Abstract
Ribonucleases play key, often essential, roles in cellular metabolism. Nineteen ribonuclease activities, from 22 different proteins, have so far been described in bacteria, the majority of them from either Escherichia coli or Bacillus subtilis. Here we examine the phylogenetic distribution of all of these ribonucleases in 50 eubacterial and archaeal species whose genomes have been completely sequenced, with particular emphasis on the endoribonucleases. Although some enzymes are very highly conserved throughout evolution, there appears to be no truly universal ribonuclease. While some organisms, like E.coli, have a large selection of ribonucleases, many with overlapping functions, others seem to have relatively few or have many that remain to be discovered.
INTRODUCTION
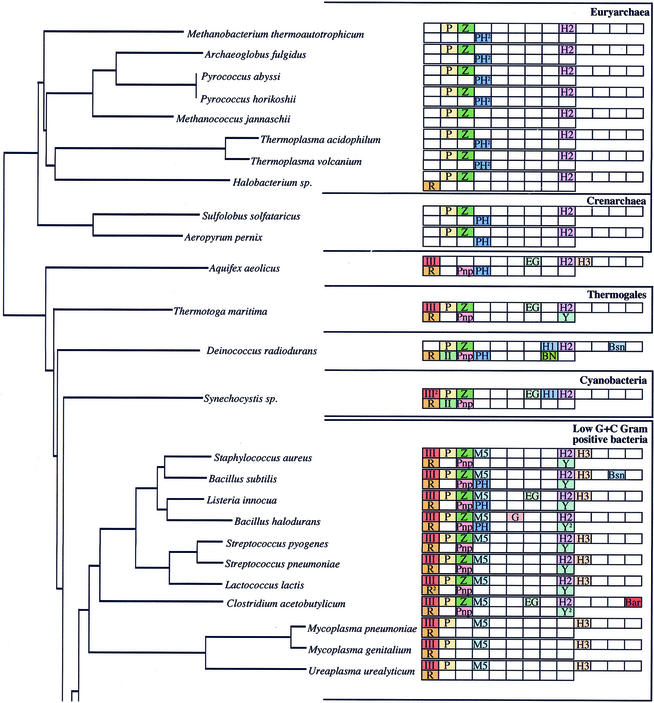
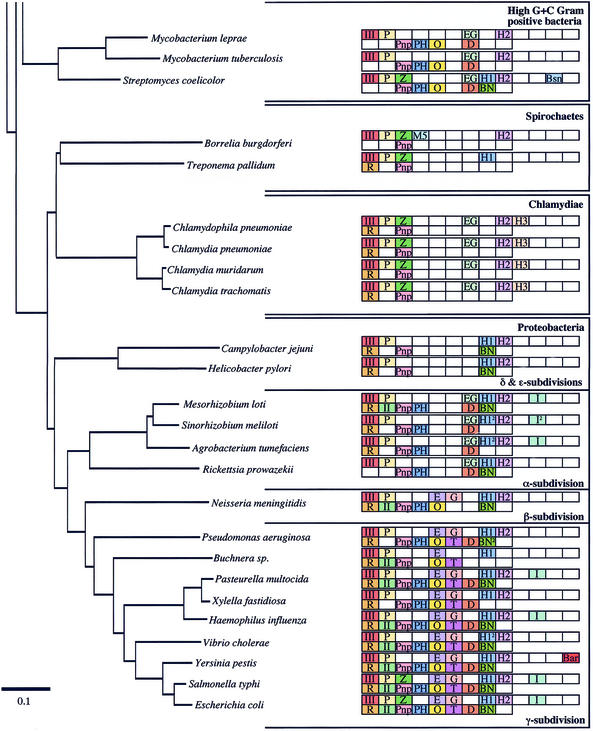
Ribonucleases (RNases) play very important roles in cellular metabolism by permitting scavenging of ribonucleotides from the extracellular environment, recycling them from cellular RNAs of diminished utility, and in the control of gene expression by altering the stability of specific mRNAs. Two general types of RNases have been described: exoribonucleases, which degrade RNAs one nucleotide at a time from the ends, and endoribonucleases, which cleave RNAs at internal sites. Bacterial RNases have been most extensively studied in Escherichia coli and Bacillus subtilis, the paradigms of Gram-negative and Gram-positive eubacteria, with a total of 17 different RNase activities (from 20 different proteins) having been identified between them. Two additional RNases have been identified from other species. A comparison of the genomes of E.coli and B.subtilis has shown that only six of their characterised RNases are shared, suggesting that, on a superficial level at least, Gram-negative and Gram-positive organisms have evolved with very different solutions to the problem of RNA maturation and decay. Since the evolutionary separation of E.coli and B.subtilis occurred over a billion years ago, even before the separation of plants and animals (1,2), we believe that the RNases identified in these two organisms will represent a very high proportion of all of the RNases to be found in eubacteria. With the explosion of genome sequencing has come the possibility of doing much more extensive and meaningful comparisons of the types of RNases available to different bacteria by homology searches. At the time of writing, 38 fully sequenced and annotated eubacterial genomes are available on the NCBI Microbial Genomes BLAST Database (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html). To these we have added the genomes of Listeria innocua (http://genolist.pasteur.fr/ListiList/) and Streptomyces coelicolor (http://www.sanger.ac.uk/Projects/S_coelicolor/blast_server.shtml), also available for BLAST search on the Internet, and examined the phylogenetic distribution of the 22 eubacterial endo- and exoribonucleases throughout the bacterial kingdom by BLAST analysis (Fig. 1). We have also included the 10 fully sequenced archaeal genomes available on the NCBI site for comparison. Orthologues for each of the RNases