Abstract
Juvenile primates develop myopia when their visual experience is degraded by lid fusion. In response to this abnormal visual input, retinal neural networks cause an excessive growth of the postequatorial segment of the eye, but the mechanism underlying this axial elongation is unknown. After fusion of the lids in one eye of juvenile rhesus macaques and green monkeys, we combined cDNA subtractions, microarray profiling, and real-time PCR to compare gene expression in the retinas of the closed and open eyes. This molecular analysis showed up-regulation of a number of genes associated with cell division in the retina of the closed eye and differential expression of six genes localized to chromosomal loci linked to forms of human hereditary myopia. In addition, it substantiated a previous observation, based on immunocytochemistry, that synthesis of vasoactive intestinal polypeptide was increased upon lid fusion. Injection of 5-bromo-2′-deoxyuridine and immunocytochemistry showed that the primate retinal periphery harbors mitotically active neuroprogenitor cells that increase in number when the visual experience is altered by lid fusion. Furthermore, the number of dividing cells is highly correlated with axial elongation of the eye and the resulting myopic refractive error. Thus, the retina undergoes active growth during the postnatal development of the primate eye. This growth is modulated by the visual input and accelerates considerably when the eye develops axial myopia. Vasoactive intestinal polypeptide may be the molecule that stimulates retinal growth.
Keywords: BrdU, gene expression, lid fusion, vasoactive intestinal polypeptide, hereditary myopia
From birth to adult age, the human eye increases ≈3-fold in size, but the development of its optical components is so well coordinated that in the normal (emmetropic) adult eye, the image of distant objects falls with great precision on the photosensitive layer of the retina in absence of accommodation. Failure of this process leads to errors of refraction, most commonly myopia, in which images of distant objects fall in front of the retinal photoreceptors in the unaccomodated eye. The cause of myopia is unclear, although population surveys indicate that both genetic and environmental factors are involved (1).
The experimental manipulation of the refractive state became possible when it was shown that juvenile primates develop myopia when their visual experience is degraded by fusion of the lids (2, 3); subsequently, myopia was experimentally produced in other species, including the chicken (4) and tree shrew (5). It is now clear that appropriate environmental cues, acting through the visual system, ensure the harmonic growth of the eye after birth (6).
Neonatal lid fusion is an expedient procedure for induction of axial elongation and myopia in primates. Commonly referred to as form deprivation, the distortion of the visual input caused by this procedure is complex, because the lids are converted into a translucent diaphragm that eliminates high spatial frequencies, alters the chromatic composition of the incoming light, and attenuates retinal illumination by about half a log unit (3). In the rhesus macaque (Macaca mulatta), the initial event that leads to excessive elongation of the eye globe takes place in the retina (3), but the mechanism by which the retinal neural networks alter the growth of the eye in response to the abnormal visual input is unknown. In this study, we gained insight into the events that take place in the primate retina upon lid fusion by combining analysis of gene expression with injection of the thymidine analog BrdU and immunocytochemistry.
Results
The lids of the right eye were fused in four rhesus macaques and eight green monkeys (Chlorocebus aethiops), 0.3 to 1.2 months of age, and kept closed for 2.1 to 3.9 months. At the end of this period of lid fusion, the rate of growth of the closed eye is still very high (3), and interocular differences in size are reliably measured. After the palpebral fissure was reestablished, accommodation was paralyzed, both eyes were refracted with a streak retinoscope, and A-scan ultrasound measurements were obtained. Tables 1 and 2 list the differences in axial dimension (depth) of the vitreous chamber between closed and open eye in all lid-fused animals. Because the effects of lid fusion are confined to the postequatorial segment of the eye, vitreous chamber depth is the most precise indicator of axial elongation (3).
Table 1.
Effects of lid fusion on depth of the vitreous chamber of juvenile monkeys
| Monkey no. | Age at closure, months | Duration of closure, months | OS vitreous chamber depth,† mm | OD vitreous chamber depth,† mm | OD–OS, mm |
|---|---|---|---|---|---|
| 296-95* | 0.4 | 3.2 | 9.9 ± 0.09 (7) | 11.25 ± 0.07 (8) | 1.35 |
| 204-95* | 0.3 | 2.8 | 9.56 ± 0.07 (14) | 10.47 ± 0.19 (12) | 0.91 |
| 298-95* | 0.3 | 3.2 | 9.98 ± 0.14 (7) | 10.48 ± 0.05 (7) | 0.50 |
| 185-03* | 0.3 | 3.9 | 9.37 ± 0.07 (6) | 10.14 ± 0.23 (10) | 0.77 |
| P047** | 0.7 | 2.1 | 9.72 ± 0.16 (12) | 10.17 ± 0.12 (10) | 0.44 |
| P049** | 0.7 | 2.1 | 9.68 ± 0.04 (8) | 9.61 ± 0.09 (12) | −0.07 |
| R071** | 1.5 | 2.5 | 10.11 ± 0.08 (11) | 11.05 ± 0.04 (9) | 0.94 |
One millimeter in depth of the vitreous chamber is equal to 4.02 diopters in M. mulatta (r = 0.957, P < 0.001, n = 21) and 3.53 diopters in C. aethiops (r = 0.752, P < 0.001, n = 36). ∗, M. mulatta; ∗∗, C. aethiops.
