Abstract
Genome-wide identification of RNAs associated with RNA-binding proteins is crucial for deciphering posttranscriptional regulatory systems. PUMILIO is a member of the evolutionary conserved Puf-family of RNA-binding proteins that repress gene expression posttranscriptionally. We generated transgenic flies expressing affinity-tagged PUMILIO under the control of an ovary-specific promoter, and we purified PUMILIO from whole adult flies and embryos and analyzed associated mRNAs by using DNA microarrays. Distinct sets comprising hundreds of mRNAs were associated with PUMILIO at the two developmental stages. Many of these mRNAs encode functionally related proteins, supporting a model for coordinated regulation of posttranscriptional modules by specific RNA-binding proteins. We identified a characteristic sequence motif in the 3′-untranslated regions of mRNAs associated with PUMILIO, and the sufficiency of this motif for interaction with PUMILIO was confirmed by RNA pull-down experiments with biotinylated synthetic RNAs. The RNA motif strikingly resembles the one previously identified for Puf3p, one of five Saccharomyces cerevisiae Puf proteins; however, proteins encoded by the associated mRNAs in yeast and Drosophila do not appear to be related. The results suggest extensive posttranscriptional regulation by PUMILIO and uncover evolutionary features of this conserved family of RNA-binding proteins.
Keywords: DNA microarray, posttranscriptional gene regulation, RNA-binding protein
Posttranscriptional regulation of gene expression plays important roles in diverse cellular processes. This regulation is often mediated by specific RNA-binding proteins (RBPs) that bind to elements in the UTRs of mRNAs and regulate the stability, translation, or localization of the mRNA (1–3). Whereas many classical studies explored the cellular role of RBPs with specific mRNA substrates, the recent development of genome-wide analysis tools enables systematic identification of mRNA substrates of RBPs and the study of posttranscriptional gene regulation on a global scale (4).
We recently used DNA microarrays to systematically identify RNAs associated with each of the five Pumilio-Fem 3-binding factor (Puf) proteins from Saccharomyces cerevisiae (5). Each Puf protein bound to a large set of mRNAs encoding functionally and cytotopically related proteins, and characteristic sequence elements in the 3′ UTRs of the target mRNAs were identified (5). These and other results suggest that there is extensive coordinate regulation of RNAs by RNA-binding proteins (4, 6, 7).
Puf proteins are an evolutionary conserved family of RBPs that are implicated in diverse physiological processes in eukaryotes (8, 9). They are defined by the presence of a structurally conserved RNA-binding domain, termed the Pum-homology domain (PumHD), composed of eight repeats of 36 aa (10–12). Puf proteins have been reported to bind to 3′ UTR sequences encompassing a “UGUR” tetranucleotide motif in the RNA, and in concert with other proteins repress gene expression by affecting mRNA translation or stability (8).
PUMILIO (Pum), the founding Puf-family member and sole member in the fruit fly Drosophila melanogaster, is implicated in diverse developmental processes. Nevertheless, only a few mRNA substrates are known to date. Pum was first identified as a maternal-effect gene required for posterior patterning in the embryo (13). Pum, Nanos (Nos), and Brain tumor (Brat) repress translation of maternally derived hunchback (hb) mRNA in the posterior of the embryo. This regulation depends on two bipartite nanos-response elements (NREs), each composed of box A (GUUGU) and box B (AUUGUA) sequences, located in the 3′ UTR of hb mRNA (11, 14, 15). The current model is that Pum binds to sequences in the NRE and recruits Nos and Brat, forming a quaternary RNA–protein complex that causes deadenylation of hb mRNA and translational repression (15). This inhibition of expression of the transcription factor Hb in the posterior of the embryo is essential for formation of the abdomen. Pum mutants also show abnormal temporal expression of bicoid (bcd), the main determinant in the anterior patterning system (16).
Pum and Nos have also been implicated in the regulation of germ cell development and maintenance. During embryogenesis, Pum and Nos accumulate in pole cells, the germ-line progenitor cells. They are required for proper pole-cell migration to the gonad and are thought to coordinate cell division in migrating pole cells by repressing translation of maternally derived cyclin B (cycB) mRNA (17). During oogenesis, Pum regulates asymmetric cell division of germ-line stem cells (18–20).
Several recent studies have implicated Pum in neuronal development and function. In a genetic screen, several pum alleles were found to affect long-term memory formation in adult flies (21). Pum apparently effects morphogenesis of larval peripheral sensory neurons into dendritic cells (22) and negatively regulates expression of the cap-binding protein eIF4E at the larval neuromuscular junction (23). Notably, the putative regulatory sequences in the 3′ UTRs of eIF4E and cycB mRNAs lack box A motifs and hence only partially match NREs present in hb and bcd, suggesting alternative strategies for regulation of these mRNAs, perhaps involving combinatorial binding with other protein factors.
Using DNA microarrays to identify mRNAs associated with Pum in Drosophila adult ovaries and embryos, we found hundreds of mRNAs that likely represent mRNA targets subject to regulation by Pum. The sets of mRNAs associated with Pum differ in adults and embryos and code for subsets of functionally related proteins. Most of the adult mRNA targets contain a conserved RNA sequence motif in their 3′ UTRs; this motif is sufficient for interaction with Pum. These results suggest extensive posttranscriptional coordination by Pum in Drosophila, providing further evidence for a highly organized and multifaceted posttranscriptional regulatory system in multicellular organisms.
