Abstract
Nearly all mammalian species like sweet-tasting foods and drinks, but there are differences in the degree of ‘sweet tooth’ both between species and among individuals of the same species. Some individual differences can be explained by genetic variability. Polymorphisms in a sweet taste receptor (Tas1r3) account for a large fraction of the differences in consumption of sweet solutions among inbred mouse strains. We wondered whether mice and rats share the same Tas1r3 alleles, and whether this gene might explain the large difference in saccharin preference among rats. We conducted three experiments to test this. We examined DNA sequence differences in the Tas1r3 gene among rats that differed in their consumption of saccharin in two-bottle choice tests. The animals tested were from an outbred strain (Sprague–Dawley; experiment 1), selectively bred to be high- or low-saccharin consumers (HiS and LoS; experiment 2), or from inbred strains with established differences in saccharin preference (FH/Wjd and ACI; experiment 3). Although there was considerable variation in saccharin preference among the rats there was no variation in the protein-coding regions of theTas1r3 gene. DNA variants in intronic regions were detected in 1 (of 12) outbred rat with lower-than-average saccharin preference and in the ACI inbred strain, which also has a lower saccharin preference than the FH/Wjd inbred partner strain. Possible effects of these intronic nucleotide variants on Tas1r3 gene expression or the presence of T1R3 protein in taste papillae were evaluated in the ACI and FH/Wjd strains. Based upon the results of these studies, we conclude that polymorphisms in the protein-coding regions of the sweet receptor gene Tas1r3 are uncommon and do not account for individual differences in saccharin preference for these strains of rats. DNA variants in intron 4 and 5 are more common but appear to be innocuous.
Introduction
Most animals prefer sugars that humans perceive as sweet, and avoid substances that humans perceive as bitter. The acceptance of sweet tastes and the rejection of bitter tastes is present at birth in humans and other primates (Steiner, 1979; Steiner et al., 2001). This observation suggests that taste preferences are innate. Differences exist in the degree to which individual humans (and members of other species) detect sweet or bitter substances (Blakeslee and Salmon, 1935), and an early hypothesis suggested that variability in sweet preferences was due to inherited differences in the ‘sensory apparatus’ (Nachman, 1959). This hypothesis has recently received support. Three members of a family of genes were discovered (Tas1r1-3), and in vivo and in vitro experiments have suggested that two members of this family (Tas1r2 and Tas1r3) dimerize and form a sweet receptor (Nelson et al., 2001; Li et al., 2002). Alleles of the Tas1r3 gene account for much of the difference among inbred mouse strains in saccharin intake and preference (Bachmanov et al., 2001a,b; Kitagawa et al., 2001; Max et al., 2001; Montmayeur et al., 2001; Nelson et al., 2001; Sainz et al., 2001; Inoue et al., 2004; Reed et al., 2004).
The observation that genetic variation in a sweet receptor gene influences sweet preference in mice has a parallel in humans for bitter perception. In most human populations tested, 10–30% of people do not perceive the bitterness of a class of chemical compounds (sometimes called nontasters), whereas other people find them extremely bitter (tasters; Mourant et al., 1976; Guo and Reed, 2001). Because both tasters and nontasters are also found within some closely related non-human primate species, investigators speculated that the alleles that produce nontasters might be shared between humans and some primates (Fisher, 1939). This idea, that specific alleles of taste genes might be preserved across species, led us to wonder whether the allelic variation found in the mouse sweet taste receptor gene Tas1r3 might also be present in a related species (rats) and explain the large variation among individuals and rat strains in saccharin preference and intake (Nachman, 1959; Vartiainen, 1967; Overstreet et al., 1993; Dess and Minor, 1996; Giza et al., 1996). To test this hypothesis, we conducted three experiments. In all experiments, we examined DNA sequence differences in the Tas1r3 gene among rats that differed in their intake and preference for saccharin. The studies used rats with diverse genetic histories: in experiment 1, the subjects were genetically heterogeneous, outbred Sprague–Dawley rats, in experiment 2, they were partially inbred strains that have been selectively bred for high or low saccharin intake (Dess and Minor, 1996), and in experiment 3, they were inbred strains that were known to differ in saccharin intake (Overstreet et al., 1999). When DNA sequence differences were found, they were evaluated in several ways to determine whether the polymorphism might influence gene function and saccharin preference. First, the DNA variants in the rat were aligned against the mouse sequence to determine whether mice with high or low saccharin preferences were allelic in the same locations as the rat. Second, in silico methods were used to determine whether the differences in DNA sequence resulted in a predicted change in amino acid, a prematurely truncated protein or would interfere with intron–exon splicing. Finally, because DNA sequence variation outside protein coding regions might influence the amount or distribution of Tas1r3 mRNA, tissues from four regions of the rat tongue (fungiform, foliate, circumvallate papillae and non-taste epithelium) were compared among rats with Tas1r3 DNA sequence variation to assess their influence on gene expression and the presence of T1R3 protein.
Materials and methods
Obtaining the rat genomic sequence for Tas1r3
When this study began, neither the rat cDNA nor the genomic sequence for Tas1r3 was known and therefore had to be determined. One genomic DNA sample from the Sprague–Dawley strain (Clontech, Palo Alto, CA) was purchased and served as the reference sequence. Primers were designed based upon the mouse Tas1r3 sequence (Table 1) and amplified using the following conditions: 94°C, 2 min denaturation; 94°C 45 s, 56–66°C 45 s, 72°C 2 min for 35 cycles; 72°C 7 min extension. All DNA sequencing was done by the University of Pennsylvania Sequencing Core Facility. The obtained sequences were aligned with the mouse and human genomic DNA, as well as the rat cDNA, and the predicted amino acid sequence was compared with the mouse (Accession #AF389853) and human (Accession #AC026283; Figure 1). During the project, the rat cDNA sequence became available (Accession #AY032620). This allowed us to check against the sequence we obtained (a match) and to compare the intron and exon structure with the new genomic DNA sequence we obtained.