are proposed based on strong overall sequence homology and size considerations in all cases. In some cases, particularly those near the limit of detection, conservation of particular amino acids was also examined. Unless otherwise stated, the search sequences used were either of E.coli or B.subtilis origin, or both, in the case of RNases held in common between the two organisms. Because a comprehensive phylogenetic survey of the eight exoribonucleases found in E.coli has recently been published (3), this paper focuses primarily on the endoribonucleases; the exoribonucleases are only included for the sake of completeness. The distribution of the different RNases is depicted on a 16S ribosomal RNA (rRNA) phylogenetic tree that shows the relationships between the bacterial species examined most clearly. This method also gives a good picture of the level of variation found within the different bacterial clades. To facilitate discussion, we have sometimes grouped the different RNases into structurally or functionally related families.
Figure 1.
(Opposite and above) The phylogenetic distribution of eubacterial RNases. The endonucleases are shown in the top row and the exonucleases in the bottom row of blocks for each species. The phylogenetic relationship between the different organisms was calculated by comparing 16S rRNA sequences using Clustal X (64). The phylogenetic tree was drawn using Phylodendron (http://iubio.bio.indiana.edu/soft/molbio/java/apps/trees/) and arranged according to Olsen et al. (65). The abbreviations for the different RNases are as follows: III, RNase III; P, RNase P; Z, RNase Z; M5, RNase M5; E, RNase E; G, RNase G; EG, RNase E/G; H1, RNase HI; H2, RNase HII; H3, RNase HIII; I, RNase I; Bsn, RNase Bsn; Bar, Barnase; R, RNase R; II, RNase II; Pnp, polynucleotide phosphorylase; PH, RNase PH; O, oligoribonuclease; T, RNase T; D, RNase D; BN, RNase BN; Y, RNase YhaM. 2Two potential orthologues present.
THE ENDORIBONUCLEASES
Ribonuclease III
Escherichia coli RNase III is a dimeric endoribonuclease of ∼26 kDa molecular weight, with specificity for double-stranded RNA (4,5). One of its major roles in this organism is the processing of 16S and 23S rRNA from its large 30S precursor transcript (6). It is also involved in the initial cleavage of several mRNAs (7–11), often producing segments of the primary transcript with different stabilities and con sequences for gene expression. RNase III is found in all eubacteria except Deinococcus radiodurans, while Synechocystis sp. has two copies of an RNase III gene. Many homologues with additional domains have been identified in eukaryotic species, the best known being the Dicer enzyme of the RNA interference pathway (12,13). Interestingly, RNase III is totally absent from archaeal species. While it is possible that the conservation of RNase III in eubacteria is related to its role in rRNA processing, functional maturation of rRNA can occur via alternate pathways in both B.subtilis and E.coli (14,15). It thus seems likely that it plays a more fundamentally important role in bacterial RNA metabolism. Indeed RNase III is an essential gene in B.subtilis, independent of rRNA processing (14).
