Abstract
Helicobacter pylori, bacteria that colonize the human gastric mucosa, possess a large number of genes for restriction-modification (R-M) systems, and essentially, every strain possesses a unique complement of functional and partial R-M systems. Nearly half of the H.pylori strains studied possess an active type IIs R-M system, HpyII, with the recognition sequence GAAGA. Recombination between direct repeats that flank the R-M cassette allows for its deletion whereas strains lacking hpyIIRM can acquire this cassette through natural transformation. We asked whether strains lacking HpyII R-M activity can acquire an active hpyIIRM cassette [containing a 1.4 kb kanamycin resistance (aphA) marker], whether such acquisition is DNase sensitive or resistant and whether restriction barriers limit acquisition of chromosomal DNA. Our results indicate that natural transformation and conjugation-like mechanisms may contribute to the transfer of large (4.8 kb) insertions of chromosomal DNA between H.pylori strains, that inactive or partial R-M systems can be reactivated upon recombination with a functional allele, consistent with their being contingency genes, and that H.pylori R-M diversity limits acquisition of chromosomal DNA fragments of ≥1 kb.
INTRODUCTION
Restriction-modification (R-M) systems defend bacteria against invasion by foreign DNA such as conjugative plasmids and bacteriophages (1). The type II family of R-M systems consists of paired enzymes that recognize identical DNA sequences but have contrasting enzymatic functions. The restriction endonuclease (ENase) cleaves DNA within the recognition site while the modification enzyme (MTase) methylates adenosyl or cytosyl residues within the recognition sequence, thereby protecting the host chromosome from cognate restriction activity (1).
Helicobacter pylori are Gram-negative curved bacteria that colonize the human stomach and increase the risk of development of peptic ulcer disease and gastric adenocarcinoma (the major form of stomach cancer in the world) (2). For H.pylori, several studies have shown significant interstrain variation in R-M system activity (3–7), a diversity that influences strain transformability (8,9). Lack of conservation in R-M activity could be due to the absence or partial absence of, or point mutations in, the R-M systems, and comparison of the genome sequences of strain 26695 and J99 provide evidence for all three phenomena (3,4,10–12).
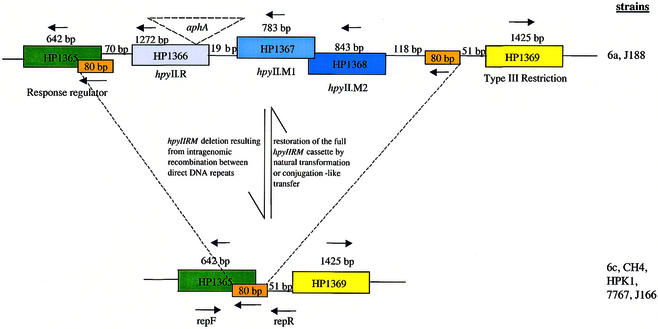
Previous work in our laboratory has shown that repetitive DNA sequences flanking the H.pylori hpyIIRM system facilitate deletion of the R-M cassette from the chromosome (13). The full hpyIIRM cassette can be re-acquired through natural transformation using chromosomal DNA from the parental strain (Fig. 1) (13). Comparison of the two available H.pylori genome sequences identified long repeat sequences flanking many strain-specific R-M systems (14) suggesting that the deletion/re-acquisition model might represent a general mechanism through which H.pylori strains may vary R-M content and that H.pylori R-M systems may act as ‘transposon-like’ mobile genetic elements (14).
Figure 1.
Spontaneous deletion and horizontal re-acquisition of the hpyIIRM system. Sequence analysis of the hpyIIRM system in H.pylori strain 26695 identified 80 bp repeats flanking the R-M system. The presence of direct repeat sequences facilitates generation of deletions through mechanisms involving sister-strand exchange or strand slippage. These deletions result in excision of the intervening region and one copy of the repeat. Helicobacter pylori strains lacking the hpyIIRM cassette, but containing one copy of the repeat unit, may re-acquire a functional cassette through transformation. Primers repF and repR flanking the 80 bp repeats were used in PCR to confirm acquisition of hpyIIRM::aphA cassettes by recipient strains. An aphA cassette was inserted into hpyIIR of strain 6a to create strain RA1.
Strain-specific R-M system activity in H.pylori presents interstrain barriers to the transfer of plasmid DNA (8); however, the role of R-M diversity in restricting chromosomal DNA uptake and transformation has not been defined. For a naturally competent organism such as H.pylori (15), the ability to restrict incoming chromosomal DNA is an efficient means of preventing competing strains from subverting the genome of a co-colonizing strain through natural transformation.