†Data are shown as mean ± SD. Number of measurements is indicated in parentheses. OS, left (open) eye; OD, right (closed) eye.
Table 2.
Relation between depth of the vitreous chamber and number of proliferating retinal cells in C. aethiops
| Monkey no. | Age at closure, months | Duration of closure, months | OS vitreous chamber depth,* mm | OD vitreous chamber depth,* mm | OD–OS, mm | BrdU-positive cells in OS† | BrdU-positive cells in OD† | |
|---|---|---|---|---|---|---|---|---|
| O032 | 1.2 | 2.9 | 10.12 ± 0.09 (5) | 11.24 ± 0.11 (5) | 1.11 | 59.13 | 193.63 | 3.27 |
| O033 | 0.8 | 2.9 | 9.98 ± 0.36 (4) | 9.37 ± 0.13 (5) | 0.61 | 26.71 | 41.61 | 1.56 |
| Q011 | 0.7 | 3.0 | 10.20 ± 0.00 (3) | 10.42 ± 0.05 (7) | 0.22 | 15.25 | 20.22 | 1.33 |
| Q009 | 0.8 | 3.0 | 9.75 ± 0.03 (4) | 9.27 ± 0.18 (9) | −0.48 | 17.49 | 11.63 | 0.66 |
| Q013 | 0.3 | 3.0 | 10.80 ± 0.06 (4) | 9.55 ± 0.16 (6) | −1.25 | 39.69 | 12.81 | 0.32 |
*Data are shown as mean ± SD. Number of measurements is indicated in parentheses.
†Number of cells mm−2. BrdU-positive cells were counted in a 1-mm strip at the extreme retinal periphery.
Analysis of Gene Expression.
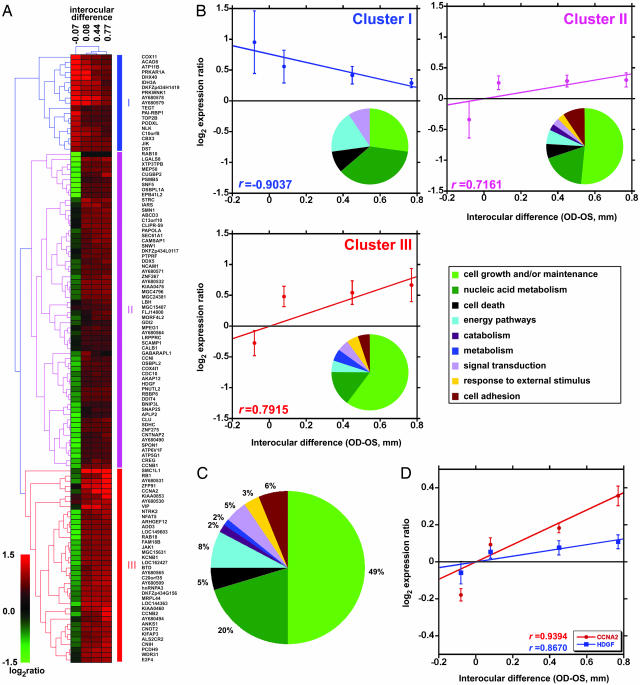
In three rhesus macaques (nos. 204-95, 296-95, and 298-95) the closed eye developed a myopic refractive error ranging from 2.5 to 4.5 diopters, caused by an increase in depth of the vitreous chamber ranging from 0.5 to 1.35 mm (Table 1; see also Fig. 6, which is published as supporting information on the PNAS web site). To compare gene expression in the retinas of the closed and open eye, we carried out cDNA subtractions (7) that resulted in two cDNA libraries: one enriched in clones potentially up-regulated and the other in clones potentially down-regulated in the retina of the closed eye. To identify differentially expressed genes, 1,000 clones from each library were sequenced, redundant clones were eliminated, and the remaining 535 cDNAs were spotted onto polyL-lysine-coated glass slides (five copies of each cDNA per slide). To obtain probes for this microarray experiment, we fused the lids of the right eye in one rhesus macaque and two green monkeys. The green monkey is ideally suited to correlate gene expression with eye growth and refractive state, because in ≈20% of individuals, lid fusion has no effect on refraction or causes hyperopia (data not shown). Indeed, the procedure induced axial elongation in the closed eye of the rhesus macaque (no. 185-03; Table 1) and one green monkey (no. P047), whereas in the other green monkey, it caused a slight decrease in the vitreous chamber depth with respect to the open eye (no. P049). As an additional control, we included in this experiment a juvenile rhesus macaque in which both eyes were left open. Cy3- and Cy5-labeled cDNA probes obtained from the retinas of these animals were hybridized to the custom-made microarrays. We carried out two hybridizations with color swap for each monkey; therefore, we had 10 measurements for each cDNA. For 119 genes (listed in Table 3, which is published as supporting information on the PNAS web site), we observed a statistically significant difference in expression between the retinas of the closed and open eyes (P < 0.05, n = 10). Three distinct clusters of genes with coordinated expression profiles could be identified when results were analyzed by hierarchical clustering (Fig. 1A and B). A group of 19 genes, found in cluster I, showed negative correlation with the difference in depth of the vitreous chamber between closed and open eye (rclusterI = −0.904), whereas 62 genes in cluster II and 38 genes in cluster III showed a positive correlation (rclusterII = 0.7161 and rclusterIII = 0.7915). Six genes were localized