Abstract
Ran is a multifunctional small GTPase that is involved in nucleocytoplasmic transport, mitotic spindle assembly, and nuclear envelope formation. Nuclear import of Ran relies on a small RanGDP-binding protein, Nuclear Transport Factor 2 (NTF2). Three proteins are expressed in Arabidopsis (Arabidopsis thaliana) that show significant sequence similarity to human and yeast (Saccharomyces cerevisiae) NTF2. Here, we demonstrate that two of them, AtNTF2a and AtNTF2b, can functionally replace the essential NTF2 gene in yeast. Consistent with this finding, both AtNTF2a and AtNTF2b interact with yeast and Arabidopsis Ran. The third NTF2-related protein, AtNTL, does not functionally replace NTF2 in yeast. Similar to yeast NTF2-green fluorescent protein (GFP), AtNTF2a-GFP and AtNTF2b-GFP accumulate at the nuclear rim. The AtNTF2a E38K and E91K mutants, which fail to bind Ran, are not functional in yeast, indicating conservation of the requirement for these key amino acids in plants and yeast. AtNTF2a overexpression, but not AtNTF2aE38K overexpression, blocks nuclear import of a plant transcription factor in Nicotiana benthamiana leaves, indicating that excess AtNTF2a disrupts nuclear import in a Ran-binding-dependent manner. On the basis of these results, we propose that AtNTF2a and AtNTF2b function in Ran import in Arabidopsis and that nuclear import of Ran is functionally conserved in plants.
Ran is an evolutionarily highly conserved small GTP-binding protein of the Ras superfamily (Bischoff and Ponstingl, 1991a, 1991b). It plays roles in nucleocytoplasmic transport, mitotic spindle formation, and nuclear envelope assembly (Carazo-Salas et al., 1999; Zhang et al., 1999; Hetzer et al., 2000). Like other small GTPases, Ran exists as both GTP- and GDP-bound forms (Avis and Clarke, 1996). The intrinsic GTPase activity of Ran is low but can be greatly stimulated by RanGTPase-activating protein (RanGAP) and its accessory factor RanBP1 (Matunis et al., 1996). The only Ran guanine nucleotide exchange factor, RCC1, charges Ran with GTP. The RanGDP versus RanGTP abundance is regulated by the compartmentalized localization of RanGAP and RanBP1 outside, and RCC1 inside, the nucleus (Gorlich and Mattaj, 1996).
Importin β family transport receptors, which play roles in the transport of proteins and RNAs between the nucleus and the cytoplasm, use RanGTP binding to regulate their affinities for cargo molecules. They enter the nucleus in the absence of RanGTP and leave the nucleus as RanGTP complexes, thereby continuously depleting Ran from the nucleus. To replenish the nuclear pools of Ran, RanGDP is imported into the nucleus by the small homodimeric protein Nuclear Transport Factor 2 (NTF2; Ribbeck et al., 1998; Quimby et al., 2000a, 2000b).
NTF2 was originally identified as a factor required for efficient import of nuclear localization signal (NLS)-containing proteins into the nucleus (Moore and Blobel, 1994). It binds specifically to RanGDP and has no detectable affinity for RanGTP (Clarkson et al., 1996; Wong et al., 1997; Stewart et al., 1998a). NTF2 also interacts with the xFxFG repeats of nucleoporins and localizes to the nuclear rim at steady state (Clarkson et al., 1996; Corbett and Silver, 1996). The yeast (Saccharomyces cerevisiae) NTF2 homolog is an essential protein (Corbett and Silver, 1996), and temperature-sensitive NTF2 mutants are defective in nuclear protein import. Moreover, temperature-sensitive mutants of the yeast Ran homolog Gsp1p can be suppressed by overexpression of NTF2, suggesting that Ran-NTF2 interaction is required for effective nuclear protein import in vivo (Wong et al., 1997). Whether NTF2 is also required for nuclear export is less clear (Corbett and Silver, 1996).
In plants, Ran, RanBP1, and RanGAP have been identified (Ach and Gruissem, 1994; Haizel et al., 1997; Rose and Meier, 2001; Pay et al., 2002). While plant Ran and RanBP1 are similar to their yeast and animal counterparts, plant RanGAP requires a unique N-terminal domain for its nuclear envelope localization, which is different from the C-terminal localization domain of its animal counterpart (Matunis et al., 1996). In addition, plant RanGAP is targeted to the cell plate, a mitotic structure unique to plants, during cell division (Jeong et al., 2005). To our knowledge, no plant homolog of RCC1 has been identified. Here, we report identification of two functional NTF2 homologs from Arabidopsis (Arabidopsis thaliana). Like Ran and RanBP1, and unlike RanGAP, plant NTF2s appear to be well conserved in their structure and function compared to their counterparts in yeast and animals.
RESULTS
Identification of Putative Plant NTF2 Homologs
Using the yeast NTF2 sequence (GenBank accession no. NP_010925), a protein BLAST search was performed against the translated Arabidopsis genome. The two hits with the lowest e values were encoded by the genes At1g27310 and At1g27970 (1e-31 and 1e-29, respectively). The third hit had a slightly lower e value (At1g11570; 3e-17), but the predicted protein was approximately the same size as other NTF2 proteins. All other hits represented larger proteins containing NTF2-like domains adjacent to additional functional domains (data not shown).
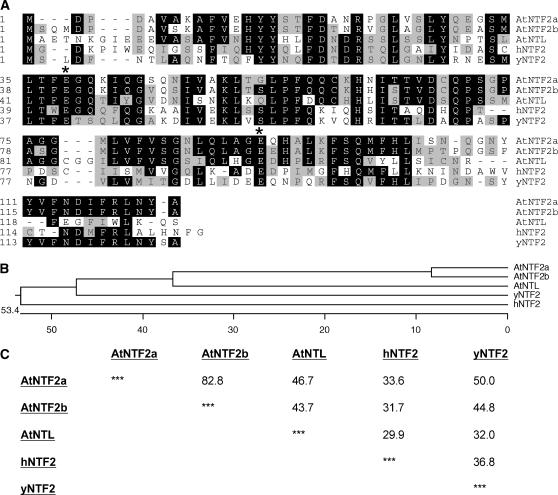
The predicted gene products of At1g27310 and At1g27970 were named AtNTF2a and AtNTF2b, respectively, and the gene product of At1g11570 was named AtNTL for Arabidopsis NTF2-like protein (see below). Figure 1A shows the alignment of the Arabidopsis proteins with human and yeast NTF2. While all three gene products share an overall similar degree of conservation, an otherwise well-conserved 12-amino acid stretch at the extreme C terminus is absent in AtNTL (Fig. 1A). AtNTF2a and AtNTF2b are 83% identical at the amino acid level, while AtNTF2a and yeast NTF2 (yNTF2) share 50% amino acid identity and AtNTF2b and yNTF2 45% amino acid identity (Fig. 1C). AtNTL shares approximately 45% sequence identity with AtNTF2a and AtNTF2b. All three Arabidopsis proteins are more closely related to each other than to the yeast or human proteins, indicating that they are paralogs (Fig. 1, B and C). AtNTF2a and AtNTF2b are more closely related to yeast NTF2 than to human NTF2, with which they share 33% and 32% identity, respectively. This level of amino acid conservation is comparable to the 37% identity between yeast and human NTF2.