Table 1.
Primers used to amplify genomic fragments of rat Tas1r3
| Name | Forward | Reverse | Annealing temperature (°C) |
|---|---|---|---|
| RSac1 | GGGTAGAAGCCAGTGCTAGCTAAGCG | CTGTCCCAATACCTATACATGCCCAG | 60 |
| RSac4 | CCCAGTGACCGGGTGCAGCT | GGCACCTGGACATGGAGCCT | 66 |
| RSac5 | AGGATCCTGAGGAGCTTGAG | TGCAACAGTGTCACAACGGC | 60 |
| RSac7 | CCTGCACACACCTTCTATTG | 56 | |
| RSac12 | TAGGGCTGGGTTCACTAAAG | 56 |
Figure 1.

Alignment of predicted mouse (strain C57BL/6ByJ), rat (strains Sprague–Dawley, HiS, LoS, ACI and FH/Wjd strains), and human VTIR3 protein. The amino acid sequence was aligned with ClustalW and plotted with BOXSHADE. Black boxes indicate regions of identity, and gray boxes indicate conservative amino acid substitutions. In mice, higher or lower saccharin preferences are associated with a polymorphism at aa position 60 (isoleucine to threonine).
Animals and saccharin intake
In experiment 1, 12 male CD™IGS (Sprague–Dawley) rats ~40 days old were purchased from Charles River (Wilmington, MA) and acclimated to laboratory conditions for 4 weeks. The daily water and saccharin intakes were measured using a two-bottle choice test, with water as one choice and 3.7 mM sodium saccharin (Sigma Chemical Corp., St Louis, MO) as the second choice. This test was conducted for four consecutive days. The position of the bottles was reversed daily to control for the preference of some rats for solutions on a particular side. Total intakes were averaged for each solution and expressed as the number of milliliters consumed per day. A preference score was computed by dividing the amount of saccharin consumed per day by the total fluid consumed per day. After this measurement was complete, small pieces of tail were obtained from each rat for DNA extraction. Experiment 1 was conducted in the laboratory of Michael G. Tordoff, at the Monell Chemical Senses Center.
In experiment 2, a second group of rats was tested from strains selectively bred to be either high-saccharin (HiS) or low-saccharin (LoS) drinkers (Dess and Minor, 1996). Both the selective breeding and the behavioral testing were conducted in the laboratory of Nancy K. Dess at Occidental College. The selective breeding project was initiated upon the incidental discovery of a male rat that was averse to saccharin solution. This male and an avid saccharin-drinking male were each mated with different pairs of females with average saccharin preference to create the Occidental Low-Saccharin (LoS) and High-Saccharin (HiS) lines. Rats in each litter with the most extreme phenotypes were selected for breeding, excluding rats with extreme baseline water intake. No matings occurred between rats that were quarter-sibs or closer in genetic relatedness, and ~15% of the rats bred every four or five generations are founders from the original stock (formerly Holtzman, now Harlan Sprague–Dawley Holtzman). The rats used in the present study were from five litters (two HiS, three LoS) in generations 19 and 20. They were phenotyped with a 2 day water-only baseline period followed by a 24 h two-bottle choice between water and 4.5 mM saccharin solution. Preference scores were computed by dividing the amount of saccharin consumed by the total amount of fluid consumed. After the behavioral testing was completed, the tail of every subject was clipped for DNA extraction.
In experiment 3, FH/Wjd and ACI rats were bred at the Monell Chemical Senses Center from groups of founder rats provided by Drs Amir Rezvani and David Overstreet at the Center for Alcohol Studies, University of North Carolina–Chapel Hill. (The FH/Wjd inbred strain is sometimes referred to as the Fawn Hooded strain.) Eight male and female rats from the inbred FH/Wjd strain and eight male and eight female rats from the ACI strain were offered ascending concentrations of saccharin to drink, with each solution offered for 2 days. Group means were compared by ANOVA, using sex, strain and concentration as factors. The 3.2 mM saccharin solution corresponds most closely to the concentrations used in experiments 1 and 2. Genomic DNA from an FH/Wjd and an ACI rat were obtained for DNA sequencing. Because the FH/Wjd and ACI strains are inbred, all rats should be genetically identical, and therefore only one DNA sample per strain was needed to detect genetic differences within the Tas1r3 gene. Experiment 3 was conducted in the laboratory of Michael G. Tordoff at the Monell Chemical Senses Center.
Note that due to procedural differences across laboratories, slightly different concentrations were used to test the rats’ preference for saccharin in experiments 1–3 (3.7, 4.5 and 3.2 mM respectively). These concentrations were chosen because they were clearly detectable; they are classically considered to fall close to but slightly below the saccharin concentration most preferred by rats (Young and Schulte, 1963). Although there is some evidence that sweetner intake of mice is under differential genetic control at threshold versus suprathreshold concentrations (Bachmanov et al., 1997), this is unlikely to be a factor for the saccharin concentrations used here.
Sequencing of rat genomic DNA samples
Genomic DNA was extracted from tails (Gentra Systems, Minneapolis, MN), and primers were designed from the mouse sequence and then modified, based on amplified rat sequence, to overlap the gene: primer sets were selected to amplify large fragments of rat Tas1r3 (RSac5F and RSac5R, 1100 bp; RSac4F and RSac4R, 1142 bp; RSac1F and RSac1R, 1773 bp; RSac7F and RSac12R, 3236 bp), and then nested primers were used to close sequencing gaps (Table 1). To reduce the likelihood that errors in the fidelity of Taq polymerase could account for sequence differences among strains, we used a Taq polymerase from Pyrococcus furiosus, which has the lowest error rate of known thermophilic DNA polymerases. Polymerase chain reaction (PCR) products were purified (Qiagen, Valencia, CA) and sequenced. Strands were aligned using Sequencher (Genecodes, Ann Arbor, MI). Nucleotide bases were compared and polymorphisms identified. When polymorphisms were found, they were confirmed by additional PCR reactions followed by sequencing.