at chromosomal susceptibility loci for human hereditary myopia (Table 4, which is published as supporting information on the PNAS web site). Expression of five of them, LOC157627, ARHGEF12, APLP2, PNUTL2, and ZNF275 (clusters II and III), was positively correlated with axial elongation whereas that of gene DHX40 (cluster I) was negatively correlated. An additional gene, AY680578 (cluster I), which was negatively correlated with axial elongation, mapped at locus 15q11.2, which is affected in the Prader-Willi and Angelman syndromes (8). The results of hierarchical clustering were confirmed by gene set enrichment analysis (GSEA), which identified a group of 100 genes exhibiting statistically significant correlation with axial elongation (Fig. 7, which is published as supporting information on the PNAS web site). This group included the 19 genes of cluster I that were negatively correlated with axial elongation, and 81 genes from clusters II and III that were positively correlated with axial elongation (the GSEA ranking scores are reported in Table 3). Gene ontology analysis revealed that 69% of all differentially expressed genes were involved in cell proliferation and nucleic acid metabolism (Fig. 1C; see also Tables 5 and 6, which are published as supporting information on the PNAS web site). Expression of 84% of these genes was positively correlated with axial elongation (clusters II and III), and a number of their products were well characterized regulators of the cell cycle (9–11) or associated with increased cell proliferation (12–15), such as cyclin A2 (CCNA2), cyclin B1, cyclin B2, E2F4, and hepatoma-derived growth factor (HDGF). The expression levels of two of these genes, cyclin A2 and cyclin B1, are known to be positively correlated with the proliferation rate of two types of tumors (16); thus, their up-regulation is an indicator of increased proliferation. Finally, expression of the gene for vasoactive intestinal polypeptide (VIP, cluster III) was positively correlated with axial elongation. To confirm the results of the microarray study, we used real-time PCR to analyze the expression of the genes coding for CCNA2 (cluster III) and HDGF (cluster II) in the same animals that were used for the microarray experiment. Expression of both genes was highly correlated with the axial dimension of the vitreous chamber (rCCNA2 = 0.9394 and rHDGF = 0.8670; Fig. 1D).
Fig. 1.
Gene expression analysis indicates that lid fusion caused increased cell proliferation in the retina. (A) Hierarchical cluster analysis of genes differentially regulated in the retina of the closed eye vs. the retina of the open eye. Interocular differences in depths of the vitreous chamber [right (closed) eye (OD)–left (open) eye (OS) in millimeters] are shown above the clusters. The two monkeys in which closure caused axial elongation (nos. 185-03 and P047) and the control monkey (No. 151-03) were clustered together, whereas the monkey in which closure caused a small decrease in vitreous length (No. P049) exhibited a different expression profile. Red represents genes that are up-regulated, and green represents genes that are down-regulated. Genes can be divided into two groups: a first group of 19 (cluster I) negatively correlated and a second group of 100 (clusters II and III) positively correlated with the depth of the vitreous chamber. The pie charts show the distribution of the differentially regulated genes according to the gene ontology terms for biological function in each cluster (B) and in all clusters combined (C). (D) Real-time PCR confirmed that expression of the genes for CCNA2 (cluster III) and HDGF (cluster II) was highly correlated with the axial dimension of the vitreous chamber.