Figure 1.
The Arabidopsis genome encodes three NTF2-like proteins. A, Alignment of the three Arabidopsis NTF2-like proteins (AtNTF2a, AtNTF2b, and AtNTL) with human NTF2 (hNTF2) and yeast NTF2 (yNTF2) is shown. Black shading indicates amino acids that match the majority (at least three out of five). Gray shading indicates amino acids similar to the majority. Asterisks indicate the position of two highly conserved Glu residues (E38 and E91 in Arabidopsis). B, Phylogenetic tree of the alignment shown in A. C, Percentage of similarity between the three Arabidopsis NTF2-like proteins and human and yeast NTF2.
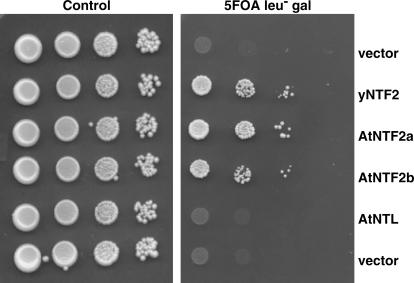
Human NTF2 can functionally replace yeast NTF2 (Corbett and Silver, 1996). Here, we tested whether AtNTF2a, AtNTF2b, and AtNTL are also functional homologs of yeast NTF2. A plasmid shuffle assay was performed to determine whether the three Arabidopsis proteins could replace the essential yeast NTF2 protein. This assay tests whether yeast cells that express AtNTF2a, AtNTF2b, and AtNTL as the only copy of NTF2 are viable. Cell growth is assessed by serial diluting and spotting. Each of the three putative Arabidopsis NTF2 cDNAs was cloned into a yeast expression plasmid under the control of the Gal-inducible GAL1/10 promoter. These plasmids were then transformed into yeast cells where the genomic copy of NTF2 had been deleted, but the cells were maintained by a plasmid-borne copy of yeast NTF2. As controls, the cells were also transformed with a vector alone and yeast NTF2 cloned into the same yeast expression vector and expressed from the Gal-inducible promoter. As shown in Figure 2 (left), all the cells grew equally well on the control plates where the yeast NTF2 maintenance plasmid was present. When cells were plated on 5-fluoroorotic acid (5-FOA) to eliminate the yeast NTF2 maintenance plasmid (right), the results indicated that both AtNTF2a and AtNTF2b can function as the sole copy of NTF2 in vivo. Growth of cells expressing each of these plasmids was similar to the growth of cells that express the control yeast NTF2 plasmid. In contrast, AtNTL did not support cell growth. Similar results were obtained when we assessed whether the AtNTF2b proteins could suppress the temperature-sensitive phenotype of two NTF2 mutant alleles, ntf2-1 and ntf2-2 (Corbett and Silver, 1996). Both AtNTF2a and AtNTF2b could suppress the temperature-sensitive growth phenotype of these mutants, whereas AtNTL could not (data not shown).
Figure 2.
AtNTF2a and AtNTF2b, but not AtNTL, can functionally replace yeast NTF2. Yeast cells deleted for the genomic copy of the essential NTF2 gene but maintained by an NTF2 genomic plasmid (see “Materials and Methods”) were transformed with plasmids expressing AtNTF2a, AtNTF2b, or AtNTL under the control of a Gal-inducible promoter. Cultures were grown to saturation, serially diluted, and spotted on control (left) or 5-FOA (right) plates to eliminate the NTF2 maintenance plasmid. The control plate indicates that similar numbers of cells were spotted for each sample. The 5-FOA plate, where each of the indicated plasmids constitutes the sole source of NTF2, shows that both AtNTF2a and AtNTF2b can function in vivo, as cells expressing these proteins grow as well as the control cells that express yNTF2. In contrast, AtNTL cannot function in place of yeast NTF2, as cells that express this plasmid show no growth (similar to the vector-only control).
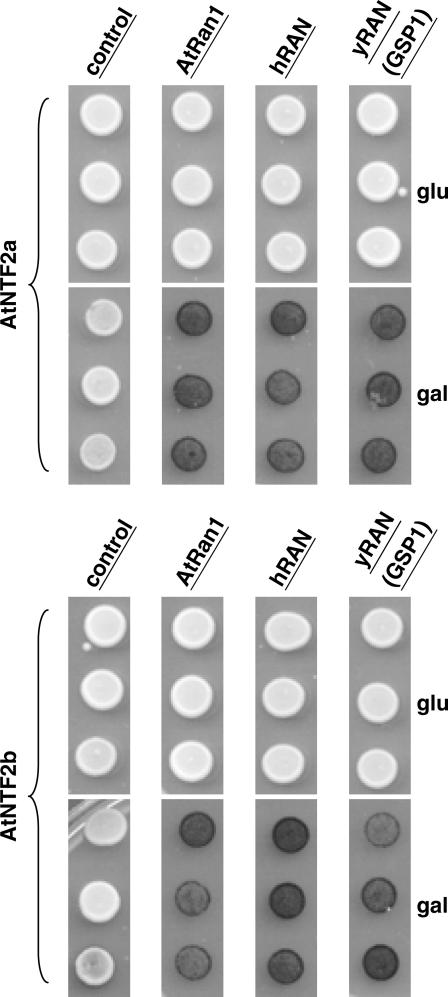
The role of NTF2 is to transport RanGDP into the nucleus (Ribbeck et al., 1998; Smith et al., 1998). Thus, any bona fide NTF2 protein should interact with Ran. Since the yeast two-hybrid method has been used previously to examine the interaction between Ran and NTF2 (Clarkson et al., 1997), this method was used here to analyze the interaction of Ran with AtNTF2a and AtNTF2b. Figure 3 shows that both proteins interact with yeast, human, and Arabidopsis Ran, but not with a control protein. Taken together, we conclude that AtNTF2a and AtNTF2b are both true orthologs of their yeast counterpart, whereas AtNTL is not a functional NTF2.
Figure 3.
