45 μm syringe filter. Absorption spectra between 200 to 450 nm were obtained using a duel beam spectrophotometer (Spectronic Genesys 5; Thermo Electron).
Microscope Images
Fluorescent images were obtained of living unfixed seedlings 24 h post BL or ABA treatment as described in the “Plant Growth and Preparation of Tissue” section. Optical sectioning was achieved by using a Zeiss Axiovert 200M microscope (Carl Zeiss), equipped with ApoTome (collected by grid projection), and a digital camera and the 4′,6-diamino-phenylindole, fluorescein isothiocyanate, and Texas Red filter sets (Chroma). Photographs of whole cotyledon fluorescence were snapped on the same microscope set up, minus the apoTome setting.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AAC73018.1.
Acknowledgments
The authors gratefully acknowledge assistance from Teresa Orenic (microscopy) and Jan Euverink (PD1 assays), and Kevin Folta for helpful reading of the manuscript.
This work was supported by the U.S. Department of Agriculture (grant no. CREES 2005–02389 to L.S.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Lon Seth Kaufman (lkaufman@uic.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071282.
References
- Agati G, Galardi C, Gravano E, Romani A, Tattini M (2002) Flavonoid distribution in tissues of Phillyrea latifolia L. leaves as estimated by microspectrofluorometry and multispectral fluorescence microimaging. Photochem Photobiol 76: 350–360 [DOI] [PubMed] [Google Scholar]
- Agati G, Pinelli P, Ebner SCS, Romani A, Cartelat A, Cerovic ZG (2005) Nondestructive evaluation of anthocyanins in olive (Olea europaea) fruits by in situ chlorophyll fluorescence spectroscopy. J Agric Food Chem 53: 1354–1363 [DOI] [PubMed] [Google Scholar]
- Ahmad M (1999) Seeing the world in red and blue: insight into plant vision and photoreceptors. Curr Opin Plant Biol 2: 230–235 [DOI] [PubMed] [Google Scholar]
- Alonzo JM, Stepanova AN, Leisse TJ, Kim CJ, Huaming Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson MB, Folta KM, Warpeha KM, Gibbons J, Gao J, Kaufman LS (1999) Blue light-directed destabilization of the pea Lhcb1*4 transcript depends upon sequences within the 5′-UTR. Plant Cell 11: 1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apone F, Alyeshmerni N, Wiens K, Chalmers D, Chrispeels MJ, Colucci G (2003) The G-protein-coupled receptor GCR1 regulates DNA synthesis through activation of phosphatidylinositol-specific phospholipase C. Plant Physiol 133: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM (2002) Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell (Suppl) 14: S355–S373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Huala E (1999) Blue light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15: 33–62 [DOI] [PubMed] [Google Scholar]
- Bruns B, Hahlbrock K, Schafer E (1986) Fluence dependence of the ultraviolet light induced accumulation of chalcone synthase mRNA and effects of blue and far-red light in cultured parsley cells. Planta 169: 393–398 [DOI] [PubMed] [Google Scholar]
- Burchard P, Bilger W, Weissenbock G (2000) Contribution of hydroxycinnamates and flavonoids to epidermal shielding of UV-A and UV-B radiation in developing rye primary leaves as assessed by ultraviolet-induced chlorophyll fluorescence measurements. Plant Cell Environ 23: 1373–1380 [Google Scholar]
- Bussotti F, Agati G, Desotgiu R, Matteini P, Tani C (2005) Ozone foliar symptoms in woody plant species assessed with ultrastructural and fluorescence analysis. New Phytol 166: 941–955 [DOI] [PubMed] [Google Scholar]
- Casal JJ (2000) Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem Photobiol 71: 1–11 [DOI] [PubMed] [Google Scholar]
- Casati P, Walbot V (2005) Differential accumulation of maysin and rhamnosylisoorientin in leaves of high-altitude landraces of maize after UV-B exposure. Plant Cell Environ 28: 788–799 [Google Scholar]
- Chapple CCS, Shirley BW, Zook M, Hammerschmidt R, Somerville SC (1994) Secondary metabolism in Arabidopsis. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. CSH Laboratory Press, Plainview, NY, pp 989–1030