Proliferating Neuroprogenitor Cells at the Retinal Periphery.
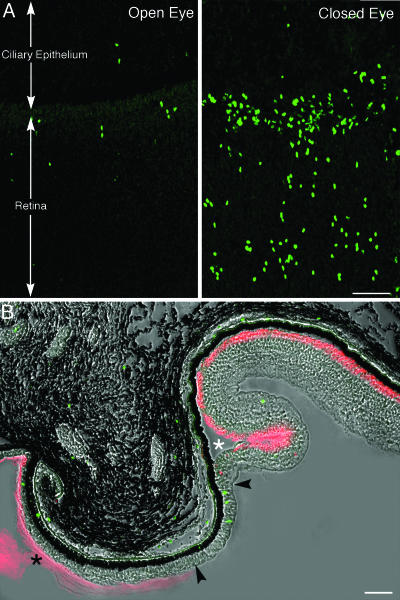
To address the issue of increased proliferation in the retina of the closed eye, we made use of injections of BrdU. In one green monkey (no. R071; Table 1), we adopted a protocol of BrdU administration aimed at labeling a large number of cells and observing the initial stages of their differentiation. This animal received six BrdU injections (100 mg·kg−1, i.p.) over a period of 3 days, 2.5 months after lid fusion. Nineteen days later (2.6 months after lid fusion), ultrasonography showed that the axial length of the vitreous chamber in the closed eye was 0.94 mm larger than in the open eye. Examination of whole mounts of the neural retina after immunostaining with an antibody to BrdU showed that in the closed eye a large number of nuclei at the extreme periphery of the retina had incorporated the tracer. BrdU-positive nuclei were also present in the peripheral retina of the open eye, but they were considerably less numerous (Fig. 2A). In both open and closed eyes, the proliferating cells occupied a region ≈250 μm wide adjacent to the ora serrata (Fig. 8, which is published as supporting information on the PNAS web site), which is the dentate boundary between retina and pars plana of the ciliary body. Very few labeled cells were present in the ciliary epithelium, in front of the ora serrata.
Fig. 2.
The peripheral retina harbors a population of proliferating cells that increase in number upon lid fusion. (A) In an animal that responded to lid closure with remarkable axial elongation, examination of whole-mounted retinas after staining with an antibody to BrdU (green) showed that the number of proliferating cells was conspicuously increased in the peripheral retina of the closed eye. The boundary (ora serrata) between neural retina and the ciliary epithelium of the pars plana of the ciliary body is indicated. These confocal micrographs represent orthogonal projections of Z series through the entire thickness of the retina. (B) A meridional section through retina and choroid of the closed eye in the region of the ora serrata was stained with both an antibody to BrdU (green) and rhodamine-conjugated peanut agglutinin (red) that binds to the surface of the outer segment of cone cells. Unexpectedly, this agglutinin also stained the ciliary zonule (filled asterisk), thus clearly marking the vitreal surface of the bistratified ciliary epithelium. A band of columnar, pseudostratified epithelium containing BrdU-positive cells (arrowheads) intervenes between the ciliary epithelium and the retina. Peanut agglutinin-positive material first appears sporadically in the thickness of the rudimentary peripheral retina. It becomes organized into a continuous layer with the differentiation of cone cell outer segments (white asterisk). (Scale bars: 50 μm.)
After staining with antibodies to BrdU and rhodamine-conjugated peanut agglutinin, which labeled both the surface of the outer segments of cone cells and the ciliary zonule, the transition between the multilayered retina and the ciliary epithelium appeared much more gradual in the closed than in the open eye. The marginal retina became progressively thinner toward the ora serrata and appeared to recapitulate in reverse its embryonic development, losing first the outer, then the inner, plexiform layer. Ultimately, it continued as a pseudostratified, columnar epithelium that resembled primitive neuroepithelium, distinct from but continuous anteriorly with the bistratified epithelium of the pars plana of the ciliary body (Fig. 2B). BrdU-positive nuclei were scattered throughout this transition zone but were especially numerous within the most peripheral, rudimentary retina. Cone outer segments began to appear just posterior to the transition between columnar epithelium and retina proper.