AtNTF2a and AtNTF2b bind to Arabidopsis, human, and yeast Ran. A yeast two-hybrid assay was used to examine the interaction between AtNTF2a, AtNTF2b, and Ran. Each of the AtNTF proteins was expressed as an activation domain fusion as described in “Materials and Methods.” Ran proteins were expressed as DNA-binding domain fusion proteins. Interactions with Arabidopsis (AtRAN1), human (hRAN), and yeast (yRAN) Ran were examined for AtNTF2a (top) and AtNTF2b (bottom). As a negative control, interaction with vector alone was also examined. In this two-hybrid system, activation domain fusion proteins are expressed under a Gal-inducible promoter; thus, interactions should only occur in the presence of Gal. As indicated by the blue color indicative of β-galactosidase expression, both AtNTF2a and AtNTF2b interact with Ran from all species tested.
The Interaction with Ran Is Critical for the Function of AtNTF2a and AtNTF2b
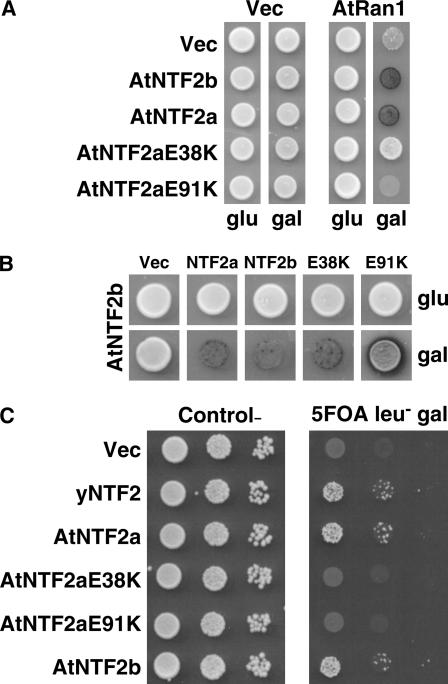
For NTF2 to fulfill its essential cellular role, it must bind to RanGDP. The interaction between NTF2 and Ran has been extensively characterized, including the resolution of the crystal structure of the complex (Stewart et al., 1998a). These analyses have revealed key residues within NTF2 that are absolutely required for Ran binding. Amino acid changes in yeast NTF2 (E42K and D91G) and rat NTF2 (E42K and D92N/D94N) lead to loss of interaction with Ran and consequent loss of NTF2 function in vivo (Clarkson et al., 1997; Quimby et al., 2000b). Both AtNTF2a and AtNTF2b contain a conserved Glu in position 38 corresponding to E42 in yeast and rat, and a conserved Glu in position 91 adjacent to the conserved Asp mutated in the yeast D91G and the rat D92G mutations (Fig. 1). Corresponding amino acid changes were incorporated into AtNTF2a to create AtNTF2aE38K and AtNTF2aE91K.
If AtNTF2a and AtNTF2b are functional homologs of NTF2, then these key conserved amino acid residues should also be critical for their function. We used yeast two-hybrid analysis to determine whether these amino acid changes within the Ran/NTF2-binding interface disrupt the interaction between AtNTF2a and AtRan1. As shown in Figure 4A, both AtNTF2aE38K and AtNTF2aE91K show significantly decreased interaction with AtRan1 as compared to wild-type AtNTF2a or AtNTF2b. This loss of interaction is not due to a loss of expression of the mutant proteins, as we can readily detect homodimerization of both of the mutant proteins with AtNTF2b in a two-hybrid assay (Fig. 4B). Figure 4C shows that neither AtNTF2aE38K nor AtNTF2aE91K is able to functionally replace yeast NTF2, indicating that their ability to bind Ran is required for their function in yeast. This analysis confirms that, like other known NTF2 homologs, the AtNTF2a/b proteins must bind Ran in vivo to function and provides further evidence that these are the Arabidopsis NTF2 homologs.
Figure 4.
E38K and E91K mutations in AtNTF2a disrupt Ran binding and AtNTF2a function in vivo. A, Yeast two hybrid was used to assay the interaction between wild-type and mutant AtNTF2a and Arabidopsis Ran (AtRAN1) or a vector control (Vec). Neither AtNTF2aE38K nor AtNTF2aE91K interacted with AtRAN, as indicated by loss of the blue color indicative of β-galactosidase expression in these spots. As controls, wild-type AtNTF2a and AtNTF2b both interacted with AtRAN in this experiment. No interaction was observed with the vector control. B, AtNTF2a mutants interact with AtNTF2b. The interaction of AtNTF2b with itself and with both wild-type and mutant AtNTF2a was examined by two-hybrid assay. The blue color indicative of β-galactosidase expression reveals that AtNTF2b interacts with all versions of NTF1/2 tested, indicating that the mutant AtNTF2a proteins are expressed and capable of interacting in the two-hybrid assay. C, The function of AtNTF2aE38K and AtNTF2aE91K was examined by a plasmid shuffle assay. Yeast cells deleted for the genomic copy of the essential NTF2 gene but maintained by an NTF2 genomic plasmid (see “Materials and Methods”) were transformed with plasmids expressing AtNTF2a, AtNTF2aE38K, AtNTF2aE91K, or as controls, vector, yNTF2, or AtNTF2b under the control of a Gal-inducible promoter. Cultures were grown to saturation, serially diluted, and spotted on control (left) or 5-FOA (right) plates to eliminate the NTF2 maintenance plasmid. Only wild-type AtNTF2a and AtNTF2b supported cell growth, as did the control (yNTF2).
AtNTF2a and AtNTF2b Are Ubiquitously Expressed in Arabidopsis
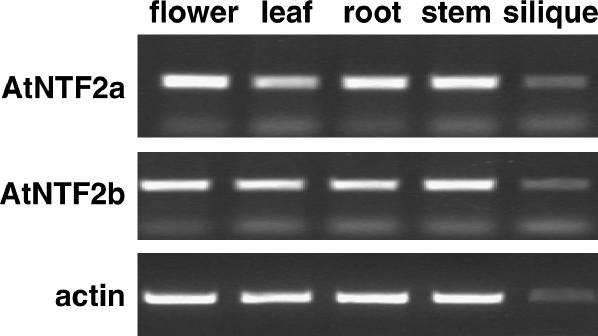
The steady-state level of mRNA expression in different tissues of flowering Arabidopsis plants was assayed by reverse transcription (RT)-PCR for AtNTF2a and AtNTF2b. For both AtNTF2a and AtNTF2b, a transcript of approximately 500 bp, which corresponds to the expected size of the open reading frame plus an approximately 100-bp 3′-untranslated region (see “Materials and Methods”), was detected in all tissues tested (Fig. 5). The Actin-related protein 6 (At3g33520) levels were tested using approximately the same amount of RNA as used in the AtNTF2a and AtNTF2b reactions (Fig. 5). For all reactions, PCR without reverse transcriptase was performed as a negative control and yielded no signal (data not shown).
