Abstract
Cargo transfer from trans-Golgi network (TGN)-derived transport carriers to endosomes involves a still undefined set of tethering/fusion events. Here we analyze a molecular interaction that may play a role in this process. We demonstrate that the GGAs, a family of Arf-dependent clathrin adaptors involved in selection of TGN cargo, interact with the Rabaptin-5–Rabex-5 complex, a Rab4/Rab5 effector regulating endosome fusion. These interactions are bipartite: GGA-GAE domains recognize an FGPLV sequence (residues 439–443) in a predicted random coil of Rabaptin-5 (a sequence also recognized by the γ1- and γ2-adaptin ears), while GGA-GAT domains bind to the C-terminal coiled-coils of Rabaptin-5. The GGA–Rabaptin-5 interaction decreases binding of clathrin to the GGA-hinge domain, and expression of green fluorescent protein (GFP)–Rabaptin-5 shifts the localization of endogenous GGA1 and associated cargo to enlarged early endosomes. These observations thus identify a binding sequence for GAE/γ-adaptin ear domains and reveal a functional link between proteins regulating TGN cargo export and endosomal tethering/fusion events.
Keywords: clathrin/endosomes/GGA/Rabaptin-5/Rabex-5
Introduction
The GGAs constitute a family of proteins peripherally associated with the cytosolic face of the trans-Golgi network (TGN) (Boman et al., 2000; Hirst et al., 2000; Dell’Angelica et al., 2000; Poussu et al., 2000; Takatsu et al., 2000). Three GGAs (termed GGA1, GGA2 and GGA3) exist in humans, two in Saccharomyces cerevisiae (Gga1p and Gga2p) and one each in Drosophila melanogaster and Caenorhabditis elegans. The GGAs are monomeric proteins organized into four distinct domains: a VHS domain (found in Vps27, Hrs and Stam), a GAT domain (found in GGAs and TOM1), a hinge domain rich in prolines and serines, and a GAE domain (similar to the γ-adaptin ear domain) (Figure 1A). The VHS domain binds acidic cluster dileucine signals present in the cytosolic tails of mannose-6-phosphate receptors (MPRs) that sort lysosomal hydrolases from the TGN to the endosomal–lysosomal system (Puertollano et al., 2001a; Takatsu et al., 2001; Zhu et al., 2001). The structural bases for this recognition have been elucidated recently (Misra et al., 2002; Shiba et al., 2002a). The GAT domain binds to the activated form of members of the Arf family of GTP-binding proteins (Boman et al., 2000; Dell’Angelica et al., 2000) and stabilizes them by preventing access of specific GTPase-activating proteins (Puertollano et al., 2001b). The hinge region binds clathrin via type I clathrin-binding motifs (Costaguta et al., 2001; Mullins and Bonifacino, 2001; Puertollano et al., 2001b; Zhu et al., 2001) and also contains autoinhibitory peptide sequences that control the accessibility of the binding site for acidic cluster dileucine signals on the VHS domain (Doray et al., 2002). As a result of these multiple interactions, the GGAs behave as clathrin adaptors that associate with the TGN in an Arf-dependent manner and mediate the sorting of MPRs.
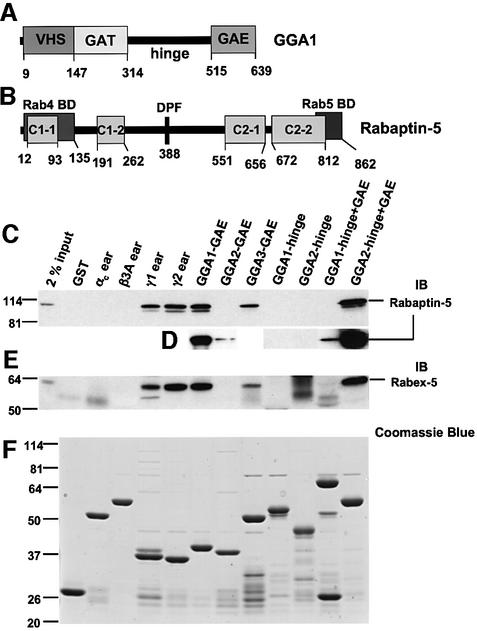
Fig. 1. Binding of GST fusion proteins to Rabaptin-5 and Rabex-5 from bovine brain cytosol. (A and B) The domains and relevant motifs in human GGA1 and human Rabaptin-5 respectively. The Rabaptin-5 scheme shows the Rab4- and Rab5-binding domains along with a DPF motif (codons 388–390) and the four regions predicted to form coiled-coil structures (C1-1, C1-2, C2-1 and C2-2). Various GST fusion proteins were immobilized on glutathione–Sepharose and subsequently incubated with bovine brain cytosol. Bound proteins were subjected to SDS–PAGE and immunoblotting (IB) using a mouse monoclonal antibody to Rabaptin-5 (C and D) or a rabbit polyclonal antiserum to Rabex-5 (E). (D) Blots from a separate experiment where membranes were overexposed to allow comparison of signals corresponding to GAE, hinge and hinge + GAE constructs of GGA1 and GGA2. (F) The Coomassie Blue staining of a gel loaded with the various GST fusion proteins. The first lanes in (C) and (E) show the signals given by 2% of the cytosol used in the binding step. The positions of molecular mass markers (in kDa) are indicated on the left.
By analogy to the ear domain of the α-adaptins (Slepnev and De Camilli, 2000), the GAE domain of the GGAs would be expected to bind accessory factors regulating the function of GGA-containing coats or of the resulting GGA-coated carriers. Indeed, recent observations have demonstrated the interaction of GGA-GAE domains with γ-synergin (Hirst et al., 2000; Takatsu et al., 2000) and Rabaptin-5 (Hirst et al., 2000; Zhu et al., 2001), two binding partners that are shared with the γ1-adaptin and γ2-adaptin subunits of the AP-1 clathrin adaptor (Page et al., 1999; Hirst et al., 2000; Zhu et al., 2001; Shiba et al., 2002b). Whereas the function of γ-synergin is unknown, Rabaptin-5 has been implicated in endosomal fusion events (Lippé et al., 2001). Rabaptin-5 is a ubiquitous 100 kDa protein originally identified in a yeast two-hybrid screen using GTPase-deficient Rab5 as a bait (Stenmark et al., 1995). Rabaptin-5 contains N- and C-terminal sequences that bind to the Rab4 and Rab5 GTPases, respectively. These Rab-binding sequences partially overlap with four regions of coiled-coils that are responsible for dimerization of Rabaptin-5 (Vitale et al., 1998) (Figure 1B). In addition to binding to and stabilizing Rab5-GTP, Rabaptin-5 forms a complex with the Rab5-specific guanine nucleotide exchange factor (GEF), Rabex-5 (Horiuchi et al., 1997; Lippé et al, 2001). The Rabaptin-5–Rabex-5 complex catalyzes the formation of Rab5-GTP on membranes, in turn increasing the affinity of Rab5 for Rabaptin-5 (Horiuchi et al., 1997). These interactions cooperate with other factors to promote endosome fusion. Along with other divalent Rab effectors such as Rabenosyn-5, Rabaptin-5 may also coordinate the movement of cargo between different subdomains of the endosomes by virtue of its ability to bind both Rab4 and Rab5 (De Renzis et al., 2002). This central role of Rabaptin-5 makes it a likely target for additional input regulating the transport of cargo in the endosomal system.
In this study, we analyze the GGA–Rabaptin-5 interaction. We show that the GGAs interact in vitro not just with Rabaptin-5 but with the Rabaptin-5–Rabex-5 complex. In addition, we show that these interactions occur in situ by demonstrating co-immunoprecipitation of the endogenous proteins from cell lysates. Molecular dissection of these interactions reveals that Rabaptin-5 interacts with both the GAE and GAT domains of the GGAs, and that an FGPLV sequence in a predicted random coil of Rabaptin-5 functions as the GAE-binding site. This FGPLV sequence is also recognized by γ1- and γ2-adaptin ears. Rabaptin-5 inhibits the binding of clathrin to the GGAs, suggesting that it may play a role in detachment of clathrin from GGA-coated intermediates. Expression of green fluorescent protein (GFP)–Rabaptin-5 causes redistribution of endogenous GGA1 to enlarged endosomes containing both cation-independent MPR (CI-MPR) and transferrin receptor (TfR). These observations suggest that the GGA–Rabaptin-5 interactions may play a role in the docking or fusion of TGN-derived intermediates to endosomes.