Results
Systematic Identification of RNAs Associated with PUMILIO.
To identify RNAs interacting with Pum in Drosophila, we generated transgenic flies expressing a tandem-affinity purification (TAP)-tagged fragment of Pum (24). A TAP-tagged full-length Pum construct could not be obtained because expression constructs were toxic to Escherichia coli. Therefore, we used the C-terminal part of Pum which includes the RNA-binding region (PumHD) and an additional 24 C-terminal amino acids that likely mediate interaction with NANOS and BRAT (12, 25). Moreover, this fragment has been shown to be sufficient for partial rescue of pum mutant phenotypes (26). TAP-PumHD was expressed in Drosophila by using the UAS-GAL4 system, which allows tissue-specific expression of transgenes (27, 28). We used maternal α-tubulin promoter-regulated GAL4 to drive TAP-PumHD expression in the female ovary. Maternally synthesized protein is loaded into the egg, as is endogenous Pum protein (29) (see Fig. 5, which is published as supporting information on the PNAS web site). Moreover, we estimated the intracellular concentration of TAP-PumHD in embryos and found it similar to that of endogenous Pum (≈40 nM) (30).
We identified RNAs associated with Pum expressed in ovaries of adult flies and from embryos by applying a method similar to one established in Saccharomyces cerevisiae that uses the TAP-tag to purify RNA-protein complexes (5, 31). We prepared cell-free extracts from adults and embryos and recovered tagged PumHD (≈5 nM in the extracts) by affinity selection on IgG beads and subsequent cleavage with TEV protease (Fig. 5). RNA was isolated from the eluate after TEV protease cleavage, and ≈250 ng of RNA per gram of transgenic adult flies and 1 μg of RNA per gram of embryos were obtained. To control for nonspecifically enriched RNAs, the same experiments were performed with wild-type flies and embryos lacking a tagged protein, yielding ≈40 ng of RNA per gram of adult flies, and 700 ng of RNA per gram of embryos. Total RNA from extracts and affinity-purified RNAs were then used to prepare cDNA probes labeled with different fluorophores, which were comparatively hybridized to a Drosophila DNA microarray. The ratio of hybridization signals from the two RNA populations at a given array element, representing a specific gene, reflects the enrichment of the respective RNA by the Pum affinity purification (5).
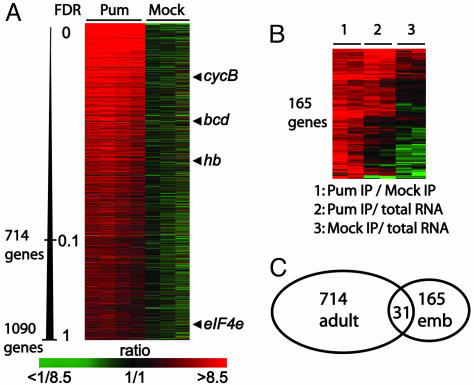
To generate a list of mRNAs that were consistently enriched by Pum affinity purification and, hence, likely targets of Pum, we compared association of transcripts from the TAP-PumHD affinity isolations to the mock isolates by two-class Significance Analysis of Microarrays (SAM) and determined false discovery rates (FDRs) for each mRNA (see Supporting Text, which is published as supporting information on the PNAS web site). In flies, 714 genes were consistently associated with Pum with a FDR of <0.1%, and 1,090 genes with a FDR of <1%. Among these 1,090 genes are the four previously identified Pum target mRNAs (hb, bcd, cycB, eIF4E) (Fig. 1A, for a complete list, see Table 2, which is published as supporting information on the PNAS web site). The data from embryos yielded no genes with a FDR of <0.1% and only 192 with a FDR of <1%. Although the smaller number of genes identified at these levels of specificity may indicate that fewer mRNAs associate with Pum in embryos, the high amounts of unspecifically bound RNAs evident in mock control isolates may have hampered our analysis and hence true mRNA targets may have been missed. Therefore, we directly compared total RNA obtained from TAP-Pum affinity isolations from embryos and mock isolates by comparative hybridization to microarrays. To define a list of potential Pum mRNA targets in embryos, we combined both types of analyses and selected 165 genes that had a FDR of <3.2% and that were at least 3-fold enriched in TAP-Pum affinity purifications compared to mock control experiments (Fig. 1B, for a complete list, see Table 3, which is published as supporting information on the PNAS web site).
Fig. 1.
Specific RNAs selectively associated with Pum in Drosophila adults and embryos. Rows represent genes (unique DNA elements) and columns represent individual experiments. The color code indicates the degree of enrichment (green–red ratio scale). (A) Relative enrichments of mRNA targets of Pum expressed in adult ovaries. Four experiments with affinity-tagged Pum proteins and three mock experiments are shown. Genes were ordered from top to bottom according to increasing FDRs determined by significance analysis of microarrays (SAM) analysis. Arrowheads depict enrichment of previously known Pum targets. (B) Pum RNA targets selected in embryos. A total of 174 transcripts representing 165 genes were clustered based on Euclidean distances with smd (50). (C) Diagram showing overlap of adult and embryo Pum associated mRNAs.