In silico analysis of DNA sequence variants
Using the reference intron–exon structure of Tas1r3, alterations of DNA sequence variants were assessed for predicted protein-coding changes, stop codons or splice junction variants (Discovery Gene Studio version 1.5, San Diego CA). Next the polymorphic site and its flanking region from each rat strain was aligned with the homologous region from the Tas1r3 gene from strains of mice chosen for higher and lower saccharin intake (Reed et al., 2004). The rationale for this procedure is twofold. First, the degree of DNA sequence conserved across species may be commensurate with its degree of biological importance. Second, if identical polymorphisms were found in mice with higher and lower saccharin preference and these polymorphisms segregated with saccharin intake, it would suggest that the rat polymorphisms might also have an influence on saccharin intake. Mouse sequences from the Tas1r3 gene were obtained by PCR amplification of genomic templates from six inbred strains, three with higher and three with lower saccharin preferences. Both forward and reverse strands were sequenced and aligned with Sequencher (GeneCodes, Ann Arbor, MI), and rat and mouse sequences were aligned with ClustalW (Chenna et al., 2003), and plotted with BOX-SHADE (version 3.21) (Figure 4). Percent identity among the aligned sequences was computed using the algorithm bl2seq (Tatusova and Madden, 1999). These mouse Tas1r3 DNA sequences have been published previously (Reed et al., 2004).
Figure 4.
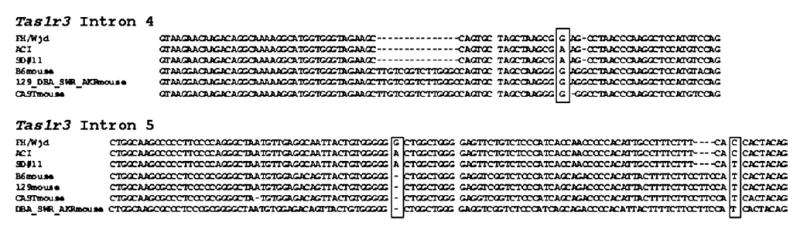
Alignment of the Tas1r3 sequence (introns 4 and 5) from the FH/Wjd and ACI inbred strains, as well as from SD rat 11, and six inbred strains of mice with high (C57BL/6J, SWR/J, CAST/Ei) and low (129P3/J, DBA/2J, AKR/J) saccharin preference. All other rats from experiments 1 and 2 had the same DNA sequence as the rats from the FH/Wjd strain. For the mice, inbred mouse strains with identical DNA sequence are shown as a single sequence. The abbreviations for the mouse strains are as follows, C57BL/6J (B6), SWR/J (SWR), CAST/Ei (CAST), 129P3/J (129), DBA/2J (DBA) and AKR/J (AKR). The rat polymorphic sites (+1873 nt, intron 4; +2130 and +2164, intron 5), are shown in the rectangular boxes.
Real-time PCR
It is possible that intronic sequence variants might lead to markedly reduced gene expression that in turn might result in fewer receptors and an altered taste preference. To determine whether Tas1r3 gene expression differed among inbred strains of rats with different genotypes, we compared the relative abundance of Tas1r3 gene expression in fungiform, foliate, circumvallate and non-taste epithelium [corrected for gustducin gene expression in one analysis and for a ubiquitously expressed control gene expression (B2m) in a second analysis]. Lingual tissues used for real-time PCR and immuncytochemistry (below) were dissected as previously described (Huang et al., 1999). The original mouse protocol was modified for rats by raising the volume of proteinase enzyme injected underneath the lingual epithelium (6 μl), and incubating the tongue for 60 min. Total RNA was extracted from the lingual tissues of four rats (two adult male rats from each inbred strain) using the Stratagene Absolutely RNA™ Microprep kit (La Jolla, CA). We could not study SD rat 11 (which also had intronic DNA sequence variants) because the tissue from this rat was not available. First strand synthesis of cDNA was conducted following the directions of the manufacturer (Invitrogen, SuperScript™ III). Transcribed cDNA (1 μl/well) was transferred to a PCR 96-well optical reaction plate. Each well was supplemented with predeveloped TaqMan assay reagents (primer and fluorescent probe, for location of probe, see Figure 3) and Universal PCR Master Mix™ to a final volume of 50 μl. The PCR product was heated to 50°C for 2 min and 95°C for 10 min and then 40 amplification cycles were conducted at 95°C for 15 s and 60°C for 60 s. DNA amplification was performed in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems), detecting fluorescence (500–560 nm) after laser light excitation. PCR cycle threshold (CT) values represent the time at which the emitted fluorescence increased above threshold. The Tas1r3 primers spanned the intron between exons 4 and 5 (Accession # NM_130818, assay ID # Rn_00590759; Figure 3). Tas1r3 gene expression was normalized as previously described (Livak and Schmittgen, 2001) to a gene that is selectively expressed in taste receptor cells (gustducin, Gnat3; Accession # NM_173139) or an ubiquitously expressed control gene (B2m; Accession # NM_012512) relative gene expression was calculated using non-taste tongue epithelium was used as a calibrator tissue. Few animals were available for study and statistical analysis of the strain differences were therefore not possible.
Figure 3.