Figure 5.
AtNTF2a and AtNTF2b are ubiquitously expressed in Arabidopsis. RT-PCR was performed on total RNA isolated from flowers, leaves, roots, stems, and siliques of 30-d-old Arabidopsis plants. As a control, the amount of Actin-related protein 6 (At3g33520; actin) is also shown for each sample.
AtNTF2a and AtNTF2b Are Located at the Plant Nuclear Envelope
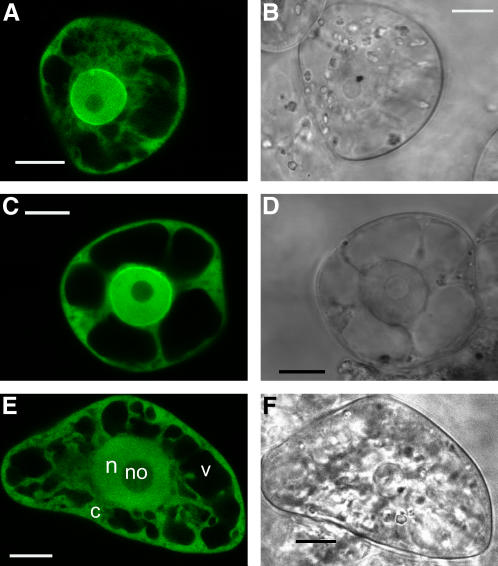
To investigate whether AtNTF2a and ATNTF2b are concentrated at the nuclear envelope, similar to what has been observed for yeast and mammalian NTF2 (Corbett and Silver, 1996; Ribbeck et al., 1998), green fluorescent protein (GFP) was fused to the C terminus of both AtNTF2a and AtNTF2b. The two GFP-fusion proteins and free GFP were transiently expressed in tobacco BY-2 cells under the control of the 35S promoter. Figure 6 shows that AtNTF2a-GFP and AtNTF2b-GFP are concentrated at the nuclear envelope as well as distributed in the cytoplasm and the nucleus, excluding the nucleolus. This pattern is very similar to the localization patterns of both yeast and mammalian NTF2 (Corbett and Silver, 1996). In addition, we tested the localization pattern of AtNTF2a-GFP and AtNTF2b-GFP in yeast and detected an accumulation at the nuclear envelope, consistent with the pattern observed for yNTF2 (data not shown).
Figure 6.
AtNTF2a and AtNTF2b accumulate at the nuclear envelope. AtNTF2a-GFP (A and B), AtNTF2b-GFP (C and D), and free GFP (E and F) were transiently expressed in tobacco BY-2 protoplasts, and GFP fluorescence was imaged by confocal laser scanning microscopy. A, C, and E, GFP fluorescence; B, D, and F, transmitted light images of protoplasts. Cellular compartments are indicated in E: n, nucleus; no, nucleolus; c, cytoplasm; and v, vacuole. Size bars = 10 μm.
Overexpression of AtNTF2a in Plants Disrupts Nuclear Import in a Ran-Binding-Dependent Manner
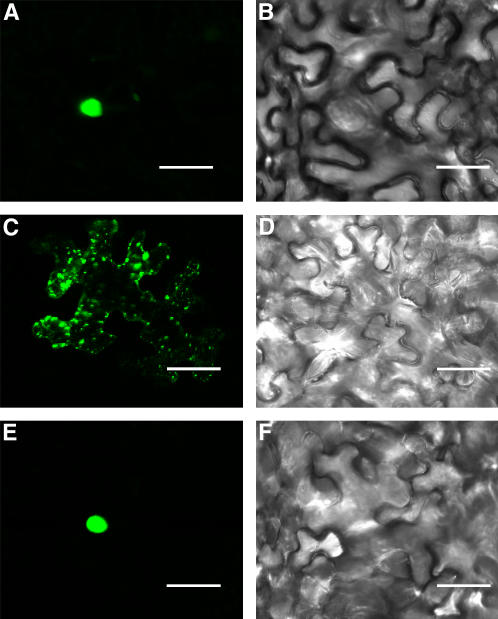
Overexpression of NTF2 in mammalian cells blocks nuclear import (Tachibana et al., 1996). In yeast, overexpression of wild-type NTF2 is slightly dominant negative (Quimby et al., 2001). To investigate the role of AtNTF2a in plant nuclear import, we tested the effect of AtNTF2a overexpression on the localization of a nuclear protein. We used the maize (Zea mays) transcription factor R fused to GFP as a reporter for nuclear import (Shieh et al., 1993) and transient expression based on infiltration of Nicotiana benthamiana leaves with Agrobacterium as an assay (Scofield et al., 1996; Tang et al., 1996). To confirm the nuclear localization of R-GFP in N. benthamiana, we agroinfiltrated a plasmid containing a 35S promoter-driven R-GFP. Figure 7A shows that R-GFP localizes exclusively in the nucleus. Next, we tested the localization of R-GFP when coexpressed with AtNTF2a under control of the 35S promoter. As shown in Figure 7C, under these conditions, R-GFP is located in cytoplasmic aggregates and no nuclear accumulation is observed. The cytoplasmic aggregation is similar to the localization of truncated R without its NLS (A. Feller and E. Grotewold, unpublished data). The localization of R-GFP is not affected by coexpressing AtNTF2aE38K, which cannot interact with Ran (Fig. 7E). In addition, coexpression of two unrelated, 35S-driven proteins did not alter R-GFP localization (data not shown). Together, these results suggest that overexpression of AtNTF2a blocks nuclear import and that this effect depends on the ability of AtNTF2a to bind Ran.
Figure 7.
Overexpression of AtNTF2a in plants disrupts nuclear import in a Ran-binding-dependent manner. A, GFP fluorescence of an R-GFP fusion protein expressed in N. benthamiana leaf epidermis cells; B, corresponding transmitted light image; C, GFP fluorescence of R-GFP coexpressed with AtNTF2a; D, corresponding transmitted light image; E, GFP fluorescence of R-GFP coexpressed with AtNTF2aE38K; and F, corresponding transmitted light image. Size bars = 10 μm.
DISCUSSION
An important component of the Ran cycle is the nuclear import receptor for RanGDP, NTF2. Without this import receptor, nuclear Ran would be depleted by the export of RanGTP complexed with exportin and importin β transport receptors. NTF2 binds RanGDP in the cytoplasm and transports it through the pore, thereby replenishing the nuclear Ran pool. While all other organisms investigated have a single gene encoding NTF2, we have identified three putative NTF2-like genes in Arabidopsis. Here, we show that two of them encode bona fide NTF2 proteins, which we call AtNTF2a and AtNTF2b, while the third gene encodes an NTF2-like protein (AtNTL) that does not function as a Ran import factor.