Ribonucleases P and Z
RNase P is an essential enzyme in both E.coli (16,17) and B.subtilis (C.Condon and H.Putzer, unpublished data). Its main role in these organisms is the maturation of the 5′ end of tRNAs. It is a two-component enzyme, the catalytic subunit being a ∼400 nt RNA molecule. A small, ∼14 kDa protein is associated with RNase P RNA, and although it plays a role in substrate recognition (18), it is not necessary for enzyme activity (19). Escherichia coli and B.subtilis RNase P RNAs are paradigms of the A-type and B-type RNAs, respectively, based on variations in secondary structure (20). M-type structures are found in some members of the Archaea, such as Methanococcus jannaschii and Archaeoglobus fulgidus. All but eight of the bacterial and archaeal species examined here have their RNase P RNA sequences and secondary structures catalogued on the RNase P database (http://www.mbio.ncsu.edu/RNaseP/home.html). The RNase P RNAs of seven of the remaining eight species were identified by homology to their closest relatives. Only Aquifex aeolicus failed to show an RNase P homologue. The apparent absence of RNase P in this organism is supported by data showing that A.aeolicus cell extracts lack RNase P activity (21). To our knowledge, this is the only known organism lacking RNase P. Homologues to the protein component of RNase P are difficult to find in some eubacterial species. In Archaea, as in eukaryotes, multiple proteins are associated with RNase P RNA (22), none of which are homologous to the eubacterial RNase P protein and none of which are found in bacteria.
Ribonuclease Z is a recently identified endonuclease that cleaves the 3′ end of eukaryotic and archaeal tRNAs at the discriminator base (23). Clear homologues to M.jannaschii RNase Z are identifiable throughout the eubacterial kingdom, and although these have not yet been shown to possess RNase Z activity, we have decided to include them here. The E.coli homologue, ElaC, is 304 amino acids in length and has 48% similarity (33% identity) to its M.jannaschii equivalent. Like RNase P, RNase Z recognises the tRNA structure; pre-tRNAs missing one or more arms of the tRNA are cleaved with drastically reduced efficiency, and pre-tRNAs missing the T-arm are not processed at all (24). Potential orthologues of RNase Z are scattered throughout the eubacteria and in all 10 of the Archaea examined. A second protein of unknown function, slightly smaller in size and of significantly lower homology than RNase Z, can also be found on many eubacterial and archaeal chromosomes.
Ribonuclease M5
5S ribosomal RNA maturation in B.subtilis is catalysed by RNase M5 (25). This is a small ∼20 kDa enzyme that cleaves 5S rRNA precursor on either side of a double-stranded RNA helix to yield mature 5S rRNA in one step. RNase M5 requires a cofactor, ribosomal protein L18, which binds to 5S rRNA and is thought to help fold the precursor into the correct conformation for cleavage (26). High concentrations of ribosomal protein L5, which also binds 5S rRNA, inhibit the cleavage reaction (26). RNase M5 does not appear to have any other substrates in B.subtilis besides 5S rRNA precursor (27), making it a highly specific enzyme, similar to RNase P. In the relatively few bacterial species that have both RNase M5 and the other 5S rRNA maturase, RNase E (see below), it is likely that both enzymes can participate in this process (28). The N-terminal half of RNase M5 consists of a Toprim domain, also found in the topoisomerases, archaeal reverse gyrases and DNA primases (29). Topoisomerases cleave double-stranded DNA to allow strand passage while relaxing supercoils in the DNA and have been shown to be capable of cleaving RNA under certain circumstances (30,31). This suggests that the cleavage mechanisms of RNase M5 and the topoisomerases may be related. RNase M5 is confined to the low G+C Gram-positive bacteria and, curiously, to the Spirochaete, Borrelia burgdorferi (28).
Ribonucleases E, G and E/G
The rate-limiting step in the degradation of most E.coli mRNAs is thought to be an endonucleolytic cleavage by RNase E. Escherichia coli RNase E is a large protein of 1061 amino acids. The N-terminal half is required for catalytic activity, while the C-terminal half acts as a scaffold for the assembly of a large multi-protein complex known as the degradosome, by binding to polynucleotide phosphorylase (PNPase), RhlB helicase and enolase (32–34). RNase E is also involved in stable RNA processing; 16S, 5S rRNA and the majority of tRNAs are processed initially by RNase E (15,35–37). Escherichia coli has a close homologue of the catalytic portion of RNase E on its chromosome, known as RNase G (or CafA). RNase G has substrate specificity and enzymatic properties very similar to RNase E (38). It is known to co-operate with RNase E in 16S rRNA maturation (15). Although RNase E is essential in E.coli, it is absent from a surprisingly large number of bacterial species. Only the β- and γ-subdivisions of the Proteobacteria have both RNase E and RNase G. Even within these subdivisions, however, the homology in the C-terminal half of RNase E becomes less and less obvious with evolutionary distance from E.coli.