To test the hypothesis that H.pylori interstrain R-M diversity restricts transformation by chromosomal DNA from other H.pylori strains, we conducted interstrain recombination experiments between isogenic or non-isogenic strains using chromosomal DNA, containing markers ranging in size from 1 to 4.8 kb. Further, we examined the role of R-M activity in restricting chromosomal DNA acquired through natural transformation or via conjugation-like mechanisms. Our findings support the hypothesis that, although both natural transformation and conjugation-like mechanisms contribute to the transfer of large (4.8 kb) fragments of chromosomal DNA, diversity in functional R-M systems represents a barrier to the acquisition of chromosomal DNA fragments of at least 1.0 kb. These results suggest that there is a double-stranded DNA intermediate in the H.pylori chromosomal transformation process.
MATERIALS AND METHODS
Bacterial strains
Helicobacter pylori strains used in this study (Table 1) were obtained from the NYU Helicobacter/Campylobacter reference strain collection. To select for spontaneous streptomycin- or spectinomycin-resistant H.pylori strains, ∼1010 cells were incubated on brucella agar (BA) plates containing streptomycin (10 µg/ml) or spectinomycin (10 µg/ml), respectively. The plates were incubated at 37°C (5% CO2) for 6–7 days and antibiotic-resistant colonies were harvested.
Table 1. Helicobacter pylori strains used in this study.
| Strain designation | Relevant genotype | hpyIIRM phenotype | Antibiotic resistance markera | Reference |
|---|---|---|---|---|
| 6a | hpyIIRM+, cagA+, vacA s1m1 | R+M+ | None | (13) |
| 6c | hpyIIRM–, cagA+, vacA s1m1 | R–M– | None | (13) |
| CH4 | hpyIIRM–, cagA+, vacA s1am1 | R–M– | None | (5) |
| HPK1 | hpyIIRM–, cagA+, vacA s1m1 | R–M– | None | (8) |
| 7767 | hpyIIRM–, cagA+, vacA s2m2 | R–M– | None | (8) |
| J166 | hpyIIRM–, cagA+, vacA s1bm1 | R–M– | None | (8) |
| J188 | hpyIIRM+, cagA–, vacA s1m1 | R–M–b | None | (8) |
| RA1 (fomerly 6aRK) | 6a (hpyIIR::aphA) | R–M+c | Kan | (13) |
| RA2 | 6a (hpyIIR::aphA, rpoB15) | R–M+c | Kan, Rif | This study |
| RA3 | 6a (vacA::aphA) | R+M+ | Kan | This study |
| RA4 | 6a (rpsL1, rrn16S1) | R+M+ | Strep, Spec | This study |
| RA5 | 6c (rpsL1, rrn16S1) | R–M– | Strep, Spec | This study |
| RA6 | J188 (rpsL1, rrn16S1) | R–M–b | Strep, Spec | This study |
| RA7 | J166 (rpsL1, rrn16S1) | R–M– | Strep, Spec | This study |
Helicobacter pylori interstrain recombination
Each interstrain recombination experiment involved two strains (A and B) with mutually exclusive antibiotic resistance properties, as described previously (13). After 48 h of growth on appropriate selective BA plates, cells of each strain were harvested and suspended in 1 ml of saline. The cells were centrifuged, the supernatant discarded, the cells resuspended in 175 µl of saline and then 25 µl aliquots spotted onto trypticase soy agar (TSA) plates as follows: strain A alone, strain B alone, strain A + B, strain A + B + DNase I (250 µg/µl). The plates were incubated overnight at 37°C in an atmosphere with 5% CO2, after which bacteria were harvested in 1 ml of saline, centrifuged for 5 min at 6000 g, supernatant discarded and cells resuspended in 1 ml of saline. The suspensions from strain A alone and strain B alone were serially diluted, and 100 µl of 10–5, 10–6 and 10–7 dilutions was inoculated on TSA plates without antibiotics. For all suspensions, 50 and 250 µl were inoculated onto BA plates containing 10% newborn calf serum and either kanamycin (25 µg/ml), streptomycin (20 µg/ml) and spectinomycin (20 µg/ml) (BA-KStSp plates); or kanamycin, streptomycin and rifampin (5 µg/ml) (BA-KStR plates). All plates were incubated for 96 h, then colonies counted and transformation frequencies calculated. Calculations were based on at least three independent experiments.
Natural transformation
Transformation of H.pylori was performed as described previously (9). In short, H.pylori cells were scraped from 36 h cultures grown on TSA with 5% sheep blood and resuspended in 175 µl saline. Then, 25 µl aliquots of cell suspension were spotted onto a TSA plate and 100 ng of the transforming chromosomal DNA or PCR product added. After incubating the mixture at 37°C (5% CO2) for 12 h, cell spots were transferred to BA plates containing 5% fetal calf serum and appropriate selective antibiotics, and incubated at 37°C (5% CO2) for 4–5 days.