Results
Binding of the Rabaptin-5–Rabex-5 complex to GGA-GAE domains
In vitro interactions of Rabaptin-5 with the GAE domains of GGA1 and GGA2 were first reported by Hirst et al. (2000), although they were found to be much weaker than those of Rabaptin-5 with the γ1-adaptin ear and, therefore, of uncertain physiological significance. Because of the possible implications of these interactions for the function of the GGAs, however, we decided to analyze them in more detail. We performed GST pull-down experiments to examine the interaction of Rabaptin-5 from bovine brain cytosol with various GGA domains. For comparison, the experiments were also performed with GST fusions to the γ1- and γ2-adaptin ears as positive controls, along with GST and GST fusions to the αc- and β3A-adaptin ears as negative controls (Figure 1C). In agreement with previous reports (Hirst et al., 2000; Zhu et al., 2001; Shiba et al., 2002b), we observed binding of Rabaptin-5 to the γ1 and γ2 ears and to the GGA-GAE domains, but not to the αc or β3A ears (Figure 1C). The relative order of avidity of the GGA-GAE domains for Rabaptin-5 was GGA1> GGA3>>GGA2. The interaction with the GGA2-GAE domain could only be observed after long exposures (Figure 1D). We did not detect interaction of Rabaptin-5 with the hinge domains of GGA1 and GGA2 (Figure 1C). The order of avidities for Rabaptin-5 was reversed when GST fusions comprising both the hinge and GAE domains were used (GGA2>>GGA1, Figure 1C; binding to the latter was also observed after prolonged exposures; Figure 1D). The differences in the signals obtained with the GAE and hinge + GAE constructs of GGA1 and GGA2 were not due to problems with the expression of the recombinant GST fusions in Escherichia coli (Figure 1F). Instead, these differences could be due to folding of the constructs or accessibility of the binding site on GAE domains depending on the presence or absence of the hinge regions. GST fusions to the hinge and hinge + GAE domains of GGA3 could not be tested because of degradation of the recombinant proteins. It is worth noting that the signals obtained with some GGA constructs (particularly GST–GGA1-GAE and GST–GGA2-hinge + GAE) were equal to or greater in intensity than those observed for the γ1 and γ2 ear constructs, suggesting that GGA–Rabaptin-5 interactions are stronger than originally appreciated.
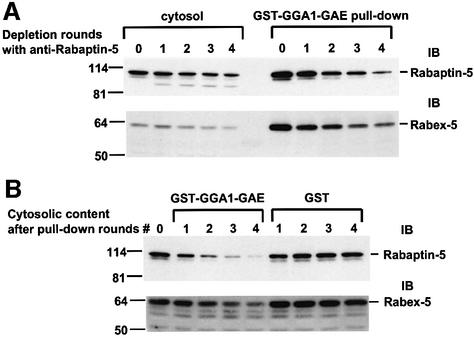
The relevance of the in vitro interactions between Rabaptin-5 and the GGAs was substantiated further by the demonstration that Rabex-5, a Rabaptin-5 partner that functions as a Rab5-GEF, was also present in the GST–GGA pull-down materials containing Rabaptin-5 (Figure 1E). Importantly, immunodepletion of Rabaptin-5 by multiple rounds of incubation with anti-Rabaptin-5 caused a parallel decrease in Rabex-5 in the cytosol, as well as in the levels of both Rabaptin-5 and Rabex-5 brought down by GST–GGA1-GAE (Figure 2A). In addition, multiple rounds of incubation with GST–GGA1-GAE, but not GST, caused a significant decrease of both Rabaptin-5 and Rabex-5 from the cytosol (Figure 2B). Taken together, these findings indicate that GGA-GAE domains interact with Rabaptin-5 as part of a complex with Rabex-5.
Fig. 2. Interaction of GGA-GAE with the Rabaptin-5–Rabex-5 complex. (A) Bovine brain cytosol was subjected to four consecutive incubations with anti-Rabaptin-5 immobilized on protein G–Sepharose. Samples of the untreated cytosol (round 0) and of the materials collected after each round of consecutive immunodepletions (rounds 1–4) were analyzed by SDS–PAGE and immunoblotting (IB) with anti-Rabaptin-5 or anti-Rabex-5 antibodies (blots on the left). The cytosol fractions corresponding to rounds 0–4 were also used in pull-down experiments with GST–GGA1-GAE immobilized to glutathione– Sepharose. The material pulled-down by the GST–GGA1-GAE was analyzed by SDS–PAGE and immunoblotting (blots on the right). (B) Bovine brain cytosol was subjected to four consecutive rounds of treatment with either GST–GGA1-GAE or GST immobilized to glutathione–Sepharose. The control cytosol (round 0) and the fractions corresponding to rounds 1–4 were analyzed by SDS–PAGE and immunoblotting.
Co-immunoprecipitation of Rabaptin-5 and GGAs
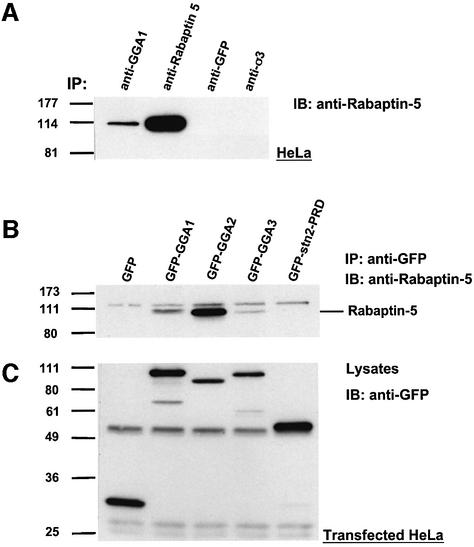
The occurrence of the GGA–Rabaptin-5 interaction in vivo was evaluated by immunoprecipitation of endogenous proteins. We observed specific co-immunoprecipitation of Rabaptin-5 with GGA1 from lysates of HeLa cells using an anti-GGA1 antiserum (Figure 3A). Due to the unavailability of antisera for immunoprecipition of endogenous GGA2 and GGA3, we analyzed the interaction of these proteins with Rabaptin-5 using GFP-tagged full-length GGAs. Immunoprecipitation of transfected HeLa cell lysates with anti-GFP revealed co-immunoprecipitation of Rabaptin-5 with the three GFP–GGAs, but not with GFP or the GFP-tagged proline-rich domain of stonin 2 (Martina et al., 2001) (Figure 3B). The differences in the extent of co-immunoprecipitation of Rabaptin-5 with the three GGA constructs were not the result of different levels of expression, as evaluated by immunoblotting of cell lysates with anti-GFP (Figure 3C). The GFP–GGA2 construct displayed the highest binding activity towards Rabaptin-5 relative to the other two GFP–GGAs. The results from these experiments thus demonstrated that Rabaptin-5–GGA interactions can take place not only in vitro but also within cells.
Fig. 3. Co-immunoprecipitation of GGAs and Rabaptin-5. (A) HeLa cell lysates were subjected to immunoprecipitation (IP) using either rabbit anti-GGA1, mouse monoclonal anti-Rabaptin-5, mouse monoclonal anti-GFP or a rabbit antiserum against the σ subunit of AP-3 (anti-σ3). The immunoprecipitated materials were analyzed subsequently by SDS–PAGE and immunobotting (IB) with mouse monoclonal anti-Rabaptin-5. (B and C) Lysates from HeLa cells transiently transfected with the indicated constructs were subjected to IP using mouse monoclonal anti-GFP followed by SDS–PAGE and IB with anti-Rabaptin-5 (B). Samples of the lysates were also analyzed by SDS–PAGE and immunoblotting with anti-GFP, in order to assess the levels of expression of the different constructs (C).