Transcripts for only 31 genes were shared between the 714 (4.3%) adult and the 165 (18.8%) embryo Pum targets. Thus, the vast majority were detected only in adults or in embryos, suggesting that Pum associates with distinct sets of mRNAs at the two developmental stages (Fig. 1C); this could be due to different patterns of expression of the target genes at the different developmental stages, to different Pum protein complexes that lead to altered RNA-binding specificities, or to different accessibility of the mRNA targets in different tissues. To estimate the bias from differential gene expression, we compared mRNA levels in adults and embryos using DNA microarrays, and then compared this to our lists of Pum mRNA targets in adults and embryos (Fig. 6, which is published as supporting information on the PNAS web site). Many mRNAs are present at both developmental stages, and the mRNAs selected with Pum from adults were not biased for genes preferentially expressed during this stage. In contrast, embryo-selected Pum targets were generally more highly expressed in embryos (Fig. 6). This trend could, at least in part, result from the higher RNA background in the embryo experiments.
Few mRNAs Have Altered Steady-State mRNA Levels in pum Mutant Flies.
We compared mRNA levels of pum13 mutant and wild-type adult female flies with DNA microarrays. Of the ≈12,500 transcripts for which data were obtained from two independent analyses, 48 (130) transcripts were at least 3-fold (2-fold) more abundant, and 59 (243) transcripts were at least 3-fold (2-fold) less abundant in pum mutant compared to wild-type flies. Notably, 13 of the 48 genes present at elevated levels in pum13 mutants (27%) encode proteins involved in the antibacterial/fungal immune response (P < 10−17); among them, five (AttB, AttC, AttD, CecC, CG13422) of the 20 mRNAs encoding antibacterial peptides (P < 10−8).
Expression levels of the “adult” Pum mRNA targets were slightly increased in Pum mutants (median, 1.1 fold; t test statistics, P < 10−17; see Fig. 7, which is published as supporting information on the PNAS web site). This finding suggests the possibility that Pum might promote accelerated degradation of mRNA targets as shown for the homologous yeast Puf3 protein regulating Cox17 mRNA stability (5, 32, 33).
Functionally Related Sets of mRNAs Are Associated with Pum.
To identify functional themes among the mRNAs associated with Pum, we searched for shared Gene Ontology (GO) annotations in the lists of Pum target mRNAs (Table 4, which is published as supporting information on the PNAS web site). The mRNAs associated with Pum expressed in adult ovaries could be grouped in two major classes. One contains mRNAs encoding nuclear proteins involved in nucleotide metabolism and transcriptional regulation. This set includes two of the previously known targets, hb and bcd mRNAs. CycB mRNA has also been shown to be regulated by Pum, and 7 of the 10 cyclin mRNAs (A, B, D, G, J, K, T) were in this set (P < 10−6; cyclins C, E, and H were analyzed but not enriched). The second group contains transcripts encoding proteins localized to organelle membranes. For example, it includes mRNAs encoding most of the subunits of the vacuolar proton-translocating V-type ATPase (Fig. 2A). This complex consists of two domains: Pum associated with mRNAs encoding six (Vha13, Vha26, Vha44, Vha55, Vha68–1/Vha68–2, VhaSFD) of the eight subunits comprising the catalytic V1 domain or the head group (P < 5 × 10−6) and four (Vha16, VhaAC39, VhaM9.7–1/7–2, VhaPPA1–1) of the five subunits of the V0 domain (P < 2 × 10−4), which forms the transmembrane channel (34).
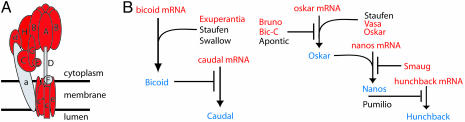
Fig. 2.
mRNAs associated with Pum encode proteins of specific macromolecular complexes and regulatory pathways. (A) Subunits of the vacuolar ATPase. Red, subunits whose mRNAs associated with Pum; gray, subunits whose mRNAs were not enriched with a FDR of <1%. Subunits of the V1 domain are labeled with capital letters (34): A, Vha68; B, Vha55; C, Vha44; D, Vha36; E, Vha26; F, Vha14; G, Vha13; H, VhaSFD. Subunits of the V0 domain are indicated by small letters: a, Vha100; c, Vha16; c″, PPA1; e, VhaM9.7; d, VhaAC39. (B) Components of anterior–posterior pattering systems associated with Pum in embryos and/or adults. mRNAs associated with Pum are shown in red and their protein products in blue. Proteins whose mRNAs were not found to be associated are in black.