PCR and restriction endonuclease digestion
To confirm acquisition of the H.pylori hpyIIRM cassette, PCR, using primers repF (nt 1427401–1427424; www.tigr.org/hpylori) and repR (nt 1430845–1430867) that flank hpyIIRM, was performed using 100 ng of chromosomal DNA from the recipient strain as template, as described previously (13). PCR, using primers vacAF (5′-GTGAAA GCGAAAAACAAGAAATTG-3′) and vacR (5′-CGTGCC ATCGGCTTTAGTGTTG-3′) were performed to amplify, from plasmid pCTB8 (13), vacA::aphA cassettes used in transformation experiments described above. Unless otherwise noted, all reactions were run for 35 cycles in mixtures containing 100 ng template DNA, 200 ng of each primer, 0.5 U Taq polymerase (Qiagen, Valencia, CA) in a 50 µl volume. DNA was digested with 10 U of MboII overnight and bands were resolved in 1% agarose gels. To assess M.HpyII activity amongst H.pylori strains, 1 µg of chromosomal DNA was digested with 5 U of MboII (NEB) at 37°C for 2 h and products visualized by agarose gel electrophoresis.
RESULTS
Effect of DNase I on natural transformation of H.pylori cells by chromosomal DNA
To examine mechanisms for chromosomal DNA transfer in H.pylori, we conducted transformation experiments between strains containing mutually exclusive antibiotic resistance markers (Table 1), in the presence and absence of DNase I. Helicobacter pylori strain RA4 was incubated with chromosomal DNA from strain RA2 and transformants were selected on BA plates containing kanamycin, streptomycin and spectinomycin (BA-KStSp). In the absence of DNase I, strain RA4 acquired a functional aphA cassette at a frequency of (1.4 × 10–4) ± (2.0 × 10–4), however, when DNase I was mixed with RA2 DNA, no transformants were observed in ∼109 recipient cells. These experiments confirm, with these strains and the aphA cassette, that DNase treatment abrogates natural transformation (16).
Horizontal transfer of a chromosomal point mutation between isogenic H.pylori strains
Next, we sought to measure the frequency of transfer of a point mutation by natural transformation or via conjugation-like mechanisms. The recipient strain was strain RA2 and strain RA4 or RA5, which have the same clonal origin but differ in the presence of the hpyIIRM locus (13), was used as the donor of the Strepr-conferring point mutation. In interstrain recombination experiments, strain RA2 cells acquired the Strepr point mutation from RA4 cells at a frequency of (2.6 × 10–5) ± (1.4 × 10–5), but the addition of DNase I decreased this rate by 95.4% (Table 2). Similarly, the point mutation conferring Strepr in RA5 cells was transferred to RA2 cells at a frequency of (5.0 × 10–5) ± (3.5 × 10–5) with a 94.5% reduction in the number of recombinant cells in the presence of DNase I (Table 2). That transformation experiments using RA4 DNA incubated with DNase yielded no RA2 transformants suggests that Strepr transformants observed during interstrain recombination experiments did not result from natural transformation of recipient cells with DNA that escaped DNase activity, consistent with earlier studies (16). All KRSt-resistant colonies also were spectinomycin sensitive, indicating that the observed transformants did not result from concomitant transfer of kanamycin and rifampin resistance from strain RA2 to strain RA4 or RA5. That incubation of strain RA2 on streptomycin-containing plates yielded no Strepr transformant indicated that spontaneous mutation was not responsible for the observed transformants. These results support the earlier finding that although natural transformation accounts for most of the observed genetic exchange between H.pylori strains, a Strepr-conferring point mutation can be transferred between H.pylori strains via a DNase-resistant mechanism (16).
Table 2. Helicobacter pylori matings to compare transfer of detectable alleles.
| H.pylori strain A | H.pylori strain Ba | Treatment with DNase (250 µg/µl) | Recombination frequency (× 10–7)b | |
|---|---|---|---|---|
| Transfer of a point mutation to strain Ac | Transfer of an altered hpyIIM allele to strain Bd | |||
| RA2e | RA4 | – | 262 ± 144 | 79 ± 20 |
| + | 12 ± 6 | 9 ± 2 | ||
| RA2 | RA5 | – | 496 ± 354 | 88 ± 72 |
| + | 27 ± 23 | 4 ± 4 | ||
| RA2 | RA6 | – | 513 ± 262 | 3 ± 2 |
| + | 29 ± 22 | <0.66 | ||
| RA2 | RA7 | – | 319 ± 225 | <0.18 |
| + | 16 ± 5 | <0.19 | ||
aEach of these strains have point mutations in rpsL and rrn16S resulting in Strepr, Specr phenotypes.
bEach result shown represents the mean ± SD of three independent matings.
cHorizontal transfer of a point mutation was determined by the ability of strain A to acquire Strepr from strain B producing a KanrRifrStrepr strain. No spontaneous mutations to Strepr were detected.
dTransfer of the hpyIIRM system was determined by the ability of strain B to acquire Kanr from strain A (hpyIIR::aphA) producing a KanrStreprSpecr strain.
eRA2 is Kanr (aphA in hpyIIRM) and Rifr.