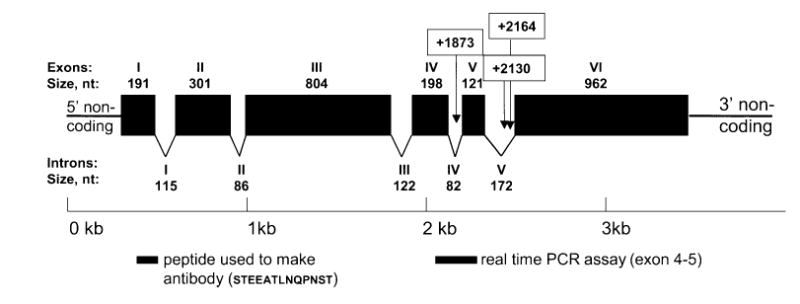
Intron–exon structure of the rat Tas1r3 gene. Above the gene structure, exons are labeled in Roman numerals, with their size (in nucleotides) shown underneath. Below the gene structure, introns are labeled in Roman numerals, and their size is shown underneath. The nucleotide positions are numbered such that the first nucleotide of the putative methionine initiation codon is +1. Polymorphisms are denoted by the changed nucleotide position and are shown in open boxes. Intron and exon structure was determined by aligning cDNA with genomic DNA. Filled boxes underneath the gene structure show the locations of the real time PCR probe and the peptide used to produce the antibody.
Immunocytochemistry
Polyclonal antisera against a hemocyanin-conjugated T1R3 peptide (aa 45–62) were raised in rabbits (see Figure 3 for the location of aa sequence). Frozen sections of rat lingual tissue (previously fixed in 4% paraformaldehyde and cryoprotected in 20% sucrose) were blocked in 3% BSA, 0.3% Triton X-100, 2% goat serum and 0.1% sodium azide in PBS for 1 h at room temperature and then incubated for 16 h at 4°C with antisera against T1R3 (1:800). The secondary antibodies were Cy3-conjugated goat-anti-rabbit Ig for T1R3. T1R3 immunoreactivity was blocked by preincubation of the antisera with the corresponding synthetic peptide at 10 μM. Preimmune serum did not show any immunoreactivity.
Results
Construction of the rat sequence of Tas1r3
We obtained 3912 bp of DNA sequence that composed and flanked the rat Tas1r3 gene. The rat gene is highly homologous at the amino acid level to the mouse and human gene (Figure 1). Similar to the mouse gene, the rat gene has six exons, interrupted by small introns. We found several polymorphisms, as described below, but none within the protein coding regions. Specifically, the I60T polymorphism in the mouse protein coding region of the Tas1r3 gene associated with saccharin preference in mice was the ‘taster’ form of the allele (i.e. isoleucine at aa position 60) in all rats studied.
Experiment 1
Spraque-Dawley rats
On average, rats drank 5.1 ± 6.3 ml (mean ± SD) of water and 66.7 ± 26.8 ml of 3.7 mM saccharin per day (Table 2). With one exception (rat 6), all rats drank more saccharin solution than water (i.e. percent preference scores were >50%). Some rats drank prodigious amounts of saccharin, and one rat (rat 1) consumed, on average, almost half of its body weight in saccharin solution daily. Eleven of the 12 Sprague–Dawley rats had a sequence of the Tas1r3 gene that was the same as the reference sequence (Table 2). Three heterozygous sequence variants were detected within the Tas1r3 gene for rat 11 (Table 5). This rat drank ~20 ml less than the group average but consumed >90% of its fluid as saccharin.
Table 2.
Experiment 1, saccharin intake of outbred Sprague-Dawley rats
| Rat no. | Body weight (g) | Water intake (ml) | 3.7 mM saccharin intake (ml) | Preference(%) | Genotype |
|---|---|---|---|---|---|
| SD 6 | 258 | 21 | 16 | 43 | reference |
| SD 8 | 229 | 16 | 74 | 83 | reference |
| SD 11 | 236 | 4 | 43 | 91 | three variantsa |
| SD 10 | 231 | 5 | 68 | 94 | reference |
| SD 12 | 292 | 3 | 81 | 96 | reference |
| SD 2 | 213 | 2 | 57 | 97 | reference |
| SD 3 | 236 | 2 | 66 | 97 | reference |
| SD 4 | 266 | 2 | 53 | 97 | reference |
| SD 7 | 210 | 2 | 58 | 97 | reference |
| SD 5 | 243 | 2 | 97 | 98 | reference |
| SD 9 | 234 | 1 | 81 | 98 | reference |
| SD 1 | 251 | 1 | 122 | 99 | reference |
Table 5.
Description of polymorphisms within the rat Tas1r3 gene
| Intron
|
||||
|---|---|---|---|---|
| Experiment | Rat | 4 | 5 | 5 |
| +1873 nt | +2130 nt | +2164 nt | ||
| 1 | SD #11 | G/G to G/A | G/G to G/A | C/C to C/T |
| 3 | ACI | G/G to A/A | G/G to A/A | reference (C/C) |
Nucleotides are numbered with A in the putative methionine start codon as +1.
Experiment 2
Rats selectively bred to differ in saccharin intake
Rats of the LoS strain had low intakes of saccharin and drank between 58 and 63 ml/day, whereas rats of the HiS strain drank between 212 and 243 ml/day. All rats showed a preference for saccharin, but the LoS preference was lower compared with the HiS strain (Mann–Whitney U = 0.50, P = 0.026; Table 3). However, all rats, bred from either the LoS or the HiS line, had the same Tas1r3 genotypes as the reference sequence (Table 3).
Table 3.