In embryos, some PumHD-bound mRNAs encode proteins involved in germ cell development or anterior–posterior axis patterning. Genes required for germ cell development included four (nos, vas, orb, out) of the eight genes with GO annotations for germ-line cyst formation (P < 2 × 10−6). Eleven of 16 mRNAs (P < 10−8) that encode proteins acting in the cascade that mediate anterior-posterior axis patterning were associated with Pum in adults and embryos (Fig. 2B). These include bcd mRNA, which encodes the anterior morphogen, the mRNA of its transcriptional target gene hb, as well as oskar, nos, and caudal mRNAs, key components of the posterior patterning system. In addition, we found mRNAs of regulators of this process: bruno-2 and bicaudal-C, which repress oskar mRNA translation, smaug, which represses translation of nos mRNA, and vasa mRNA, which encodes an ATP-dependent RNA helicase that promotes oskar and nanos mRNA translation. The striking identification of mRNAs that encode many of the proteins required for posterior patterning suggests a general role for Pum in the coordination of this process, in addition to its previously identified role in translational regulation of hb.
Pum also associated with mRNAs encoding regulators of dorso–ventral axis formation, namely gastrulation defective (gd), spaetzle, and possibly easter. These genes are also linked to the antimicrobial immune response in Drosophila, which also involves the Toll signaling pathway. No array data were obtained for snake, the fourth component of a protease cascade leading to Toll activation (35).
A Common Sequence Motif in the 3′ UTR of Pum mRNA Targets.
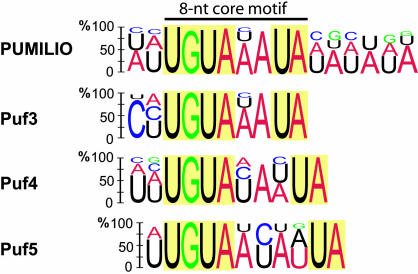
Characteristic sequence motifs have been found in the 3′ UTRs of the mRNA targets of diverse Puf family members (5, 8, 36, 37). Therefore, we examined the sets of mRNAs that associate with Pum for the presence of common motifs using multiple expectation maximization for motif elicitation (MEME) as a motif discovery tool (38). The 3′ UTR sequence information could be retrieved (FlyBase) for 113 of the 150 genes most highly enriched in Pum affinity isolates from adult flies, and MEME analysis revealed a 16-nt consensus sequence (Fig. 3; see also Table 5, which is published as supporting information on the PNAS web site). The consensus includes a highly conserved 8-nt core motif UGUA(A/U/C)AUA that contains the UGUA tetranucleotide found in previously identified mRNAs that interact with Puf proteins (5, 26, 30, 36, 37, 39). The core is flanked by less conserved AU-rich sequences that could determine the recruitment of other transacting factors or modulate binding affinities, as shown recently for Fbf-1, a Puf family member from Caenorhabditis elegans (36).
Fig. 3.
RNA consensus motif in 3′ UTR sequences associated with Pum and yeast Puf3, Puf4 and Puf5 proteins (5). Height of the letter indicates the probability of appearing at the position in the motif. Nucleotides with <10% appearance were omitted.
The UGUA tetranucleotide motif was found in 80% of the 3′ UTRs in our list of adult and embryo Pum mRNA targets, a significant enrichment relative to its occurrence (67%) among all of the available 3′ UTR sequences from D. melanogaster (Table 1). Enrichment of the 8-nt core motif UGUA(A/U/C)AUA was more striking. This motif was present in 54% of the adult (P < 10−106) and in 22% of the embryo (P < 0.02) Pum target 3′ UTRs, compared to its genome-wide occurrence in 3′ UTRs (15%) (Table 1; for a list of all Drosophila 3′ UTRs containing this motif, see Table 6, which is published as supporting information on the PNAS web site). The reason for the relatively rare occurrence of the 8-nt motif among the embryo Pum targets is not known. False positives from Pum affinity purifications, additional elements in the RNA, or other transacting proteins that bind RNA may have contributed to the measured RNA associations.
Table 1.
Sequence motifs enriched in Pum targets
| Consensus motif | Search option | 3′ UTRs available | 3′ UTRs with motif (%) | P value |
|---|---|---|---|---|
| UGUA | Genome | 9,777 | 6,207 (67) | |
| Adult (0.1% FDR) | 577 | 473 (82) | 10−26 | |
| Embryo | 134 | 104 (78) | 0.0003 | |
| UGUAHAUA | Genome | 9,777 | 1,431 (15) | |
| Adult (0.1% FDR) | 577 | 311 (54) | 10−106 | |
| Embryo | 134 | 29 (22) | 0.02 |
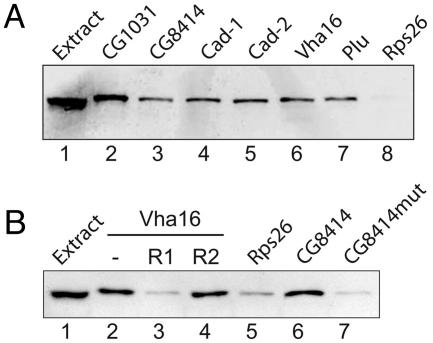
To test putative mRNA targets for interaction with Pum and to evaluate our predicted RNA recognition motif, we performed RNA pull-down experiments using synthetic biotinylated transcripts added to Drosophila extracts expressing TAP-PumHD. We made biotinylated 3′ UTR sequences of five Pum targets (Vha16, caudal, CG1031, CG8414, plutonium) and one negative control (Rps26) that was not found to be associated with Pum. All of the five potential target mRNAs bound Pum protein, whereas the Rps26 control 3′ UTR sequence did not (Fig. 4A). To further map sequences required for Pum-RNA interactions, we mutated the highly conserved UGU trinucleotide within the core consensus motif of the 3′ UTR of CG8414 to ACA. Assays with the mutated biotinylated RNA showed no specific interaction with Pum (Fig. 4B). Likewise, addition of a 10-nt competitor RNA comprising the consensus sequence prevented interaction of biotinylated Vha16 3′ UTR RNA with Pum, but no such competition was seen with a control RNA in which the conserved core UGU was mutated to ACA (Fig. 4B). The same results were obtained in assays performed with extracts prepared from embryos expressing TAP-PumHD (data not shown). Thus, RNA sequences encompassing the computationally inferred core motif are sufficient for association with PumHD in Drosophila extracts.
