Abstract
Molecular chaperones play crucial roles in various aspects of the biogenesis and maintenance of proteins in the cell. The heat shock protein 70 (HSP70) chaperone system, in which HSP70 proteins act as chaperones, is one of the major molecular chaperone systems conserved among a variety of organisms. To shed light on the evolutionary history of the constituents of the chordate HSP70 chaperone system and to identify all of the components of the HSP70 chaperone system in ascidians, we carried out a comprehensive survey for HSP70s and their cochaperones in the genome of Ciona intestinalis. We characterized all members of the Ciona HSP70 superfamily, J-proteins, BAG family, and some other types of cochaperones. The Ciona genome contains 8 members of the HSP70 superfamily, all of which have human and protostome counterparts. Members of the STCH subfamily of the HSP70 family and members of the HSPA14 subfamily of the HSP110 family are conserved between humans and protostomes but were not found in Ciona. The Ciona genome encodes 36 J-proteins, 32 of which belong to groups conserved in humans and protostomes. Three proteins seem to be unique to Ciona. J-proteins of the RBJ group are conserved between humans and Ciona but were not found in protostomes, whereas J-proteins of the DNAJC14, ZCSL3, FLJ13236, and C21orf55 groups are conserved between humans and protostomes but were not found in Ciona. J-proteins of the sacsin group seem to be specific to vertebrates. There is also a J-like protein without a conserved HPD tripeptide motif in the Ciona genome. The Ciona genome encodes 3 types of BAG family proteins, all of which have human and protostome counterparts (BAG1, BAG3, and BAT3). BAG2 group is conserved between humans and protostomes but was not found in Ciona, and BAG4 and BAG5 groups seem to be specific to vertebrates. Members for SIL1, UBQLN, UBADC1, TIMM44, GRPEL, and Magmas groups, which are conserved between humans and protostomes, were also found in Ciona. No Ciona member was retrieved for HSPBP1 group, which is conserved between humans and protostomes. For several groups of the HSP70 superfamily, J-proteins, and other types of cochaperones, multiple members in humans are represented by a single counterpart in Ciona. These results show that genes of the HSP70 chaperone system can be distinguished into groups that are shared by vertebrates, Ciona, and protostomes, ones shared by vertebrates and protostomes, ones shared by vertebrates and Ciona, and ones specific to vertebrates, Ciona, or protostomes. These results also demonstrate that the components of the HSP70 chaperone system in Ciona are similar to but simpler than those in humans and suggest that changes of the genome in the lineage leading to humans after the separation from that leading to Ciona increased the number and diversity of members of the HSP70 chaperone system. Changes of the genome in the lineage leading to Ciona also seem to have made the HSP70 chaperone system in this species slightly simpler than that in the common ancestor of humans and Ciona.
INTRODUCTION
Molecular chaperones play crucial roles in various aspects of the biogenesis and maintenance of proteins in the cell (Frydman 2001; Soti et al 2005). They are responsible for the folding of nascent proteins in the cytosol, translocation of proteins into organelles, assembly and disassembly of protein complexes, control of the biological activity of regulatory proteins, renaturation of denatured proteins after stress, and degradation of misfolded or harmful proteins (Hartl and Hayer-Hartl 2002; Craig et al 2003). The heat shock protein 70 (HSP70) chaperone system, in which HSP70 family proteins act as chaperones, is one of the major molecular chaperone systems conserved among a variety of organisms (Bukau and Horwich 1998). In the process of the folding of nascent proteins, for example, HSP70s bind to and release hydrophobic regions of substrate proteins in an ATP-dependent manner. HSP70s in the adenosine 5′-diphosphate (ADP)-bound state bind to the substrate proteins stably, whereas HSP70s in the adenosine triphosphate (ATP)-bound state release them rapidly. The folding of the substrate proteins is thought to be attained through the repetition of this cycle (Fan et al 2003; Mayer and Bukau 2005).
The activity of HSP70s is regulated by many types of cochaperones (Caplan 2003; Mayer and Bukau 2005). For example, members of J-domain–containing protein (J-protein) family stimulate ATP hydrolysis by HSP70s (Wall et al 1994; Tsai and Douglas 1996). BAG-1, a member of the BAG domain–containing protein family (BAG family), acts as a nucleotide exchange factor for HSP70s and accelerates ADP release from HSP70s (Alberti et al 2003). The actions of these cochaperones also affect the substrate specificity of HSP70s and link the HSP70 chaperone system with other chaperone systems and/or other biological systems in the cell, such as the protein degradation pathway (Caplan 2003).
Generally, there are multiple HSP70s and J-proteins in the genomes of eukaryotes, and the number is larger in higher than in lower eukaryotes. For example, the yeast genome contains 14 and 22 genes encoding HSP70s and J-proteins, respectively, whereas the human genome contains 21 and 50 genes, respectively. Although the evolution of HSP70s has been studied extensively (Gupta and Golding 1993; Boorstein et al 1994; Rensing and Maier 1994; Feder and Krebs 1998; Karlin and Brocchieri 1998; Easton et al 2000; Bettencourt and Feder 2001; Nikolaidis and Nei 2004), the details of the changes in the repertoire of HSP70s during the early phases of chordate evolution are unknown. Furthermore, little is known about the evolution of the J-protein family in chordates.
Because the molecular chaperones were originally found as stress-inducible factors, much attention was initially given to their roles in stress responses of the cell. Subsequently, their roles under normal conditions, such as the quality control of proteins in the cell, were investigated intensively. More recently, several studies have focused on the roles of the molecular chaperones in animal development. With regard to the HSP70 chaperone system, for example, it has been reported that some HSP70s are required for spermatogenesis in mouse embryos (Christians et al 2003). DnaJB6, one of the J-proteins, is required for placenta formation in mouse embryos (Hunter et al 1999). lethal(2) tumorous imaginal discs is a J-protein-encoding gene required for the differentiation of imaginal disc cells in Drosophila (Kurzik-Dumke et al 1995). The results of these studies suggest that the HSP70 chaperone system is involved in various aspects of animal development, the details of which remain to be clarified.