- Chapple CCS, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Colucci G, Apone F, Alyeshmerni N, Chalmers D, Chrispeels MJ (2002) GCR1, the putative Arabidopsis G protein-coupled receptor gene is cell cycle-regulated, and its overexpression abolishes seed dormancy and shortens time to flowering. Proc Natl Acad Sci USA 99: 4736–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Lewis NG, Umezawa T (1992) Phenylpropanoid metabolism: biosynthesis of monolignols, lignans and neolignans, lignins and suberins. In HA Stafford, RK Ibrahim, eds, Recent Advances in Phytochemistry, Vol 26. Plenum Press, New York, pp 325–375
- Dunwell JM, Purvis A, Khuri S (2004) Cupins: the most functionally diverse protein superfamily? Phytochemistry 65: 7–17 [DOI] [PubMed] [Google Scholar]
- Eberhard J, Ehrler TT, Epple P, Felix G, Raesecke HR, Amrhein N, Schmid J (1996) Cytosolic and plastidic chorismate mutase isozymes from Arabidopsis thaliana: molecular characterization and enzymatic properties. Plant J 10: 815–821 [DOI] [PubMed] [Google Scholar]
- Euverink GJW, Wolters DJ, Dijkhuizen L (1995) Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of wild type and deregulated mutant proteins. Biochem J 308: 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Kaufman LS (1994) Blue-light regulation of the Arabidopsis thaliana Cab1 gene. Plant Physiol 104: 1251–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DG, Kosuge T (1980) Aromatic amino acid biosynthesis and its regulation. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 5. BJ Miflin, Academic Press, New York, pp 507–531
- Haslam E (1993) Shikimic Acid Metabolism and Metabolites. John Wiley and Sons, New York
- Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C (2004) Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J 38: 765–778 [DOI] [PubMed] [Google Scholar]
- Hudson G, Davidson BE (1984) Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol 180: 1023–1051 [DOI] [PubMed] [Google Scholar]
- Jones AM, Assmann SM (2004) Plants: the latest model system for G-protein research. EMBO Rep 5: 572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E, Zamir LO, Jensen RA (1986) Chloroplasts of higher plants synthesize L-phenylalanine via L-arogenate. Proc Natl Acad Sci USA 83: 7231–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ (2004) Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant Cell Environ 27: 675–684 [Google Scholar]
- Lapik Y, Kaufman LS (2003) The Arabidopsis cupin domain protein AtPirin1 and AtGPA1, the Arabidopsis Gα subunit interact with each other and regulate seed germination and early seedling development. Plant Cell 15: 1578–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E (1989) Tannins: their place in plant metabolism. In RW Hemingway, JJ Karchesy, eds, Chemistry and Significance of Condensed Tannins. Plenum Press, New York, pp 23–47
- Lin C (2000) Plant blue-light receptors. Trends Plant Sci 5: 337–341 [DOI] [PubMed] [Google Scholar]
- Long JC, Jenkins GI (1998) Involvement of a plasma membrane redox activity and calcium homeostasis in the UV-B UV-A blue light induction of gene expression in Arabidopsis. Plant Cell 10: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM (1990) Molecular cloning and characterization of GPA1, a G protein alpha subunit gene from Arabidopsis thaliana. Proc Natl Acad Sci USA 87: 3821–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA, Kaufman LS (1991) Rapid transcriptional regulation of the Cab and pEA207 gene families by blue light in the absence of cytoplasmic protein synthesis. Planta 183: 327–333 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Kaufman LS (1999) Cloning and characterization of PGA1 and PGA2, two G protein alpha-subunits from pea that promote growth in the yeast Saccharomyces cerevisiae. Plant J 19: 237–247 [DOI] [PubMed] [Google Scholar]
- Mobley EM, Kunkel BN, Keith B (1999) Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana. Gene 240: 115–123 [DOI] [PubMed] [Google Scholar]