Fig. 4.
Validation of Pum mRNA targets. RNA–protein complexes formed between biotin-labeled 3′ UTRs and extracts of adult Drosophila expressing TAP-PumHD were purified on streptavidin magnetic beads and monitored for the presence of TAP-PumHD by immunoblot analysis with Peroxidase-Anti-Peroxidase Soluble Complex (Sigma). (A) Biotin-labeled 3′ UTR sequences for indicated genes (lanes 2 to 8) were incubated with Drosophila extract (input, lane 1). Rps26 3′ UTR was used as a negative control probe RNA (lane 8). (B) Validation of the core sequence motif identified in 3′ UTRs sequences of Pum target mRNAs. Lane 1, input (Drosophila extract); lanes 2–4, biotin-labeled RNA corresponding to the Vha16 3′ UTR was combined with Drosophila extract (lane 2) and 100-fold excess of competitor RNA (R1; AUUGUAAAUA; lane 3) or control RNA where the core motif is mutated (R2; AUACAAAAUA; lane 4). The conserved trinucleotide motif UGU in the 3′ UTR of CG8414 was left intact (lane 6) or was mutated to ACA (lane 7). Rps26 3′ UTR is the negative control probe RNA (lane 5).
Binding Sequence Motif, but Not Gene Function, Conserved Between Yeast and Drosophila Puf mRNA Targets.
The Pum core motif is strikingly similar to the previously determined RNA recognition sequence for Puf3 and less similar to sequences recognized by the Puf4 and Puf5 proteins from S. cerevisiae (Fig. 3) (5). This finding correlates with amino acid sequence conservation within the respective PumHD domains of the orthologous proteins: Puf3p is 46%, Puf4p and Puf5p are 30%, and Puf1 and Puf2 proteins are 20% identical to Drosophila Pum (9). Furthermore, the three amino acid residues in each of the eight PumHD repeats that make direct contact with one RNA base (12) are conserved between Pum and Puf3 but differ at three positions in the Puf4 and Puf5 protein sequences (for a multiple sequence alignment of PumHDs from Puf-family members and respective RNA recognition sequences see Fig. 8, which is published as supporting information on the PNAS web site). The differences in these PumHD residues may contribute to the altered RNA recognition specificities of Puf4 and Puf5 compared to Pum and Puf3. Further investigation will be required to define precise rules for RNA recognition by this protein family.
We wondered whether the apparent conservation of protein structure and RNA recognition sequences extended to the functional level, i.e., whether the set of proteins encoded by the target mRNAs of Puf proteins have been evolutionary conserved between Drosophila and S. cerevisiae. Twenty-eight percent of all Drosophila proteins match a homologous S. cerevisiae protein sequence (40). A similar fraction (29%) of proteins encoded by the Pum mRNA targets in embryos have a yeast homolog (Table 7, which is published as supporting information on the PNAS web site; for proteinblast results, see Tables 8 and 9, which are published as supporting information on the PNAS web site). However, a significantly greater fraction of proteins encoded by the adult Pum mRNA targets, 39% (P < 2 × 10−8), have a yeast homolog, which may indicate a role of Pum in regulating evolutionarily conserved cellular processes. We also determined the fraction of the yeast homologs to the Pum targets whose mRNAs associated with at least one of the five yeast Puf proteins. Only 15% were targets for a yeast Puf protein (5), suggesting that the Drosophila Pum targets are not particularly conserved in yeast.
These differences are further underscored by comparison of the targets of yeast Puf3 and Drosophila Pum, the most highly conserved family members. Yeast Puf3p binds almost exclusively to transcripts of nuclear genes that encode mitochondrial proteins (5). In contrast, only 49 (4%) of the Drosophila Pum mRNA targets encode mitochondrial proteins. Nevertheless, a large fraction of those targets (23 mRNAs) contain in their 3′ UTRs our computationally defined 8-nt Pum core motif; this motif is present in only 31 (8%) of the 400 Drosophila nuclear genes encoding mitochondrial proteins (for a list of the compiled 400 mitochondrial proteins, see Table 10, which is published as supporting information on the PNAS web site). Thus, Puf3p and Pum both appear to interact with nuclear-encoded mRNAs encoding mitochondrial proteins. Whereas Puf3p interacts predominantly with these mRNAs (87% of targets), Pum interacts with only a small fraction of this group (4%). Conversely, most of the Pum targets are mRNAs encoding for nonmitochondrial proteins. It is possible that the need for greater complexity in gene regulation and coordination in more complex, multicellular organisms has been addressed, in part, by breaking up functionally related mRNAs into smaller subgroups. In addition, the ability to use combinatorial binding of RBPs would allow coordination of and differentiation between these subgroups.