Ascidians are primitive chordates that belong to the subphylum Urochordata of the phylum Chordata. The genome and a total of more than 680 000 expressed sequence tags (ESTs) of Ciona intestinalis have been sequenced and it has been shown that the 159-Mb genome contains 15 852 genes that comprise a basic set of chordate-type genes with fewer gene duplications than are observed for vertebrate genes (Dehal et al 2002; Satou et al 2002a, 2002b, 2005). Through rapid and well-described embryogenesis, ascidians form tadpole-like larvae, which are much simpler than but similar to vertebrate embryos. These features make C intestinalis one of the excellent model animals to explore the evolutionary trails of vertebrate genes and to investigate the roles of genes in development (Corbo et al 2001; Satoh, 2003; Satoh et al 2003).
In this study, we carried out a comprehensive survey for HSP70s and their cochaperones in the genome of C intestinalis with the following 2 aims. One was to shed light on the evolutionary history of the HSP70 chaperone system in chordates by comparing the repertoires of components of the HSP70 chaperone system in C intestinalis with those in other animals. The other aim was to identify all of the components of the HSP70 chaperone system in C intestinalis to set up a starting point for studies on the roles of the HSP70 chaperone system in ascidian development. We characterized all of the members of HSP70 superfamily, J-proteins, BAG family, and some other types of cochaperones (SIL1, UBQLN, UBADC1, TIMM44, GRPEL, and Magmas) in this ascidian species.
MATERIALS AND METHODS
Searching for C intestinalis genes
Search for C intestinalis genes was carried out as described in Satou et al (2003). The genome database (http://genome.jgi-psf.org/ciona4/) and cDNA/EST database (http://ghost.zool.kyoto-u.ac.jp/indexr1.html) of C intestinalis were screened with all members of HSP70 superfamily, J-protein family, BAG family, SIL1, HSPBP1, UBQLN, UBADC1, TIMM44, GRPEL1/2, and Magmas groups in humans and Drosophila as queries using the TBLASTN program with default parameters. Sequences of candidate genes were selected from cDNA sequences (eg, citb001h23), sequences of predicted gene models in the assembled genome sequences (eg, ci0100123456) or EST sequences (eg, cilv003o45EST), giving priority to cDNA sequences. When the predicted gene model seemed to lack part(s) of the gene but the EST(s) covered the region that the gene model lacked, assembled sequence of the gene model and the EST(s) was used for analyses (eg, ciad067d89EST_ci0100123456). All Ciona sequences used for analyses are listed in supplementary Table S1.1 The sequences from other organisms used for analyses are designated by the accession number registered in public databases, abbreviation of the species (HS for human, MM for mouse, CI for C intestinalis, DM for Drosophila melanogaster, AG for Anopheles gambiae, CE for Caenorhabditis elegans, SC for Saccharomyces cerevisiae, and SP for Schizosacchromyces pombe), and gene name. For example, human DNAJA2 (accession number NP_005871) is represented as “NP_005871_HS_DNAJA2.” Domains and motifs in the deduced proteins were searched with the SMART (http://smart.embl-heidelberg.de/) and Pfam (http://www.sanger.ac.uk/Software/Pfam/search.shtml), and were summarized in supplementary Tables S2–S5.
Molecular phylogenetic analysis
Molecular phylogenetic analysis was carried out as described in Satou et al (2003). The sequences were aligned using the ClustalX program with default parameters (Thompson et al 1997). As for the tree in supplementary Figure S3, the J-domain sequences were used for the alignment. As for other trees, the full-length sequences were used. The problematic regions that would disturb the alignment (eg, long flanking sequence that is not conserved among most members of a given group) were removed from the sequences used for the alignment before the alignment was performed. The alignment was refined and all gaps were removed using the BioEdit program (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The phylogenetic trees were constructed based on the neighbor-joining method with 1000 bootstrap replicates using MEGA2 (Saitou and Nei 1987; Kumar et al 2001).
Best-hit analysis
In addition to molecular phylogenetic analysis, the orthology of Ciona and human proteins was tested by a method that we call best-hit analysis, as described in Satou et al (2003). The given Ciona proteins were compared with the human proteome by the BLASTP program using the NCBI web site. The best-hit human protein obtained by this comparison was then used for the TBLASTN search against the Ciona genome. In the case that the best-hit sequence of the TBLASTN search coincided with the genomic region encoding the initial Ciona protein, the relationship between the 2 proteins suggests their orthology and was called the “bidirectional best-hit relationship.” In the other cases, the relationship between the 2 proteins was called the “unidirectional best-hit relationship.”
RESULTS
HSP70 superfamily
The HSP70 superfamily consists of the HSP70 family, which contains prototypical HSP70 proteins (Gupta and Golding 1993; Boorstein et al 1994; Rensing and Maier 1994; Karlin and Brocchieri 1998), and the HSP110 family, which is composed of larger, distantly related Hsp110 and homologous proteins (Easton et al 2000). Most members of the HSP70 family in eukaryotes belong to the DnaK subfamily, whereas the remaining, divergent members belong to the STCH subfamily. Members of the DnaK subfamily are classified further into distinct clusters that correspond to their intracellular localization (the cytoplasm, endoplasmic reticulum [ER], mitochondria, and chloroplasts) (Boorstein et al 1994; Lin et al 2001). Similarly to the HSP70 family, the HSP110 family consists of cytoplasmic members and ER-resident members (Easton et al 2000; Nikolaidis and Nei 2004). Cytoplasmic members of the HSP110 family are subdivided into the HSP110/SSE subfamily and HSPA14 subfamily. The ER-resident group of the HSP110 family is called the GRP170 subfamily. Human HSPA12A and HSPA12B form a group (HSPA12 family) that belongs to the HSP70 superfamily but is distinct from the HSP70 family and HSP110 family (Han et al 2003).