- Mol J, Jenkins GI, Schafer E, Weiss D (1996) Signal perception, transduction, and gene expression involved in anthocyanin biosynthesis. CRC Crit Rev Plant Sci 15: 525–557 [Google Scholar]
- Noh B, Spalding EP (1998) Anion channels and the stimulation of anthocyanin accumulation by blue light in Arabidopsis seedlings. Plant Physiol 116: 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl S, Hahlbrock K, Schafer E (1989) A stable blue-light-derived signal modulates ultraviolet-light-induced activation of the chalcone-synthase gene in cultured parsley cells. Planta 177: 228–236 [DOI] [PubMed] [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang H, Bartlam M, Zeng QH, Miyatake H, Hisano T, Miki K, Wong LL, Gao GF, Rao ZH (2004) Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J Biol Chem 279: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Patel N, Pierson DL, Jensen RA (1977) Dual enzymatic routes to L-Tyrosine and L-Phenylalanine via pre-tyrosine in Pseudomonas aeroginosa. J Biol Chem 252: 5839–5846 [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM (2004) Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr Opin Plant Biol 7: 719–731 [DOI] [PubMed] [Google Scholar]
- Quinones MA, Lu ZM, Zeiger E (1998) Genetic variation of stomatal conductance, blue light sensitivity and zeaxanthin content in guard cells of Pima cotton (Gossypium barbadense). Physiol Plant 103: 560–566 [Google Scholar]
- Quinones MA, Zeiger E (1994) A putative role of the xanthophyll, zeaxanthin, in blue light photoreception of corn coleoptiles. Science 264: 558–561 [DOI] [PubMed] [Google Scholar]
- Razal RA, Ellis S, Singh S, Lewis NG, Towers GHN (1996) Nitrogen recycling in phenylpropanoid metabolism. Phytochemistry 41: 31–35 [Google Scholar]
- Razal RA, Lewis NG, Towers GHN (1994) Pico-Tag analysis of arogenic acid and related free amino acids from plant and fungal extracts. Anal Biochem 5: 98–104 [Google Scholar]
- Romero RM, Roberts MF, Phillipson JD (1995) Chorismate mutase in microorganisms and plants. Phytochemistry 40: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Chapple C (2001) Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159: 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J, Amrhein N (1995) Molecular organization of the shikimate pathway in higher plants. Phytochemistry 39: 737–749 [Google Scholar]
- Ullah H, Chen JG, Wang SC, Jones AM (2002) Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol 129: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Wade HK, Bibikova TN, Valentine WJ, Jenkins GI (2001) Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. Plant J 6: 675–685 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Warpeha KMF, Hamm HE, Rasenick MM, Kaufman LS (1991) A blue-light activated GTP binding protein in the plasma membrane of etiolated pea. Proc Natl Acad Sci USA 88: 8925–8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KMF, Kaufman LS (1990. a) Two distinct blue-light responses regulate epicotyl elongation in pea. Plant Physiol 92: 495–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha KMF, Kaufman LS (1990. b) Two distinct blue-light responses regulate the levels of transcripts of specific nuclear-coded genes in pea. Planta 182: 553–558 [DOI] [PubMed] [Google Scholar]
- Winkel BSJ (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Xia TH, Ahmad S, Zhao GS (1991) A single cyclohexadienyl dehydratase specifies the prephenate dehydratase and arogenate dehydratase components of one of 2 independent pathways to L-phenylalanine in Erwinia herbicola. Arch Biochem Biophys 286: 461–465 [DOI] [PubMed] [Google Scholar]
- Zhao J, Wang XM (2004) Arabidopsis phospholipase D alpha 1 interacts with the heterotrimeric G-protein alpha-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem 279: 1794–1800 [DOI] [PubMed] [Google Scholar]
- Zurawski G, Brown K, Killingly D, Yanofsky C (1977) Nucleotide sequence of the leader region of the phenylalanine operon of Escherichia coli. Proc Natl Acad Sci USA 75: 4271–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]