Delineation of the structural determinants of Rabaptin-5–GGA interactions
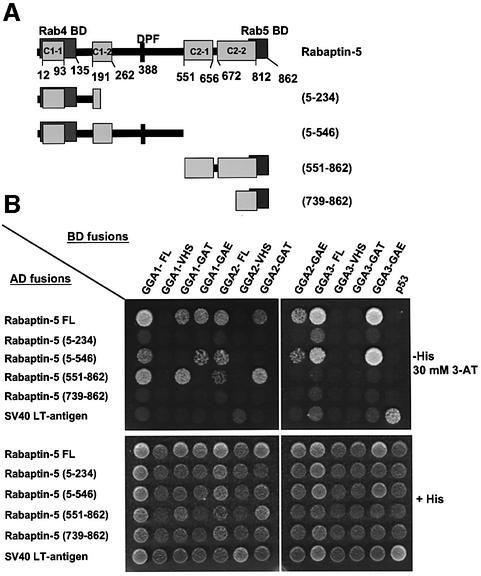
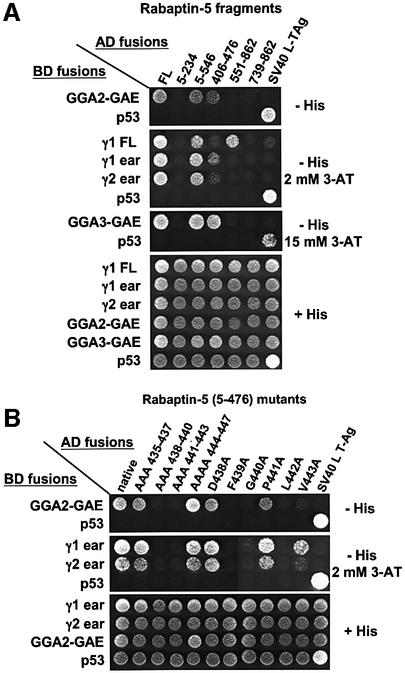
The structural determinants of the Rabaptin-5–GGA interactions were analyzed using the yeast two-hybrid system. The experiments were performed by generating fusions of the Gal4 activation domain (AD) and Gal4 DNA-binding domain (BD) with fragments of Rabaptin-5 and GGAs, respectively. The specificity of the interactions was determined by co-transformation of AD–Rabaptin-5 constructs with a BD–p53 construct, and of BD–GGA constructs with an AD–SV40 large T-antigen construct, used as negative controls. Co-transformation with vectors encoding the BD–p53 and AD–SV40 large T-antigen fusions provided a positive control for the assays. These experiments confirmed the interaction between the full-length GGAs and Rabaptin-5, and helped define the domains that are involved in these interactions. We observed that full-length Rabaptin-5 interacted not only with each of the three full-length GGAs, but also with the GAT and GAE domains of GGA1 and GGA2, as well as with the GAE domain of GGA3 (Figure 4). No interaction was detected with GGA-VHS domains (Figure 4). Interestingly, two regions in Rabaptin-5 appeared responsible for interactions with the GGAs: the Rabaptin-5 (5–546) fragment containing the two N-terminal coiled-coils (C1-1 and C1-2) and the Rab4-binding domain interacted with the three GGA-GAE domains, whereas the Rabaptin-5 (551–862) fragment containing the two C-terminal coiled-coils (C2-1 and C2-2) and the Rab5-binding domain interacted with the GAT domains of GGA1 and GGA2 but not with that of GGA3 (Figure 4). No interactions were observed with the constructs expressing the Rabaptin-5 (5–234) and (739–862) fusions, suggesting that the regions responsible for interactions with GAE and GAT domains were contained within residues 234–546 and 551–739, respectively (Figure 4).
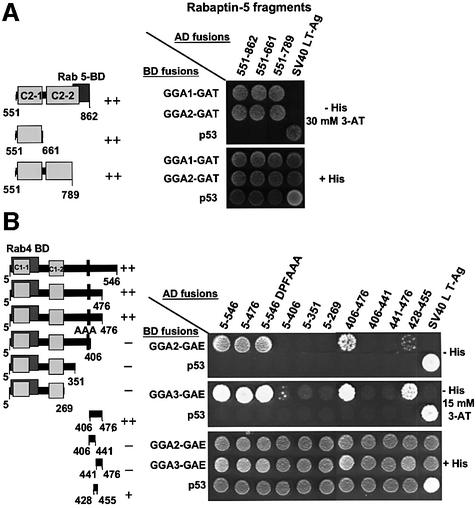
Fig. 4. Different regions of Rabaptin-5 are responsible for interaction with GAT and GAE domains of GGAs. The AH109 yeast strain was co-transformed with the indicated constructs (GGA domains fused to the GAL4 binding domain and Rabaptin-5 fragments fused to the GAL4 activation domain). (A) The various Rabaptin-5 constructs that were assayed. After selection, co-transformants were plated on medium without histidine (–His), to monitor HIS3 reporter gene activation due to interaction of fusion proteins, and on medium containing histidine (+His) to control growth/loading of the co-transformants (B). The –His plates were supplemented with 30 mM 3-AT (a competitive inhibitor of the His3 protein) to suppress background growth and minimize non-specific interactions between constructs.
The region of Rabaptin-5 responsible for interactions with the GAT domains of GGA1 and GGA2 was delineated further by constructing two additional deletion mutants: Rabaptin-5 (551–789) [comprising the C2-1 domain and a larger fraction of the C2-2 domain interrupted in Rabaptin-5 (739–862), but excluding the Rab5-binding domain] and Rabaptin-5 (551–661) (containing only the C2-1 domain) (see schemes in Figure 5A). Two-hybrid analyses revealed a similar interaction of the three C-terminal fragments of Rabaptin-5 (551–862, 551–661 and 551–789) with either GGA1-GAT or GGA2-GAT (Figure 5A). Thus, the C-terminal coiled-coil regions of Rabaptin-5 bind to the GAT domains of GGA1 and GGA2. The C2-1 domain is sufficient for this interaction, while the Rab5-binding domain is dispensable.
Fig. 5. C-terminal coiled-coils in Rabaptin-5 bind the GAT domains of GGA1 and GGA2, while residues in the Rabaptin-5 428–455 region are important for interaction with the GGA-GAE domain. (A) The interaction between deletion mutants of the Rabaptin-5 C-terminal region and GAT domains of GGA1 and GGA2. (B) The interaction of an additional set of deletion mutants in the N-terminal half of Rabaptin-5 with GGA-GAE domains. Also shown in (B) is the interaction with a Rabaptin-5 (5–546) mutant containing a DPF388–390AAA substitution. Experiments were performed as indicated in the legend to Figure 4, with the exception that co-transformants were plated on –His medium with or without 30 mM (A) or 15 mM 3-AT (B). The ++, + or – symbols indicate the relative strength of interactions between GGA domains and Rabaptin-5 mutants.
The region of Rabaptin-5 binding to the GGA-GAE domains was mapped using a series of truncations of the Rabaptin-5 (5–546) construct (see scheme in Figure 5B). Two-hybrid analyses showed that the 5–546 and 5–476 fragments bound to the GAE domains of GGA2 and GGA3, whereas the 5–406, 5–351 and 5–269 fragments did not, indicating that the 406–476 segment is necessary for these interactions (Figure 5B). Testing of the 406–476 fragment revealed that it was sufficient for interactions with GAE domains of GGA2 and GGA3 (Figure 5B). Further subdivision of the 406–476 segment into halves (406–441 and 441–476 fragments) resulted in loss of activity, whereas removal of residues from both ends of this construct (428–455 fragment) still allowed for interactions with GGA-GAE domains, albeit with reduced avidity. Similar results were obtained with a GGA1-GAE construct (data not shown). These observations indicated that the minimal GAE-binding determinant is contained within residues 428–455 of Rabaptin-5.
The Rabaptin-5 (428–455) segment does not contain any known binding motifs. A single DPF motif is present at positions 388–390 of Rabaptin-5, outside the GAE-binding segment. Since DPF and DPW motifs bind to the α-adaptin ear (Owen et al., 1999), it nonetheless was of interest to examine the effect of mutating these residues on the interactions with GGA-GAE domains. We observed that mutation of this motif had no effect on the interactions (Figure 5B). This observation, together with the mapping of the GAE-binding determinant to residues 428–455 of Rabaptin-5, suggested that GGA-GAE and α-adaptin ear domains recognize distinct structural determinants.
Identification of a novel sequence within Rabaptin-5 that mediates interactions with GGA-GAE domains
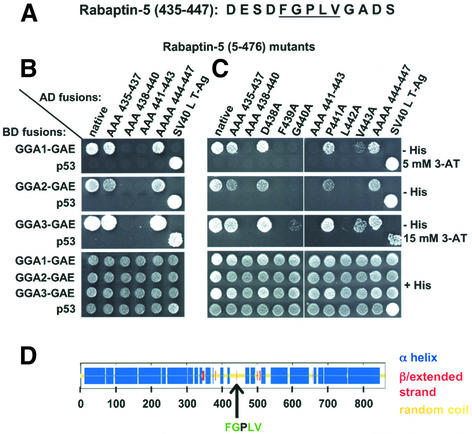
The delineation of the GAE-binding determinant to residues 428–455 made it feasible to use alanine scan mutagenesis to identify residues critical for interactions. This was performed by substituting blocks of 2–4 residues in this region by alanine residues. The substitutions were introduced in Rabaptin-5 (428–455) and Rabaptin-5 (406–476). Yeast two-hybrid analyses demonstrated the importance of the central portion of the 428–455 segment and the dispensability of residues at its two boundaries (428–434 and 448–455) (data not shown). Based on these observations, a more limited set of alanine substitutions in the 435–447 region (Figure 6A) subsequently was introduced into Rabaptin-5 (5–476). Analysis of these mutants revealed that the residues critical for interaction with the GAE domains reside within the 438–443 segment of Rabaptin-5 (Figure 6B).
Fig. 6. Alanine scan mutagenesis in Rabaptin-5 (5–476) defines a GGA-GAE-binding sequence: FGXLV. (A) Sequence of the Rabaptin-5 (435–447) segment. (B and C) Interaction of GGA-GAE domains with Rabaptin-5 (5–476) mutants: either blocks of 3–4 contiguous amino acids corresponding to codons 435–447 (B) or individual residues in the 438–443 region (C) were introduced into Rabaptin-5 (5–476). Experiments were performed as indicated in the legend to Figure 5, with the exception that the co-transformants were plated on –His plus 5 mM 3-AT, –His medium, and –His medium plus 15 mM 3-AT for evaluation of interactions with GAE domains of GGA1, GGA2 and GGA3, respectively. (D) The consensus secondary structure prediction for Rabaptin-5 (Network Protein Sequence Analysis of the Pole Bio-informatique Lyonnais), as well as the position of the GGA-GAE-binding sequence.
