Abstract
Once the plant coenzyme A (CoA) biosynthetic pathway has been elucidated by comparative genomics, it is feasible to analyze the physiological relevance of CoA biosynthesis in plant life. To this end, we have identified and characterized Arabidopsis (Arabidopsis thaliana) T-DNA knockout mutants of two CoA biosynthetic genes, HAL3A and HAL3B. The HAL3A gene encodes a 4′-phosphopantothenoyl-cysteine decarboxilase that generates 4′-phosphopantetheine. A second gene, HAL3B, whose gene product is 86% identical to that of HAL3A, is present in the Arabidopsis genome. HAL3A appears to have a predominant role over HAL3B according to their respective mRNA expression levels. The hal3a-1, hal3a-2, and hal3b mutants were viable and showed a similar growth rate as that in wild-type plants; in contrast, a hal3a-1 hal3b double mutant was embryo lethal. Unexpectedly, seedlings that were null for HAL3A and heterozygous for HAL3B (aaBb genotype) displayed a sucrose (Suc)-dependent phenotype for seedling establishment, which is in common with mutants defective in β-oxidation. This phenotype was genetically complemented in aaBB siblings of the progeny and chemically complemented by pantethine. In contrast, seedling establishment of Aabb plants was not Suc dependent, proving a predominant role of HAL3A over HAL3B at this stage. Total fatty acid and acyl-CoA measurements of 5-d-old aaBb seedlings in medium lacking Suc revealed stalled storage lipid catabolism and impaired CoA biosynthesis; in particular, acetyl-CoA levels were reduced by approximately 80%. Taken together, these results provide in vivo evidence for the function of HAL3A and HAL3B, and they point out the critical role of CoA biosynthesis during early postgerminative growth.
CoA is a cofactor for a multitude of enzymatic reactions, including the oxidation of fatty acids, carbohydrates, and amino acids, as well as many synthetic reactions (Begley et al., 2001). CoA is synthesized in five steps from pantothenate, and recently all the biosynthetic enzymes have been cloned in both prokaryotes and higher eukaryotes (Begley et al., 2001; Daugherty et al., 2002; Kupke et al., 2003; Leonardi et al., 2005). Indeed, both in humans and plants, the complete biosynthetic pathway from pantothenate to CoA has been reconstituted in vitro using recombinant enzymes (Daugherty et al., 2002; Kupke et al., 2003). The universal pathway for biosynthesis of CoA from pantothenate is initiated by phosphorylation of this precursor to generate 4′-phosphopantothenate, which is catalyzed by pantothenate kinase (PK). Then, the addition of Cys to 4′-phosphopantothenate gives rise to 4′-phospho-N-pantothenoyl-cysteine (PPC), which is catalyzed by PPC synthetase. In the next step, PPC is decarboxylated to 4′-phosphopantetheine by PPC decarboxylase (PPCDC). Finally, 4′-phosphopantetheine is converted to CoA by the sequential action of the enzymes 4′-phosphopantetheine adenylyltransferase and dephospho-CoA kinase. In humans, the two latter activities are encoded in a single bifunctional enzyme (Daugherty et al., 2002). In Escherichia coli, both the addition of Cys to 4′-phosphopantothenate and the subsequent decarboxylation of PPC are catalyzed by the bifunctional enzyme Dfp (mnemonic for DNA and flavoprotein; Kupke et al., 2000; Strauss et al., 2001; Kupke, 2002).
In plants, every step of CoA biosynthesis from pantothenate is catalyzed by single monofunctional enzymes, and the pathway has been reconstituted in vitro by combining the recombinant enzymes PK (AtCoaA, At1g60440), PPC synthetase (AtCoaB, At1g12350), PPCDC (HAL3A, AtCoaC, and At3g18030), 4′-phosphopantetheine adenylyltransferase (AtCoaD, At2g18250), and dephospho-CoA kinase (AtCoaE, At2g27490). However, many aspects of the pathway are not well understood and, to our knowledge, the physiological consequences for plant life of impairing CoA biosynthesis have not been addressed by genetic analysis. In particular, the subcellular location of the Arabidopsis (Arabidopsis thaliana) CoA biosynthetic enzymes has not been studied in detail. It is known that the last enzyme of the pantothenate biosynthesis pathway (pantothenate synthetase) is found in the cytosol (Ottenhof et al., 2004), whereas PK activity appears to be predominantly localized in the chloroplast in spinach (Spinacia oleracea; Falk and Guerra, 1993), with some activity observed in cytosol. However, the five Arabidopsis members of the PK family (At1g60440, At4g32180, At2g17320, At2g17340, and At4g35360) as well as AtCoaC, AtCoaD, and AtCoaE do not show chloroplast-targeting sequences according to prediction programs such as ChloroP or PSORT. In any case, CoA itself is found in all cellular compartments and this multicompartmentation implies that there must be transporters present for shuttling intermediates. However, to our knowledge, currently only one transport activity for import of CoA into mitochondria has been reported in potato (Solanum tuberosum), although the corresponding gene has not been cloned (Neuburger et al., 1984).
As CoA plays an essential role in metabolism, null mutations in CoA biosynthetic genes are presumed to be lethal, unless there is some degree of genetic redundancy in the organism. For instance, in both yeast (Saccharomyces cerevisiae) and fly (Drosophila melanogaster), each with only one PK gene, the null mutant is nonviable (Winzeler et al., 1999; Afshar et al., 2001). To identify plant mutants impaired in CoA biosynthesis, we took advantage of the fact that some steps of this pathway are catalyzed by more than one gene product in Arabidopsis (Kupke et al., 2003). Plant mutants partially impaired in CoA biosynthesis offer the possibility to genetically test the function of CoA in plant biology. For instance, in oilseed plants the use of storage lipids during early seedling establishment is a key process for plant survival that is CoA dependent (Graham and Eastmond, 2002). Additionally, as CoA biosynthesis appears to be a sensitive step in plants under salt stress (Espinosa-Ruiz et al., 1999; Yonamine et al., 2004), an improved knowledge of this pathway might help our understanding of how plants cope with abiotic stresses. Finally, as the CoA biosynthetic pathway is evolutionarily conserved, its study in plants might have clinical relevance, as defects in this pathway lead to a human neurodegenerative disease (Zhou et al., 2001).
The plant CoA biosynthetic pathway has been recently defined in plants (Kupke et al., 2003), and the biochemistry of one of the biosynthetic enzymes, HAL3A (AtCoaC, PPCDC), has been studied in detail (Albert et al., 2000; Kupke et al., 2001; Hernandez-Acosta et al., 2002; Steinbacher et al., 2003). HAL3A is a flavoprotein that catalyzes the decarboxylation of PPC to 4′-phosphopantetheine (Kupke et al., 2001; Hernandez-Acosta et al., 2002), and overexpression of this enzyme leads to improved plant tolerance to salt and osmotic stress (Espinosa-Ruiz et al., 1999; Yonamine et al., 2004). Two highly homologous genes, HAL3A (At3g18030) and HAL3B (At1g48605), are present in the Arabidopsis genome. Expression of HAL3A and HAL3B mRNAs was previously analyzed in seeds and different organs of adult plants (root, shoot, leaf, flower, and silique), as well as in 12-d-old seedlings (Espinosa-Ruiz et al., 1999). As a general result, transcript level of HAL3B mRNA was found to be lower than HAL3A mRNA. Therefore, according to their relative transcript levels, HAL3A appears to play a predominant role over HAL3B. For instance, HAL3B mRNA was expressed to very low level in seeds, whereas strong expression was observed for HAL3A mRNA. During embryogenesis, in situ hybridization of HAL3A mRNA revealed that the transcript was mainly detected in the cotyledons and hypocotyl of mature seeds, and to a lower level in the seed coat outer integument (Espinosa-Ruiz et al., 1999). Finally, according to northern-blot analysis, HAL3B mRNA was expressed 3- to 4-fold less than HAL3A mRNA in 12-d-old seedlings (Espinosa-Ruiz et al., 1999).