Discussion
We have systematically analyzed mRNAs associated with Pum, an important posttranscriptional regulator of gene expression in Drosophila by a method that combines RNA copurification with an affinity-tagged RNA-binding protein and DNA microarray analysis of the associated RNAs (5, 31, 41–46). We identified >1,000 distinct Pum-associated mRNAs, many of which encode functionally related proteins and contain characteristic 3′ UTR sequence elements sufficient for interaction with Pum. This finding represents a tremendous increase in potential mRNA targets that may be subject to translational or other posttranscriptional regulation by Pum, and highlights the potential importance of posttranscriptional regulation in multicellular organisms (8, 9). The roles of Pum in embryonic development, stem cell biology, and the function of the nervous system were discovered by classical forward-genetic approaches; although these approaches uncovered essential functions of Pum and identified several important target mRNAs, our genomic analysis points to many hitherto unrecognized targets mRNAs whose products may be involved in processes less readily accessed by classical genetic approaches. However, our assay is unlikely to exclusively and completely uncover target mRNAs that are associated with Pum in vivo: nontarget mRNAs may associate with Pum and true mRNA targets may dissociate during the affinity-isolation procedure (47). Moreover, our assay does not reveal whether Pum interacts directly with its target mRNA or indirectly via another protein. Further biochemical and functional experiments are required to verify and dissect regulation of particular mRNAs by Pum. Nevertheless, the identification of a sequence motif for Pum and the striking functional links among mRNA targets strongly suggest an underlying biological role for many of the interactions we have identified.
Not only was the number of mRNAs associated with Pum remarkably large, but the protein products of these mRNAs shared functional links, including function in the anterior–posterior patterning system, most cyclins, and most subunits of the vacuolar H-ATPase. These functional links add to the growing evidence for an extensive posttranscriptional regulatory system and support recent models for functionally related posttranscriptional modules organized by RBPs (5–7). Perhaps Pum, in concert with other proteins, coordinates the temporal or spatial pattern of translation of a large set of mRNAs. For instance, Pum may help ensure that maternally derived mRNAs, which are stored in the unfertilized egg, are translated at the correct developmental stage. Translational repression of maternally derived mRNAs before fertilization is an important mechanism to control the onset of expression of anterior–posterior patterning genes (48). Another role of Pum is to regulate mitosis of migrating pole cells by inhibition of cycB expression (17). Our finding that most cyclin mRNAs were associated with Pum introduces the possibility of a more general role for Pum in the coordination of the cell-cycle, although cyclins can also have non-cell-cycle-related functions (49). Regulation of cyclins by Pum may also have a role in sustaining proliferation of stem cells, a proposed ancestral function of Puf-family proteins (8).
In this work, as in the previous work on yeast Puf proteins (5), a consensus RNA-binding element was defined by a genome-wide unbiased search for common sequence motifs among mRNAs selected by a biochemical procedure (Fig. 3). The 8-nt core motif [UGUA(A/C/U)AUA] defined here is remarkably similar to the sequences in and surrounding box B of the hb NRE, and it resembles core motifs bound by diverse Puf family proteins (5, 26, 30, 36, 39). It is also in agreement with a crystal structure of human PumHD in complex with hb RNA, which revealed that each of the eight repeats comprising the PumHD interacts with one of eight bases in the bound RNA and suggested that RNA recognition is highly modular (12). Interestingly, the three amino acid residues in each repeat that directly interact with one RNA base are conserved between Pum and yeast Puf3p (Fig. 8, which is published as supporting information on the PNAS web site) paralleling the almost identical core sequences bound by these proteins (Fig. 3). Other Puf proteins that bear the same critical amino acid residues for RNA base contacts may bind to highly similar RNA consensus sequences.
The definition of core motifs allows us to search for additional potential mRNA targets that were not identified in our affinity isolation procedure. About 10% of all genes in Drosophila contain our computationally defined 8-nt core motif in their 3′ UTR; a search for GO terms overrepresented among these 1431 annotated genes found that an unexpectedly large fraction encode proteins involved in morphogenesis or organ development (243 genes, P < 10−29), neurogenesis (153 genes, P < 10−27), transcriptional regulation (172 genes, P < 10−17), or proteins that are localized to membranes (264 genes, P < 5 × 10−5), in particular the plasma membrane (116 genes, P < 2 × 10−12). Interestingly, many of the mRNAs that have the putative Pum binding site but were not enriched in our assays encode proteins with neuronal functions; e.g., Complexin (Cpx; CG32490), which bears a cluster of 10 core motifs in its 3′ UTR. Because TAP-Pum was specifically expressed in the ovaries of flies, neuron-specific mRNA targets would not have been accessible to TAP-Pum in vivo and therefore were not expected to be identified. It will be important to extend this analysis to other tissues and organs including the nervous system by the use of tissue-specific drivers available in Drosophila. In addition to identification of tissue-specific potential mRNA targets of Pum, these experiments will also allow the determination of whether and to what extent exchange of Pum-associated mRNAs occurs after cell lysis.