Experiment 2, rats selectively bred for high (HiS) and low (LoS) saccharin intake
| Rat no. and group | Body weight (g) | Water intake (ml) | 4.5 mM saccharin intake (ml) | Preference (%) | Genotype |
|---|---|---|---|---|---|
| 5 LoS | 319 | 6 | 58 | 91 | reference |
| 6 LoS | 288 | 8 | 60 | 88 | reference |
| 1 LoS | 523 | 13 | 63 | 83 | reference |
| 4 LoS | 450 | 1 | 71 | 99 | reference |
| Mean ± SD | 395 ± 111 | 7 ± 5 | 63 ± 6 | 90 ± 7 | |
| 8 HiS | 277 | 1 | 212 | 100 | reference |
| 2 HiS | 485 | 1 | 230 | 100 | reference |
| 3 HiS | 529 | 1 | 234 | 100 | reference |
| 7 HiS | 283 | 2 | 243 | 99 | reference |
| Mean ± SD | 394 ± 132 | 1 ± 0.4 | 230 ± 13 | 100 ± 0 |
Water intake and saccharin intake are given as the total amount consumed during a 2 day test.
Experiment 3
ACI
FH/Wjd rats differ in sequence of the Tas1r3 gene. Saccharin intake was significantly higher in the FH/Wjd than in the ACI rats, F(1,30) = 41.56, P < 0.000001 (Fig. 2), which confirms earlier observations that the FH/Wjd strain is a saccharin-preferring inbred strain. The FH/Wjd strain had the reference sequence and the ACI strain had two sequence variants (Tables 4 and 5; Figure 3): a substitution of G to A at nt 1873 was found in intron 4, and a G to A substitution at nt 2130 was found in intron 5. Neither of these polymorphisms was within the splice junction and would not therefore be predicted to interfere with the creation of the correct exon order and length (Figure 3).
Figure 2.
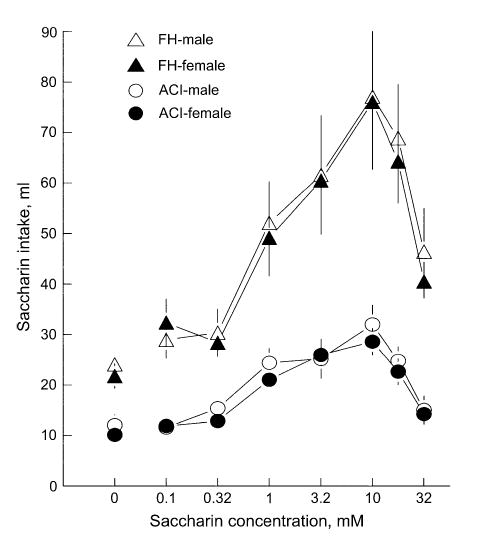
Mean (±SEM) saccharin intakes of male and female FH/Wjd and ACI rats. Intakes are daily averages of 48 h tests. Water intake for all groups is shown as 0 mM saccharin. Water intake during tests with saccharin available was minimal (not shown).
Table 4.
Experiment 3, 3.2 mM saccharin intake of two inbred rat strains
| Strain | Water intake (ml) | Saccharin intake (ml) | Preference (%) | Genotype |
|---|---|---|---|---|
| ACI | 3.6 | 25 ± 4 | 83 ± 9 | two variants* |
| FH/Wjd | 4.8 | 62 ± 12 | 90 ± 5 | reference |
Comparison of the intronic rat DNA sequence variants with DNA sequences from inbred mouse strains with high and low saccharin preference
The DNA sequence flanking the rat intronic variants was aligned with homologous sequence of mice selected for high and low saccharin intake (Bachmanov et al., 2001a,b; Reed et al., 2004) to determine whether the nucleotides were conserved and whether a pattern of polymorphism was shared between rats and mice with similar saccharin preference (Figure 4). The homology between rat and mouse sequence from intron 4 and 5 was high (>80%), and was higher for intron 4 (75/82 identity; 91%) than for intron 5 (140/172; 81%). The intron 4 rat polymorphism (G to A at position +1873) was a G in all mice regardless of saccharin preference. For the two polymorphisms in intron 5, the first polymorphism (G to A at +2130) was not conserved between mouse and rats. This nucleotide was deleted in all mouse strains studied. The second polymorphism in intron 5 was a C at position +2164 for both the FH/Wjd and ACI rat strains, whereas the SD rat 11 and the six mouse strains shared a T at this position. Of the three Tas1r3 polymorphisms identified in the rats, none shared a common pattern of polymorphism with mice strains selected for high and low saccharin preference.
Tas1r3 gene expression
Gene expression in the foliate papillae was more abundant relative to the fungiform or circumvallate papillae, and was somewhat higher in the FH/Wjd strain compared with the ACI strain (Figure 5). There was little or no Tas1r3 gene expression in non-taste epithelium. The pattern of gene expression in tongue tissue is consistent with previous reports of the expression pattern of Tas1r2 (Hoon et al., 1999), a gene that co-localizes with Tas1r3 in taste receptor cells (Li et al., 2002). Similar patterns of Tas1r3 expression were found whether the abundance was expressed relative to gustducin, a taster-receptor cell-specific gene (Huang et al., 1999), or B2m, a ubiquitously expressed control gene.
Figure 5.
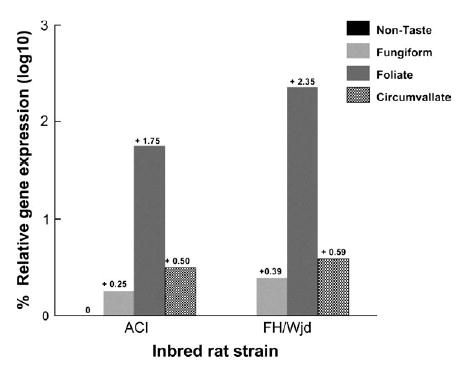
Expression of the Tas1r3 gene in the fungiform, foliate, circumvallate papillae calibrated to expression in non-taste epithelium of the rat tongue from inbred strains with higher (FH/Wjd) or lower (ACI) saccharin intakes. Relative Tas1r3 gene expression, either expressed as a proportion of control gene expression (B2m, as shown) or gustducin (data not shown) was similar between strains, although the expression in foliate papillae was somewhat higher in the FH/Wjd than the ACI strain.