Even though the function of HAL3B has not been experimentally addressed (Leonardi et al., 2005), taking into account the high sequence similarity between both genes, we hypothesized that HAL3B might partially play a redundant role to HAL3A. Keeping in mind the crucial role of CoA biosynthesis for plant life, a certain degree of genetic redundancy would allow the identification of viable hal3a and hal3b mutants. Thus, T-DNA-disrupted alleles of HAL3A and HAL3B were identified in Arabidopsis T-DNA collections, and the corresponding homozygous mutants were isolated and found to be viable. Indeed, the single hal3a and hal3b mutants showed similar phenotypes to wild-type plants; however, a hal3a hal3b double mutant was embryo lethal, proving the expected crucial role for CoA biosynthesis. Unexpectedly, a Suc-dependent seedling establishment phenotype was found for hal3a plants that were heterozygous for the T-DNA-disrupted hal3b allele (aaBb individuals), which were unable to surpass the heterotrophic growth phase that occurs upon germination.
RESULTS
Isolation of T-DNA Insertional Mutations in the Arabidopsis HAL3A and HAL3B Genes
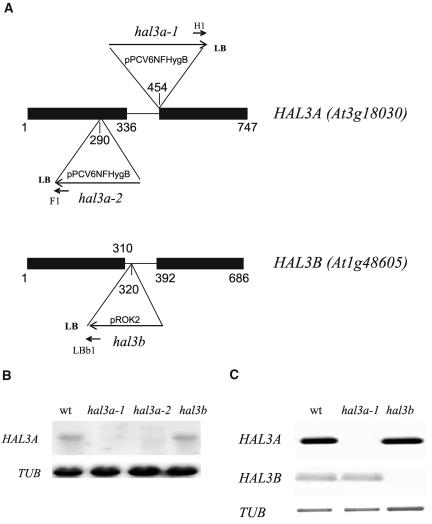
Two T-DNA-disrupted alleles of HAL3A were identified from the T-DNA collection of the Max-Planck Institute (Cologne, Germany) through PCR-based screening of Arabidopsis plants containing random T-DNA insertions (Rios et al., 2002; Fig. 1A). The first allele, hal3a-1, is a T-DNA insertion in the intron of the gene, and it is localized 454 nucleotides downstream from the ATG start codon (Fig. 1A). The second one, hal3a-2, is a T-DNA insertion in the first exon of the gene, and it is localized 290 nucleotides downstream from the ATG start codon. No HAL3A transcript was detected in seedlings of either mutant (Fig. 1B). In the case of HAL3B, no T-DNA-disrupted allele could be recovered from the Max-Planck collection. Moreover, only one T-DNA line from the currently available collections leads to disruption of the transcription unit (http://signal.salk.edu/cgi-bin/tdnaexpress), corresponding to donor stock number SALK_045607 (Fig. 1A). This hal3b allele is a T-DNA insertion in the intron of the gene, and it is localized 320 nucleotides downstream from the ATG start codon (Fig. 1A).
Figure 1.
Molecular characterization of hal3a and hal3b mutants. A, Scheme of the HAL3A and HAL3B genes and localization of the respective T-DNA insertions in the different alleles. The numbering begins at the ATG translation start codon. The T-DNA left border (LB) primers that were used to localize the T-DNA insertion are indicated. B, Northern-blot analysis of wild-type, hal3a-1, hal3a-2, and hal3b mRNAs probed with a cDNA probe specific for HAL3A transcript. C, RT-PCR analysis of wild-type, hal3a-1, and hal3b mRNAs prepared from 5-d-old seedlings. Primers specific for HAL3A, HAL3B, and TUB transcripts were used.
Reverse transcription (RT)-PCR analysis was performed both for HAL3A and HAL3B mRNA expression in 5-d-old seedlings from wild-type, hal3a-1, and hal3b mutants (Fig. 1C). The expression of HAL3B was found to be lower than HAL3A in wild type (Fig. 1C), which confirms that HAL3A is the predominant isoform of HAL3 present during seedling establishment (Espinosa-Ruiz et al., 1999). Indeed, 5-d-old seedlings showed a 4-fold higher expression level for HAL3A compared to HAL3B, as measured by RT-quantitative-PCR analyses (data not shown). Expression of HAL3A and HAL3B was abolished in the hal3a-1 and hal3b mutants, respectively (Fig. 1C). RT-quantitative-PCR analyses of HAL3A in hal3b or HAL3B in hal3a-1 revealed a similar expression level to the one observed in wild type for each gene (data not shown).
hal3a-1 hal3b Double Mutant Is Embryo Lethal
Transgenic Arabidopsis plants (two lines) that express an antisense full-length cDNA of HAL3A were reported to show a delayed growth rate and impaired osmotic stress tolerance compared to wild-type plants (Espinosa-Ruiz et al., 1999). In contrast, the null mutant hal3a-1 failed to show those phenotypes, as growth rate (Fig. 2A), salt sensitivity (see later Fig. 4C), and osmotic stress tolerance (data not shown) were quite comparable to wild-type plants. Similar results to those of hal3a-1 were obtained for the allelic hal3a-2 mutant (data not shown). Whereas a decrease in HAL3A transcript amount was measured in the two antisense lines constructed by Espinosa-Ruiz et al. (1999), HAL3B expression was not investigated. Antisense expression of full-length HAL3A cDNA might lead to a decreased HAL3B transcript amount because of the high sequence identity between both genes at the nucleotide level. This explanation might reconcile the discrepancy between our results for hal3a-1 and hal3a-2 mutants, and the antisense approach of Espinosa-Ruiz et al. (1999).
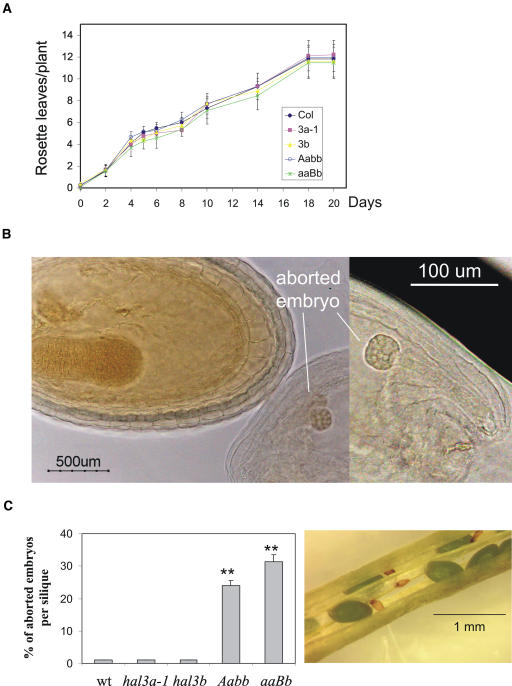
Figure 2.
A, Growth rate of wild-type, hal3a-1, hal3b, Aabb, and aaBb plants. Values are averages ± sd (n = 20). B, The hal3a-1 hal3b double mutant is embryo lethal. Nomarski image of chemically cleared seed progeny from aaBb plants showing a viable embryo at the torpedo stage and an aborted embryo arrested at the early/midglobular stage. C, Percentage of aborted embryos per silique. Siliques of wild-type, hal3a-1, hal3b, Aabb, and aaBb plants were examined under a Nikon SMZ800 binocular glass. A representative silique from aaBb individuals is shown. Values are averages ± sd (n = 20). Double asterisks (**) indicate P < 0.03 (Student's t test) with respect to wild type (wt).