Horizontal transfer of the hpyIIRM system between isogenic H.pylori strains
Next, to determine whether larger DNA fragments could be mobilized by conjugation-like mechanisms between isogenic strains, we examined horizontal transfer of a 1.3 kb aphA cassette within the hpyIIRM locus (from strain RA2 to RA4) or the entire 4.8 kb hpyIIRM::aphA cassette (which contains aphA in hpyIIR) from strain RA2 to RA5, in the presence or absence of DNase I. Co-culture of strains RA2 and RA4 and selection for transformants on BA-StSpK plates indicated that strain RA4 acquired the aphA cassette at a frequency of (7.9 × 10–6) ± (2.0 × 10–6), and that addition of DNase I decreased this frequency by 88.9% (Table 2). Similarly, co-culture of strains RA2 and RA5 and selection for transformants on BA-StSpK plates resulted in the transfer of the entire 4.8 kb hpyIIRM::aphA cassette from RA2 cells to RA5 cells at a frequency of (8.8 × 10–6) ± (7.2 × 10–6) (Table 2), confirming that a strain lacking hpyIIRM (6c) can re-acquire an R-M system through horizontal gene transfer (13). Addition of DNase I decreased this recombination frequency by 96.0%. All observed transformants were spectinomcyin sensitive, indicating that they did not result from the concomitant transfer of streptomycin and spectinomycin resistance from strain RA4 or RA5 to strain RA2. Similarly, that strain RA4 or RA5 incubated on kanamycin-containing plates yielded no kanamycin-resistant transformants indicated that the observed transformants did not result from spontaneous kanamycin resistance. For strains RA2, RA4 and RA5, there were no significant differences in the frequency of transfer of the Strepr point mutation (1 bp), the aphA cassette (∼1.3 kb) or the hpyIIRM::aphA cassette (∼4.8 kb) between clonal variants. These results indicate that chromosomal DNA fragments of ≤4.8 kb can be transferred without significant barriers between strains of the same clonal origin through both natural transformation and DNase-resistant methods (13).
Genetic exchange of chromosomal DNA between non-isogenic H.pylori strains
Previous studies have shown that plasmid DNA uptake by H.pylori from non-isogenic strains is limited by restriction barriers (8,9). To examine now whether barriers against transfer of chromosomal DNA exist between non-isogenic H.pylori strains, we conducted interstrain recombination experiments between strain RA2 and the hpyIIRM+ strain RA6 or the hpyIIRM– strain RA7. Co-culture of strains RA2 and RA6 and selection on BA-KRSt plates indicated that strain RA2 acquired Strepr at a frequency of (5.1 × 10–5) ± (2.6 × 10–5) with a 94.3% decrease in transformation frequency in the presence of DNase I (Table 2). That all transformants were spectinomycin sensitive, and that strain RA2 incubated alone yielded no Strepr transformants, indicated that strain RA2 had acquired the Strepr-conferring point mutation from a non-isogenic H.pylori strain (RA6) at a frequency similar to that of strains of the same clonal origin (RA4, RA5). Parallel results were obtained from co-culture of strain RA2 with strain RA7 and selection on BA-KRSt plates, confirming that there are no significant barriers against DNase-sensitive or -resistant transfer of a chromosomal point mutation between non-isogenic H.pylori strains.
Transfer of the aphA cassette between non-isogenic strains was measured by co-culture of strains RA2 and RA6 and selection on BA-StSpK plates. Strain RA6 acquired the aphA cassette at a frequency of (3.4 × 10–7) ± (2.1 × 10–7) and no transformants were observed in the presence of DNase I (Table 2). That strain RA6 acquired the aphA cassette from strain RA2 at a significantly lower frequency (P = 0.02) than either strain RA4 or RA5 indicates that barriers between non-isogenic H.pylori strains affect the frequency of insertion of the 1.3 kb cassette. Co-culture of strain RA2 and RA7 and selection on BA-StSpK plates yielded no transformants in the presence or absence of DNase I (Table 2). Failure of the 4.8 kb hpyIIRM::aphA cassette to be transferred from RA2 to RA7 at a detectable frequency further defines the extent of the barriers existing between non-isogenic H.pylori strains that limit integration of larger chromosomal DNA insertions.