Redundancy appears to be a common theme of Ran cycle components in Arabidopsis. There are three genes for Ran itself (Ran1, Ran2, and Ran3; Haizel et al., 1997), two for RanGAP (RanGAP1 and RanGAP2; Pay et al., 2002), and two for RanBP1 (RanBP1a and RanBP1b; Haizel et al., 1997). No RCC1 homolog has yet been identified in Arabidopsis. Currently, there is little evidence for specific functions of individual gene family members of Ran cycle proteins in Arabidopsis. The three Rans have highly similar sequences and show very similar and largely overlapping expression patterns (Haizel et al., 1997). A knockout T-DNA insertion in RanGAP1 leads to no phenotype, indicating redundancy between RanGAP1 and RanGAP2 (Q. Zhao and I. Meier, unpublished data). In line with these findings, we show here that AtNTF2a and AtNTF2b can form both homodimers and heterodimers, and we have found that a knockout T-DNA insertion in AtNTF2a has no phenotype (data not shown), suggesting functional redundancy of the two NTF2 copies as well.
AtNTL is only slightly less similar to human NTF2 than AtNTF2a and AtNTF2b, but it is significantly less similar to yNTF2 (Fig. 1). This difference is mainly due to a stretch of sequence at the very C terminus that is conserved between yNTF2, AtNTF2a, and AtNTF2b, but not AtNTL and human NTF2. Although the C-terminal similarity between the two Arabidopsis NTF2 orthologs and yeast NTF2 is striking, this is unlikely the reason for the lacking function of AtNTL in yeast, as human NTF2 can functionally replace yeast NTF2 (Corbett and Silver, 1996) but also has limited sequence similarity at the very C terminus. While it is currently not known why NTL is not functional as an NTF2, one notable difference is a reduction in the number of bulky hydrophobic amino acids compared to AtNTF2a and AtNTF2b, which are involved in interaction with the xFxFG repeats and Ran (Stewart et al., 1998a; Quimby et al., 2001).
A highly conserved Glu residue (E38 in Arabidopsis) and a small cluster of acidic residues are crucial for Ran binding of NTF2. The crystal structure of the RanGDP/NTF2 complex has been resolved, and this structure reveals that these residues contact Lys-71 and Arg-76 in the switch II loop of Ran (Stewart et al., 1998b). The Glu residue is conserved in all three Arabidopsis proteins and the acidic cluster is represented by a conserved Glu, followed by Glu, Asp, or Gln. Both Glu-38 and Glu-91 were replaced by a Lys residue in AtNTF2b and shown to be necessary both for binding to Ran and function in yeast. Lys-71 and Arg-76 are conserved in all three plant Rans as Lys-74 and Arg-79. On the basis of these results, we assume that these crucial salt bridges are made between plant Ran and NTF2 family members, too.
It has been demonstrated previously in a semi-in vitro assay using permeabilized mammalian cells that an excess of exogenous NTF2 can block nuclear import of an NLS-containing reporter protein (Tachibana et al., 1996). In yeast, overexpression of NTF2 is slightly dominant negative (Quimby et al., 2001). Here, we have demonstrated that overexpressing AtNTF2a in agroinfiltrated N. benthamiana leaf epidermis cells inhibits nuclear import of a plant transcription factor. These data imply that AtNTF2a is functional in plant nuclear import and that excess protein has a deleterious effect on nuclear import, similar to the results in yeast and mammalian cells. In contrast to wild-type AtNTF2a, the E38K mutant has no effect on nuclear import, indicating that Ran binding is required for the observed effect.
One model for the negative effect of overabundant NTF2 is a nuclear competition for binding of RanGDP between NTF2 and RCC1. This would imply that the excess of NTF2 can enter the nucleus, which is indicated by the subcellular distribution of overexpressed NTF2-GFP (Fig. 6). Comparison of the crystal structures of the NTF2/Ran complex and the Ran/RCC1 complex indicates that NTF2 and RCC1 cannot bind RanGDP simultaneously (Renault et al., 2001). Thus, RanGDP has to dissociate from NTF2 for RCC1-mediated nucleotide exchange to occur (Yamada et al., 2004). A mathematical model analyzing the role of the known components of the Ran cycle suggested that the delivery of Ran to RCC1 is one of the two particularly sensitive parameters (Riddick and Macara, 2005). Import receptors release their substrate upon direct interaction with RanGTP (Chi et al., 1996; Gorlich and Mattaj, 1996; Izaurralde et al., 1997; Siomi et al., 1997) and exit the nucleus as RanGTP complex. Depletion of nuclear RanGTP by competitive binding between an excess of nuclear NTF2 and endogenous RCC1 might therefore inhibit nuclear import. This is consistent with the finding that Ran binding is required for the overexpression effect.
An alternative model was proposed by Tachibana et al. (1996), who suggested that excess NTF2 might “clog” the nuclear pore by occupying an excess of binding sites at xFxFG repeat nucleoporins and that the inhibition occurs at the translocation step. This model is more difficult to reconcile with our finding of Ran-binding dependence. It has been clearly demonstrated both by crystallography and by mutant analyses that the binding sites for RanGDP and nucleoporins are located at the two different poles of NTF2 (Clarkson et al., 1996; Quimby et al., 2001; Bayliss et al., 2002), making it unlikely that the E38K mutation affects nucleoporin interaction per se. However, it has been shown that binding of NTF2 to the nuclear envelope is enhanced by Ran (Chaillan-Huntington et al., 2000). Therefore, there remains the possibility that Ran-bound NTF2 has a higher affinity for the nuclear pore, leading to a stronger “clogging” effect for the wild-type protein.
The data presented here clearly indicate that AtNTF2a and AtNTF2b are bona fide plant NTF2 orthologs. Like the Ran and RanBP1 orthologs, they occur in likely redundant gene families in Arabidopsis, but are otherwise structurally and functionally very similar to their yeast and mammalian counterparts. So far, RanGAP is the only protein in the plant Ran cycle that has a diverse structure as well as a demonstrated unique localization pattern (Rose and Meier, 2001; Jeong et al., 2005). We might expect a somewhat similar situation for RCC1, which cannot be identified by methods of sequence similarity searches and is therefore likely to be relatively diverged from yeast and mammalian RCC1. A yeast complementation screen might finally allow cloning of the gene for RCC1, the now last missing player in the plant Ran cycle at the Arabidopsis nuclear pore.