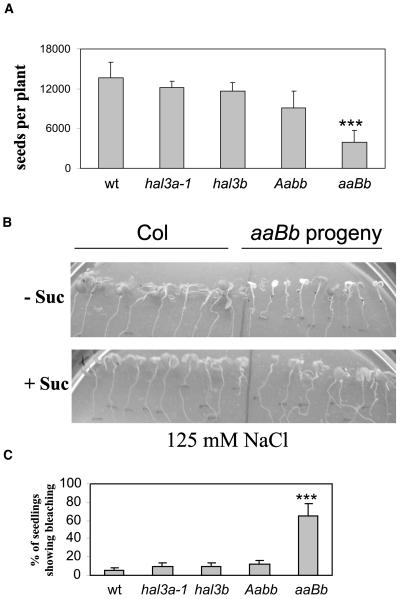
Figure 4.
Reduced seed production and enhanced salt sensitivity of aaBb plants. A, Seed production of wild type, hal3a-1, hal3b, Aabb, and aaBb plants. Values are averages ± sd (n = 20). Triple asterisks (***) indicate P < 0.01 (Student's t test) with respect to wild type. B, Salt hypersensitivity of aaBb seedlings. Seedlings grown in medium containing 1% Suc were transferred to medium supplemented with 125 mm NaCl and lacking (−Suc) or containing 1% Suc (+Suc). The photograph was taken after 7 d in medium containing NaCl. C, Percentage of seedlings showing bleaching when transferred to medium supplemented with 125 mm NaCl and lacking Suc. Values are averages ± sd for three independent experiments (40 seedlings each). Triple asterisks (***) indicate P < 0.01 (Student's t test) with respect to wild type (wt).
No visible phenotype for the single hal3a or hal3b mutant was observed under our experimental conditions (Figs. 2, 3, and 4). As the predicted HAL3A and HAL3B gene products show 90% amino acid similarity, we reasoned that some functional redundancy might exist between both genes. To establish whether HAL3A function is partially redundant with HAL3B, we tried to generate a double mutant by crossing hal3a-1 and hal3b homozygous mutants in both directions. We could not recover double-homozygous mutants in spite of genotyping more than 200 plants of the F2 progeny. However, it was possible to identify hal3a-1 plants that were heterozygous for the T-DNA-disrupted hal3b allele (aaBb genotype). Chemically cleared preparations from young fruits of aaBb plants showed that embryogenesis was arrested in approximately one quarter of the seeds, which degenerated into brown aborted seeds during maturation (Fig. 2, B and C). Indeed, no aabb double mutant was obtained in the progeny of self-fertilized aaBb plants, and the ratio of heterozygous HAL3B/hal3b to homozygous HAL3B/HAL3B plants was close to 2:1 (188:95, χ2 = 0.012, P > 0.9). This finding suggests that viable embryos of the aaBb seed progeny must represent either the aaBB or aaBb genotype, whereas the aborted embryo must correspond to a putative aabb double mutant (Fig. 2B). The aabb embryo was arrested at the early/midglobular stage (Fig. 2B), and the number of aborted embryos per silique was in agreement with the expected lethality of the aabb genotype (Fig. 2C). These results indicate that homozygous hal3b embryos are not viable in the hal3a-1 background. Additionally, the fact that one or two wild-type copies of HAL3B (in a hal3a-1 background) support the growth of viable embryos reflects a partial functional redundancy between HAL3A and HAL3B genes.
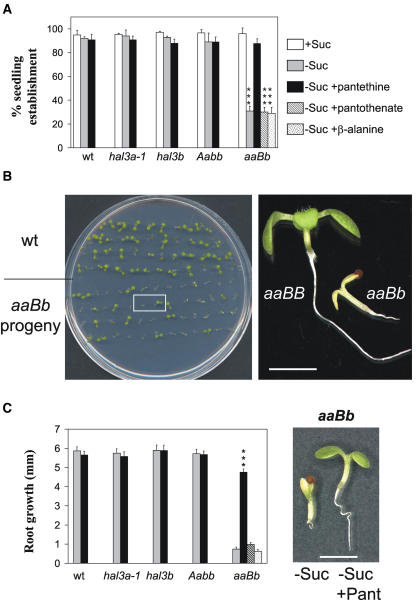
Figure 3.
Suc-dependent phenotype of aaBb seedlings. Genetic and chemical complementation. A, Seedling establishment of the seed progeny from wild-type, hal3a-1, hal3b, Aabb, and aaBb plants in medium supplemented with 1% Suc (+Suc), lacking Suc (−Suc), lacking Suc and supplemented with 50 μg/mL of pantethine (−Suc + pantethine), pantothenate (−Suc + pantothenate), or β-Ala (−Suc + β-Ala). Values are averages ± sd for three independent experiments (200 seeds each). Triple asterisks (***) indicate P < 0.01 (Student's t test) with respect to aaBb + Suc. B, Segregating phenotype of the seed progeny from aaBb plants in medium lacking Suc. Representative aaBB and aaBb seedlings were removed from the medium at 7 d after sowing and they were photographed under a Nikon SMZ800 binocular glass. Bar = 2 mm. C, Root growth at 7 d after sowing from wild-type, hal3a-1, hal3b, Aabb, and aaBb individuals in medium lacking Suc (gray bars) or lacking Suc but supplemented with 50 μg/mL of pantethine (black bars), pantothenate (cross-hatched bar), or β-Ala (stippled bar). Root growth in medium with 1% Suc was not statistically different for the seedlings. Values are averages ± sd for two independent experiments (40 seedlings each). Triple asterisks (***) indicate P < 0.01 (Student's t test) with respect to aaBb in medium lacking Suc. Representative aaBb seedlings were removed from medium lacking Suc (−Suc) or lacking Suc but supplemented with 50 μg/mL pantethine (−Suc + Pant) and rearranged on agar plates (right). Photograph was taken at 5 d after sowing. Bar = 2 mm. wt, Wild type.
During the analysis of the F2 progeny described above, we also identified hal3b plants that were heterozygous for the T-DNA-disrupted hal3a-1 allele (Aabb genotype). Siliques of these plants also revealed approximately one quarter of aborted embryos (Fig. 2C), likewise indicating that hal3a-1 embryos are not viable in the hal3b homozygous background. Growth rate of Aabb plants was quite similar to wild type, whereas aaBb plants showed a slight delay with respect to wild type (Fig. 2A).
aaBb Plants Require Exogenous Suc for Seedling Establishment
According to the main reserve compound, Arabidopsis qualifies as an oilseed plant, and mutants severely impaired in the ability to catabolize storage lipid require an exogenous supply of Suc for seedling establishment (Hayashi et al., 1998, 2002; Germain et al., 2001; Zolman et al., 2001; Footitt et al., 2002; Fulda et al., 2004). We reasoned that mutants impaired in CoA biosynthesis might have compromised fatty acid β-oxidation, leading to a requirement for Suc supplementation in the heterotrophic growth phase of the plant. Therefore, we analyzed seedling establishment of the different mutant backgrounds described above on media supplemented with or lacking exogenous Suc (Fig. 3A). Wild-type seeds germinated and grew normally, regardless of the presence or absence of Suc in the growth medium. Similar behavior was observed for seeds of hal3a-1 and hal3b single mutants (Fig. 3A). In contrast, seedling establishment of the progeny of aaBb plants was notably impaired in the absence of Suc (Fig. 3A), whereas germination was not severely compromised under the conditions used.