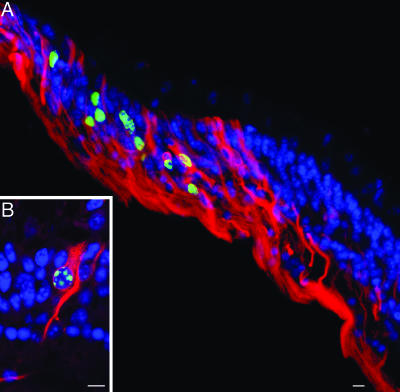
To determine whether the proliferating cells at the retinal margin had the characteristics of neuroprogenitor cells, we examined the expression of the neural precursor cell marker nestin (17) in retinal sections double-labeled with anti-BrdU antibody. A large proportion of the proliferating cells in the retinal periphery contained filament bundles immunoreactive for nestin (Fig. 3A and B). They commonly exhibited the elongate shape typical of embryonic neuroepithelial precursors and spanned the entire thickness of the neural retina.
Fig. 3.
Proliferating cells are neuroprogenitors. (A) Sections of the retinal periphery were stained with antibodies to nestin (red) and BrdU (green), and nuclei were counterstained with DAPI (blue). Numerous cells contained nestin-positive bundles of cytoskeletal filaments and exhibited the elongate shape typical of embryonic neuroepithelial precursors. In many of these cells, the nucleus was BrdU-positive. (B) At higher magnification, BrdU-stained nucleus and nestin-positive cytoplasmic filaments were colocalized in the same cell. (Scale bars: 10 μm.)
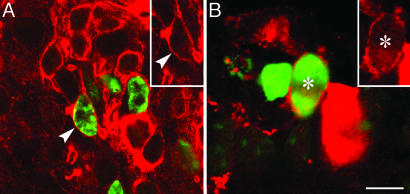
Finally, to establish whether the proliferating cells were capable of differentiating into retinal neurons, we investigated the colocalization of BrdU with two photoreceptor cell markers, rhodopsin, the photopigment of rod cells, and cone-specific arrestin, a cytosolic protein of cone cells. In the peripheral retina, BrdU-positive cells deeply situated in the developing outer nuclear layer were immunoreactive for rhodopsin. The staining was confined to the plasma membrane, because the cells did not possess yet an outer segment (Fig. 4A). A smaller number of cells with BrdU-positive nuclei contained cytoplasmic arrestin; they also were lacking an outer segment and were deeply situated with respect to the row of developing cone cells abutting the pigment epithelium (Fig. 4B). Thus, the proliferating cells at the retinal periphery of the myopic eye are neuroprogenitors that can give rise to photoreceptors.
Fig. 4.
Neuroprogenitors differentiate into photoreceptor cells. (A) Immunostaining with antibodies to BrdU (green) and rhodopsin (red) showed that the membrane of many BrdU-positive cells contained rhodopsin (arrowheads); the positive cells were located deeply in the developing outer nuclear layer. (B) A smaller number of neuroprogenitors (asterisks) also gave rise to precursors of cone photoreceptor cells, as revealed by double-staining with antibodies to BrdU (green) and cone-specific arrestin (red). (Scale bar: 10 μm.)
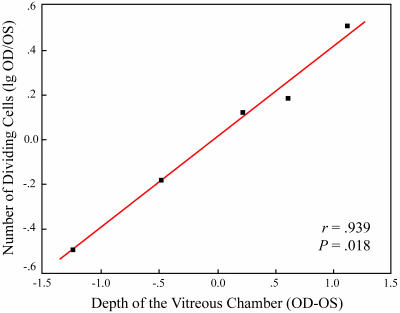
It was important to establish whether the increased proliferation of neuroprogenitor cells was correlated with the postequatorial expansion of the eye globe. To this purpose, we counted the BrdU-positive cells in five lid-sutured green monkeys in which the difference in axial length between closed and open eye varied considerably at the end of 3 months of lid fusion (nos. O032, O033, Q009, Q011, and Q013; Table 2). In these animals, BrdU (125 mg·kg−1) was slowly administered i.v. over a period of 1.5 h and the retina was harvested 2 h after the end of the infusion. Three monkeys responded to lid fusion with varying degrees of axial elongation, whereas in the remaining two animals, the vitreous depth was less in the closed than in the open eye (Table 2). In all of these animals, the number of BrdU-positive nuclei was counted in the retinal periphery in the closed and open eyes by using both whole mounts and sections. When the normalized cell counts were plotted against the difference in depth of the vitreous chambers of the closed and open eyes, it became clear that the number of proliferating cells grew exponentially in parallel with the increase in depth of the vitreous chamber. The logarithm of the normalized cell counts showed a high positive correlation with the interocular difference in axial length of the vitreous chamber (r = 0.939, P = 0.018; Fig. 5).
Fig. 5.
Interocular difference in the depth of the vitreous chamber between the OD and OS is positively correlated with the logarithm of the normalized cell counts. r, Pearson’s correlation coefficient; P, significance value calculated by ANOVA.