T1R3 immunochemistry
The results of immunocytochemistry with an anti-T1R3 antibody demonstrated that (i) a subset of taste receptor cells from all types of taste papillae contained T1R3 immunoreactivity and (ii) there was little or no T1R3 immunoreactivity in non-taste lingual epithelium (Figure 6).
Figure 6.
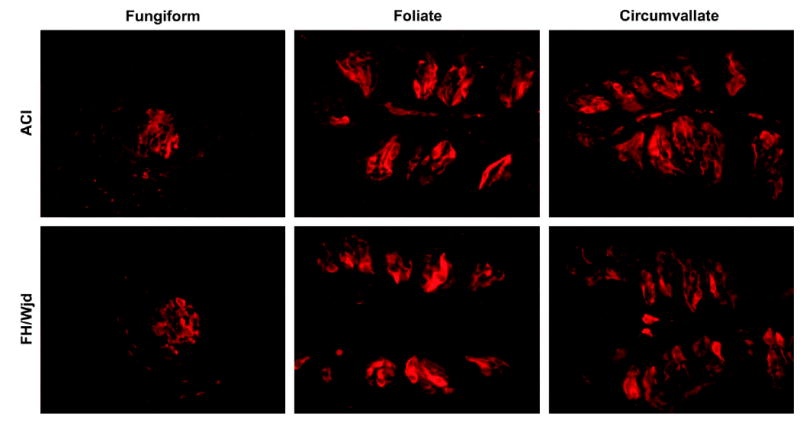
T1R3 immunoreactivity in taste tissue of FH/Wjd and ACI rats. Longitudinal sections from fungiform, foliate, circumvallate and papillae were immunostained with rabbit antiserum directed against a synthetic peptide from the N-terminal domain of T1R3, with a Cy3-conjugated anti-rabbit secondary antibody. No differences in expression levels of the receptor in all sections examined from the two rat strains were apparent. See Figure 3 for location of amino acid sequence used to generate the antibody. 1Magnification: 250×.
Discussion
Polymorphisms within the Tas1r3 protein-coding regions make no contribution to saccharin preference in the rats tested. We tested the hypothesis that polymorphisms within the protein-coding regions of the sweet taste receptor gene Tas1r3 account for individual differences in saccharin intake among rats and, further, that rats with low saccharin preference might have the same alleles that predict amino acid changes in Tas1r3 as mice with low saccharin preference (Reed et al., 2004). We found no protein-coding sequence variants among the rats tested, and no DNA sequence variation that matched alleles in mice with lower saccharin preference. While sequence variants were found in the introns of the rat Tas1r3 gene, there was little correspondence between these polymorphisms and behavior. The FH/Wjd rats may have somewhat higher Tas1r3 gene expression levels compared to the ACI strain. Both the FH/Wjd and ACI strains have T1R3 protein in the appropriate lingual tissues as measured by immunocytochemistry. Thus, our results do not support the hypothesis that Tas1r3 polymorphisms underlie the range of saccharin preferences observed in rats, although there is always the possibility that rare Tas1r3 polymorphisms may exist in the SD strain (or other outbred strains) or that inbred rat strains not tested here might have alleles in the protein-coding regions of the Tas1r3 gene.
The Tas1r3 gene codes for a receptor that is thought to work by binding sugars and other sweeteners in taste receptor cells, and it regulates the intensity of the sensory signal sent to the brain (Bachmanov et al., 1997). This observation suggests that the effect of different alleles of this gene on behavior is limited to the perception of compounds that bind to the receptor such as sucrose and saccharin (Nelson et al., 2001). However, rats selected and bred for high or low saccharin intake have many behavioral differences (e.g. brain self-stimulation, ethanol consumption) in addition to their sweetener preference (Ganchrow et al., 1981; Gosnell and Krahn, 1992; Overstreet et al., 1993; Dess and Minor, 1996; Dess et al., 1998; Kampov-Polevoy et al., 1999; Over-street et al., 1999; Dess, 2000). Therefore, genes involved in central sensory and hedonic processing, such as those underlying brain reward pathways, rather than in peripheral sensitivity, may account for differences in saccharin preference among rats.
Alternatively, or additionally, allelic differences in other genes contributing to the taste pathway may be involved. In addition to Tas1r3, two members of the same receptor gene family (Tas1r1 and Tas1r2) also transduce taste information. Tas1r1 and Tas1r2 were discovered several years before Tas1r3, and originally Tas1r1 was thought to be the Sac gene (Hoon et al., 1999). Genetic mapping studies disconfirmed this hypothesis (Li et al., 2001), and later studies have suggested that T1R1 combines with T1R3 to transduce information about amino acid rather than sweet taste (Nelson et al., 2001; Li et al., 2002). Therefore, alleles in the Tas1r1 gene are unlikely to account for differences in saccharin intake. The Tas1r2 gene is a better candidate gene because its protein product is thought to combine with T1R3 to become a sweet taste receptor. However, although alleles in the Tas1r2 gene might be involved in individual differences in sweet taste perception in some strains of rats, the search for rat quantitative trait loci for saccharin preference using genome scan methods have not implicated the region containing the Tas1r2 gene (Murphy et al., 2002). Alleles in bitter receptors or in cell signaling might also influence saccharin intake because these might enhance or diminish the bitter taste of saccharin. Finally, individual differences in central integration of taste information may influence saccharin intake, for instance, rats that prefer water instead of saccharin may perceive saccharin as bitter and salty (Giza et al., 1996).