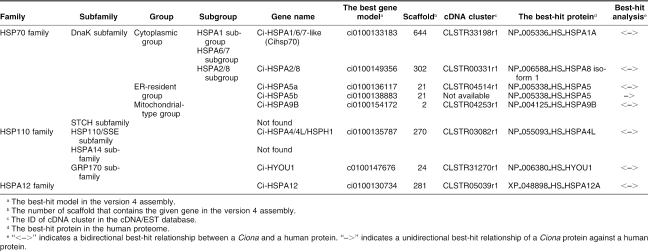
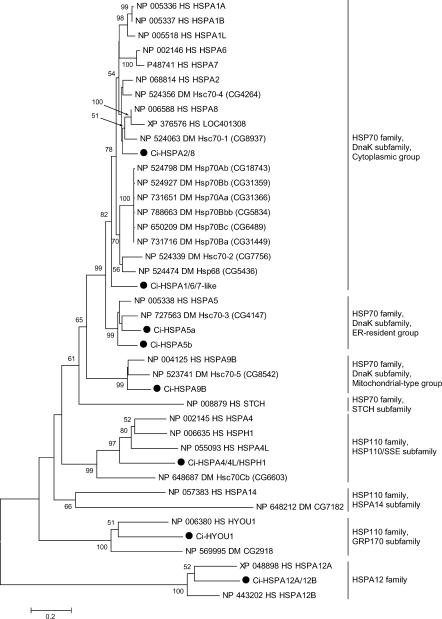
In the public databases, 21 human genes encoding HSP70 superfamily–related proteins are registered (HSPA1A, HSPA1B, HSPA1L, HSPA2, HSPA4, HSPA4L, HSPA5–8, HSPA9B, HSPA12A, HSPA12B, HSPA14, HSPH1, HYOU1, STCH, LOC401308, LOC442509, LOC402461 and LOC440490). LOC442509, LOC402461, and LOC440490 were excluded from the present analysis, because proteins encoded by them are short and correspond to a limited part of typical HSP70 superfamily proteins. The Drosophila genome contains 10 genes for HSP70 superfamily proteins. In the present survey, we identified 8 Ciona genes for the HSP70 superfamily proteins (Table 1; supplementary Table S2). The phylogenetic analysis suggests that 6 clades (the cytoplasmic group of the DnaK subfamily of the HSP70 family, ER-resident group of the DnaK subfamily of the HSP70 family, mitochondrial-type group of the DnaK subfamily of the HSP70 family, HSP110/SSE subfamily of the HSP110 family, GRP170 subfamily of the HSP110 family, and the HSPA12 family) contain both human and Ciona members (Fig 1). Every clade except for the HSPA12 family also contains Drosophila members. Notably, the cytoplasmic group of the DnaK subfamily contains only 2 Ciona proteins, whereas 10 Drosophila and 8 human proteins belong to this group (Fig 1). The cytoplasmic group of the DnaK subfamily consists of HSPA2/8 subgroup, HSPA6/7 subgroup, HSPA1 subgroup, and Drosophila-specific cytoplasmic HSP70 subgroup (supplementary Fig S1). One of the 2 Ciona proteins was assigned into HSPA2/8 subgroup and the other was located outside of the 4 clades and thus the gene for this protein was named Ci-HSPA1/6/7–like (Table 1; supplementary Fig S1). As for the STCH subfamily of the HSP70 family and HSPA14 subfamily of the HSP110 family, no candidate gene was found by the present survey, although each subfamily contains both vertebrate and protostome members.
Table 1.
Genes for HSP70 superfamily proteins in the C intestinalis genome
Fig 1.
Phylogenetic tree of HSP70 superfamily proteins constructed based on the full-length sequences of HSP70 superfamily proteins of humans, C intestinalis, and D melanogaster. Ciona proteins are indicated by large black dots. The number at each branch indicates the percentage of times that a node was supported in 1000 bootstrap pseudoreplications. Percentages <49% are omitted for simplicity. Proteins are named as explained in Materials and Methods. The unrooted tree is shown as a rooted tree for simplicity. The scale bar indicates an evolutionary distance of 0.2 amino acid substitutions per position. Bars on the right indicate gene groups. As for members of cytoplasmic group of DnaK subfamily, another tree was constructed to know the relationship among them more precisely (supplementary Fig S1)
J-proteins
Members of the J-protein family are characterized by the presence of a conserved domain called the J-domain (Walsh et al 2004). The J-domain consists of approximately 70–75 amino acid residues that form 4 helices with the invariant tripeptide HPD between the second and third helices, and is essential for J-proteins to stimulate ATP hydrolysis by HSP70s (Wall et al 1994; Tsai and Douglas 1996).
A system for classification of the J-proteins according to their structure was proposed (Cheetham and Caplan 1998) and then partly modified (Ohtsuka and Hata 2000). In the proposed system, the J-proteins are categorized into 3 groups: type A (or type I) proteins have all of the J-domain, the glycine- and phenylalanine-rich region, and the DnaJ central domain; type B (or type II) proteins have the J-domain and the glycine- and phenylalanine-rich region but not the DnaJ central domain; and type C (or type III) proteins have the J-domain but lack the other 2 domains found in type A. Members of each type are numbered according to the chronological order of registration of the sequence data (Cheetham and Caplan 1998; Ohtsuka and Hata 2000).