Figure 3B shows that approximately two thirds of the progeny were represented by stunted individuals with a very short root, whereas one third of the seedlings were similar to wild-type plants (188:95, χ2 = 0.012, P > 0.9). Stunted individuals became senescent after 2 weeks in the absence of Suc, but they could be rescued to normal growth by transfer to a medium supplemented with Suc. Genotyping of these individuals revealed they had an aaBb genotype, whereas those seedlings that did not require Suc for postgerminative growth had an aaBB genotype. Thus, a single copy of the HAL3B gene (in a hal3a-1 background) was not able to support postgerminative growth in medium lacking Suc. These results also show genetic complementation of the Suc-dependent phenotype of aaBb seedlings by an additional copy of the HAL3B gene (right section of Fig. 3B, compare aaBb and aaBB siblings).
In contrast to the phenotype reported above for aaBb, the progeny of Aabb plants did not show a segregating phenotype in medium lacking Suc (Fig. 3A). Genotyping of this progeny revealed no double hal3a-1 hal3b mutant but hal3b individuals that had either one or two wild-type copies of the HAL3A gene. This result indicates that a single copy of HAL3A, in a hal3b background, is enough to support postgerminative growth in medium lacking Suc. Thus, whereas aaBb seedlings were Suc dependent, this was not the case for Aabb genotype. This observation can be explained by the fact that HAL3A expression in seedlings is higher than HAL3B (Espinosa-Ruiz et al. 1999; Fig. 1C). Additionally, this finding confirms that HAL3A function is particularly crucial for seedling establishment.
The Suc-Dependent Phenotype of aaBb Seedlings Is Complemented by Pantethine
HAL3A catalyzes the decarboxylation of 4′-phosphopantothenoyl-cysteine to 4′-phosphopantetheine, and therefore, this step of the CoA biosynthetic pathway must be severely impaired in aaBb individuals. The 4′-phosphopantetheine compound is not commercially available and additionally, phosphorylated precursors of CoA or CoA itself are not able to efficiently cross the plasma membrane (Shibata et al., 1983). However, we could obtain pantethine, which is the dimer resulting from the oxidation of the thiol group of pantetheine and subsequent formation of a disulfide bond. Thus, we were interested in examining whether supplementation of the media with pantethine might complement the Suc-dependent seedling establishment phenotype of the aaBb mutant. Figure 3A shows that seedling establishment in medium lacking Suc was recovered in aaBb upon pantethine supplementation, which was also reflected by measurements of root growth (Fig. 3C). In contrast, supplementation of the medium with other CoA precursors such as pantothenate or β-Ala, which are upstream of HAL3A function, was not able to complement the Suc-dependent phenotype of aaBb seedlings (Fig. 3, A and C). Pantethine complementation of the Suc-dependent phenotype indicates that impaired CoA biosynthesis is responsible for the observed phenotype in the aaBb mutant.
Seed Production and Salt Tolerance Are Severely Impaired in aaBb Plants
During photoautotrophic growth both Aabb and aaBb plants did not show obvious vegetative phenotypes, suggesting that a single gene copy of either HAL3A or HAL3B provides enough CoA for this growth phase. However, reproductive growth was particularly impaired in aaBb plants, where less inflorescence stems were present, although inflorescence and floral development were comparable to those of wild-type plants (data not shown). As a result, seed production was severely impaired in aaBb plants and to a lesser extent in Aabb plants, resulting in a reduction of approximately 70% compared to wild type (Fig. 4A).
Finally, as overexpression of HAL3A improves plant tolerance to salt stress (Espinosa-Ruiz et al., 1999; Yonamine et al., 2004), we decided to test salt sensitivity of the different mutant backgrounds. To this end, 7-d-old seedlings grown in a medium containing 1% Suc were transferred to a medium lacking Suc and supplemented with 125 mm NaCl. Compared to wild-type plants, aaBb individuals were hypersensitive to salt stress, as they bleached after 7 d in medium supplemented with NaCl (Fig. 4B). However, a similar salt sensitivity as that in wild-type plants was observed after transfer to medium supplemented with 125 mm NaCl and 1% Suc (Fig. 4B). Salt sensitivity of hal3a-1, hal3b, and Aabb mutants was quite similar to wild type both in the presence (data not shown) or absence of exogenous Suc (Fig. 4C).
Fatty Acid and Acyl-CoA Profiling
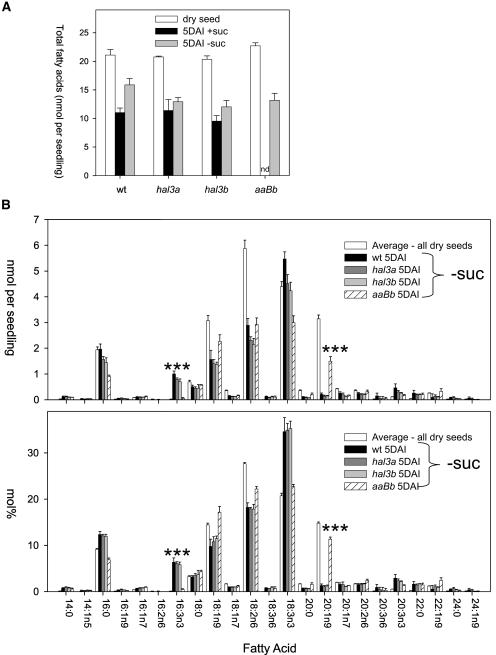
Dry seed from wild-type, aaBB, Aabb, and aaBb plants all had total fatty acid yields between 20 to 23 nmol seed−1 (30% w/w), as determined by gas chromatography-flame ionization detector analysis (Fig. 5A), with a similar molar percent distribution for individual fatty acids (Fig. 5B). Therefore, although seed yield per plant was lower for aaBb plants, normal oil deposition within individual seeds occurred during seed development. Dry seed of an aaBb plant contains, respectively, two thirds and one third of aaBb and aaBB seeds, therefore only a relative comparison with data obtained for aaBb seedlings can be made (see below).
Figure 5.
Altered fatty acid profile in aaBb seedlings. Dry seeds or seedlings grown on medium in the presence or absence of 1% Suc were extracted at 5 DAI for fatty acid profiling. Total fatty acids (A) and profile data (B) are shown. Data in B are expressed both as nmol/seedling as well as mol percentage (nmol fatty acid/total nmol fatty acids). Data in B refer to seedlings that were obtained in the absence of Suc (−Suc). Total fatty acid content of aaBb and Aabb genotypes in medium supplemented with Suc could not be determined because their seed progeny consists of a segregating population. Dry seeds of an aaBb plant contain approximately two thirds and one third of aaBb and aaBB seeds, respectively. For profile data, an average value for dry seeds from all lines is shown as these were very similar. Values are averages ± sd for five separate determinations, expressed on a per seed(ling) basis. Triple asterisks (***) indicate P < 0.01 (Student's t test) when compared to data from aaBb and wild type (wt).
In addition, total content of fatty acids was measured for wild type, hal3a-1, and hal3b in 5 d after imbibition (DAI) seedlings that were grown in medium either supplemented with or lacking Suc. Total content of fatty acids for aaBb and Aabb genotypes in medium supplemented with Suc could not be determined because their seed progeny consists of a segregating population. However, we could select (and measure total fatty acid content in) aaBb seedlings in medium lacking Suc by the stunted phenotype described above. In germinated seedlings of this line, some lipid catabolism took place. However, although total lipid levels decreased in 5-DAI aaBb seedlings germinated in the absence of Suc relative to dry seeds (Fig. 5A), the fatty acid profile of these seedlings indicated incomplete storage lipid catabolism (Fig. 5B). Notably, the level of the storage triacylglycerol-specific fatty acid, eicosenoic acid (20:1n9), decreased only approximately 50% in aaBb seedlings, whereas a 95% reduction was observed in the other lines (Fig. 5B). The accumulation of 20:1n9 in aaBb suggests that storage triacylglycerol-derived fatty acid catabolism was stalled, in contrast to the sharp decay observed in wild type and the other genetic backgrounds (Fig. 5B, mol percentage). Additionally, the photosynthetic membrane-specific hexadecatrienoic acid (16:3n3) did not increase, as would be expected in established seedlings (Fig. 5B). Therefore, this result reflects that fatty acids are not properly mobilized from the lipid body to reach the chloroplast in aaBb mutant. In contrast, the other lines all had fatty acid profiles that indicated their storage lipids were almost completely catabolized by 5 DAI, with the balance of lipids made up of membrane-specific fatty acids expected in actively growing tissue.