Acquisition of M.HpyII function
Next, we sought to determine whether the kanamycin-resistant RA5 and RA6 transformants acquired a functional M.HpyII in the original locus. Chromosomal DNA from these transformants was subjected to PCR using primers that flank the hpyIIRM cassette in strain RA2, and was also digested with MboII, an isoschizomer of HpyII. That strain RA5 DNA yielded only a 0.3 kb empty-site PCR product, and was sensitive to MboII digestion, whereas all kanamycin-resistant transformants yielded 4.8 kb PCR products consistent with the presence of the entire hpyIIRM::aphA cassette and were resistant to MboII digestion (Fig. 2), confirms that RA5 transformants acquired the full R-M cassette containing a functional hpyIIM in its original locus. The presence of empty-site products reflects the relative ease of amplifying shorter PCR products, and the existence of mixed populations (13). For strain RA6, which contains a full-length hpyIIRM cassette but lacks M.HpyII activity (13), PCR indicated that all 10 transformants examined acquired the aphA cassette in the original hpyIIR locus as expected; however, MboII digestion indicated that only seven (70%) of these 10 transformants acquired the functional hpyIIM allele. These results indicate that the recombination events in RA6 that yielded a selectable marker (aphA in hpyIIR) involved exchanges that varied in the extent of replacement of the flanking DNA containing hpyIIM.
Figure 2.
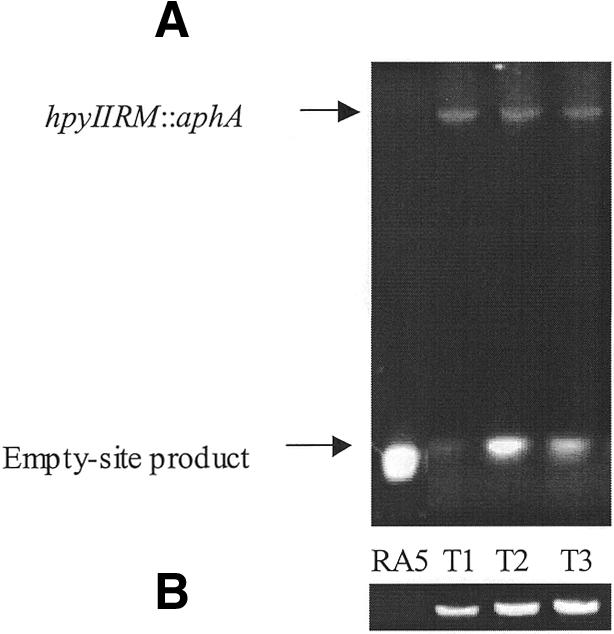
Acquisition of hpyIIM function in kanamycin-resistant RA5 transformants. (A) To determine whether kanamycin-resistant RA5 transformants acquired the hpyIIRM::aphA cassette in its original locus, PCR was performed using primers that flank the R-M cassette in strain RA2, with template chromosomal DNA from strain RA5 (control) and three transformants. (B) To determine whether kanamycin-resistant RA5 transformants acquired M.HpyII function, chromosomal DNA from strain RA5 (control) and the three transformants was digested with MboII, an isoschizomer of HpyII.
Restriction barriers limit horizontal transfer of chromosomal DNA between non-isogenic H.pylori strains
Next, we sought to determine whether the H.pylori barriers that limit acquisition of chromosomal DNA from non-isogenic strains reflect differences in natural competence. Experiments were conducted to examine the frequency of transforming the Strepr point mutation (1 bp), the aphA cassette (1.3 kb) or the hpyIIRM::aphA cassette (4.8 kb) originating from strain RA4, RA3 or RA1, respectively, into the hpyIIRM– strain 6c, which shares the same clonal origin or non-isogenic hpyIIRM– strains J166, CH4, HPK1, 7767 (Table 3). All five strains were transformed to Strepr to essentially the same extent, indicating that there were no significant differences in the competence of these strains. For control recipient strain 6c, there were also no significant differences in transformation by chromosomal DNA when selecting for the Strepr point mutation, the aphA cassette or hpyIIRM::aphA. However, for all four non-isogenic strains studied, the Strepr point mutation was acquired at significantly higher frequencies (P < 0.05) than was the 1.3 kb aphA or 4.8 kb hpyIIRM::aphA cassette. These data confirm that the limited acquisition of chromosomal DNA >1 kb from non-isogenic H.pylori strains reflects barriers rather than a lack of competence or competence signals.
Table 3. Transformation of isogenic and non-isogenic H.pylori strains by chromosomal DNA containing selectable markers of different sizes.
| Recipient H.pylori strain | Transformation frequency (× 10–7) | ||
|---|---|---|---|
| Transfer of a point mutation (1 bp)a from RA4 | Transfer of aphA cassette (1.3 kb)b from RA3 | Transfer of hpyIIRM::aphA cassette (4.8 kb) from RA1 | |
| 6cc | 141 ± 32 | 100 ± 19 | 49 ± 40 |
| J166d | 98 ± 7e | 4 ± 0.9 | <0.4 |
| CH4d | 106 ± 38e | 3 ± 2 | <0.4 |
| HPK1d | 222 ± 160e | 1 ± 0.7 | <0.5 |
| 7767d | 113 ± 67e | 3 ± 2 | <0.4 |
aStrepr point mutation.
baphA present in vacA.
cStrain 6c has the same clonal origin as strain 6a.
dStrain has different origin to strain 6a.
eStrain was transformed by the point mutation in rpsL (A128G) conferring Strepr at a significantly higher frequency (P < 0.05) than by the aphA cassette or by the hpyIIRM::aphA cassette.