Fifty human J-proteins are registered in the public databases. Among them, 5 (DNAJA1–DNAJA5), 11 (DNAJB1–DNAJB9, DNAJB11, and DNAJB12), and 16 (DNAJC1–DNAJC5, DNAJC5beta, DNAJC5gamma, and DNAJC6–DNAJC14) proteins have been categorized as type A, type B, and type C, respectively, and have been designated systematically. The other 17 proteins have not been classified into any of these 3 types and have been designated in a nonsystematic manner. Our present analysis suggested that the J-domain sequences of 4 of the J-proteins (LOC387820, FLJ14281, MGC29463, and TSARG6) show similarity to those of type B proteins; hence they were classified into type B. Another 14 proteins (GAK, DNAJD1, TIM14, SEC63, RBJ, HSC20, WBSCR18, sacsin, GNG10, ZCSL3, C21orf55, KIAA0962, FLJ13236, and FLJ10634) were regarded as type C proteins.
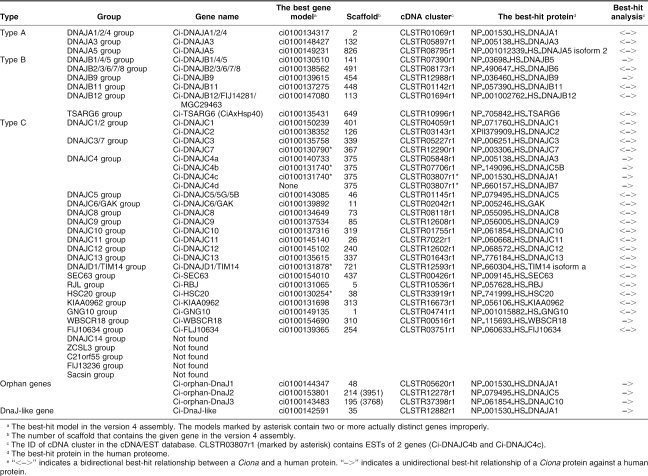
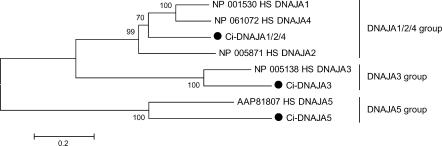
Very recently, identification of 15 J-protein–encoding genes in the C intestinalis genome was reported (Satouh et al 2005). In the present survey, we identified 36 Ciona genes for J-proteins, including all of the 15 genes reported in that paper. The phylogenetic analyses suggest that 36 Ciona and 50 human genes for J-proteins are classified into 33 groups along with 3 orphan genes specific to Ciona (see Table 2; supplementary Table S3; Fig 2; supplementary Figs S2–S4). Among the 33 groups, 28 groups contain both human and Ciona proteins, whereas the other 5 groups (DNAJC14, ZCSL3, C21orf55, FLJ13236, and sacsin groups) contain only human proteins. All of the 33 groups except for RBJ and sacsin groups contain protostome members (data not shown). Thus, the sacsin group seems to be vertebrate specific. In addition to 36 genes for true J-proteins, we identified a gene for a J-like protein that lacks the HPD tripeptide motif but showed similarity to type A J-proteins (Ci-DnaJ–like) (Table 2; Table S3; supplementary Fig S5).
Table 2.
Genes for J-proteins in the C intestinalis genome
Fig 2.
Phylogenetic tree of type A J-proteins constructed based on the full-length sequences of type A J-proteins of humans and C intestinalis. Ciona proteins are indicated by large black dots. The number at each branch indicates the percentage of times that a node was supported in 1000 bootstrap pseudoreplications. Proteins are named as explained in Materials and Methods. Bars on the right indicate gene groups. The unrooted tree is shown as a rooted tree for simplicity. The scale bar indicates an evolutionary distance of 0.2 amino acid substitutions per position
BAG family proteins
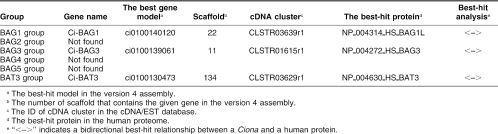
The BAG domain is a conserved domain that comprises about 50 amino acid residues and is shared by 6 human BAG family proteins: BAG1, BAG2, BAG3, BAG4, BAG5, and BAT3 (Takayama and Reed 2001; Doong et al 2002; Alberti et al 2003). The BAG domain of BAG1, BAG2, BAG3, and BAG4 has been shown to bind to HSP70s, and the BAG family proteins are thought to be involved in the regulation of HSP70s. In protostomes, BAG1, BAG2, BAG3, and BAT3 group proteins have been identified. The present survey identified 3 genes for BAG family proteins (Ci-BAG1, Ci-BAG3, and Ci-BAT3) in the genome of C intestinalis (Table 3; supplementary Table S4). The results of best-hit analyses and comparisons of protein sequences suggest that Ci-BAG1, Ci-BAG3, and Ci-BAT3 belong to BAG1, BAG3, and BAT3 groups, respectively (Table 3; supplementary Figs S6–S8). As for BAG2, BAG4, and BAG5 groups, no candidate gene was found by the present survey. Genes of BAG4 and BAG5 groups seem to be vertebrate specific.
Table 3.