Analysis of single alanine substitutions in the Rabaptin-5 (438–443) segment allowed definition of a GAE-binding sequence (Figure 6C). This analysis revealed that F439, G440, L442 and, to some extent, V443 are the residues critical for binding to GAE domains of the three GGAs. Substitution of the P residue at position 441 had no effect on the interactions. Consequently, the sequence FGPLV corresponding to codons 439–443 in Rabaptin-5 represents the GGA-GAE recognition site, and the sequence FGXLV can be considered a putative GGA-GAE-binding box given the minor contribution of the central P residue. Theoretical analysis of the secondary structure of Rabaptin-5 predicts that the GAE-binding sequence is located in an unstructured region, which probably makes this site accessible for interactions (scheme in Figure 6D).
Interaction of Rabaptin-5 with γ1- and γ2-adaptin ears
We also compared the interactions of GGA-GAE and γ-adaptin ear domains with Rabaptin-5. Full-length γ1-adaptin interacted not only with full-length Rabaptin-5 but also with the 5–546 and 551–862 fragments of this protein (Figure 7A), similarly to full-length GGA1 and GGA2 (Figure 4). In addition, we observed that the γ1 and γ2 ears interacted with full-length Rabaptin-5 as well as with the 5–546 and 406–476 fragments of Rabaptin-5. In contrast, the γ1 and γ2 ears did not interact with the 551–862 fragment of Rabaptin-5 (Figure 7A). Further analysis using the panel of alanine substitutions in Rabaptin-5 (5–476) demonstrated that, like GGA-GAE domains, the γ-adaptin ears also bind to the FGXLV motif, with the V residue being less critical for this interaction (Figure 7B).
Fig. 7. Definition of domains important for the γ-adaptin–Rabaptin-5 interaction. (A) Different Rabaptin-5 fragments (schematized in Figures 4 and 5) were used to study the interactions with full-length γ1, γ1 and γ2 ears and GAE domains of GGA2 and GGA3 by yeast two-hybrid analysis. (B) Interaction between γ-adaptin ears and Rabaptin-5 (5–476) mutants. Co-transformants were plated on +His medium and on –His medium with the indicated concentrations of 3-AT.
In vitro analysis of the γ-ear/GGA-GAE-binding sequence in Rabaptin-5
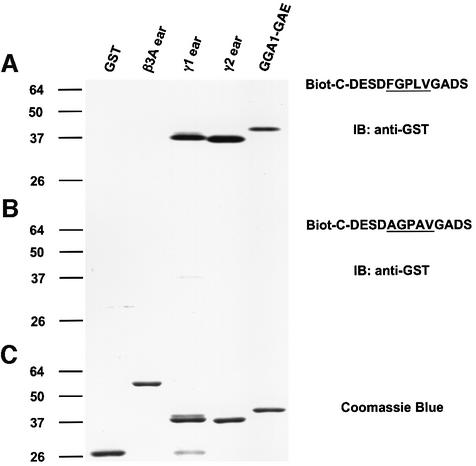
The definition of the sequence recognizing the γ-ear and GGA-GAE domains was tested by performing in vitro binding assays using peptides that were biotinylated and immobilized on streptavidin–agarose. The results summarized in Figure 8 revealed that a 14mer peptide containing the FGPLV sequence specifically binds to GST fusions to the γ-adaptin ear and GGA1-GAE domains, and that this binding is markedly reduced by substitution of the F and L residues. Similar results were obtained using GST fusions to the other GGA-GAE domains (data not shown).
Fig. 8. Binding of biotinylated Rabaptin-5 peptides to γ-adaptin ear and GGA-GAE domains. Both a biotinylated peptide corresponding to the Rabaptin-5 (435–447) sequence (A) and a variant with F439A and L442A substitutions (B) were immobilized on streptavidin–agarose and tested for their ability to bind GST fusions to the γ1- and γ2-adaptin ear and GGA1-GAE domains (GST and GST–β3A were used as negative controls). The material bound to the immobilized peptides was analyzed by SDS–PAGE and immunoblotting (IB) using anti-GST antiserum (A and B). The Coomassie Blue staining of a gel loaded with the various GST fusions is depicted in (C).
Rabaptin-5 interferes with GGA–clathrin interactions
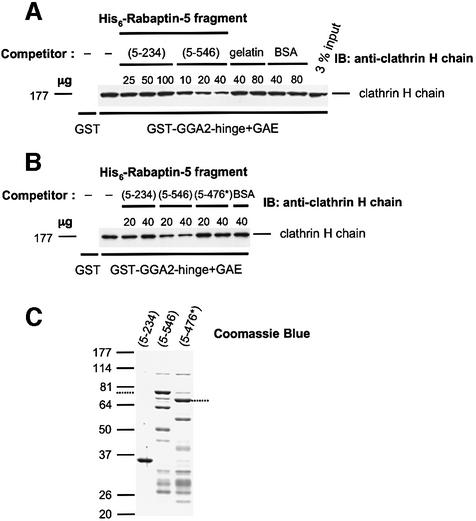
The hinge and GAE domains of the GGAs mediate binding to clathrin. GGA2, in particular, has an LIDLE sequence within its hinge domain that conforms to a type I consensus motif for clathrin binding (Dell’Angelica et al., 2000; Puertollano et al., 2001b; Zhu et al., 2001). The proximity of this motif to the Rabaptin-5 interaction site on GGA2 prompted us to test whether Rabaptin-5 interferes with clathrin binding to GGA2. To this end, we examined the effect of His6-tagged Rabaptin-5 fragments on in vitro binding of clathrin to GST–GGA2-hinge + GAE (Figure 9). We observed that His6-Rabaptin-5 (5–546) inhibited the binding of clathrin to the GST–GGA2-hinge + GAE construct in a concentration-dependent fashion, whereas even larger amounts of His6-Rabaptin-5 (5–234) or irrelevant proteins [gelatin or bovine serum albumin (BSA)] had no effect (Figure 9A). Similar observations were made when a GST–GGA1-hinge + GAE construct was used (not shown). These results were consistent with the yeast two-hybrid assays showing binding of GGA-GAE to Rabaptin-5 (5–546) but not to Rabaptin-5 (5–234) (Figures 4 and 5). Importantly, a His6-tagged Rabaptin-5 (5–476) mutant unable to interact with GGA-GAE domains (AAA438–440 in Figure 6, including the substitution of the critical F residue at position 439) did not interfere with the pull-down of clathrin by the GGA2-hinge + GAE fusion, consistent with the notion that the Rabaptin-5–GGA interaction is required for inhibition of clathrin binding (5–476* construct in Figure 9B). Clathrin binding to the GGA hinge + GAE constructs was also interfered with by His6-tagged full-length Rabaptin-5, but was unaffected by the 14mer peptide containing the FGPLV sequence depicted in Figure 8A (data not shown). The lack of effect of this short peptide suggests that interference with clathrin association upon binding of Rabaptin-5 to GGA-GAE is probably due to steric hindrance, as opposed to a more global conformational change in the GGA molecule.
Fig. 9. Rabaptin-5 interferes with GGA–clathrin interactions. (A) The effects of increasing concentrations of His6-Rabaptin-5 (5–234) and His6-Rabaptin-5 (5–546) on the binding of clathrin from bovine brain cytosol to GST–GGA2-hinge + GAE were assessed in a pull-down interference assay. Specificity controls include pull-down incubations with GST (left lane) as well as the effects of increasing concentrations of gelatin or BSA. (B) A mutant His6-Rabaptin-5 (5–476) fragment containing three alanine substitutions in codons 438–440 (5–476*) does not affect the binding of clathrin to GST–GGA2-hinge + GAE. (C) Coomassie Blue staining of gels analyzing the preparations of His6-tagged Rabaptin-5 fragments 5–234, 5–546 and 5–476*. The mobility of the full-length His6-Rabaptin-5 (5–546) and 5–476* fragments is indicated by dotted lines on the left and right of the panel, respectively; preparations of these two fragments contained two proteolytic fragments recognized by anti-Rabaptin-5 in addition to the full-length protein.