MATERIALS AND METHODS
Sequence Comparison
AtNTF2a (At1g27310), AtNTF2b (At1g27970), and AtNTL (At1g11570) were identified by sequence similarity searches using Protein BLAST. MEGALIGN protein alignment software (DNASTAR) was used for multiple sequence alignments using Clustal algorithm performed as previously described (Rose and Meier, 2001).
RT-PCR Cloning of AtNTF2a, AtNTF2b, and AtNTL
The AtNTF2a, AtNTF2b, and AtNTL cDNAs were isolated by RT-PCR with the primers NTF1-F (5′-CAC CAT GGA TCC AGA CGC TGT TGC-3′), NTF1-R (5′-TCA GGC ATA GTT CAA CCT GAA TAT GTC-3′), NTF2-F (5′-CAC CAT GTC TCA GAT GGA TCC CGA CG-3′), NTF2-R (5′-TCA GGC ATA GTT CAA CCT GAA TAT GTC-3′), NTL-F (5′-GCT CTA GAA TGG CAG AGA CAA ATA AAG GAA-3′), and NTL-R (5′-GCT CTA GAC TAA GAT TGT TTT AAC CAA ATG-3′). RNA was prepared from Arabidopsis (Arabidopsis thaliana) ecotype Columbia leaf tissue using the RNeasy Plant Mini kit (Qiagen). RT-PCR was performed using the ProSTAR HF single-tube RT-PCR system from Stratagene with 100 ng total RNA, 100 ng of each primer, and 48°C annealing temperature. The resulting RT-PCR products of approximately 400 bp in length were cloned into the pENTR TOPO vector (Invitrogen) to create pIM1011 (AtNTF2a) and pIM1012 (AtNTF2b). The AtNTL construct served as PCR template for the construction of pIM1020. All cDNA inserts were confirmed by sequencing.
Plasmid Construction
All plasmids used for this study are listed in Table I. PCR reactions were performed with PFU polymerase (Invitrogen). The sources for the AtNTF2a and AtNTF2b cDNAs for all cloning steps were pIM1011 and pIM1012, respectively. All inserts were confirmed by sequencing. To construct pIM1018 and pIM1019, the coding regions of AtNTF2a and AtNTF2b were amplified by PCR with primers introducing a KpnI site at the 5′ and 3′ ends. PCR products were cloned into pYES2 (Invitrogen). pIM1003, pIM1004, pIM1020, pIM1007, and pIM1008 were made by amplifying the coding region of AtNTF2a, AtNTF2b, AtNTL, AtNTF2aE38K, and AtNTF2aE91K. Primers were designed to introduce a SpeI site at the 5′ and 3′ ends. pIM1000 and pIM1001 were made by amplifying the coding region of AtNTF2a and AtNTF2b with flanking EcoRI sites and cloning the PCR products into pEG202 (Gyuris et al., 1993).
Table I.
List of plasmids used in this study
Abbreviations: AMPR, ampicillin resistance; Y2H, yeast two hybrid; DBD, DNA-binding domain; AD, activation domain; KanR, kanamycin resistance; SPR, spectinomycin resistance; HYGR, hygromycin resistance.
| Name | Description | Reference |
|---|---|---|
| pAC19 | pGAL 2μ LEU2 AMPR | Schlenstedt et al. (1995) |
| pEG202 (pAC20) | Y2H LexA-DBD fusion plasmid; pADH 2μ HIS3 AMPR | Gyuris et al. (1993) |
| pJG4-5 (pAC21) | Y2H LexA-AD fusion plasmid; pGAL 2μ TRP1 AMPR | Gyuris et al. (1993) |
| pAC626 (pPS883) | Yeast NTF2; CEN URA3 AMPR | Corbett and Silver (1996) |
| pAC370 | Human RAN in pAC21; pGAL 2μ TRP1 AMPR | Clarkson et al. (1997) |
| pAC373 | Yeast RAN (GSP1) in pAC21; pGAL 2μ TRP1 AMPR | Wong et al. (1997) |
| pAC410 | GSP1-GFP (yeast RAN); 2μ URA3 AMPR | Quimby et al. (2001) |
| pAC825 | C-terminal GFP fusion plasmid; pMET CEN URA3 AMPR | Niedenthal et al. (1996) |
| pIM1000/pAC1870 | AtNTF2a in pAC20; pADH 2μ HIS3 AMPR | This study |
| pIM1001/pAC1871 | AtNTF2b in pAC20; pADH 2μ HIS3 AMPR | This study |
| pIM1002/pAC1875 | AtRan1 in pAC21; pGAL 2μ TRP1 AMPR | This study |
| pIM1003/pAC1878 | AtNTF2a in pAC19; pGAL 2μ LEU2 AMPR | This study |
| pIM1004/pAC1879 | AtNTF2b in pAC19; pGAL 2μ LEU2 AMPR | This study |
| pIM1005/pAC1941 | AtNTF2aE38K in pAC20; pADH 2μ HIS3 AMPR | This study |
| pIM1006/pAC1942 | AtNTF2aE91K in pAC20; pADH 2μ HIS3 AMPR | This study |
| pIM1007/pAC1947 | AtNTF2aE38K in pAC19; pGAL 2μ LEU2 AMPR | This study |
| pIM1008/pAC1948 | AtNTF2aE91K in pAC19; pGAL 2μ LEU2 AMPR | This study |
| pIM1009/pAC1949 | AtNTF2a in pAC825; pMET CEN URA3 AMPR | This study |
| pIM1010/pAC1950 | AtNTF2b in pAC825; pMET CEN URA3 AMPR | This study |
| pK7FWG2.0 | C-terminal fusion with GFP; KanR SPR | Karimi et al. (2002) |
| pH2GW7 | Overexpression vector in plant; HYGR SPR | Karimi et al. (2002) |
| pYES2 | Inducible yeast expression vector; pGAL; URA3 AMPR | Invitrogen |
| pIM1011 | AtNTF2a in pENTR TOPO; KanR | This study |
| pIM1012 | AtNTF2b in pENTR TOPO; KanR | This study |
| pIM1013 | AtNTF2aE38K in pENTR TOPO; KanR | This study |
| pIM1014 | AtNTF2a in pK7FWG2.0; KanR SPR | This study |
| pIM1015 | AtNTF2b in pK7FWG2.0; KanR SPR | This study |
| pIM1016 | AtNTF2a in pH2GW7; HYGR SPR | This study |
| pIM1017 | AtNTF2aE38K in pH2GW7; HYGR SPR | This study |
| pIM1018 | AtNTF2a in pYES2; pGAL; URA3 AMPR | This study |
| pIM1019 | AtNTF2b in pYES2; pGAL; URA3 AMPR | This study |
| pIM1020 | AtNTL in pAC19; pGAL 2μ LEU2 AMPR | This study |
| pAC2024 | Yeast NTF2 in pAC19; pGAL 2μ LEU2 AMPR | This study |
| pENTR TOPO | Blunt-end PCR product entry vector; KanR | Invitrogen |
To construct pIM1002, the coding region of AtRan1 was first amplified by RT-PCR and cloned into a pENTR TOPO vector. It was then re-amplified with flanking EcoRI sites by PCR and cloned into pJG4-5 (Gyuris et al., 1993). pIM1009 and pIM1010 were made by cloning the EcoRI inserts from pIM1000 and pIM1001, respectively, into pAC825. pIM1013 was made by amplifying the coding region of AtNTF2aE38K by PCR from pIM1005. The PCR product was cloned into the pENTR TOPO vector. pIM1014 and pIM1015 were made by moving the inserts from pIM1011 and pIM1012 by LR recombination into pK7FWG2.0 (Karimi et al., 2002), which contains a cassette of the cauliflower mosaic virus 35S promoter and GFP flanking the recombination site leading to fusions of AtNTF2a and AtNTF2b to the N terminus of GFP. pIM1016 and pIM1017 were made by moving the inserts from pIM1011 and pIM1013 by LR recombination to pH2GW7 (Karimi et al., 2002), which contains a cassette of the cauliflower mosaic virus 35S promoter flanking the recombination site.