Genes for BAG-family proteins in the C intestinalis genome
Other types of cochaperones
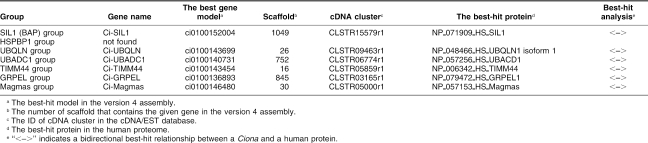
In addition to J-proteins and BAG family proteins, several proteins with different structural properties were shown to act as cochaperones of HSP70s. A significant fraction of such cochaperones is characterized by the presence of the TPR domain (Blatch and Lassle 1999). However, TPR domain–containing cochaperones for HSP70s in Ciona will be described in our upcoming paper that reports HSP90 family proteins and their cochaperones (Hamada et al, in preparation), because some of them also interact with HSP90s. Therefore, cochaperones for HSP70s without the TPR domain were subjected to the present analysis and are described in this section. We searched for genes of following groups in the Ciona genome: SIL1 (BAP) (Tyson and Stirling 2000; Chung et al 2002), HSPBP1 (Hsp70 binding protein 1) (Raynes and Guerriero 1998; Kabani et al 2002), UBQLN (Ubiquilin) (Kaye et al 2000), UBADC1 (Ubiquilin associated domain containing 1) (Li et al 2000), TIMM44, GRPEL, and Magmas (Koehler 2004; Wiedemann et al 2004). For all of these groups, there are both human and protostome members. The present survey identified single members in Ciona for each group except for HSPBP1 (Ci-SIL1, Ci-UBQLN, Ci-UBADC1, Ci-TIMM44, Ci-GRPEL, and Ci-Magmas) (see Table 4; supplementary Table S5; supplementary Figs S9 and S10). Ci-Magmas is identical to the recently reported magmas-like protein of C intestinalis (Peng et al 2005).
Table 4.
Genes for other types of cochaperones for HSP70s in the C intestinalis genome
DISCUSSION
Components of the HSP70 chaperone system are highly conserved but different among vertebrates, Ciona, and protostomes
In the present study, we searched for members of the HSP70 superfamily, J-proteins, BAG family, and other types of cochaperone (SIL1, HSPBP1, UBQLN, UBADC1, TIMM44, GRPEL, and Magmas) in C intestinalis. The results of the present study show that genes of the HSP70 chaperone system can be distinguished into groups that are shared by vertebrates, Ciona, and protostomes, ones shared by vertebrates and protostomes, ones shared by vertebrates and Ciona, and ones specific to vertebrates, Ciona, or protostomes.
Many of gene groups examined in the present study (42 groups of 54 groups) are shared by vertebrates, Ciona, and protostomes. This confirms that the components of the HSP70 chaperone system are highly conserved among metazoans and suggests that these gene groups are essential for function of the HSP70 chaperone system in metazoans.
Genes of 8 groups are found in both vertebrates and protostomes but not identified in Ciona (STCH, HSPA14, DNAJC14, ZCSL3, C21orf55, FLJ13236, BAG2, and HSPBP1 groups). Those genes may have been lost in the lineage leading to ascidians after the separation from that leading to vertebrates. Alternatively, because the published sequence of the Ciona genome was estimated to contain about 95% of genes that are present in the genome (Dehal et al 2002), those genes may be located in regions of the genome that have not been sequenced. However, if that is the case, those genes should be rarely transcribed or be unstable in cDNA libraries, because we searched for the cDNA database as well. Another possibility is that sequences of those genes have changed extensively during evolution of Ciona so that we are not able to recognize them.
Among gene groups examined in the present study, the RBJ group of J-protein family is the only group that is found in vertebrates and Ciona but not in protostomes. Thus, the RBJ group may be related to a deuterostome-specific or chordate-specific innovation. Proteins of the RBJ group belong to the Ras superfamily of GTP binding proteins and have a Ras-related GTPase domain in addition to the J-domain (Nepomuceno-Silva et al 2004). In vertebrates, RBJ group genes are predominantly expressed in the nervous system, and it has been suggested that they are involved in the development or maintenance of the sophisticated chordate nervous system (Nepomuceno-Silva et al 2004).
Three gene groups (sacsin group of the J-protein family and BAG4 and BAG5 groups of BAG family) were found in neither Ciona nor protostomes and seem to be specific to vertebrates. Although the function of BAG5 is undetermined, known functions of sacsin and BAG4 suggest that they may be related to vertebrate-specific innovations. Human sacsin has been identified as a gene responsible for a human genetic neurodegenerative disease called ARSACS (autosomal recessive spastic ataxial of Charlevoix-Saguenary), which is involved in a developmental defect in myelination of both retinal and peripheral nerve fibers (Engert et al 2000). Expression of sacsin mRNA is detected in the entire central nervous system of the developing and adult rat (Engert et al 2000). The fact that ARSACS is involved in abnormal myelination process is interesting, because myelin sheaths occur in gnathostomes but not in agnathans and invertebrates. It is possible that sacsin has emerged in gnathostome lineage during evolution in accord with the emergence of myelin sheaths.
BAG4 was identified as a protein that binds to the tumor necrosis factor receptor 1 (TNFR1) and regulates the activity of TNFR1 (Jiang et al 1999). BAG4 recognizes the death domain of TNFR1 (Jiang et al 1999). It has been suggested that vertebrate TNFR superfamily proteins have acquired the death domain during evolution (Bridgham et al 2003). In Drosophila, a single gene encoding a protein related to vertebrate TNFR superfamily proteins was identified, and this protein lacks the death domain (Kanda et al 2002; Kauppila et al 2003). In Ciona, 3 genes each encoding a protein related to vertebrate TNFR superfamily proteins were identified (Terajima et al 2003). Similar to the TNFR-related protein in Drosophila, all Ciona TNFR-related proteins lack the death domain. Therefore, it is likely that the emergence of both BAG4 and TNFR superfamily proteins with the death domain has occurred in the lineage leading to vertebrates after the separation from that leading to ascidians.