To determine specifically whether restriction affects H.pylori transformation frequencies of chromosomal DNA fragments, PCR products containing the Strepr point mutation, the aphA cassette or the hpyIIRM::aphA cassette from the strain 6a derivatives were used to transform strain 6c (Table 4). There was no significant difference in Strepr transformation frequency of 6c between chromosomal DNA and the unmethylated PCR products. However, there was significantly less transformation (P < 0.05) by the aphA or the hpyIIRM::aphA cassette when the PCR product was used compared with chromosomal DNA. That <50 bp homology is needed for successful transformation of H.pylori (S.Levine and M.J.Blaser, manuscript in preparation), suggests that the lengths of the PCR product-flanking regions (Table 4) were sufficient, and thus were not responsible for the significant reduction in transformation frequency. To determine whether the difference in transformation frequency was due to the lack of DNA methylation of the PCR products in the pattern specific for strain 6a, all of the products were premethylated using cell extracts from strain 6a, as described previously (9), and then used to transform strain 6c. For the product containing Strepr, there was little difference between the three forms of donor DNA. For both cassettes >1 kb, transformation frequency increased significantly (P < 0.05) compared with the unmethylated PCR products. To confirm that differences in transformation frequency resulted from modification of chromosomal DNA (e.g. methylation), strain 6c was transformed with donor strain (RA4, RA3 or RA1) DNA, the PCR products or the 6c-transformant DNA (Table 4) and transformation frequencies for all three markers were calculated. As expected, for the aphA and hpyIIRM::aphA markers, 6c transformant DNA transformed strain 6c at a significantly higher frequency (P < 0.05) than did PCR products, but not at a significantly different frequency than donor strain DNA (data not shown). These results provide evidence that H.pylori R-M systems represent barriers to acquisition of chromosomal DNA >1 kb from non-isogenic strains. These barriers may result from differing distributions of relevant restriction sites or differing methylation patterns in the donor DNA.
Table 4. Effect of DNA methylation status on transformation frequency of strain 6c.
| Donora DNA | Transformation frequency (× 10–7) | ||
|---|---|---|---|
| Point mutation from RA4 | aphA cassette from RA3b | Altered hpyIIRM cassette from RA1c | |
| Chromosome | 138 ± 32 | 102 ± 20 | 81 ± 30 |
| PCR productd | 193 ± 81 | 10 ± 8 | <0.4 |
| Methylated PCR producte | 94 ± 50 | 35 ± 12 | 12 ± 10 |
aStrepr point mutation, aphA cassette or hpyIIRM::aphA cassette, respectively, which were selected for during transformation experiments.
bThe PCR product contains 1577 bp flanking the aphA cassette that have homology to vacA.
cThe PCR product contains 367 bp flanking the aphA cassette that have homology to hpyIIR.
dPCR products for the Strepr point mutation, aphA cassette and hpyIIRM::aphA cassette were amplified from strains RA4, RA3 and RA1 chromosomal DNA, respectively.
ePCR products were methylated to the host strain specificities, as described previously (9).
DISCUSSION
Although H.pylori R-M systems exhibit significant interstrain variation (5–7), and R-M diversity influences plasmid transformation frequencies (8,9), its effect on transformation by H.pylori chromosomal DNA transformation was not established. Here, we show that H.pylori can transfer 4.8 kb chromosomal DNA inserts through natural transformation or conjugation-like mechanisms between clonal variants, but that restriction barriers between non-isogenic strains limit transfer of DNA fragments ≥1 kb. These findings are consistent with the observation that horizontally transferred DNA fragments in H.pylori have a median size of 417 bp, much smaller than for other bacteria studied (17). For the naturally competent H.pylori, the presence of barriers that limit incoming chromosomal DNA from other H.pylori strains may be an effective means to prevent one strain from completely transforming the genome of another strain during co-colonization of a host (8). The acquisition of relatively smaller DNA fragments is also consistent with the presence of mosaic loci (17,18) within the H.pylori chromosome.