Site-Directed Mutagenesis
Point mutations were introduced using the QuikChange XL site-directed mutagenesis kit (Stratagene) with pIM1000 as template. Primers 5′-AAT CTT CAG CTC GCT GGT AAA CAA CAC GCT CTC AAG TTC AG-3′ and 5′-CTG AAC TTG AGA GCG TGT TGT TTA CCA GCG AGC TGA AGA TT-3′ were employed to change E91 to K. Primers 5′-ATC CAT GTT GAC CTT CAA AGG CA GAA GAT CCA GGG C-3′ and 5′-GCC CTG GAT CTT CTG GCC TTT GAA GGT CAA CAT GGA T-3′ were employed to change E38 to K. NTF1 inserts were sequenced to confirm that only the desired mutations were created. The plasmids containing the E38K and E91K mutants were named pIM1005 and pIM1006, respectively.
Yeast Cell Growth Assays/Plasmid Shuffle Assays
To analyze the function of the putative Arabidopsis NTF2s, the AtNTF2a, AtNTF2b, and AtNTL open reading frames were cloned into yeast (Saccharomyces cerevisiae) expression vectors under the control of a Gal-inducible promoter. As a control, yeast NTF2 was also cloned into a Gal-inducible vector (pAC2024). Each of these plasmids, as well as the vector-only control, was transformed into yeast cells where the endogenous NTF2 gene had been deleted (ACY114) but the viability of the strain was maintained by the presence of a URA3-marked yeast NTF2 genomic plasmid (pAC626; Corbett and Silver, 1996). To assess the function of the Arabidopsis NTF2 homologs, cultures of cells carrying each of the plasmids were grown to saturation, serially diluted, and then spotted on either control plates (ura− leu− Glc) or 5-FOA leu− gal plates. On control plates, the genomic yeast NTF2 maintenance plasmid was retained. Thus, all cells expressed yeast NTF2 and grew equally well, indicating that similar numbers of cells were spotted for each sample. On the 5-FOA plates, the URA3-marked yeast NTF2 maintenance plasmid was lost (Boeke et al., 1987). These plates contained Gal, so the Arabidopsis NTF2 proteins were expressed as was the control yeast NTF2. No growth was seen when cells were spotted on 5-FOA plates lacking Gal (data not shown).
Yeast Two-Hybrid Analysis
Two-hybrid/interaction trap constructs were generated by cloning the appropriate genes into either the LexA-binding domain plasmid pEG202 or the acid-blob transcriptional activator (activation domain) plasmid pJG4-5 (Gyuris et al., 1993). The pJG4-5 plasmid is designed so that the activation domain fusion proteins are only expressed in the presence of Gal. Thus, for all experiments, plating on Glc serves as a negative control as the pJG4-5-encoded protein will not be expressed. For this reason, valid interactions should be detected only on Gal plates and not on Glc plates. For all constructs, levels of expression were examined by immunoblotting either with anti-LexA antibodies (pEG202 constructs) or with anti-hemagglutinin antibodies (pJG4-5 constructs). Levels of expression were approximately equal for all proteins, indicating that all fusion proteins had comparable stability and so the failure of some to interact with NTF2 was not due to their levels of expression or stability being substantially lower. Interactions between proteins were examined by cotransforming pEG202 (DNA-binding domain) and pJG4-5 (activation domain) plasmids into the yeast strain EGY48 (MATa ura3 trp1 his3 lexAopLEU2; see Wong et al., 1997). An interaction was scored as positive if the cells were Leu+ (data not shown) and turned blue on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside indicator plates (Gyuris et al., 1993).
Gene Expression Pattern
The cDNAs for AtNTF2a, AtNTF2b, and the control gene (actin) were synthesized and amplified by RT-PCR using specific primers for each gene. Primers 5′-CAC CAT GGA TCC AGA CGC TGT TGC-3′ and 5′-CCA CCA CTC TCT TTT CAG CTT CC-3′ were used for NTF1, and 5′-CAC CAT GTC TCA GAT GGA TCC CGA CG-3′ and 5′-CTG TTC CCA AGG TAA ATC ATC TGG G-3′ were used for NTF2. Both reverse primers are in the 3′-untranslated region to distinguish between the two genes. For Actin-related protein 6 (At3g33520) gene as control, primers 5′-AAA ACC ACT TAC AGA GTT CGT TCG-3′and 5′-GTT GAA CGG AAG GGA TTG AGA GT-3′ were used. Total RNA was prepared from flowers, leaves, roots, stems, and siliques of 30-d-old Arabidopsis ecotype Columbia using the RNeasy Plant Mini kit (Qiagen). RT-PCR was performed as described above.
Ballistic Transient Transformation of Tobacco BY-2 Cells
Transient transformation was performed as described previously (Rose and Meier, 2001) using pIM1014 and pIM1015. Transformed cells were examined for GFP by confocal laser scanning microscopy essentially as described previously (Rose and Meier, 2001).