Systematic identification of mRNAs associated with homologous RBPs in various species provides a basis for considering their evolution. Large sets of target mRNAs can now be compared with respect to their structural and functional commonalties and differences. In the case of Pum, conservation of amino acid residues in the PumHDs between homologous Puf proteins correlates with our identified core motifs in 3′ UTR of mRNA targets. However, the proteins encoded by the mRNA targets appeared not to be particularly conserved. This discordance suggests that acquisition or loss of RBP-binding motifs in UTRs of genes may provide a surprisingly fluid evolutionary mechanism to modify posttranscriptional regulatory connections.
Materials and Methods
Oligonucleotide Sequences.
See Supporting Text.
RNA Affinity Isolations.
Wild-type (yellow white, y w) or mat-α-tubGAL4-VP16(V67);UAS-TAP-PumHD(3–1-4) flies were grown in large food cages (the generation of transgenic TAP-Pum flies is described in Supporting Text). Adults (0–5 days old) were collected, frozen in liquid nitrogen, and stored at −80°C. Embryos (0–16 h) were collected from apple juice agar plates, dechorionated, and washed twice with buffer A (20 mM Tris·HCl, pH 8.0/150 mM NaCl/10 mM EDTA, pH 8.0/0.2% Nonidet P-40/0.02 mg/ml heparin) before freezing.
Five grams of adult flies or 2.5 g of embryos were used in each affinity purification. Flies or embryos were suspended in 15 ml of buffer B [buffer A plus 1.5 mM DTT/1 mM PMSF/0.5 μg/ml leupeptin/0.8 μg/ml pepstatin/20 units/ml DNase I/100 units/ml Rnasin (Promega)/0.2 mg/ml heparin) in a mortar filled with liquid nitrogen and ground with a pestle to a fine powder. The powder was transferred to a glass-dounce, thawed, and dounced until the pestle reached the bottom. The suspension was centrifuged twice at 4°C and 10,000 × g for 10 min. The fat layer on top was aspirated off after each centrifugation. Cleared extract (12.5 ml) was incubated with 600 μl of a slurry (50% vol/vol) of IgG-agarose beads (Sigma) for 90 min at 4°C. The beads were washed once with buffer B for 15 min at 4°C, and three times for 15 min at 4°C with buffer C (20 mM Tris·HCl, pH 8.0/150 mM NaCl/1 mM EDTA, pH 8.0/10% glycerol/0.01% Nonidet P-40/1 mM DTT/10 units/ml RNasin). PumHD was released from beads by incubation with 150 units of AcTEV protease (Invitrogen) for 2 h at room temperature. RNA was isolated from extracts and from the TEV eluates with TRIzol reagent (Invitrogen) followed by RNeasy Mini kit (Qiagen) purification according to the manufacturer’s instructions.
Concentration of tagged transgene products in extracts was determined by a filter affinity blot analysis using protein A as a standard for calibration (5). Intracellular protein concentrations in embryos were estimated with parameters described in ref. 30.
Microarray Analysis and Bioinformatics.
Procedures for microarray analysis, data selection, bioinformatics and gene expression profiling are described in Supporting Text. Microarray data sets are available at the Stanford Microarray Database (SMD) (50) or at the Gene Expression Omnibus at www.ncbi.nlm.nih.gov/geo (accession nos. GSE3580GSE3581–GSE3582).
Synthesis of Biotinylated RNAs and Pull-Down Experiment.
See Supporting Text.
Supplementary Material
Acknowledgments
We thank Dr. Eric Johnson for help with motif searches and Dan Hogan for discussions. We also thank Stanford Microarray Database and the Functional Genomics Center Zürich for providing microarray related infrastructure. A.P.G. and S.L. were supported by long-term fellowships from the Human Frontier Science Program Organization. P.O.B. and M.A.K. are investigators of the Howard Hughes Medical Institute. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health Grant GM49243 (to D.H.).
Abbreviations
- RBP
RNA-binding protein
- PumHD
Pum-homology domain
- NRE
nanos-response element
- FDR
false discovery rate
- GO
gene ontology
- TAP
tandem affinity purification
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE3580GSE3581–GSE3582).