Redistribution of endogenous GGA1 to Rabaptin-5- induced enlarged endosomes
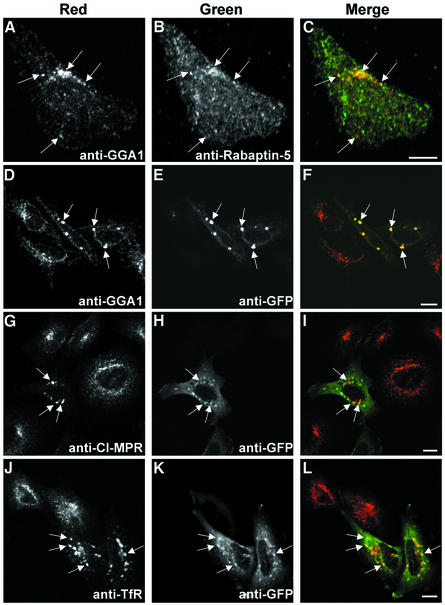
Possible sites of common intracellular localization of endogenous Rabaptin-5 and GGAs were investigated by double-indirect immunofluorescence microscopy in HeLa cells. Consistent with previous reports (Stenmark et al., 1995), labeling of Rabaptin-5 with a monoclonal antibody resulted in a diffuse cytosolic signal, while a polyclonal anti-GGA1 antibody highlighted the TGN distribution of GGA1 (data not shown). In order to reduce the cytosolic staining of Rabaptin-5, cells were subjected to a brief (1 min) permeabilization with 0.004% (w/v) saponin immediately prior to fixation. Confocal microscopy of these cells revealed discrete areas of co-localization of Rabaptin-5 and GGA1, although for the most part the patterns of staining were distinct (Figure 10A–C).
Fig. 10. Changes in the intracellular distribution of endogenous GGA1, CI-MPR and TfR upon expression of GFP–Rabaptin-5 in HeLa cells. (A–C) Confocal immunofluorescence microscopy showing discrete areas of co-localization of endogenous Rabaptin-5 and GGA-1 in untransfected HeLa cells. HeLa cells were subjected to a brief permeabilization pulse immediately before fixation with 4% formaldehyde. Fixed cells were subjected to double staining using mouse monoclonal anti-Rabaptin-5 and rabbit anti-GGA1 antibodies, followed by incubation with Cy3-conjugated anti-rabbit and Alexa 488-conjugated anti-mouse antisera. Scale bar = 10 µm. (D–L) Localization of endogenous GGA1 (D–F), CI-MPR (G–I) and TfR (J–L) to enlarged endosomes (∼2 µm) induced by transient overexpression of GFP–Rabaptin-5 in HeLa cells. Fixed cells were subjected to double immunostaining using: rabbit anti-GGA1 and mouse monoclonal anti-GFP, followed by Cy3-conjugated anti-rabbit and Alexa 488-conjugated anti-mouse secondary antisera (D–F); mouse monoclonal anti-CI-MPR and rabbit anti-GFP, followed by Alexa 568-conjugated anti-mouse and Alexa 488-conjugated anti-rabbit antisera (G–I); and mouse monoclonal anti-TfR and rabbit anti-GFP followed by Alexa 568-conjugated anti-mouse and Alexa 488-conjugated anti-rabbit antisera (J–L). The use of anti-GFP antisera allowed a more sensitive detection of cells expressing low levels of GFP–Rabaptin-5 than direct detection of GFP fluorescence. Scale bar = 10 µm.
We next analyzed changes in the intracellular distribution of endogenous GGA1, and of the associated cargo, CI-MPR, in HeLa cells that had been transiently co-transfected with GFP–Rabaptin-5. As reported previously (Stenmark et al., 1995), overexpression of GFP–Rabaptin-5 induced the formation of large (∼2 µm) TfR-positive vesicles corresponding to an enlarged endosomal compartment (Figure 10J–L). Importantly, the distribution of endogenous GGA1 and CI-MPR was shifted from a mainly juxtanuclear localization to the Rabaptin-5-stabilized enlarged endosomes (Figure 10D–I). This observation is consistent with Rabaptin-5 promoting conveyance of cargo from GGA-containing carriers into early endosomes.
Discussion
Our observations demonstrate that GGAs interact with the Rabaptin-5–Rabex-5 complex both in vitro and in vivo. We also show that γ1- and γ2-adaptins interact not just with Rabaptin-5, as previously reported (Hirst et al., 2000; Zhu et al., 2001; Shiba et al., 2002b), but with the Rabaptin-5–Rabex-5 complex. The magnitude of the interactions for some of the GST–GGA-GAE constructs (e.g. GST–GGA1-GAE or GST–GGA2-hinge + GAE) with the Rabaptin-5–Rabex-5 complex is quantitatively similar to that of the γ-adaptin ears (Figure 1), indicating that these interactions may all be equally significant. Importantly, the GGA–Rabaptin-5 interaction could also be demonstrated by co-immunoprecipitation of endogenous proteins or of full-length GGA constructs from cell extracts. Together, these observations indicate that interactions between the GGAs and Rabaptin-5 are robust and likely to be physiologically significant.
GGA–Rabaptin-5 interactions are mediated by binding of the GAT domains of GGA1 and GGA2 to the C-terminal coiled-coil domains of Rabaptin-5 (with the C2-1 module at 551–661 being sufficient for binding), and of the GAE domains of the three GGAs to an FGPLV sequence (residues 439–443) in a predicted random coil in the central region of Rabaptin-5. The GAE domains are analogous to the ear domains of the large subunits of the adaptor protein complexes AP-1, AP-2, AP-3 and AP-4 (Boehm and Bonifacino, 2001). The main function of these domains is the recruitment of accessory factors that regulate coat function. To date, however, peptide sequences that mediate interactions with the ear domains have only been defined for the α-adaptin subunit of AP-2 (i.e. DPF, DPW, FXDXF; Owen et al., 1999; Brett et al., 2002). The FGPLV sequence is thus the first binding sequence defined for an ear-like domain other than that of α-adaptin.
The FGPLV sequence is conserved in human, rat, mouse and chicken Rabaptin-5, and is also present in four variants of Rabaptin-5 named neurocrescin (Rabaptin-5 Δ1–84; Nishimune et al., 1997), Rabaptin-4 (Rabaptin-5 Δ758–790; Nagelkerken et al., 2000), Rabaptin-5γ and Rabaptin-5δ (Rabaptin-5 Δ12–54 and Rabaptin-5 Δ177–216, respectively; Korobko et al., 2002). This sequence is not conserved, however, in Rabaptin-5β, another Rab5 effector sharing 42% sequence identity with Rabaptin-5 (Gournier et al., 1998). The presence of consensus sites for cleavage by caspase-3 in Rabaptin-5, as well as its proteolysis during apoptosis, suggested that Rabaptin-5 may play a central role in the alterations in endosomal trafficking that characterize programmed cell death (Cosulich et al., 1997). Interestingly, the sequence DESD438F, one of two sites recognized by caspase-3 in human Rabaptin-5, immediately precedes the FGPLV motif (Swanton et al., 1999).
Individual alanine substitutions of residues in the FGPLV sequence revealed that the F, G, L and, to a lesser extent, V residues are important for GAE binding. On this basis, the GAE-binding motif can be defined tentatively as FGXLV (where X has been preliminarily designated as any amino acid). Further refinement of this motif will require a more detailed substitution analysis.
Two recent publications have analyzed the structural basis for the recruitment of binding partners by the γ1-adaptin ears (Kent et al., 2002; Nogi et al., 2002). The crystal structure of the unliganded domain, as well as a complementary mutational analysis, demonstrated the importance of a shallow hydrophobic groove (Kent et al., 2002) surrounded by basic residues (Nogi et al., 2002) in the β-sandwich fold of the γ1 ear for binding to γ-synergin and Rabaptin-5. Interestingly, both the residues lining the hydrophobic pocket and the cluster of basic residues in the γ1 ear are also conserved in GGA-GAE domains, suggesting that all of these modules may share a similar mechanism of ligand recognition. The FGPLV sequence in Rabaptin-5 is made up of hydrophobic amino acid residues that are likely to bind to the hydrophobic grooves on the GGA-GAE and γ-adaptin ear domains, and is surrounded by acidic residues that may stabilize ligand binding through electrostatic interactions with a basic platform. However, mutation of acidic residues surrounding the FGPLV sequence in the Rabaptin-5 (5–476) fragment did not affect the interaction with either GGA-GAE or γ-adaptin ear domains (Figures 6 and 7). However, block substitution of amino acids 435–437 in Rabaptin-5 (including D435 and E436) by alanine residues within the context of smaller peptides [Rabaptin-5 (406–476) or Rabaptin-5 (428–455)] weakened the interaction of these peptides with GGA-GAE domains (results not shown). These findings indicate that hydrophobic interactions between the Rabaptin-5 FGPLV box and a putative hydrophobic groove in GGA-GAE domains are the main contributors to binding, while electrostatic interactions between acidic residues surrounding the FGPLV sequence and basic residues in GGA-GAE may provide a less crucial stabilizing effect that is apparent only when assaying smaller peptides.
Shiba et al. (2002b) recently have reported the results of yeast two-hybrid analyses of the Rabaptin-5–γ1-adaptin interaction that are at variance with our results. These authors conducted two sets of experiments. In the first, they examined the interaction of several Rabaptin-5 fragments with full-length γ1-adaptin. They concluded that the 546–728 fragment of Rabaptin-5 (including C2-1 and a fraction of C2-2) was mainly responsible for interaction with γ1-adaptin. In the second set, they tested for interactions of full-length Rabaptin-5 with the N-terminal trunk (residues 1–594) and the C-terminal ear (residues 706–822) of γ1-adaptin. They found that full-length Rabaptin-5 bound to the γ1-adaptin ear. These results led them to conclude that the C-terminal coiled-coil region of Rabaptin-5 binds to the γ1-adaptin ear. In contrast, we find that the FGPLV sequence corresponding to codons 439–443 in Rabaptin-5 binds to the ear domains of both γ1- and γ2-adaptins (Figure 7), analogously to the interaction with the GGA-GAE domains (Figure 6).