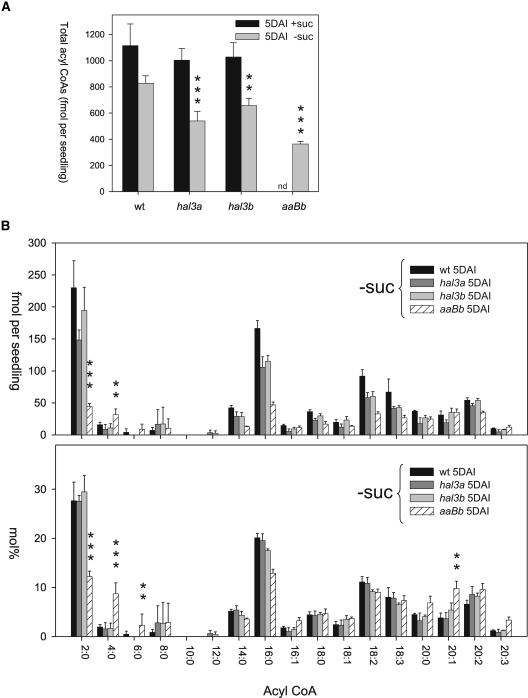
Measurement of the total acyl-CoA pool was obtained as described by Larson and Graham (2001). Total acyl-CoA content was quite similar in wild type, hal3a-1, and hal3b seedlings grown in medium supplemented with Suc (Fig. 6A). In medium lacking Suc, total acyl-CoA content was reduced in hal3a-1, hal3b, and aaBb seedlings to 65%, 79%, and 44% of wild-type levels (540, 657, 364, and 828 fmol/seedling, respectively). The reduction in total acyl-CoA content reported for hal3a-1 and hal3b did not lead to a visible phenotype, whereas the more than 50% reduction measured in aaBb led to a severe phenotype for seedling establishment (see Fig. 3, A and B). Particularly noticeable in this line was the low level of acetyl-CoA (2:0), approximately 20% of wild type (Fig. 6B, fmol/seedling), and the accumulation of 4:0, 6:0, and 20:1 CoA with respect to wild type (Fig. 6B, mol percentage). A low CoA supply in aaBb appears to limit β-oxidation and hence acetyl-CoA production as, for instance, the thiolysis step requires the input of a new CoA molecule for every 2-carbon cleavage.
Figure 6.
Altered acyl-CoA profile in aaBb seedlings. Seedlings grown on medium in the presence or absence of 1% Suc were extracted at 5 DAI for acyl-CoA profiling. Total fatty acids (A) and profile data (B) are shown. Data in B are expressed both as fmol/seedling as well as mol percentage (fmol acyl-CoA/total fmol acyl-CoAs). Data in B are from seedlings grown in the absence of Suc (−Suc). Values are averages ± sd for five separate determinations, expressed on a per seedling basis. Double (**) and triple (***) asterisks indicate P < 0.03 or P < 0.01 (Student's t test), respectively, when compared data from aaBb and wild type (wt).
DISCUSSION
Comparative genomics in both prokaryotic and eukaryotic organisms has been fruitful in the discovery of genes of universal metabolic pathways. In particular, elucidation of the human and plant genes involved in the CoA biosynthetic pathway has greatly benefited from this approach (Daugherty et al., 2002; Kupke et al., 2003). A step forward in plant physiology should be the analysis of reduction-of-function mutants impaired in CoA biosynthesis. In this work we provide an initial effort in that direction, by reporting the identification and characterization of plant knockout mutants impaired in CoA biosynthesis.
Two allelic Arabidopsis mutants with a lesion in the HAL3A (AtCoaC1, PPCDC) gene, hal3a-1 and hal3a-2, did not reveal major phenotypical differences compared to wild-type plants. The Arabidopsis genome encodes a second gene, HAL3B (AtCoaC2), whose gene product shows 86% amino acid sequence identity to that of HAL3A. Therefore, we suspected the corresponding gene products might be able to complement each other. A reverse genetics approach was used to isolate a knockout mutant for HAL3B. As it happened with hal3a-1, the hal3b mutant behaved quite similarly to wild-type plants. Analysis of the progeny of hal3a-1/+ hal3b/+ plants failed to identify a hal3a-1 hal3b double mutant, however we could identify aaBb and Aabb individuals. The percentage of aborted embryos in siliques of these plants was in agreement with the expected nonviability of hal3a-1 hal3b double mutants. Moreover, the results of the χ2 test in the progeny of aaBb plants was in agreement with the 2:1 ratio (HAL3B heterozygous to homozygous) expected if aabb embryos were lethal. The aabb embryos were arrested to the early/midglobular stage (Fig. 2B), which represents an early step of embryogenesis (36–48 h after flowering). It can be speculated that once the residual CoA present in the aabb zygote is titrated below a certain threshold by early cell divisions, further development is arrested. Additionally, this result shows that embryogenesis arrest in aabb occurs before the synthesis of fatty acids and lipid deposition take place (Baud et al., 2002), revealing a crucial role for CoA at early stages of embryo development. Moreover, de novo CoA biosynthesis by the embryo is required for embryogenesis, as either aaBb or Aabb mother plants do not support growth of aabb embryos. A maternal effect on the seed by CoA supply is unlikely, as this compound as well as CoA precursors are phosphorylated and therefore do not efficiently cross plasma membrane. Pantothenate is the most advanced CoA precursor taken up by cells (Begley et al., 2001). Indeed, whereas exogenous pantothenate complements mutants lacking de novo pantothenate biosynthesis, exogenous CoA does not complement mutants impaired in CoA biosynthesis (Begley et al., 2001; Leonardi et al., 2005).
The progeny of aaBb mother plants showed a segregating phenotype with respect to seedling establishment in medium lacking Suc, which was not present in the progeny of Aabb plants. Thus, root elongation, expansion, and greening of the cotyledons, as well as production of true leaves from the apical meristem, were severely impaired in hal3a-1 individuals that were heterozygous for HAL3B (Fig. 3B). An additional wild-type copy of HAL3B in aaBB siblings of the progeny restored normal growth (Fig. 3B), which provides genetic complementation of the phenotype and proves that the phenotype is due to impaired HAL3 function. Additionally, chemical complementation of the phenotype was obtained by supplementation of the media with pantethine (Fig. 3C), which is a direct precursor of the CoA metabolite synthesized by HAL3A. Taken together, these results indicate that impaired HAL3 function is responsible for the Suc-dependent seedling establishment phenotype and that adequate CoA biosynthesis is required for seedling establishment. Additionally, these results provide in vivo evidence on HAL3B function, and they confirm that HAL3A is predominant over HAL3B at this stage.