Interestingly, the contribution of the TNF signaling in apoptosis differs between mammals and Drosophila. In mammals, the TNF signaling activates 3 distinct pathways (FADD/caspase-8–mediated pathway, NF-κB–mediated pathway, and JNK-mediated pathway), whereas only the JNK-mediated pathway is downstream to the TNF signaling in Drosophila (Moreno et al 2002). It has been suggested that the presence of the death domain in mammalian TNFR superfamily proteins accounts for such difference (Bridgham et al 2003). Thus, the emergence of BAG4 may be related to the death domain–mediated functional diversification of the TNF signaling.
Putative evolutionary events that led to differences in the components of the HSP70 chaperone system between Ciona and humans
The results of the present study provide a light on the evolutionary process that resulted in similar but different HSP70 chaperone systems in Ciona and humans, as discussed below.
The number of HSP70 superfamily proteins in Ciona is about one-third of that in humans. This is due in part to the fact that multiple members for 3 of the HSP70 superfamily in humans are represented by a single protein in Ciona (ER-resident group of the DnaK subfamily of the HSP70 family, cytoplasmic HSP110 subfamily of the HSP110 family and HSPA12 family). Similarly, the number of proteins in the cytoplasmic group of the DnaK subfamily of the HSP70 family in Ciona is much smaller than that in humans (2 in Ciona vs 8 in humans). It seems unlikely that there is another member of this group in regions of the Ciona genome that have not been sequenced, because, as mentioned above, the published sequence was estimated to contain about 95% of genes that are present in the Ciona genome. Therefore, it is likely that the amplification of members of this group, which appears to have occurred in many lineages, including those leading to lineages of yeasts, Drosophila, C elegans, fish, and mammals, did not occur or was not retained in the Ciona lineage. It has been proposed that HSP70 genes are targets of natural selection and that the amplification of HSP70 genes in Drosophila has facilitated the evolution of thermotolerance and niche expansion of this species (Feder and Krebs 1998). In this respect, whether and how environmental factors have influenced the number of HSP70 genes in Ciona is an interesting issue for future studies.
There are 36 J-proteins in Ciona, whereas there are 50 in humans. Similar to the case of HSP70s, this difference is due in part to the fact that multiple members for 7 groups of J-proteins in humans are represented by a single protein in Ciona (DNAJA1/2/4, DNAJB1/4/5, DNAJB2/3/6/7/8, DNAJB12, DNAJC5, DNAJC6/GAK, and DNAJD1/TIM15 groups). As for the UBQLN and GRPEL groups, humans have multiple members but Ciona has a single one. These results also suggest that the amplification of genes and/or genomic regions in the lineage leading to humans after the separation from that leading to Ciona increased the number and diversity of members of the HSP70 chaperone system.
Gene amplification events seem to have occurred in 2 gene groups in Ciona as well (ER-resident group of DnaK subfamily of HSP70 family and DNAJC4 group of J-protein family). As for the ER-resident group of DnaK subfamily, the Ciona genome contains 2 genes (Ci-HSPA5a and Ci-HSPA5b), whereas the human genome contains 1 gene. Ci-HSPA5a and Ci-HSPA5b are located on the same scaffold, although the distance of the 2 genes is over 250 kb. Analysis of ESTs showed that Ci-HSPA5a is actively transcribed, whereas Ci-HSPA5b is not transcribed as far as it was examined by the cDNA project (data not shown). However, the predicted protein sequence of Ci-HSPA5b shows overall similarity to the typical members of the ER-resident group of DnaK subfamily, although it lacks the ER retention signal at the C terminus. Further analysis is required to distinguish whether Ci-HSPA5b is a pseudogene or not. Although there are number of HSP70 pseudogenes (ie, sequences that encode fragmentary HSP70 superfamily protein) in the human genome, no such sequence was found in the Ciona genome.
The Ciona genome contains 4 genes of the DNAJC4 group of J-protein family, whereas the human genome contains 1 gene of this group. Four DNAJC4 group genes in Ciona are aligned in tandem in the same scaffold, suggesting that they are products of local gene duplication events. Analysis of ESTs suggests that every gene is transcribed (data not shown). The significance of the amplification of DNAJC4 group genes in Ciona remains to be investigated.
There are 3 orphan genes encoding J-proteins and a gene encoding J-like protein in the Ciona genome. It is likely that these genes have emerged in the lineage leading to Ciona after the separation from that leading to humans, because there are no counterparts for these genes in protostomes as well. Similar to the case of gene groups that are conserved in vertebrates and protostomes but not in Ciona, there is alternative possibility that sequences of those genes have changed extensively in during evolution of both vertebrates and protostomes so that we are not able to recognize them.
In summary, the results of the present study suggest that the difference in the components of the HSP70 chaperone system between humans and Ciona has originated form the following events: the vertebrate-specific innovation of 1 group (sacsin group), the gene amplification in humans in 13 groups (cytoplasmic group of the DnaK subfamily, ER-resident group of the DnaK subfamily, cytoplasmic HSP110 subfamily of the HSP110 family, HSPA12 family, DNAJA1/2/4, DNAJB1/4/5, DNAJB2/ 3/6/7/8, DNAJB12, DNAJC5, DNAJC6/GAK, DNAJD1/ TIM15, UBQLN, and GRPEL groups), the gene loss in Ciona in 8 groups (STCH, HSPA14, DNAJC14, ZCSL3, C21orf55, FLJ13236, BAG2, and HSPBP1 groups), the gene amplification in Ciona in 2 groups (ER-resident group of DnaK subfamily of HSP70 family and DNAJC4 group of J-protein family), and the occurrence of 4 genes unique to Ciona (3 orphan genes encoding J-proteins and a gene encoding J-like protein). The difference in the variety of the components of the HSP70 chaperone system may be related to the difference in the ability of stress response between humans and Ciona. Studies on stress response in Ciona and function of Ciona genes identified in this study will be helpful to test this idea.