Discussion
We have used a combination of cDNA subtractions, microarray analysis, and real-time PCR to investigate gene expression in the retinas of juvenile rhesus macaques and green monkeys deprived of form vision by lid fusion. This molecular analysis showed a statistically significant difference in the expression of 119 genes between the retinas of the closed and open eyes. Crucial findings were up-regulation of a number of genes associated with cell division, suggesting that the abnormal visual input stimulated growth of the retina, and up-regulation of the gene for VIP, which confirms a previous observation, based on immunocytochemistry, that VIP synthesis was increased in a class of amacrine cells upon lid fusion (18). VIP stimulates neurogenesis (for review, see ref. 19), thus, it may be one of the molecules that mediate the effect of lid fusion on the proliferation of retinal cells. Particularly noteworthy was the finding that six genes differentially expressed in the retinas of the closed and open eyes were localized at chromosomal susceptibility loci for human hereditary myopia and, thus, represent potential candidate genes for the genetic anomalies identified in previous genomewide linkage studies of myopia (for reference, see Table 4). Two of these genes, ARHGEF12 and APLP2, code for known proteins, a Rho guanine nucleotide exchange factor and an amyloid beta precursor-like protein, respectively. The function of the remaining four genes, LOC157627, PNUTL2, DHX40, and one coding for a novel transcription factor ZNF275, are not known. Another potentially important finding concerns an unknown gene, AY680578, whose expression was negatively correlated with axial elongation. It is localized at 15q11.2, a locus that resides in the chromosomal regions affected in Prader-Willi and Angelman syndromes, in which multiple organ anomalies are frequently associated with high myopia (20). Thus, this gene may be important for emmetropization in juvenile individuals.
In amphibians and teleost fishes, postnatal growth of the retina results from the proliferation of stem or neuroprogenitor cells located in the germinal zone of the ciliary margin (for review, see ref. 21) and a population of mitotically active neuroprogenitor cells has been identified in the peripheral retina of newly hatched chickens (22). Stem cells, which can be induced to proliferate in vitro, have been isolated from the ciliary epithelium of adult mice (23) and from the epithelium of the ciliary body and iris of humans, including old individuals (24). It is generally accepted, however, that neurogenesis ceases in the mammalian retina shortly after birth (25, 26). In contrast with this belief, we demonstrated by injection of BrdU and immunocytochemistry that a population of proliferating cells, exhibiting the hallmarks of embryonic neuroprogenitors, persists in the extreme periphery of the retina of juvenile primates, and it increases dramatically when the visual experience is altered by lid fusion, a powerful stimulus to the elongation of the postequatorial segment of the eye. At the ora serrata, these proliferating cells are situated both in a band of pseudostratified columnar epithelium, which resembles the embryonic retinal neuroepithelium, and in the adjacent rudimentary retina, which is progressively acquiring its typical layers. In this region, the bipolar, mitotically active neuroepithelial cells contain bundles of nestin-positive intermediate filaments, a marker of neuronal precursors. Finally, the neuroprogenitors possess the ability to generate mature retinal neurons, because a proportion of the BrdU-positive cells express the photoreceptor cell markers rhodopsin and cone-specific arrestin. Thus, it appears that neurogenesis persists in the peripheral retina of all vertebrates after completion of embryonic development and, in primates, participates in the growth of the retina from birth to adult age.
The number of proliferating cells in the retinal periphery of the closed eye is highly correlated with the axial length of the eye and, therefore, with its refractive state. This finding demonstrates that in juvenile primates, as in young chickens (22), the excessive ocular growth induced by the altered visual input is accompanied by retinal growth, due to increased proliferation of neuroprogenitor cells at the retinal periphery. On the other hand, in the monkeys in which lid fusion slowed down eye growth, fewer proliferating cells were found in the retina of the closed eye than in that of the open one. Thus, increased retinal neurogenesis in the closed eye is induced by the complex alteration of the visual input that leads to axial elongation and myopia, rather than represent a trivial consequence of the light attenuation caused by the fused lids.