The Suc-dependent phenotype during postgerminative growth of aaBb is similar to that of various mutants affected in fatty acid breakdown and subsequent utilization of the resulting acetyl-CoA units. These include the pxa1/ped3/cts mutant, which shows a lesion in a peroxisomal ATP-binding cassette transporter involved in uptake of fatty acids into the peroxisome (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002) and a long-chain acyl-CoA synthetase (LACS) lacs6-1 lacs7-1 double mutant affected in peroxisomal LACS responsible for activation of fatty acids to fatty acyl-CoAs, i.e. the substrates for β-oxidation (Fulda et al., 2004). Disruption of β-oxidation itself also results in a Suc-dependent seedling establishment phenotype as demonstrated by the acx1 acx2 double mutant disrupted in the first step of the pathway (Adham et al., 2005; Pinfield-Wells et al., 2005) and the ped1/kat2 mutant, which is deficient in the last thiolytic cleavage step (Hayashi et al., 1998; Germain et al., 2001). Finally, mutations in different enzymatic steps of the peroxisomal glyoxylate cycle, which plays a central role in the conversion of acetyl units derived from β-oxidation to sugars, also gives rise to sugar-dependent seedling establishment phenotypes of varying degrees of severity (Eastmond et al., 2000; Eastmond and Graham, 2001; Cornah et al., 2004). The icl and mls mutants are devoid of the corresponding isocitrate lyase and malate synthase glyoxylate cycle enzymes, but despite this, they exhibit a relatively weak sugar-dependent seedling establishment phenotype best characterized by impaired hypocotyl elongation in dark-grown seedlings (Eastmond et al., 2000; Cornah et al., 2004). These weak phenotypes are put down due to the fact that in the absence of isocitrate lyase activity, acetyl units from β-oxidation can still be respired (Eastmond et al., 2000), and in the absence of malate synthase, acetyl-CoA can still be used by the seedling for gluconeogenesis, because the glyoxylate from isocitrate lyase can be metabolized by an alternative pathway (Cornah et al., 2004). In contrast to icl and mls mutant seedlings, the csy2 csy3 double mutant, which lacks the major peroxisomal citrate synthase isoforms involved in storage reserve mobilization, remains dormant until the seed coat is removed and is then dependent on exogenous sugar for seedling establishment (Pracharoenwattana et al., 2005). This increased dormancy phenotype is also exhibited by the cts, kat2, and acx1 acx2 mutants (Pinfield-Wells et al., 2005; Pracharoenwattana et al., 2005). The severity of the csy2 csy3 double-mutant phenotype is thought to be due to the fact that export of acetyl units from the peroxisome in Arabidopsis is absolutely dependent on their conversion to citrate.
The severity of the hal3 aaBb sugar-dependent seedling establishment phenotype (Fig. 3) is much closer to those mutants that are completely blocked in fatty acid breakdown (due either to disruption of a component of the pathway or export of product from the peroxisome) than to the mls and icl mutants, which are still able to utilize acetyl-CoA derived from fatty acid breakdown. Malate synthase and citrate synthase both use acetyl-CoA derived from the last step of β-oxidation and recycle CoA, thus maintaining the peroxisomal pool. This pool of CoA will almost certainly need to increase to meet demand with the onset of storage lipid mobilization during postgerminative seedling growth. The hal3 aaBb mutant appears to be unable to respond to this increased demand resulting in an inability to break down and utilize storage lipid derived fatty acids, which leads to a sugar-dependent seedling establishment phenotype.
The Suc rescue of seedling establishment in the hal3 aaBb mutant demonstrates that these seedlings are able to utilize sugars as a respiratory carbon source. In the presence of Suc the mitochondrial CoA pool must therefore be adequate to support the production of acetyl-CoA required for the TCA cycle. It is possible that exogenous sugar feeding, as well as providing a utilizable carbon source, could also alleviate the limitation in peroxisomal CoA brought about through increased demand, since sugars are known to delay and in some cases inhibit breakdown of storage lipid derived fatty acids (Eastmond et al., 2000; Martin et al., 2002). Furthermore, sugars such as Suc and Glc can also have regulatory roles, acting in many cases to either increase or decrease the activity of various metabolic pathways at the transcriptional and posttranscriptional level (Smeekens, 2000). It is possible that Suc could lead to a sufficient up-regulation of the CoA biosynthetic pathway to alleviate the seedling establishment phenotype of the hal3 aaBb mutant. In fact Suc feeding actually increases the total acyl-CoA pool in both wild type and the hal3a-1 and hal3b mutants, which suggests that increased levels of CoA are available in the presence of Suc (Fig. 6).
Increases in the total acyl-CoA pool are also seen in the various mutants disrupted in fatty acid breakdown (Germain et al., 2001; Footitt et al., 2002; Rylott et al., 2003; Fulda et al., 2004; Pinfield-Wells et al., 2005). In contrast, the aaBb seedling total acyl-CoA pool measured in this study was lower than that measured for the wild type on a per seedling basis. This suggests that the observed retention of storage lipid specific fatty acids in this line could at least partly be due to a decreased CoA supply to cytosolic and peroxisomal acyl-CoA synthetases, which are required to activate fatty acids to acyl-CoA esters for subsequent β-oxidation. Alternately, a bottleneck in peroxisomal β-oxidation arising as a consequence of a limiting supply of CoA provision for the last thiolytic step in the pathway could result in feedback inhibition of storage lipid breakdown as previously proposed (Graham et al., 2002). Indeed, the accumulation of C4, C6, and C20:x acyl-CoAs (on a mole percentage) in the aaBb line suggests a bottleneck operated over the entire range of acyl-CoA chain lengths that are generated during the cyclic 2C cleavage of long-chain fatty acids. That C16:0 and C18:x CoAs do not accumulate despite the fact that these are the predominant fatty acids in storage lipids most likely reflects an impairment in the acylation of fatty acids (because of a reduced supply of CoA) together with compromised de novo fatty acid synthesis.
In conclusion, the data presented in this work provide strong evidence that HAL3B plays the same catalytic role as HAL3A in CoA biosynthesis. Despite the fact that HAL3B is expressed at significantly lower levels than HAL3A (Espinosa-Ruiz et al., 1999; Fig. 1C), it can compensate for the lack of HAL3A during embryo development and the vegetative growth of the plant. HAL3B mRNA is detected (at lower level than HAL3A mRNA) in root, flower, and silique, whereas it is hardly detectable in shoot, leaf, and seed (Espinosa-Ruiz et al., 1999). Therefore, at specific stages (reproductive growth, salt stress, and seedling establishment), presumably due to increased demand for CoA, two rather than one copy of the HAL3B gene are required to compensate for the lack of HAL3A. Thus, aaBB seedlings, still containing 65% of wild-type levels of total acyl-CoAs, did not show a Suc-dependent phenotype for seedling establishment. Instead, further reduction (up to 44%) in aaBb seedlings correlated with a dependence on exogenous Suc for seedling establishment. As free CoA is only 10% to 15% of total CoAs in tissues with active fatty acid metabolism (Jackowski and Rock, 1986; Tahiliani and Beinlich, 1991), the reduced content of total acyl-CoAs likely reflects impaired CoA synthesis in the aaBb mutant. In addition, we clearly show that acetyl-CoA levels are compromised in the aaBb line with respect to hal3a-1, hal3b, or wild-type plants (approximately 80% reduction with respect to wild type), and that this correlates with compromised fatty acid breakdown. The fact that Suc rescues seedling establishment in the aaBb line demonstrates that the main cause of this phenotype is a compromised reserve mobilization, rather than other pleiotrophic effects of CoA deficiency. It will be interesting to establish whether the salt hypersensitivity observed in aaBb seedlings (Fig. 4C) correlates with similar metabolic defects or whether it is due to other roles of CoA. However, the low fresh weight of these seedlings and their growth arrest in the absence of Suc also suggest that the retention of storage lipid may have been a consequence of detrimental pleiotrophic effects of CoA deficiency (i.e. impaired amino acid biosynthesis) on cell development and expansion.