What could be the physiological role of GGA– Rabaptin-5 interactions? The GGAs function in the sorting of MPRs into clathrin-coated carriers budding from the TGN (Puertollano et al., 2001a, 2003; Takatsu et al., 2001; Zhu et al., 2001), while the Rabaptin-5–Rabex-5 complex participates in endosome fusion (Lippé et al., 2001). It is thus reasonable to hypothesize that GGA–Rabaptin-5 interactions could enable fusion of TGN-derived carriers with endosomes, allowing the transfer of MPR from the TGN to the endosomal system. This hypothesis is supported by the localization of endogenous GGA1 and CI-MPR to enlarged endosomes stabilized by expression of Rabaptin-5 (Figure 10). In the course of these interactions, the Rabaptin-5–Rabex-5 complex could affect the activities of the GGAs. Indeed, in vitro studies showed that His6-tagged Rabaptin-5 (5–546) inhibited the binding of clathrin to GST–GGAs (Figure 9). This suggests that binding of the Rabaptin-5–Rabex-5 complex may induce the release of clathrin from GGA-coated intermediates or prevent clathrin re-binding, perhaps facilitating fusion with endosomes. The identification of a second binding site for Rabaptin-5 in the GGA1- and GGA2-GAT domains also raises the possibility of interference with the binding of Arf by these domains.
Conversely, GGA binding could also regulate the GEF activity of the Rabaptin-5–Rabex-5 complex. In this context, GGAs may affect the ability of the Rabaptin-5–Rabex complex to function as a time delay mechanism, stabilizing Rab5-GTP and increasing the likelihood of fusion events (Horiuchi et al., 1997).
The presence of subdomains containing distinct Rab proteins represents a critical mechanism of compartmentalization of endosomal membranes. While early endosomes appear enriched in Rab5 and Rab4, recycling endosomes are characterized by the presence of Rab4 and Rab11 (Miaczynska and Zerial, 2002). In this context, it has been proposed that divalent Rab effectors, such as Rabaptin-5, may coordinate the communication between contiguous endosomal subdomains (De Renzis et al., 2002). The coupling of Rabaptin-5 to both Rab4 and Rab5 suggests that the GGAs may also interact with different endosomal subdomains and participate in their coordination. It is also conceivable that binding of Rab4 and Rab5 to Rabaptin-5 may differentially affect the affinity of this molecule for the GGAs, resulting in the conveyance of its associated cargo to a specific endosomal subdomain enriched in one of these GTPases. Future studies on these issues are likely to provide a better understanding of the cross-talk between coat proteins involved in cargo sorting at the TGN and regulators of endosome fusion.
Materials and methods
DNA recombinant procedures
GST fusion constructs. The GST–mouse αC-ear construct (residues 701–938; Traub et al., 1999) was a gift from L.Traub. The GST–human β3A-ear (799–1081) and GST–human GGA3-GAE (residues 494–723, comprising a portion of the hinge in addition to the GGA3-GAE domain) were described previously (Dell’Angelica et al., 1998, 2000). GST constructs encoding human GGA1-hinge + GAE (residues 315–639), human GGA1-hinge (315–514), human GGA1-GAE (515–639), human GGA2-hinge + GAE (331–613), human GGA2-hinge (331–488) and human GGA2-GAE (489–613) were described by Puertollano et al. (2001b). GST–mouse γ1-ear (707–822) and GST–human γ2-ear (670–785) were generated by PCR amplification and subcloning into EcoRI–XhoI sites of pGEX-5X-1 (Amersham-Pharmacia Biotech., Piscataway, NJ).
pEGFP constructs. Plasmids resulting from subcloning full-length human GGA1 and GGA3 into EcoRI–SalI-digested pEGFP-C2 (Clontech, Palo Alto, CA) were described previously (Puertollano et al., 2001b). A full-length human GGA2 cDNA obtained after partial EcoRI digestion of pCR3.1-GGA2 was subcloned into EcoRI-restricted pEGFP-C2. The pEGFP-C2 human stonin2 proline-rich domain was described by Martina et al. (2001). pEGFP-C3 human Rabaptin-5 was obtained by filling in with T4 DNA polymerase an ApaI–NotI fragment excised from pCI-neo-myc-human Rabaptin-5 followed by ligation into the SmaI site of pEGFP-C3.
pGBT9 constructs. EcoRI–SalI fragments encoding full-length GGA1, GGA1-VHS (residues 5–168), GGA2-VHS (residues 1–183), full-length GGA3, GGA3-VHS (residues 1–146) and GGA3-GAT (residues 147–313) obtained from the pGAD424 constructs described by Puertollano et al. (2001a), as well as EcoRI–SalI fragments encoding GAT domains of GGA1 (residues 148–314) and GGA2 (164–330), obtained from the cognate pEGFP constructs described by Puertollano et al. (2001b), were subcloned into the corresponding sites of pGBT9 (Clontech). An EcoRI fragment encoding full-length GGA2 (obtained after partial digestion) was subcloned into the corresponding site of pGBT9. Similarly, EcoRI–SalI and BamHI–SalI fragments of GGA1-GAE and GGA2-GAE, respectively, obtained from the above-described GST constructs, were subcloned into corresponding sites of pGBT9. An EcoRI–BglII fragment encoding GGA3-GAE (residues 494–723, also comprising a fraction of the hinge) obtained from the corresponding pGAD424 construct (Puertollano et al., 2001a) was subcloned into EcoRI–BamH1-restricted pGBT9. Finally, EcoRI–XhoI fragments of mouse γ1- and human γ2-ears (described in the GST fusion section) as well as full-length mouse γ1 were subcloned into EcoRI–SalI sites of pGBT9.
pGAD-human Rabaptin-5 constructs. pGAD10-myc-Rabaptin-5, pGAD10- myc-Rabaptin-5/5–234, pGAD10-myc-Rabaptin-5-/5–546, pGADGH-Rabaptin-5/551–862 and pGADGH-Rabaptin-5/739–862 were described by Vitale et al. (1998) (termed pGAD10-mycUEP, pGAD10-mycUEP-5–235, pGAD10-myc-UEPΔ547–862, pGADGH-L1_46 and pGADGH-Rabaptin-739–862, respectively in this reference).
Human Rabaptin-5 mutants. pGAD constructs encoding deletion mutants corresponding to the 5–269, 5–351, 5–406, 5–476, 551–661 and 551–789 fragments of human Rabaptin-5, as well as the 5–476 fragment containing the DPF388–390AAA substitution, were prepared from pGAD10-myc-Rabaptin-5/5–546 or pGAD10-GH-Rabaptin-5/551–862 templates using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Deletion mutants corresponding to Rabaptin-5 fragments 311–476, 406–476, 406–441, 441–476 and 428–455 were prepared by PCR amplification from pGAD-myc-Rabaptin-5/5–546, using upstream and downstream primers including EcoRI and BamHI sites, respectively, and other reagents in the Advantage–HF2-PCR system (Clontech). Amplified fragments were digested with the corresponding enzymes and subcloned into EcoRI–BamHI-restricted pGAD424 (Clontech). The alanine scanning mutagenesis, either substituting blocks of 2–4 contiguous amino acids in the Rabaptin-5 429–455 region or introducing single alanine substitutions in the Rabaptin-5 438–443 region for alanine residues, was also performed using the QuickChange Site-Directed Mutagenesis system. The effect of the substitution of blocks of 2–4 residues in the 429–455 region was analyzed in the 5–476, 406–476 and 428–455 fragments of Rabaptin-5, while single substitutions were only performed in the 5–476 fragment.
His6-tagged fusion proteins. EcoRI fragments encoding the 5–234 or 5–546 regions of myc-tagged human Rabaptin-5 (obtained from the pGAD constructs described above), as well as an EcoRI–SalI fragment encoding a Rabaptin-5/5–476 mutant with a DFG438–440AAA substitution were subcloned into corresponding sites of pET-28a(+) (Novagen, Madison, WI). The His6-tagged proteins were expressed in E.coli strain BL21(DE3) (Novagen) and purified over Ni-NTA columns (Qiagen, Valencia, CA).