It has been suggested that R-M systems represent ‘selfish’ mobile genetic elements that, once established within the genome, would lead to adverse consequences for their host cell if eliminated (19). That the full 4.8 kb hpyIIRM::aphA cassette cannot be transferred between non-isogenic H.pylori strains indicates that R-M systems within cells are barriers to acquisition of other R-M systems, since the resident restriction endonuclease activity attacks improperly methylated incoming DNA. Since elimination (or inactivation) of resident R-M systems may increase host-cell susceptibility to subversion by competing H.pylori DNA, the resident R-Ms may be considered to be ‘selfishly’ (19) improving their own security by retarding novel R-Ms from entering the chromosome. The strong avoidance of cognate R-M recognition sequences within the H.pylori genome is indicative of the strong selective pressures exerted by the competing R-M systems (20). Conversely, cognate sequence avoidance may contribute to improved acquisition of DNA fragments of H.pylori, but not of non-H.pylori origin, thereby deepening the differential between species-homologous and -heterologous DNA.
In that context, the presence of partial or inactive R-M systems (3,4,10) may serve as a new class of ‘contingency locus’ (21) that can be reactivated upon recombination with a functional allele, as illustrated by the acquisition of M.HpyII activity by strain RA6 through natural transformation using donor DNA from RA2. Such findings provide evidence for the ‘fluidity’ of the H.pylori R-M systems with the potential to move both vertically and horizontally throughout the host cell population. Recent studies of iceA1 (hpyIR) indicate four potential states: functional enzyme, single point mutations that can be fixed by endogenous repair (i.e. frameshifts, reversion point mutations), multiple mutations and complete absence of the gene; all three of the latter states can be repaired by recombination with the exogenous wild-type gene (22). That in each strain the cognate methylase (hpyIM) is fully functional (6,23,24), indicates that any H.pylori strain is capable of acquiring R.HpyI function regardless of the initial hpyIR status. That H.pylori genomes are diverse in sequence and gene content (10) suggests that the reactivation of inactive ‘contingency genes’ through acquisition of fully functional alleles may be a general paradigm for gene regulation.
Naturally competent organisms are capable of ingesting environmental DNA with genomic integration through homologous recombination (25). For Haemophilus influenzae (26) and Neisseria gonorrheae (27), but not H.pylori (28), DNA uptake sequences have been identified that permit donor DNA to bind to recipient cells before transport across outer and inner membranes and integration into the chromosome. For Gram-negative prokaryotes such as H.influenzae, transformation results in single-stranded integration (29); however, single-stranded DNA of donor origin has not been detected within H.influenzae cells. Type II ENases, including several carried by H.pylori, preferentially cleave double-stranded DNA (1,3–5,8). That pre-methylated hpyIIRM::aphA PCR products preferentially transformed H.pylori cells suggests that, after internalization by recipient cells, double-stranded donor DNA intermediates are present and thus subject to restriction. Consistent with this hypothesis are recent experimental findings that indicate that double-stranded DNA is substantially more efficient at transforming H.pylori cells than is identical single-stranded DNA (S. M. Levine and M. J. Blaser, unpublished results).
In conclusion, our findings demonstrate that H.pylori R-M diversity presents barriers to interstrain transfer of chromosomal DNA fragments of >1 kb, and extend the theory that R-M systems are ‘selfish genes’ by providing evidence that acquisition of novel R-M systems is limited by the presence of resident R-M systems. Further exploration of this highly tractable experimental system encompassing both H.pylori competence and restriction may broaden our understanding of the dynamics of gene flow in natural bacterial populations.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (R01 GM63270, R01 DK58587, 5T32 AI07180-21, and the NCI Cancer Core grant to NYU) and by the Medical Research Service of the Department of Veterans’ Affairs.
REFERENCES
- 1.Wilson G.G. (1991) Organization of restriction-modification systems. Nucleic Acids Res., 19, 2539–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peek R.M. Jr and Blaser,M.J. (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature Rev. Cancer, 2, 28–37. [DOI] [PubMed] [Google Scholar]
- 3.Kong H., Lin,L.F., Porter,N., Stickel,S., Byrd,D., Posfai,J. and Roberts,R.J. (2000) Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res., 28, 3216–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L., Posfai,J., Roberts,R.J. and Kong,H. (2001) Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA, 98, 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Q., Morgan,R., Roberts,R. and Blaser,M.J. (2000) Identification of type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc. Natl Acad. Sci. USA, 97, 9671–9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takata T., Aras,R., Tavakoli,D., Ando,T., Olivares,A.Z. and Blaser,M.J. (2002) Phenotypic and genotypic variation in methylases involved in type II restriction-modification systems in Helicobacter pylori. Nucleic Acids Res., 30, 2444–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitkute J., Stankevicius,K., Tamulaitiene,G., Maneliene,Z., Timinskas,A., Berg,D.E. and Janulaitis,A. (2001) Specificities of eleven different DNA methyltransferases of Helicobacter pylori strain 26695. J. Bacteriol, 183, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ando T., Xu,Q., Torres,M., Kusugami,K., Israel,D.A. and Blaser,M.J. (2000) Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol., 37, 1052–1065. [DOI] [PubMed] [Google Scholar]