Two crucial problems are now open to future investigation: the identification of the precise stimulus that causes proliferation of neuroprogenitor cells at the retinal periphery and the role played by retinal growth in the mechanism of axial elongation of the eye. Based on the spatial distribution of dopaminergic amacrine cells in the retina of goggle-wearing chickens (27), it was suggested that the rate of progenitor cell proliferation at the margin of the retina increases in myopia because the retina is stretched as a result of excessive ocular growth (22). Even though an effect of mechanical stretch on progenitor cell proliferation cannot be ruled out, the following issues have to be considered. First, there is no direct evidence that the tension of the wall of the postequatorial segment of the eye globe is increased in the lid-sutured eye, although the sclera at the posterior pole does become thinner as a result of remodeling of the extracellular matrix (28, 29). Second, the eye in the chicken grows much more rapidly than in the monkey (6). Finally, there is no indication that neuroprogenitor cells are capable of responding with proliferation to mechanical solicitation, such as is the case for the cells of the connective tissue. It is more likely that either the neural networks trigger retinal cell proliferation in response to the abnormal visual input, as recently shown in the adult hippocampus (30), or that unrestricted vision slows down retinal proliferation. An interesting possibility is that the distorted visual input causes an increased release of VIP by a class of amacrine cells and that this peptide, in turn, stimulates the proliferation of the progenitors at the retinal periphery. On the other hand, we cannot exclude the possibility that growth may result from signals from the pigment epithelium, choroid, or sclera that feed back onto the retina, as indicated by experiments in cold-blooded vertebrates (31).
Our findings also suggest that a population of proliferating retinal neuroprogenitors may persist in the peripheral retina of juvenile humans and be responsible for the growth of the retina from birth to the end of adolescence. In individuals susceptible to becoming nearsighted, this population could increase, as is the case for monkeys’ lid-sutured eyes. If this hypothesis is confirmed, it will have significant clinical applications both in attempts at repairing a damaged retina in children and for prevention of myopia.
Materials and Methods
Lid fusion was carried out as described in ref. 2. Ultrasound measurements were obtained by using a Digital Biometric Ruler DBR-300 (Sonometrics Systems, New York) with a 10 MHz ultrasound tranducer and a resolution of 0.0375 mm.
Subtracted cDNA Libraries, Microarray Analysis, and Real-Time PCR.
Details of the procedure for harvesting the retina are presented in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. cDNA subtractions, construction of cDNA libraries, and differential screening were carried out as described in ref. 7. For microarray hybridization with Cy3- and Cy5-labeled cDNA probes, we used the protocol described at http://cmgm.stanford.edu/pbrown/protocols. After hybridization, slides were scanned by using a GenePix 4000B microarray scanner (Axon Instruments, Union City, CA). The images were acquired and processed by using the genepix pro 5.1 software package (Axon Instruments). Analysis of the microarray data and determination of the level of CCNA2 and HDGF mRNA expression by multiplex real-time PCR are presented in Supporting Materials and Methods.
Immunocytochemistry and Cell Counts.
Details of the procedures for immunocytochemistry and cell counts are presented in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank D.E. Redmond and the staff of the St. Kitts Biomedical Research Foundation for invaluable help; R. S. Molday (University of British Columbia, Vancouver) for the rhodopsin antibody; Y. Zhang and C. M. Craft (University of Southern California, Los Angeles) for the arrestin antibody; and P. Kara for help with the statistical analysis. This work was supported by grants from the Ilfeld Macular Degeneration Fund and the Edward R. and Anne G. Lefler Center at Harvard Medical School. A.V.T. was the recipient of a Dana–Mahoney Neuroscience Fellowship Award.
Abbreviations
- HDGF
hepatoma-derived growth factor
- OD
right (closed) eye
- OS
left (open) eye
- VIP
vasoactive intestinal polypeptide.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The nucleotide sequences reported in this paper have been deposited in the GenBank database (accession nos. AY680431–AY680585), and the microarray data were deposited in Gene Expression Omnibus, www.ncbi.nlm.nih.gov/geo (accession no. GSE3300).