GST fusion pull-down assays
Bovine brain cytosol was prepared by homogenization in 25 mM HEPES pH 7.7, 250 mM sucrose, 2 mM EDTA supplemented with protease inhibitors (EDTA-free Complete™, Roche, Indianapolis, IN). The bovine brain was homogenized at a 3:1 buffer:tissue ratio using a Polytron-Brinkmann (Westbury, NY) (four bursts of 20 s, position 10). The homogenate was centrifuged for 10 min at 12 000 g and 4°C, and the corresponding supernatant subjected to further centrifugation for 60 min at 105 000 g and 4°C. The 105 000 g supernatant (cytosolic fraction) was aliquoted and stored at –80°C. Thawed cytosolic fractions were centrifuged for 15 min at 300 000 g and 4°C in a Beckman TL-100 desktop ultracentrifuge immediately before assays. Samples containing 30 µg of GST fusion proteins were incubated for 2 h at 4°C with 40 µl of glutathione–Sepharose (Pharmacia Biotech.) in a final volume of 1 ml of phosphate-buffered saline (PBS)/0.25% Triton X-100 supplemented with protease inhibitors. Beads containing immobilized GST fusions were washed once by resuspension in 1.0 ml of PBS/0.25% Triton X-100 and microfuge centrifugation (3 min, 2000 g). Washed beads were subsequently incubated for 2 h at 4°C with bovine brain cytosol (1.5 mg of protein) in a final volume of 1 ml of 15 mM HEPES pH 7.0, 75 mM NaCl, 0.25% Triton X-100 (binding buffer), supplemented with protease inhibitors. Following incubation, beads were washed twice by resuspension in 1 ml of binding buffer and centrifugation as described above. After removal of the last wash supernatant, the beads were resuspended in 50 µl of 2× Laemmli buffer, followed by incubation for 10 min at 90°C and microfuge centrifugation (2 min, 16 000 g). The supernatants with the eluted proteins were subjected to SDS–PAGE (10% acrylamide) and immunoblotting.
Effect of His6-Rabaptin-5 fragments on GGA–clathrin interaction
GST fusion pull-down assays were performed as described in the above paragraph with the following modifications: (i) 20 µg of GST–GGA1-hinge + GAE or GST–GGA2-hinge + GAE were used; (ii) following immobilization of the GST fusion proteins, and prior to the addition of cytosol, the washed beads were incubated for 30 min at 4°C with or without competitors (His6-Rabaptin-5 fragments, gelatin or BSA) in a final volume of 650 µl of binding buffer; (iii) at the end of this period, 250 µl of cytosol (protein concentration 6 mg/ml) was added and the incubation at 4°C continued for an additional 90 min.
Binding of biotinylated peptides to γ-adaptin ear and GGA-GAE domains
Peptides CDESDFGPLVGADS and CDESDAGPAVGADS were obtained from New England Peptide (Fitchburg, MA) and subjected to biotinylation using EZ-Link PEO-maleimide-activated biotin (Pierce, Rockford, IL) as described by Aguilar et al. (2001). Biotinylated peptides were immobilized to streptavidin–agarose by incubation for 2 h at 20°C (30 nmol of peptides and 50 µl of washed beads in 1 ml of PBS pH 7.0).
Peptide-bound beads were washed three times by resuspension in 1 ml of 15 mM HEPES pH 7.0, 75 mM NaCl, 0.25% Triton X-100 supplemented with 0.1% BSA and protease inhibitors (peptide binding buffer) and centrifugation for 3 min at 2000 g. Washed beads were subsequently incubated for 2 h at 4°C with 3 µg of GST fusion proteins in a final volume of 1 ml of peptide binding buffer. Following incubation, beads were washed three times with 1 ml of binding buffer without BSA as described above. Bound proteins were eluted as described for the GST pull-down assays and subjected to SDS–PAGE followed by immunoblotting with rabbit anti-GST antiserum.
Cell transfection
HeLa cells (American Type Culture Collection, Manassas, VA) were cultured on 100 mm dishes at 37°C under a humidified atmosphere (95:5 air:CO2) in Dulbecco’s modified Eagle’s medium/high glucose (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin. When cells reached ∼30% confluence, they were transfected with 15–20 µg of DNA constructs using the Fugene™ reagent (Roche, Indianapolis, IN).
Immunoprecipitation
HeLa cells cultured on 100 mm dishes (either wild-type, or 15–24 h after transfection) were washed twice with cold PBS and immediately resuspended in 1 ml of 50 mM Tris–HCl pH 7.4, 0.5% Triton X-100, 75 mM NaCl (lysis buffer), supplemented with protease inhibitors. After a 60 min treatment at 4°C, lysates were centrifuged for 20 min at 16 000 g and 4°C. Supernatants were pre-cleared by incubation (60 min at 4°C) with 20 µl of protein G–Sepharose and centrifugation (10 min at 16 000 g and 4°C). Pre-cleared lysates were subsequently incubated for 2–4 h at 4°C with 1 µg BSA and 20 µl of protein–G Sepharose (Amersham-Pharmacia Biotech.) bound to either rabbit anti-GGA1 (M.S.Robinson, University of Cambridge), mouse monoclonal anti-Rabaptin-5 (Transduction Laboratories), mouse monoclonal anti-GFP (Roche) or rabbit anti-σ3 (Dell’Angelica et al., 1997). The IgG-bound protein G–Sepharose was prepared previously by incubation with 2.5 µg of antibody in 500 µl of PBS/0.01% Triton X-100 for 2 h at 4°C, followed by two washes with lysis buffer. Following immunoprecipitation, samples were centrifuged for 3 min at 2000 g and 4°C and washed twice with 1 ml of lysis buffer containing a reduced (0.1%) concentration of Triton X-100. Washed beads were subsequently processed as described for GST pull-down assays.
Depletion of Rabaptin-5–Rabex-5 complexes from bovine brain cytosol
A 1.5 ml sample of bovine brain cytosol (protein concentration 6 mg/ml) was incubated for 90 min at 4°C with protein G–Sepharose-immobilized anti-Rabaptin-5 (prepared as described in the Immunoprecipitation section). At the end of this period, the sample was centrifuged for 2 min at 16 000 g and 4°C. The supernatant with the immunodepleted cytosol was subsequently subjected to three additional rounds of incubation with new aliquots of protein G–Sepharose–anti-Rabaptin-5. Samples of untreated cytosol and of the fractions obtained after each immunodepletion step were analyzed by SDS–PAGE and immunoblotting, and were also used in pull-down experiments with GST–GGA1-GAE. In a second experimental set, a 1 ml sample of cytosol was incubated for 90 min at 4°C with GST–GGA1-GAE immobilized on glutathione–Sepharose (see GST pull-down section). Following incubation, the treated cytosol was separated by centrifugation for 2 min at 16 000 g and 4°C and subjected to three additional rounds of treatment with fresh aliquots of GST–GGA1-GAE. Samples of untreated cytosol, and of the fractions obtained after each treatment, were analyzed by SDS–PAGE and immunoblotting.
Antisera used in immunoblotting
The following antisera dilutions were used for immunoblotting: 1:500 mouse monoclonal anti-Rabaptin-5 (Transduction Laboratories, Lexington, KY); 1:1000 rabbit anti-Rabex-5 (Horiuchi et al., 1997); 1:1000 mouse monoclonal anti-clathrin heavy chain (Transduction Laboratories); 1:1250 mouse monoclonal anti-GFP (Roche); and 1:5000 rabbit anti-GST (Dell’Angelica et al., 1998). Immunoblots were developed using horseradish peroxidase-coupled secondary antisera and an enhanced chemiluminiscence reagent (Western Lightning, Perkin Elmer Life Sciences, Boston, MA).
Yeast two-hybrid analysis
The AH109 yeast reporter strain was maintained on YPD agar plates. Transformation of AH109 cells with pGAD- and pGBT9-based constructs by the lithium acetate method was performed following the guidelines in the Matchmaker two-hybrid system (Clontech). Double transformants were isolated on synthetic defined medium lacking leucine and tryptophan. Interaction of fusion proteins was monitored by activation of HIS3 gene transcription following plating on medium lacking histidine, leucine and tryptophan, in the presence or absence of the indicated concentrations of the competitive inhibitor of the HIS3 protein 3-amino-1,2,4-triazole (3-AT).
Immunofluorescence microscopy
HeLa cells were plated on 12 mm coverslips and grown to ∼30% confluency in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. Where indicated, coverslips were washed quickly by immersion in PBS and subjected to a short (1 min) permeabilization pulse in 25 mM HEPES/HCl, 150 mM potassium glutamate, 25 mM potassium chloride, 2.5 mM potassium acetate, 5 mM EDTA and 0.00375% saponin. Coverslips were washed quickly by immersion in PBS and immediately fixed for 12 min in a 4% formaldehyde solution in PBS. Following fixation, coverslips were washed twice for 10 min in PBS and incubated for 1 h at room temperature with the indicated combinations of primary antisera diluted in 0.1% BSA, 0.1% saponin, 0.02% sodium azide in PBS (immunofluorescence buffer): rabbit anti-GGA1 (M.S.Robinson, University of Cambridge), mouse monoclonal anti-Rabaptin-5 (Transduction Laboratories), mouse monoclonal anti-CI-MPR (Susan Pfeffer, Stanford University), mouse monoclonal anti-TfR (B3/25), mouse monoclonal anti-GFP (Roche) and rabbit anti-GFP (Molecular Probes, Eugene, OR). At the end of this period, coverslips were washed twice with PBS followed by incubation for 45 min with the indicated combinations of secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA) also diluted in immunofluorescence buffer (see legend to Figure 10). Following incubation, coverslips were again washed twice with PBS and mounted on slides using Fluoromount-G (Southern Biotech. Associates, Birmingham, AL). Images were obtained in an inverted confocal laser scanning microscope (LSM410; Carl Zeiss Inc., Thornwood, NY).