Acknowledgments
The authors thank Kazuko Hirayama, Shigeki Fujiwara, Nobuo Yamaguchi, and all of the members of the Maizuru Fisheries Research Station of Kyoto University; the Field Science Center, Graduate School of Agricultural Science, Tohoku University; the International Coastal Research Center, Ocean Research Institute, University of Tokyo; and Marine Biological Laboratory, Graduate School of Science, Hiroshima University, for collecting and culturing of C intestinalis. The present study was a project of the CREST program (Development, Differentiation, and Regeneration), JST, Japan.
Footnotes
Supplementary tables and figures available online at: http://dx.doi.org/10.1379/CSC-137R.s1.
REFERENCES
- Alberti S, Esser C, Hohfeld J. BAG-1—a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones. 2003;8:225–231. doi: 10.1379/1466-1268(2003)008<0225:bnefoh>2.0.co;2.1466-1268(2003)008[0225:BNEFOH]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt BR, Feder ME. Hsp70 duplication in the Drosophila melanogaster species group: how and when did two become five? Mol Biol Evol. 2001;18:1272–1282. doi: 10.1093/oxfordjournals.molbev.a003912.0737-4038(2001)018[1272:HDITDM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N.0265-9247(1999)021[0932:TTRASM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490.0022-2844(1994)038[0001:MEOTHM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Wilder JA, Hollocher H, Johnson AL. All in the family: evolutionary and functional relationships among death receptors. Cell Death Differ. 2003;10:19–25. doi: 10.1038/sj.cdd.4401174.1350-9047(2003)010[0019:AITFEA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9.0092-8674(1998)092[0351:THAHCM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Caplan AJ. What is a co-chaperone? Cell Stress Chaperones. 2003;8:105–107. doi: 10.1379/1466-1268(2003)008<0105:wiac>2.0.co;2.1466-1268(2003)008[0105:WIAC]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2.1466-1268(1998)003[0028:SFAEOD]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians ES, Zhou Q, Renard J, Benjamin IJ. Heat shock proteins in mammalian development. Semin Cell Dev Biol. 2003;14:283–290. doi: 10.1016/j.semcdb.2003.09.021.1084-9521(2003)014[0283:HSPIMD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Chung KT, Shen Y, Hendershot LM. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem. 2002;277:47557–47563. doi: 10.1074/jbc.M208377200.0021-9258(2002)277[47557:BAMBPI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Corbo JC, Di Gregorio A, Levine M. The ascidian as a model organism in developmental and evolutionary biology. Cell. 2001;106:535–538. doi: 10.1016/s0092-8674(01)00481-0.0092-8674(2001)106[0535:TAAAMO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Craig EA, Eisenman HC, Hundley HA. Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr Opin Microbiol. 2003;6:157–162. doi: 10.1016/s1369-5274(03)00030-4.1369-5274(2003)006[0157:RMCTFL]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, and Campbell RK. et al. 2002 The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 298:2157–2167. [DOI] [PubMed] [Google Scholar]
- Doong H, Vrailas A, Kohn EC. What's in the “BAG”?—a functional domain analysis of the BAG-family proteins. Cancer Lett. 2002;188:25–32. doi: 10.1016/s0304-3835(02)00456-1.0304-3835(2002)188[0025:WITBFD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2.1466-1268(2000)005[0276:THAGSP]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert JC, Berube P, and Mercier J. et al. 2000 ARSACS, a spastic ataxia common in northeastern Quebec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat Genet. 24:120–125. [DOI] [PubMed] [Google Scholar]
- Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2.1466-1268(2003)008[0309:MFROHF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder M, Krebs RA. Natural and genetic engineering of thermotolerance in Drosophila melanogaster. Am Zool. 1998;38:503–517.0003-1569(1998)038[0503:NAGEOT]2.0.CO;2 [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603.0066-4154(2001)070[0603:FONTPI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gupta RS, Golding GB. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743.0022-2844(1993)037[0573:EOHGAI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Han Z, Truong QA, Park S, Breslow JL. Two Hsp70 family members expressed in atherosclerotic lesions. Proc Natl Acad Sci U S A. 2003;100:1256–1261. doi: 10.1073/pnas.252764399.0027-8424(2003)100[1256:THFMEI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408.0193-4511(2002)295[1852:MCITCN]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hunter PJ, Swanson BJ, Haendel MA, Lyons GE, Cross JC. Mrj encodes a DnaJ-related co-chaperone that is essential for murine placental development. Development. 1999;126:1247–1258. doi: 10.1242/dev.126.6.1247.1011-6370(1999)126[1247:MEADCT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Woronicz JD, Liu W, Goeddel DV. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science. 1999;283:543–546. doi: 10.1126/science.283.5401.543.0193-4511(1999)283[0543:POCTRS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002;531:339–342. doi: 10.1016/s0014-5793(02)03570-6.0014-5793(2002)531[0339:HAHOTY]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kanda H, Igaki T, Kanuka H, Yagi T, Miura M. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J Biol Chem. 2002;277:28372–28375. doi: 10.1074/jbc.C200324200.0021-9258(2002)277[28372:WAMOTD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution. J Mol Evol. 1998;47:565–577. doi: 10.1007/pl00006413.0022-2844(1998)047[0565:HSPFMS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kauppila S, Maaty WSA, and Chen P. et al. 2003 Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene. 22:4860–4867. [DOI] [PubMed] [Google Scholar]