References
- 1.Dreyfuss G., Kim V. N., Kataoka N. Nat. Rev. Mol. Cell. Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 2.Gebauer F., Hentze M. W. Nat. Rev. Mol. Cell. Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez de Heredia M., Jansen R. P. Curr. Opin. Cell. Biol. 2004;16:80–85. doi: 10.1016/j.ceb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Hieronymus H., Silver P. A. Genes Dev. 2004;18:2845–2860. doi: 10.1101/gad.1256904. [DOI] [PubMed] [Google Scholar]
- 5.Gerber A. P., Herschlag D., Brown P. O. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keene J. D., Tenenbaum S. A. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 7.Keene J. D., Lager P. J. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- 8.Wickens M., Bernstein D. S., Kimble J., Parker R. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 9.Spassov D. S., Jurecic R. IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- 10.Zamore P. D., Williamson J. R., Lehmann R. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 11.Sonoda J., Wharton R. P. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., McLachlan J., Zamore P. D., Hall T. M. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann R., Nusslein-Volhard A. Nature. 1987;329:167–170. [Google Scholar]
- 14.Wharton R. P., Struhl G. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- 15.Sonoda J., Wharton R. P. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamberi C., Peterson D. S., He L., Gottlieb E. Development (Cambridge, U.K.) 2002;129:2699–2710. doi: 10.1242/dev.129.11.2699. [DOI] [PubMed] [Google Scholar]
- 17.Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K., Kobayashi S. Nat. Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- 18.Lin H., Spradling A. C. Development (Cambridge, U.K.) 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 19.Forbes A., Lehmann R. Development (Cambridge, U.K.) 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- 20.Szakmary A., Cox D. N., Wang Z., Lin H. Curr. Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Dubnau J., Chiang A. S., Grady L., Barditch J., Gossweiler S., McNeil J., Smith P., Buldoc F., Scott R., Certa U., et al. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 22.Ye B., Petritsch C., Clark I. E., Gavis E. R., Jan L. Y., Jan Y. N. Curr. Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 23.Menon K. P., Sanyal S., Habara Y., Sanchez R., Wharton R. P., Ramaswami M., Zinn K. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 25.Edwards T. A., Pyle S. E., Wharton R. P., Aggarwal A. K. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 26.Wharton R. P., Sonoda J., Lee T., Patterson M., Murata Y. Mol. Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- 27.Brand A. H., Perrimon N. Development (Cambridge, U.K.) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 28.Rorth P. Mech. Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 29.Macdonald P. M. Development (Cambridge, U.K.) 1992;114:221–232. doi: 10.1242/dev.114.1.221. [DOI] [PubMed] [Google Scholar]
- 30.Zamore P. D., Bartel D. P., Lehmann R., Williamson J. R. Biochemistry. 1999;38:596–604. doi: 10.1021/bi982264s. [DOI] [PubMed] [Google Scholar]
- 31.Shepard K. A., Gerber A. P., Jambhekar A., Takizawa P. A., Brown P. O., Herschlag D., DeRisi J. L., Vale R. D. Proc. Natl. Acad. Sci. USA. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivas W., Parker R. EMBO J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson J. S., Jr., Houshmandi S. S., Lopez Leban F., Olivas W. M. RNA. 2004;10:1625–1636. doi: 10.1261/rna.7270204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allan A. K., Du J., Davies S. A., Dow J. A. Physiol. Genomics. 2005;22:128–138. doi: 10.1152/physiolgenomics.00233.2004. [DOI] [PubMed] [Google Scholar]
- 35.Dissing M., Giordano H., DeLotto R. EMBO J. 2001;20:2387–2393. doi: 10.1093/emboj/20.10.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein D., Hook B., Hajarnavis A., Opperman L., Wickens M. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Opperman L., Hook B., Defino M., Bernstein D. S., Wickens M. Nat. Struct. Mol. Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- 38.Bailey T. L., Elkan C. Proc. Int. Conf. Intell. Syst. Mol. Biol.; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 39.White E. K., Moore-Jarrett T., Ruley H. E. RNA. 2001;7:1855–1866. [PMC free article] [PubMed] [Google Scholar]
- 40.Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R., Hariharan I. K., Fortini M. E., Li P. W., Apweiler R., Fleischmann W., et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenenbaum S. A., Carson C. C., Lager P. J., Keene J. D. Proc. Natl. Acad. Sci. USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takizawa P. A., DeRisi J. L., Wilhelm J. E., Vale R. D. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 43.Roy P. J., Stuart J. M., Lund J., Kim S. K. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- 44.Hieronymus H., Silver P. A. Nat. Genet. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- 45.Inada M., Guthrie C. Proc. Natl. Acad. Sci. USA. 2004;101:434–439. doi: 10.1073/pnas.0307425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Guisbert K., Duncan K., Li H., Guthrie C. RNA. 2005;11:383–393. doi: 10.1261/rna.7234205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mili S., Steitz J. A. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickens M., Goodwin E. B., Kimble J., Strickland S., Hentze M. W. In: Translational Control of Gene Expression. Sonenberg N., Hershey J. W. B., Mathews M. B., editors. Plainview, New York: Cold Spring Harbor Lab. Press; 2000. pp. 295–370. [Google Scholar]
- 49.Edgar B. A., Lehner C. F. Science. 1996;274:1646–1652. doi: 10.1126/science.274.5293.1646. [DOI] [PubMed] [Google Scholar]
- 50.Ball C. A., Awad I. A., Demeter J., Gollub J., Hebert J. M., Hernandez-Boussard T., Jin H., Matese J. C., Nitzberg M., Wymore F., et al. Nucleic Acids Res. 2005;33:D580–D582. doi: 10.1093/nar/gki006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.