Finally, the Suc-dependent phenotype of aaBb seedlings was complemented upon media supplementation with pantethine. Reduction and phosphorylation of this compound by the cell metabolism must have occurred to generate 4′-phosphopantetheine, which is the CoA precursor generated by HAL3A enzyme. This result suggests that it might be possible to therapeutically deliver pantethine or an alternative intermediary compound to bypass certain enzymatic defects in CoA biosynthesis. In the case of PK-associated neurodegeneration, the only human illness currently associated to a defect in CoA biosynthesis (Zhou et al., 2001), pantethine supplementation might not be effective, as presumably PK activity is required to generate 4′-phosphopantetheine from pantethine. However, for a different illness associated to a defect in other steps of the CoA pathway (CoaB or CoaC), treatment with pantethine might prove to be useful. In general, this result serves to illustrate that a defect in the CoA biosynthesis pathway might be complemented by nonphosphorylated downstream intermediates, provided a functional PK is present.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were routinely grown under greenhouse conditions in pots containing a 1:3 vermiculite-soil mixture. For in vitro culture, seeds were surface sterilized by treatment with 70% ethanol for 20 min, followed by commercial bleach (2.5% sodium hypochlorite) containing 0.05% Triton X-100 for 10 min, and, finally, four washes with sterile distilled water. Stratification of the seeds was conducted in the dark at 4°C for 4 d. Then (0 DAI), seeds were sowed on Murashige and Skoog plates (Murashige and Skoog, 1962) composed of Murashige and Skoog basal salts, 0.1% 2-[N-morpholino]ethanesulfonic acid, 1% agar, and pH adjusted to 5.7 with potassium hydroxide before autoclaving. When stated, 1% Suc was included in the media. Plates were sealed and incubated in a controlled environment growth chamber at 22°C under a 16-h light, 8-h dark photoperiod at 80 to 100 μE m−2 s−1.
Mutant Isolation by PCR Screening
An Arabidopsis insertion mutant collection of 90,000 lines (Columbia background) that carry the T-DNA of vector pPCV6NFHyg was screened as described by Rios et al. (2002). The PCR screen was performed with the T-DNA left border primers HOOK1 and FISH1 (Rios et al., 2002) and the following HAL3A primers: 5′-CCAACGGTTTTAGCAGGTCGACTCTTAC and 5′-CAGAGTGGAGCTAGTAGTGCAAATGGTC. Finally, two plants corresponding to donor stock numbers 13332 and 32247 were found to contain independent single T-DNA insertions in the HAL3A gene, and they were, respectively, named hal3a-1 and hal3a-2. To identify individuals homozygous for the T-DNA insertion, genomic DNA was obtained from hygromicin-resistant seedlings and submitted to PCR genotyping using the above-described primers.
A line (Columbia background) containing a single T-DNA insertion in HAL3B was identified from the SALK T-DNA collection (http://signal.salk.edu/cgi-bin/tdnaexpress), corresponding to donor stock number SALK_045607. To identify individuals homozygous for the T-DNA insertion, genomic DNA was obtained from kanamycin-resistant (25 μg/mL) seedlings and submitted to PCR genotyping using the following HAL3B primers: 5′-TGTGACTGGGTCATAGTCTTACTGAACAC and 5′-TACTCGAGTCGTTGTGCCACATAAAACC. As T-DNA left border primer of the pROK2 vector, we used the following one: 5′-GCCGATTTCGGAACCACCATC.
The T-DNA tagged mutant lines were backcrossed once with the wild type, verified by DNA gel-blot hybridization and also by segregation analysis of the encoded antibiotic resistance gene (for the SALK line, the partially silenced selectable marker was scored under a 25 μg/mL kanamycin concentration).
Cytological Techniques
Green and mature siliques were fixed in an ethanol/acetic acid mixture and then cleared using the following solution: chloral hydrate/glycerol/water 8:1:2 (w/v/v) according to the protocol of Weigel and Glazebrook (2002). Seeds, usually cleared for 12 to 16 h, were examined with an Eclipse E600 microscope (Nikon) equipped with Nomarski optics.
Seedling Establishment, Complementation, and Salt Tolerance Assays
Seedling establishment of the different genetic backgrounds was scored as the percentage of seeds that developed green expanded cotyledons and the first pair of true leaves. Complementation of the Suc-dependent phenotype of aaBb individuals was assayed supplementing the medium with 50 μg/mL of bis[N-pantothenylamidoethyl] disulfide (pantethine, Sigma P2125). Salt tolerance assays were performed by transferring 7-d-old seedlings grown in medium containing Suc to a medium supplemented with 125 mm NaCl and lacking or containing 1% Suc. Previous assays for testing salt resistance of 35S:AtHAL3A transgenic lines were done by testing plant growth in Murashige and Skoog medium supplemented with 3% Suc and 100 mm NaCl (Espinosa-Ruiz et al., 1999). In tobacco (Nicotiana tabacum), salt resistance assays were performed in calli derived of Bright Yellow 2 cells that were transformed with a 35S:NtHAL3A construct (Yonamine et al., 2004).
RNA Analyses
Seedlings were collected and frozen in liquid nitrogen. Total RNA was extracted using a Qiagen RNeasy plant mini kit, separated on formaldehyde-agarose gels, and blotted to a nylon membrane. Blots were hybridized with random-priming 32P-labeled probes. A specific cDNA probe for HAL3A was prepared as described previously (Espinosa-Ruiz et al., 1999). RNA samples for RT-PCR analysis were treated with DNase (RNase free) and, after precipitation in ethanol, they were dissolved to a final concentration of 1 μg/μL. One microgram of the RNA solution obtained was reverse transcribed using 0.1 μg oligo(dT)15 primer and Moloney meurine leukemia virus reverse transcriptase (Roche), to finally obtain a 20-μL cDNA solution. PCR reactions were performed on 1-μL cDNA template using the following primers: primers HAL3A, 5′-ATG GAG AAT GGG AAA AGA GAC and 5′-AAG ATT ATC ACA AAG CCC ACC; primers HAL3B, 5′-GATTCAGATAGAGAAGAAGATGA and 5′-CATGCTCTTACTATACATGTC; primers TUB, 5′-CCTGATAACTTCGTCTTTGG and 5′-GTGAACTCCATCTCGTCCAT.
Fatty Acid and Acyl-CoA Profiling
Lipids from 50 dry seeds were extracted and transmethylated to their fatty acid methyl esters (FAMEs) together with tripentadecanoin as an internal standard using a one-step procedure (Browse et al., 1986). FAMEs were dissolved in hexane, and 2-μL aliquots injected for gas chromatography-flame ionization detector analysis using a BPX70 60 m × 0.25 mm i.d. × 0.25 μm film thickness capillary column (SGE) and a CE instruments GC8000 Top GC (Thermoquest). Injection was made into a hydrogen carrier gas stream at 1.3 mL min−1 (average linear velocity 35 cm s−1) at a 30:1 split ratio. Temperature was ramped as follows: 110°C isothermal 1 min; 7.5°C min−1 to 260°C; cool down 70°C min−1 to 110°C; total analysis time 23 min. FAMEs were identified by comparison to a 37 FAME mix (Supelco). Ten seedlings (approximately 10 mg) were extracted for quantitative acyl-CoA analysis by HPLC with fluorescence detection of acyl etheno CoA derivatives (Larson and Graham, 2001). The lipid portion of the acyl-CoA extracts were used for fatty acid determinations as described above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF166262 and U80192.