Acknowledgments
Acknowledgements
We thank Rubén C.Aguilar, José A.Martina and Rosa Puertollano for valuable discussions and DNA constructs, as well as Suzanne Pfeffer, Linton Traub and Margaret S.Robinson for generous gifts of reagents.
References
- Aguilar R.C., Boehm,M., Gorshkova,I., Crouch,R.J., Tomita,K., Saito,T., Ohno,H. and Bonifacino,J.S. (2001) Signal-binding specificity of the µ4 subunit of the adaptor protein complex AP-4. J. Biol. Chem., 276, 13145–13152. [DOI] [PubMed] [Google Scholar]
- Boehm M. and Bonifacino,J.S. (2001) Adaptins: the final recount. Mol. Biol. Cell, 12, 2907–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman A.L., Zhang,C., Zhu,X. and Kahn,R.A. (2000) A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell, 11, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett T.J., Traub,L.M. and Fremont,D.H. (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure, 10, 797–809. [DOI] [PubMed] [Google Scholar]
- Costaguta G., Stefan,C.J., Bensen,E.S., Emr,S.D. and Payne,G.S. (2001) Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell, 12, 1885–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosulich S.C., Horiuchi,H., Zerial,M., Clarke,P.R. and Woodman,P.G. (1997) Cleavage of Rabaptin-5 blocks endosome fusion during apoptosis. EMBO J., 16, 6182–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica E.C., Ohno,H., Ooi,C.E., Rabinovich,E., Roche,K.W. and Bonifacino,J.S. (1997) AP-3: an adaptor-like protein complex with ubiquitous expression. EMBO J., 16, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica E.C., Klumperman,J., Stoorvogel,W. and Bonifacino,J.S. (1998) Association of the AP-3 adaptor complex with clathrin. Science, 280, 431–434. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E.C., Puertollano,R., Mullins,C., Aguilar,R.C., Vargas, J.D., Hartnell,L.M. and Bonifacino,J.S. (2000) GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol., 149, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzis S., Sonnichsen,B. and Zerial,M. (2002) Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat. Cell Biol., 2, 124–133. [DOI] [PubMed] [Google Scholar]
- Doray B., Bruns,K., Ghosh,P. and Kornfeld,S.A. (2002) Autoinhibition of the ligand-binding site of GGA1/3 VHS domains by an internal acidic cluster-dileucine motif. Proc. Natl Acad. Sci. USA, 99, 8072–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournier H., Stenmark,H., Rybin,V., Lippé,R. and Zerial,M. (1998) Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J., 17, 1930–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Lui,W.W., Bright,N.A., Totty,N., Seaman,M.N. and Robinson,M.S. (2000) A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol., 149, 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H. et al. (1997) A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell, 90, 1149–1159. [DOI] [PubMed] [Google Scholar]
- Kent H.M., McMahon,H.T., Evans,P.R., Benmerah,A. and Owen,D.J. (2002) γ-Adaptin appendage domain: structure and binding site for Eps15 and γ-synergin. Structure, 10, 1139–1148. [DOI] [PubMed] [Google Scholar]
- Korobko E.V., Kiselev,S.L. and Korobko,I.V. (2002) Multiple Rabaptin-5-like transcripts. Gene, 292, 191–197. [DOI] [PubMed] [Google Scholar]
- Lippé R., Miaczynska,M., Rybin,V., Runge,A. and Zerial,M. (2001) Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol. Biol. Cell, 12, 2219–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Bonangelino,C.J., Aguilar,R.C. and Bonifacino,J.S. (2001) Stonin 2: an adaptor-like protein that interacts with components of the endocytic machinery. J. Cell Biol., 153, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M. and Zerial M. (2002) Mosaic organization of the endocytic pathway. Exp. Cell Res., 272, 8–14. [DOI] [PubMed] [Google Scholar]
- Misra S., Puertollano,R., Kato,Y., Bonifacino,J.S. and Hurley,J.H. (2002) Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature, 415, 933–937. [DOI] [PubMed] [Google Scholar]
- Mullins C. and Bonifacino,J.S. (2001) Structural requirements for function of yeast GGAs in vacuolar protein sorting, α-factor maturation and interactions with clathrin. Mol. Cell Biol., 23, 7981–7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelkerken B., Van Anken,E., Van Raak,M., Gerez,L., Mohrmann,K., Van Uden,N., Holthuizen,J., Pelkmans,L. and Van Der Sluijs,P. (2000) Rabaptin4, a novel effector of the small GTPase rab4a, is recruited to perinuclear recycling vesicles. Biochem. J., 346, 593–601. [PMC free article] [PubMed] [Google Scholar]
- Nishimune H., Uyeda,A., Nogawa,M., Fujimori,K. and Taguchi,T. (1997) Neurocrescin: a novel neurite-outgrowth factor secreted by muscle after denervation. Neuroreport, 8, 3649–3654. [DOI] [PubMed] [Google Scholar]
- Nogi T. et al. (2002) Structural basis for the accessory protein recruitment by the γ-adaptin ear domain. Nat. Struct. Biol., 9, 527–531. [DOI] [PubMed] [Google Scholar]
- Owen D.J., Vallis,Y., Noble,M.E., Hunter,J.B., Dafforn,T.R., Evans,P.R. and McMahon,H.T. (1999) A structural explanation for the binding of multiple ligands by the α-adaptin appendage domain. Cell, 97, 805–815. [DOI] [PubMed] [Google Scholar]
- Page L.J., Sowerby,P.J., Lui,W.W. and Robinson,M.S. (1999) γ-Synergin: an EH domain-containing protein that interacts with γ-adaptin. J. Cell Biol., 146, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poussu A., Lohi,O. and Lehto,V.P. (2000) Vear, a novel Golgi-associated protein with VHS and γ-adaptin ‘ear’ domains. J. Biol. Chem., 275, 7176–7183. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Aguilar,R.C., Gorshkova,I., Crouch,R.J. and Bonifacino,J.S. (2001a) Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science, 292, 1712–1716. [DOI] [PubMed] [Google Scholar]
- Puertollano R., Randazzo,P.A., Presley,J.F., Hartnell,L.M. and Bonifacino,J.S. (2001b) The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell, 105, 93–102. [DOI] [PubMed] [Google Scholar]
- Puertollano R., van der Well,N.N., Greene,L.E., Eisenberg,E., Peters,P.J. and Bonifacino,J.S. (2003) Morphology and dynamics of clathrin/GGA1-coated carrier budding from the trans-Golgi network. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T. et al. (2002a) Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature, 415, 937–941. [DOI] [PubMed] [Google Scholar]
- Shiba Y., Takatsu,H., Shin,H.W. and Nakayama,K. (2002b) γ-Adaptin interacts directly with Rabaptin-5 through its ear domain. J. Biochem., 131, 327–336. [DOI] [PubMed] [Google Scholar]
- Slepnev V.I. and De Camilli,P. (2000) Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat. Rev. Neurosci., 1, 161–172. [DOI] [PubMed] [Google Scholar]
- Stenmark H., Vitale,G., Ullrich,O. and Zerial,M. (1995) Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell, 83, 423–432. [DOI] [PubMed] [Google Scholar]
- Swanton E., Bishop,N. and Woodman,P. (1999) Human Rabaptin-5 is selectively cleaved by caspase-3 during apoptosis. J. Biol. Chem., 274, 37583–37590. [DOI] [PubMed] [Google Scholar]
- Takatsu H., Yoshino,K. and Nakayama,K. (2000) Adaptor γ ear homology domain conserved in γ-adaptin and GGA proteins that interact with γ-synergin. Biochem. Biophys. Res. Commun., 271, 719–725. [DOI] [PubMed] [Google Scholar]
- Takatsu H., Katoh,Y., Shiba,Y. and Nakayama,K. (2001) Golgi-localizing, γ-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem., 276, 28541–28545. [DOI] [PubMed] [Google Scholar]
- Traub L.M., Downs,M.A., Westrich,J.L. and Fremont,D.H. (1999) Crystal structure of the α appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc. Natl Acad. Sci. USA, 96, 8907–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale G., Rybin,V., Christoforidis,S., Thornqvist,P., McCaffrey,M., Stenmark,H. and Zerial,M. (1998) Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J., 17, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Doray,B., Poussu,A., Lehto,V.P. and Kornfeld,S. (2001) Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science, 292, 1716–1718. [DOI] [PubMed] [Google Scholar]