Polymorphisms in the flanking regulatory sequence may contribute to the difference in saccharin preference. Intronic DNA variants were associated with reduced saccharin intake of the ACI compared with the FH/Wjd strain, and it is possible that these variants have functional consequences on gene expression levels or alternative splicing. The immunocytochemistry and measures of gene expression presented here suggest that there are no large differences between the ACI and FH/Wjd strains in Tas1r3 gene expression or T1R3 protein abundance in the taste tissues tested. However, the somewhat higher expression of the Tas1r3 gene in the foliate papillae of the FH/Wjd strain compared with the ACI strain may be worthy of further study. Because the probes used to measure gene expression span exons 4 and 5, it is unlikely that intronic variants in the rats studied cause a disruption in the correct intron–exon splice for these nearby exons. Because the promoter and enhancer of Tas1r3 are not characterized, we did not look for polymorphisms in the flanking regulatory sequence. The possibility exists that sequence variants among these regions contribute to the difference in saccharin preference among the rats tested.
In addition to the sequencing of candidate genes, two other approaches are worth further consideration to understand genetic variation and sweetness perception in the rat. One approach would be to survey inbred and outbred rat strains for individual or strain differences in response to other sweeteners such as sucralose (Sclafani and Clare, 2004) or fructose (De Francisco and Dess, 1998) and to assess heritability for these traits. Second, when and if high heritability is found, genome scans (a method whereby all genomic regions are assayed for the presence of genes that contribute to behavioral variation) could be a next step in identifying genes involved in sweet taste perception. Several rat genome scans have been conducted using saccharin preference as a trait but the investigators were primarily interested in ethanol and therefore the rats strains selected for study differed in ethanol as well as saccharin preference (Foroud et al., 2002; Terenina-Rigaldie et al., 2003). None of the linkage peaks were near the Tas1r3 gene. Along with a candidate gene method as reported here, these other approaches are likely to lead to a better understanding of taste and genetics, which in turn may increase our understanding of the mammalian sweet tooth and ultimately have implications for human health.
Acknowledgments
This work was supported by National Institutes of Health grants R01DC04188 and R01DK55853 (D.R.R.), R01DC00882 (G.K.B.), R01AA011028 (A.A.B.), R03 DC05154 (L.H.) and R01AA11028 (M.G.T). Dr David Overstreet kindly provided the ACI and FH/Wjd rats used in the molecular experiments. The T1R3 antibody was developed in collaboration with Dr Kazumi Taniguchi (Kitasato University). The technical assistance of Diane Pilchak, Lynn Vo and Patricia J. Watson are gratefully acknowledged. Stuart McCaughey provided valuable comments on an earlier draft of this manuscript.
References
- Bachmanov AA, Li X, Reed DR, Ohmen JD, Li S, Chen Z, Tordoff MG, de Jong PJ, Wu C, West DB, Chatterjee A, Ross DA, Beauchamp GK. Positional cloning of the mouse saccharin preference (Sac) locus. Chem Senses. 2001;26:925–933. doi: 10.1093/chemse/26.7.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov AA, Reed DR, Ninomiya Y, Inoue M, Tordoff MG, Price RA, Beauchamp GK. Sucrose consumption in mice: major influence of two genetic loci affecting peripheral sensory responses. Mamm Genome. 1997;8:545–548. doi: 10.1007/s003359900500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov AA, Tordoff MG, Beauchamp GK. Sweetener preference of C57BL/6ByJ and 129P3/J mice. Chem Senses. 2001;26:905–913. doi: 10.1093/chemse/26.7.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee AF, Salmon TN. Genetics of sensory thresholds: individual taste reactions for different substances. Proc Natl Acad Sci U S A. 1935;21:84–90. doi: 10.1073/pnas.21.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Francisco JC, Dess NK. Aspartame consumption in rats selectively bred for high versus low saccharin intake. Physiol Behav. 1998;65:393–396. doi: 10.1016/s0031-9384(98)00215-7. [DOI] [PubMed] [Google Scholar]
- Dess NK. Responses to basic taste qualities in rats selectively bred for high versus low saccharin intake. Physiol Behav. 2000;69:247–257. doi: 10.1016/s0031-9384(99)00246-2. [DOI] [PubMed] [Google Scholar]
- Dess NK, Badia-Elder NE, Thiele TE, Kiefer SW, Blizard DA. Ethanol consumption in rats selectively bred for differential saccharin intake. Alcohol. 1998;16:275–8. doi: 10.1016/s0741-8329(98)00010-x. [DOI] [PubMed] [Google Scholar]
- Dess NK, Minor TR. Taste and emotionality in rats selectively bred for high versus low saccharin intake. Animal Learning & Behavior. 1996;24:105–115. [Google Scholar]
- Fisher R. Taste-testing the Anthropoid apes. Nature. 1939;144:750. [Google Scholar]
- Foroud T, Bice P, Castelluccio P, Bo R, Ritchotte A, Stewart R, Lumeng L, Li TK, Carr L. Mapping of QTL influencing saccharin consumption in the selectively bred alcohol-preferring and -nonpreferring rat lines. Behav Genet. 2002;32:57–67. doi: 10.1023/a:1014459912935. [DOI] [PubMed] [Google Scholar]
- Ganchrow JR, Lieblich I, Cohen E. Consummatory responses to taste stimuli in rats selected for high and low rates of self-stimulation. Physiol Behav. 1981;27:971–6. doi: 10.1016/0031-9384(81)90356-5. [DOI] [PubMed] [Google Scholar]