- 9.Donahue J.P., Israel,D.A., Peek,R.M., Blaser,M.J. and Miller,G.G. (2000) Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol., 37, 1066–1074. [DOI] [PubMed] [Google Scholar]
- 10.Alm R.A., Ling,L.S., Moir,D.T., King,B.L., Brown,E.D., Doig,P.C., Smith,D.R., Noonan,B., Guild,B.C., de Jonge,B.L. et al. (1999) Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature, 397, 176–180. [DOI] [PubMed] [Google Scholar]
- 11.Tomb J.F., White,O., Kerlavage,A.R., Clayton,R.A., Sutton,G.G., Fleischmann,R.D., Ketchum,K.A., Klenk,H.P., Gill,S., Dougherty,B.A. et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature, 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 12.Nobusato A., Uchiyama,I. and Kobayashi,I. (2000) Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene, 259, 89–98. [DOI] [PubMed] [Google Scholar]
- 13.Aras R.A., Takata,T., Ando,T., van der Ende,A. and Blaser,M.J. (2001) Regulation of the HpyII restriction-modification system of Helicobacter pylori by gene deletion and horizontal reconstitution. Mol. Microbiol., 42, 369–382. [DOI] [PubMed] [Google Scholar]
- 14.Nobusato A., Uchiyama,I., Ohashi,S. and Kobayashi,I. (2000) Insertion with long target duplication: a mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene, 259, 99–108. [DOI] [PubMed] [Google Scholar]
- 15.Tsuda M., Karita,M. and Nakazawa,T. (1993) Genetic transformation in Helicobacter pylori. Microbiol. Immunol., 37, 85–89. [DOI] [PubMed] [Google Scholar]
- 16.Kuipers E.J., Israel,D.A., Kusters,J.G. and Blaser,M.J. (1998) Evidence for a conjugation-like mechanism of DNA transfer in Helicobacter pylori. J. Bacteriol., 180, 2901–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falush D., Kraft,C., Taylor,N.S., Correa,P., Fox,J.G., Achtman,M. and Suerbaum,S. (2001) Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl Acad. Sci. USA, 98, 15056–15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atherton J.C., Cao,P., Peek,R.M.,Jr, Tummuru,M.K., Blaser,M.J. and Cover,T.L. (1995) Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem., 270, 17771–17777. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi I. (2001) Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res., 29, 3742–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pride D.T. and Blaser,M.J. (2002) Identification of horizontally acquired genetic elements in Helicobacter pylori and other prokaryotes using oligonucleotide difference analysis. Genome Lett., 1, 2–15. [Google Scholar]
- 21.Bayliss C.D., Field,D. and Moxon,E.R. (2001) The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Invest., 107, 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Q., Morgan,R.D., Roberts,R.J., Xu,S.Y., Van Doorn,L.J., Donahue,J.P., Miller,G.G. and Blaser,M.J. (2002) Functional analysis of iceA1, a CATG-recognizing restriction endonuclease gene in Helicobacter pylori. Nucleic Acids Res., 30, 3839–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Q., Peek,R.M.,Jr, Miller,G.G. and Blaser,M.J. (1997) The Helicobacter pylori genome is modified at CATG by the product of hpyIM. J. Bacteriol., 179, 6807–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q. and Blaser,M.J. (2001) Promoters of the CATG-specific methyltransferase gene hpyIM differ between iceA1 and iceA2 Helicobacter pylori strains. J. Bacteriol., 183, 3875–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubnau D. (1999) DNA uptake in bacteria. Annu. Rev. Microbiol., 53, 217–244. [DOI] [PubMed] [Google Scholar]
- 26.Danner D.B., Deich,R.A., Sisco,K.L. and Smith,H.O. (1980) An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene, 11, 311–318. [DOI] [PubMed] [Google Scholar]
- 27.Elkins C., Thomas,C.E., Seifert,H.S. and Sparling,P.F. (1981) Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J. Bacteriol., 173, 3911–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders N.J., Peden,J.F. and Moxon,E.R. (1999) Absence in Helicobacter pylori of an uptake sequence for enhancing uptake of homospecific DNA during transformation. Microbiology, 145, 3523–3528. [DOI] [PubMed] [Google Scholar]
- 29.Notani N. and Goodgal,S.H. (1966) On the nature of recombinants formed during transformation in Hemophilus influenzae. J. Gen. Physiol., 49, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heep M., Beck,D., Bayerdorffer,E. and Lehn,N. (1999) Rifampin and rifabutin resistance mechanism in Helicobacter pylori. Antimicrob. Agents Chemother., 43, 1497–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dennis P.P. (1979) Transcription and translation in a pleiotropic streptomycin-resistant mutant of Escherichia coli. J. Bacteriol., 137, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigmund C.D., Ettayebi,M. and Morgan,E.A. (1984) Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res., 12, 4653–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]