- Kaye FJ, Modi S, Ivanovska I, Koonin EV, Thress K, Kubo A, Kornbluth S, Rose MD. A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett. 2000;467:348–352. doi: 10.1016/s0014-5793(00)01135-2.0014-5793(2000)467[0348:AFOUPB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Koehler CM. New developments in mitochondrial assembly. Annu Rev Cell Dev Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057.1081-0706(2004)020[0309:NDIMA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244.1367-4803(2001)017[1244:MMEGAS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kurzik-Dumke U, Gundacker D, Renthrop M, Gateff E. Tumor suppression in Drosophila is causally related to the function of the lethal(2) tumorous imaginal discs gene, a dnaJ homolog. Dev Genet. 1995;16:64–76. doi: 10.1002/dvg.1020160110.0192-253X(1995)016[0064:TSIDIC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Li C, Rodriguez M, Adamson JW, Banerjee D. Identification of a glialblastoma cell differentiation factor-related gene mRNA in human microvascular endothelial cells. Genomics. 2000;65:243–252. doi: 10.1006/geno.2000.6176.0888-7543(2000)065[0243:IOAGCD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6:201–208. doi: 10.1379/1466-1268(2001)006<0201:gaoths>2.0.co;2.1466-1268(2001)006[0201:GAOTHS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6.1420-682X(2005)062[0670:HCCFAM]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol. 2002;12:1263–1268. doi: 10.1016/s0960-9822(02)00954-5.0960-9822(2002)012[1263:EOTSMJ]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nepomuceno-Silva JL, de Melo LD, Mendonca SM, Paixao JC, Lopes UG. RJLs: a new family of Ras-related GTP-binding proteins. Gene. 2004;327:221–232. doi: 10.1016/j.gene.2003.11.010.0378-1119(2004)327[0221:RANFOR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nikolaidis N, Nei M. Concerted and nonconcerted evolution of the Hsp70 gene superfamily in two sibling species of nematodes. Mol Biol Evol. 2004;21:498–505. doi: 10.1093/molbev/msh041.0737-4038(2004)021[0498:CANEOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ohtsuka K, Hata M. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2.1466-1268(2000)005[0098:MDHCON]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Huang C-H, Short MK, Jubinsky PT. Magmas gene structure and evolution. In Silico Biol. 2005;5:0024.1386-6338(2005)005[0024:MGSAE]2.0.CO;2 [PubMed] [Google Scholar]
- Raynes DA, Guerriero V. Inhibition of Hsp70 ATPase activity and protein renaturation by a novel Hsp70-binding protein. J Biol Chem. 1998;273:32883–32888. doi: 10.1074/jbc.273.49.32883.0021-9258(1998)273[32883:IOHAAA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Maier UG. Phylogenetic analysis of the stress-70 protein family. J Mol Evol. 1994;39:80–86. doi: 10.1007/BF00178252.0022-2844(1994)039[0080:PAOTSP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454.0737-4038(1987)004[0406:TNMANM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Satoh N. The ascidian tadpole larva: comparative molecular development and genomics. Nat Rev Genet. 2003;4:285–295. doi: 10.1038/nrg1042.1471-0056(2003)004[0285:TATLCM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Satoh N, Satou Y, Davidson B, Levine M. Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet. 2003;19:376–381. doi: 10.1016/S0168-9525(03)00144-6.0168-9525(2003)019[0376:CIAEMF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Levine M, Kohara Y, Rokhsar D, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. I. Genes for bHLH transcription factors. Dev Genes Evol. 2003;213:213–221. doi: 10.1007/s00427-003-0319-7.0949-944X(2003)213[0213:AGSODR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Satou Y, Kawashima T, Shoguchi E, Satoh N. An integrated database of the ascidian Ciona intestinalis: towards functional genomics. Zool Sci. 2005;22:837–843. doi: 10.2108/zsj.22.837.0289-0003(2005)022[0837:AIDOTA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Satou Y, Takatori N, and Fujiwara S. et al. 2002a Ciona intestinalis cDNA projects: expressed sequence tag analyses and gene expression profiles during embryogenesis. Gene. 287:83–96. [DOI] [PubMed] [Google Scholar]
- Satou Y, Yamada L, and Mochizuki Y. et al. 2002b A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 33:153–154. [DOI] [PubMed] [Google Scholar]
- Satouh Y, Padma P, Toda T, Satoh N, Ide H, Inaba K. Molecular characterization of radial spoke subcomplex containing radial spoke protein 3 and heat shock protein 40 in sperm flagella of the ascidian Ciona intestinalis. Mol Biol Cell. 2005;16:626–636. doi: 10.1091/mbc.E04-09-0784.1059-1524(2005)016[0626:MCORSS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soti C, Pal C, Papp B, Csermely P. Molecular chaperones as regulatory elements of cellular networks. Curr Opin Cell Biol. 2005;17:210–215. doi: 10.1016/j.ceb.2005.02.012.0955-0674(2005)017[0210:MCAREO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:E237–E241. doi: 10.1038/ncb1001-e237.1465-7392(2001)003[E237:MCTARB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Terajima D, Shida K, Takada N, Kasuya A, Rokhsar D, Satoh N, Satake M, Wang H-G. Identification of candidate genes encoding the core components of the cell death machinery in the Ciona intestinalis genome. Cell Death Differ. 2003;10:749–753. doi: 10.1038/sj.cdd.4401223.1350-9047(2003)010[0749:IOCGET]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876.0305-1048(1997)025[4876:TCXWIF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Douglas MG. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347.0021-9258(1996)271[9347:ACHSOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tyson JR, Stirling CJ. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440.1460-2075(2000)019[6440:LASPAL]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Zylicz M, Georgopoulos C. The NH2-terminal. 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem. 1994;269:5446–5451.0021-9258(1994)269[5446:TNAAOT]2.0.CO;2 [PubMed] [Google Scholar]
- Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172.1469-221X(2004)005[0567:TJFMPA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Frazier AE, Pfanner N. The protein import machinery of mitochondria. J Biol Chem. 2004;279:14473–14476. doi: 10.1074/jbc.R400003200.0021-9258(2004)279[14473:TPIMOM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]