References
- 1.Curtin B. J. The Myopias: Basic Science and Clinical Management. Philadelphia: Harper & Row; 1985. [Google Scholar]
- 2.Wiesel T. N., Raviola E. Nature. 1977;266:66–68. doi: 10.1038/266066a0. [DOI] [PubMed] [Google Scholar]
- 3.Raviola E., Wiesel T. N. N. Engl. J. Med. 1985;312:1609–1615. doi: 10.1056/NEJM198506203122505. [DOI] [PubMed] [Google Scholar]
- 4.Wallman J., Turkel J., Trachtman J. Science. 1978;201:1249–1251. doi: 10.1126/science.694514. [DOI] [PubMed] [Google Scholar]
- 5.Sherman S. M., Norton T. T., Casagrande V. A. Brain Res. 1977;124:154–157. doi: 10.1016/0006-8993(77)90872-1. [DOI] [PubMed] [Google Scholar]
- 6.Wallman J., Winawer J. Neuron. 2004;43:447–468. doi: 10.1016/j.neuron.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Tkatchenko A. V., Le Cam G., Leger J. J., Dechesne C. A. Biochim. Biophys. Acta. 2000;1500:17–30. doi: 10.1016/s0925-4439(99)00084-8. [DOI] [PubMed] [Google Scholar]
- 8.Nicholls R. D. Curr. Opin. Genet. Dev. 1993;3:445–456. doi: 10.1016/0959-437x(93)90119-a. [DOI] [PubMed] [Google Scholar]
- 9.Girard F., Strausfeld U., Fernandez A., Lamb N. J. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 10.Pagano M., Pepperkok R., Verde F., Ansorge W., Draetta G. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pines J., Hunter T. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 12.Kishima Y., Yamamoto H., Izumoto Y., Yoshida K., Enomoto H., Yamamoto M., Kuroda T., Ito H., Yoshizaki K., Nakamura H. J. Biol. Chem. 2002;277:10315–10322. doi: 10.1074/jbc.M111122200. [DOI] [PubMed] [Google Scholar]
- 13.Klagsbrun M., Sasse J., Sullivan R., Smith J. A. Proc. Natl. Acad. Sci. USA. 1986;83:2448–2452. doi: 10.1073/pnas.83.8.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepourcelet M., Tou L., Cai L., Sawada J.-I., Lazar A. J. F., Glickman J. N., Williamson J. A., Everett A. D., Redston M., Fox E. A., et al. Development (Cambridge, U.K.) 2005;132:415–427. doi: 10.1242/dev.01579. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Z., Yamamoto Y., Sugai F., Yoshida K., Kishima Y., Sumi H., Nakamura H., Sakoda S. J. Biol. Chem. 2004;279:27320–27326. doi: 10.1074/jbc.M308650200. [DOI] [PubMed] [Google Scholar]
- 16.Whitfield M. L., Sherlock G., Saldanha A. J., Murray J. I., Ball C. A., Alexander K. E., Matese J. C., Perou C. M., Hurt M. M., Brown P. O., Botstein D. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lendahl U., Zimmerman L. B., McKay R. D. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 18.Stone R. A., Laties A. M., Raviola E., Wiesel T. N. Proc. Natl. Acad. Sci. USA. 1988;85:257–260. doi: 10.1073/pnas.85.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moody T. W., Hill J. M., Jensen R. T. Peptides. 2003;24:163–177. doi: 10.1016/s0196-9781(02)00290-5. [DOI] [PubMed] [Google Scholar]
- 20.Fox R., Sinatra R. B., Mooney M. A., Feurer I. D., Butler M. G. J. Pediatr. Ophthalmol. Strabismus. 1999;36:331–336. doi: 10.3928/0191-3913-19991101-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reh T. A., Levine E. M. J. Neurobiol. 1998;36:206–220. [PubMed] [Google Scholar]
- 22.Fischer A. J., Reh T. A. Dev. Biol. 2000;220:197–210. doi: 10.1006/dbio.2000.9640. [DOI] [PubMed] [Google Scholar]
- 23.Tropepe V., Coles B. L. K., Chiasson B. J., Horsford D. J., Elia A. J., McInnes R. R., van der Kooy D. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- 24.Coles B. L. K., Angènieux B., Inoue T., Del Rio-Tsonis K., Spence J. R., McInnes R. R., Arsenijevic Y., van der Kooy D. Proc. Natl. Acad. Sci. USA. 2004;101:15772–15777. doi: 10.1073/pnas.0401596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubota R., Hokoc J. N., Moshiri A., McGuire C., Reh T. A. Brain. Res. Dev. Brain Res. 2002;134:31–41. doi: 10.1016/s0165-3806(01)00287-5. [DOI] [PubMed] [Google Scholar]
- 26.Young R. W. Brain Res. 1985;353:229–239. doi: 10.1016/0165-3806(85)90211-1. [DOI] [PubMed] [Google Scholar]
- 27.Teakle E. M., Wildsoet C. F., Vaney D. I. Vision Res. 1993;33:2383–2396. doi: 10.1016/0042-6989(93)90117-f. [DOI] [PubMed] [Google Scholar]
- 28.Guggenheim J. A., McBrien N. A. Invest. Ophthalmol. Vis. Sci. 1996;37:1380–1395. [PubMed] [Google Scholar]
- 29.Rada J. A., Nickla D. L., Troilo D. Invest. Ophthalmol. Vis. Sci. 2000;41:2050–2058. [PubMed] [Google Scholar]
- 30.Deisseroth K., Singla S., Toda H., Monje M., Palmer T. D., Malenka R. C. Neuron. 2004;42:535–552. doi: 10.1016/s0896-6273(04)00266-1. [DOI] [PubMed] [Google Scholar]
- 31.Mitsuda S., Yoshii C., Ikegami Y., Araki M. Develop. Biol. 2005;280:122–132. doi: 10.1016/j.ydbio.2005.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.