Acknowledgments
We thank the Max-Planck Institute (Cologne, Germany) for providing access to its Arabidopsis T-DNA collection, Joseph Ecker and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, and Arabidopsis Biological Resource Center (Ohio State University, Columbus)/Nottingham Arabidopsis Stock Centre for distributing these seeds. We thank Stuart Graham (University of York, UK) for extensive technical assistance in the acyl-CoA analyses.
This work was supported by the Ministerio de Educación y Ciencia (grant nos. BIO2002–03090 and BIO2005–01760, and fellowship to M.G.-G.), Fondo Europeo de Desarrollo Regional, and Consejo Superior de Investigaciones Científicas (fellowship to S.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Pedro L. Rodriguez (prodriguez@ibmcp.upv.es).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.072066.
References
- Adham AR, Zolman BK, Millius A, Bartel B (2005) Mutations in Arabidopsis acyl CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. Plant J 41: 859–874 [DOI] [PubMed] [Google Scholar]
- Afshar K, Gonczy P, DiNardo S, Wasserman SA (2001) fumble encodes a pantothenate kinase homolog required for proper mitosis and meiosis in Drosophila melanogaster. Genetics 157: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert A, Martinez-Ripoll M, Espinosa-Ruiz A, Yenush L, Culianez-Macia FA, Serrano R (2000) The x-ray structure of the FMN-binding protein AtHal3 provides the structural basis for the activity of a regulatory subunit involved in signal transduction. Struct Fold Des 8: 961–969 [DOI] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Begley TP, Kinsland C, Strauss E (2001) The biosynthesis of coenzyme A in bacteria. Vitam Horm 61: 157–171 [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Cornah JE, Germain V, Ward JL, Beale MH, Smith SM (2004) Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J Biol Chem 279: 42916–42923 [DOI] [PubMed] [Google Scholar]
- Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, Crecy-Lagard V, Osterman A (2002) Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J Biol Chem 277: 21431–21439 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA (2000) Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA 97: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Graham IA (2001) Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci 6: 72–78 [DOI] [PubMed] [Google Scholar]
- Espinosa-Ruiz A, Belles JM, Serrano R, Culianez-Macia FA (1999) Arabidopsis thaliana AtHAL3: a flavoprotein related to salt and osmotic tolerance and plant growth. Plant J 20: 529–539 [DOI] [PubMed] [Google Scholar]
- Falk KL, Guerra DJ (1993) Coenzyme A biosynthesis in plants: partial purification and characterization of pantothenate kinase from spinach. Arch Biochem Biophys 301: 424–430 [DOI] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M, Schnurr J, Abbadi A, Heinz E, Browse J (2004) Peroxisomal acyl CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde JP, Bryce JH, Graham IA, Smith SM (2001) Requirement for 3-ketoacyl CoA thiolase-2 in peroxisome development, fatty acid beta-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28: 1–12 [DOI] [PubMed] [Google Scholar]
- Graham IA, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41: 156–181 [DOI] [PubMed] [Google Scholar]
- Graham IA, Li Y, Larson TR (2002) Acyl CoA measurements in plants suggest a role in regulating various cellular processes. Biochem Soc Trans 30: 1095–1099 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M (2002) Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid beta-oxidation. Plant Cell Physiol 43: 1–11 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Acosta P, Schmid DG, Jung G, Culianez-Macia FA, Kupke T (2002) Molecular characterization of the Arabidopsis thaliana flavoprotein AtHAL3a reveals the general reaction mechanism of 4′-phosphopantothenoylcysteine decarboxylases. J Biol Chem 277: 20490–20498 [DOI] [PubMed] [Google Scholar]
- Jackowski S, Rock CO (1986) Consequences of reduced intracellular coenzyme A content in Escherichia coli. J Bacteriol 166: 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupke T (2002) Molecular characterization of the 4′-phosphopantothenoylcysteine synthetase domain of bacterial dfp flavoproteins. J Biol Chem 277: 36137–36145 [DOI] [PubMed] [Google Scholar]
- Kupke T, Hernandez-Acosta P, Culianez-Macia FA (2003) 4′-Phosphopantetheine and coenzyme A biosynthesis in plants. J Biol Chem 278: 38229–38237 [DOI] [PubMed] [Google Scholar]
- Kupke T, Hernandez-Acosta P, Steinbacher S, Culianez-Macia FA (2001) Arabidopsis thaliana flavoprotein AtHAL3a catalyzes the decarboxylation of 4′-phosphopantothenoylcysteine to 4′-phosphopantetheine, a key step in coenzyme A biosynthesis. J Biol Chem 276: 19190–19196 [DOI] [PubMed] [Google Scholar]
- Kupke T, Uebele M, Schmid D, Jung G, Blaesse M, Steinbacher S (2000) Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J Biol Chem 275: 31838–31846 [DOI] [PubMed] [Google Scholar]
- Larson TR, Graham IA (2001) Technical advance: a novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J 25: 115–125 [DOI] [PubMed] [Google Scholar]
- Leonardi R, Zhang YM, Rock CO, Jackowski S (2005) Coenzyme A: back in action. Prog Lipid Res 44: 125–153 [DOI] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Neuburger M, Day DA, Douce R (1984) Transport of coenzyme A in plant mitochondria. Arch Biochem Biophys 229: 253–258 [DOI] [PubMed] [Google Scholar]
- Ottenhof HH, Ashurst JL, Whitney HM, Saldanha SA, Schmitzberger F, Gweon HS, Blundell TL, Abell C, Smith AG (2004) Organisation of the pantothenate (vitamin B5) biosynthesis pathway in higher plants. Plant J 37: 61–72 [DOI] [PubMed] [Google Scholar]
- Pinfield-Wells H, Rylott EL, Gilday AD, Graham S, Job K, Larson TR, Graham IA (2005) Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J 43: 861–872 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17: 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios G, Lossow A, Hertel B, Breuer F, Schaefer S, Broich M, Kleinow T, Jasik J, Winter J, Ferrando A, et al (2002) Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J 32: 243–253 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA (2003) Arabidopsis mutants in short- and medium-chain acyl CoA oxidase activities accumulate acyl CoAs and reveal that fatty acid beta-oxidation is essential for embryo development. J Biol Chem 278: 21370–21377 [DOI] [PubMed] [Google Scholar]
- Shibata K, Gross CJ, Henderson LM (1983) Hydrolysis and absorption of pantothenate and its coenzymes in the rat small intestine. J Nutr 113: 2207–2215 [DOI] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Steinbacher S, Hernandez-Acosta P, Bieseler B, Blaesse M, Huber R, Culianez-Macia FA, Kupke T (2003) Crystal structure of the plant PPC decarboxylase AtHAL3a complexed with an ene-thiol reaction intermediate. J Mol Biol 327: 193–202 [DOI] [PubMed] [Google Scholar]
- Strauss E, Kinsland C, Ge Y, McLafferty FW, Begley TP (2001) Phosphopantothenoylcysteine synthetase from Escherichia coli: identification and characterization of the last unidentified coenzyme A biosynthetic enzyme in bacteria. J Biol Chem 276: 13513–13516 [DOI] [PubMed] [Google Scholar]
- Tahiliani AG, Beinlich CJ (1991) Pantothenic acid in health and disease. Vitam Horm 46: 165–228 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J, editors (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 104–105
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Yonamine I, Yoshida K, Kido K, Nakagawa A, Nakayama H, Shinmyo A (2004) Overexpression of NtHAL3 genes confers increased levels of proline biosynthesis and the enhancement of salt tolerance in cultured tobacco cells. J Exp Bot 55: 387–395 [DOI] [PubMed] [Google Scholar]
- Zhou B, Westaway SK, Levinson B, Johnson MA, Gitschier J, Hayflick SJ (2001) A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat Genet 28: 345–349 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]