- Giza BK, McCaughey SA, Zhang L, Scott TR. Taste responses in the nucleus of the solitary tract in saccharin-preferring and saccharin-averse rats. Chem Senses. 1996;21:147–157. doi: 10.1093/chemse/21.2.147. [DOI] [PubMed] [Google Scholar]
- Gosnell BA, Krahn DD. The relationship between saccharin and alcohol intake in rats. Alcohol. 1992;9:203–206. doi: 10.1016/0741-8329(92)90054-e. [DOI] [PubMed] [Google Scholar]
- Guo SW, Reed DR. The genetics of phenylthiocarbamide perception. Ann Hum Biol. 2001;28:111–142. doi: 10.1080/03014460151056310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJ, Zuker CS. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- Huang L, Shanker YG, Dubauskaite J, Zheng JZ, Yan W, Rosenzweig S, Spielman AI, Max M, Margolskee RF. Ggamma13 colocalizes with gustducin in taste receptor cells and mediates IP3 responses to bitter denatonium. Nat Neurosci. 1999;2:1055–62. doi: 10.1038/15981. [DOI] [PubMed] [Google Scholar]
- Inoue M, Reed DR, Li X, Tordoff MG, Beauchamp GK, Bachmanov AA. Allelic variation of the Tas1r3 taste receptor gene selectively affects behavioral and neural taste responses to sweeteners in the F2 hybrids between C57BL/6ByJ and 129P3/J mice. J Neurosci. 2004;24:2296–303. doi: 10.1523/JNEUROSCI.4439-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampov-Polevoy AB, Garbutt JC, Janowsky DS. Association between preference for sweets and excessive alcohol intake: a review of animal and human studies. Alcohol Alcohol. 1999;34:386–395. doi: 10.1093/alcalc/34.3.386. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Kusakabe Y, Miura H, Ninomiya Y, Hino A. Molecular genetic identification of a candidate receptor gene for sweet taste. Biochem Biophys Res Commun. 2001;283:236–242. doi: 10.1006/bbrc.2001.4760. [DOI] [PubMed] [Google Scholar]
- Li X, Inoue M, Reed DR, Huque T, Puchalski RB, Tordoff MG, Ninomiya Y, Beauchamp GK, Bachmanov AA. High-resolution genetic mapping of the saccharin preference locus (Sac) and the putative sweet taste receptor (T1R1) gene (Gpr70) to mouse distal Chromosome 4. Mamm Genome. 2001;12:13–16. doi: 10.1007/s003350010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proceedings of the National Academy of Sciences. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Max M, Shanker YG, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee RF. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- Montmayeur JP, Liberles SD, Matsunami H, Buck LB. A candidate taste receptor gene near a sweet taste locus. Nat Neurosci. 2001;4:492–498. doi: 10.1038/87440. [DOI] [PubMed] [Google Scholar]
- Mourant, A.E., Kopec, A.C. and Domaniewska-Sobczak, K. (1976) The distribution of human blood groups and other polymorphisms London, Oxford University Press.
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, Lumeng L, Li TK. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32:363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Nachman M. The inheritance of saccharin preference. Journal of Comp Physiol Psychol. 1959;52:451–457. doi: 10.1037/h0048853. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Overstreet DH, Kampov-Polevoy AB, Rezvani AH, Murrelle L, Halikas JA, Janowsky DS. Saccharin intake predicts ethanol intake in genetically heterogeneous rats as well as different rat strains. Alcohol Clin Exp Res. 1993;17:366–369. doi: 10.1111/j.1530-0277.1993.tb00777.x. [DOI] [PubMed] [Google Scholar]
- Overstreet DH, Rezvani AH, Parsian A. Behavioural features of alcohol-preferring rats: focus on inbred strains. Alcohol Alcohol. 1999;34:378–385. doi: 10.1093/alcalc/34.3.378. [DOI] [PubMed] [Google Scholar]
- Reed DR, Li S, Li X, Huang L, Tordoff MG, Starling-Roney R, Taniguchi K, West DB, Ohmen JD, Beauchamp GK, Bachmanov AA. Polymorphisms in the taste receptor gene (Tas1r3) region are associated with saccharin preference in 30 mouse strains. J Neurosci. 2004;24:938–946. doi: 10.1523/JNEUROSCI.1374-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz E, Korley JN, Battey JF, Sullivan SL. Identification of a novel member of the T1R family of putative taste receptors. J Neurochem. 2001;77:896–903. doi: 10.1046/j.1471-4159.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- Sclafani A, Clare RA. Female Rats show a Bimodal Preference Response to the Artificial Sweetener Sucralose. Chem Senses. 2004;29:523–528. doi: 10.1093/chemse/bjh055. [DOI] [PubMed] [Google Scholar]
- Steiner, J.E. (1979) Human facial expressions in response to taste and smell stimulation Adv Child Dev Behav Reese, H and Lipsitt, L New York, Academic Press. 13: 257–295. [DOI] [PubMed]
- Steiner JE, Glaser D, Hawilo ME, Berridge KC. Comparative expression of hedonic impact: affective reactions to taste by human infants and other primates. Neurosci Biobehav Rev. 2001;25:53–74. doi: 10.1016/s0149-7634(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Terenina-Rigaldie E, Jones BC, Mormede P. Pleiotropic effect of a locus on chromosome 4 influencing alcohol drinking and emotional reactivity in rats. Genes Brain Behav. 2003;2:125–131. doi: 10.1034/j.1601-183x.2003.00018.x. [DOI] [PubMed] [Google Scholar]
- Vartiainen I. The inheritance of craving for sugar in rats. Ann Med Int Fenn. 1967;56:155–171. [PubMed] [Google Scholar]
- Young PT, Schulte RH. Isohedonic contours and tongue activity in three gustatory areas of the rat. J Comp Physiol Psychol. 1963;56:465–475. doi: 10.1037/h0045768. [DOI] [PubMed] [Google Scholar]