Agroinfiltration of Nicotiana benthamiana
The constructs pIM1016 and pIM1017 were transformed into Agrobacterium tumefaciens (strain ABI), and transformants were selected with 50 μg/mL spectinomycin, 50 μg/mL kanamycin, and 17 μg/mL chloramphenicol. As a reporter gene for nuclear import, maize (Zea mays) R fused to GFP in pGWB5 was used (R-GFP; gift from Dr. Erich Grotewold, The Ohio State University [OSU]; Shieh et al., 1993). R-GFP was transformed into A. tumefaciens GV3101, and transformants were selected with 40 μg/mL gentamicin. A construct expressing the gene-silencing suppressor P19 under control of the 35S promoter (gift from Dr. Desh Pal Verma, OSU) was transformed into A. tumefaciens C58 with selection on tetracycline (5 μg/mL final) and kanamycin (50 μg/mL final). Agrobacterium growth and infiltration conditions were essentially as described (Scofield et al., 1996; Tang et al., 1996). R-GFP, P19, and either AtNTF2a or AtNTF2aE38K containing Agrobacterium cultures were mixed at a ratio of 1:1:1. For the control, R-GFP- and P19-containing strains were mixed at a 1:1 ratio. Transformed cells were examined for GFP by confocal laser scanning microscopy essentially as described (Rose and Meier, 2001).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_174051 (AtNTF2a), NP_174118 (AtNTF2b), and NP_172623 (AtNTL).
Acknowledgments
We thank Dr. Erich Grotewold (OSU) for the R-GFP vector, Dr. David Bisaro (OSU) for the P19 vector, Xianfeng Xu (OSU) for the AtRan1 cDNA, and the Meier and Corbett Laboratories and Dr. Murray Stewart for helpful discussions.
This work was supported by grants from the National Science Foundation (to I.M.) and the National Institutes of Health (to A.H.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Iris Meier (meier.56@osu.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.075499.
References
- Ach RA, Gruissem W (1994) A small nuclear GTP-binding protein from tomato suppresses a Schizosaccharomyces pombe cell-cycle mutant. Proc Natl Acad Sci USA 91: 5863–5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avis JM, Clarke PR (1996) Ran, a GTPase involved in nuclear processes: its regulators and effectors. J Cell Sci 109: 2423–2427 [DOI] [PubMed] [Google Scholar]
- Bayliss R, Leung SW, Baker RP, Quimby BB, Corbett AH, Stewart M (2002) Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J 21: 2843–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H (1991. a) Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354: 80–82 [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H (1991. b) Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci USA 88: 10830–10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J, Trueheart J, Natsoulis G, Fink G (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW (1999) Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400: 178–181 [DOI] [PubMed] [Google Scholar]
- Chaillan-Huntington C, Braslavsky CV, Kuhlmann J, Stewart M (2000) Dissecting the interactions between NTF2, RanGDP, and the nucleoporin XFXFG repeats. J Biol Chem 275: 5874–5879 [DOI] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Visser GD, Adam SA (1996) RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import. J Cell Biol 135: 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson WD, Corbett AH, Paschal BM, Kent HM, McCoy AJ, Gerace L, Silver PA, Stewart M (1997) Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J Mol Biol 272: 716–730 [DOI] [PubMed] [Google Scholar]
- Clarkson WD, Kent HM, Stewart M (1996) Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J Mol Biol 263: 517–524 [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA (1996) The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J Biol Chem 271: 18477–18484 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Mattaj IW (1996) Nucleocytoplasmic transport. Science 271: 1513–1518 [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75: 791–803 [DOI] [PubMed] [Google Scholar]
- Haizel T, Merkle T, Pay A, Fejes E, Nagy F (1997) Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J 11: 93–103 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW (2000) GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell 5: 1013–1024 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 16: 6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Joseph J, Dasso M, Meier I (2005) Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J 42: 270–282 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G (1994) Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA 91: 10212–10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hegemann JH (1996) Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 12: 773–786 [DOI] [PubMed] [Google Scholar]
- Pay A, Resch K, Frohnmeyer H, Fejes E, Nagy F, Nick P (2002) Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J 30: 699–709 [DOI] [PubMed] [Google Scholar]
- Quimby BB, Lamitina T, L'Hernault SW, Corbett AH (2000. a) The mechanism of ran import into the nucleus by nuclear transport factor 2. J Biol Chem 275: 28575–28582 [DOI] [PubMed] [Google Scholar]
- Quimby BB, Leung SW, Bayliss R, Harreman MT, Thirumala G, Stewart M, Corbett AH (2001) Functional analysis of the hydrophobic patch on nuclear transport factor 2 involved in interactions with the nuclear pore in vivo. J Biol Chem 276: 38820–38829 [DOI] [PubMed] [Google Scholar]
- Quimby BB, Wilson CA, Corbett AH (2000. b) The interaction between Ran and NTF2 is required for cell cycle progression. Mol Biol Cell 11: 2617–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L, Kuhlmann J, Henkel A, Wittinghofer A (2001) Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell 105: 245–255 [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Gorlich D (1998) NTF2 mediates nuclear import of Ran. EMBO J 17: 6587–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, Macara I (2005) A systems analysis of importin-{alpha}-{beta} mediated nuclear protein import. J Cell Biol 168: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Meier I (2001) A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA 98: 15377–15382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Saavedra C, Loeb JD, Cole CN, Silver PA (1995) The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and appearance of poly(A)+ RNA in the cytoplasm. Proc Natl Acad Sci USA 92: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ (1996) Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274: 2063–2065 [DOI] [PubMed] [Google Scholar]
- Shieh MW, Wessler SR, Raikhel NV (1993) Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol 101: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G (1997) Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol 138: 1181–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, Brownawell A, Macara I (1998) Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol 8: 1403–1406 [DOI] [PubMed] [Google Scholar]
- Stewart M, Kent HM, McCoy AJ (1998. a) Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J Mol Biol 277: 635–646 [DOI] [PubMed] [Google Scholar]
- Stewart M, Kent HM, McCoy AJ (1998. b) The structure of the Q69L mutant of GDP-Ran shows a major conformational change in the switch II loop that accounts for its failure to bind nuclear transport factor 2 (NTF2). J Mol Biol 284: 1517–1527 [DOI] [PubMed] [Google Scholar]
- Tachibana T, Hieda M, Sekimoto T, Yoneda Y (1996) Exogenously injected nuclear import factor p10/NTF2 inhibits signal-mediated nuclear import and export of proteins in living cells. FEBS Lett 397: 177–182 [DOI] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Wong DH, Corbett AH, Kent HM, Stewart M, Silver PA (1997) Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol 17: 3755–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Mattaj IW, Yoneda Y (2004) An ATP-dependent activity that releases RanGDP from NTF2. J Biol Chem 279: 36228–36234 [DOI] [PubMed] [Google Scholar]
- Zhang C, Hughes M, Clarke PR (1999) Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J Cell Sci 112: 2453–2461 [DOI] [PubMed] [Google Scholar]