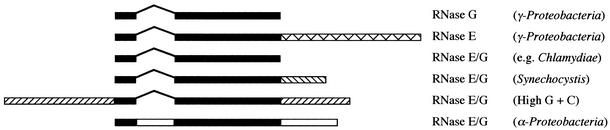
A single RNase E/G family member is found on the chromosomes of about half of the eubacterial species outside of the β- and γ-Proteobacteria. In most cases, this protein is similar to RNase G in size (∼500 amino acids), but is equally homologous to both RNase G and the N-terminal half of RNase E. This seems likely to be the ancestral protein, giving rise to RNase E and RNase G by gene duplication and subsequent divergent evolution. Variants of RNase E/G are found in the α-Proteobacteria, Synechocystis sp. and in the high G+C Gram-positive bacteria (Fig. 2). The α-Proteobacteria have an insertion of 100–200 amino acids at N82 of the E.coli RNase E sequence and they have a C-terminal tail of 130–320 amino acids that shows no significant homology to RNase E from the β- and γ-Proteobacteria. The RNase E/G domain of this group of proteins shows more homology to that of RNase E than RNase G. However, given that the organisation and amino acid sequence of the additional domains bear little resemblance to those of RNase E, we have preferred to call these proteins RNase E/G in Figure 1. Synechocystis sp. RNase E/G has a 180 amino acid C-terminal extension, which has no homology to the degradosome scaffold of E.coli RNase E and, indeed, has been shown to be incapable of promoting degradosome assembly (39). The high G+C Gram-positives, exemplified by S.coelicolor and its close relatives, have a 320–560 amino acid N-terminal extension and a 180–370 amino acid C-terminal extension to the RNase E/G domain. As in the previous examples, none of the extensions is homologous to the degradosome-forming portion of E.coli RNase E, even though the overall size of these enzymes is similar to that of RNase E. The function of these various insertions and extensions is not known, but they presumably give additional properties to the enzyme.
Figure 2.
RNase E/G protein families. The ∼500 amino acid core RNase E/G domain is shown in black. This domain is continuous in all RNase E/G proteins except those of the α-Proteobacteria, which have an insertion. N-terminal and C-terminal extensions (hatched or white bars) are depicted as the average size of those of the different family members examined. The different fillings reflect the lack of sequence homology.
There does not seem to be much pattern to the distribution of RNase E/G within the low G+C Gram-positive group. Bacillus subtilis lacks RNase E/G, for example, while some of its closest relatives, Bacillus halodurans and L.innocua, each have an RNase E/G family member. The 500 amino acid B.halodurans protein is intriguing in that it is the only short RNase E/G family member we have come across that is not equally homologous to RNase E and G, but shows a much closer relationship to RNase G. It is thus labelled as such in Figure 1.
Ribonucleases HI, HII and HIII
Ribonuclease H cleaves the RNA of RNA–DNA hybrids produced during the process of chromosome replication by primases. Its primary function in E.coli appears to be the prevention of aberrant chromosome replication at sites other than oriC (40,41). Three RNase H activities have been characterised between E.coli and B.subtilis, termed RNase HI, HII and HIII. These enzymes can be distinguished at the sequence level, although RNases HII and HIII show some homology at their C-termini. Ninety-five percent of RNase H activity in E.coli is provided by RNase HI and the remainder by RNase HII (42). RNase HI is largely confined to the Proteobacteria, although some examples are scattered among the high G+C Gram-positives, the Spirochaetes and the Cyanobacteria. Some members of the Proteobacteria, namely Vibrio cholerae, Sinorhizobium meliloti and Agrobacterium tumefaciens, actually have two potential orthologues of the RNase HI gene. The second copy generally has lower homology scores and may thus have been acquired from another source by horizontal gene transfer. Although some members of the low G+C Gram-positive family and Halobacterium sp. have a protein with some homology to RNase HI, this protein, YpdQ, does not appear to have RNase H activity in B.subtilis (43,44). They have thus not been included in Figure 1. RNase H activity in B.subtilis is rather provided by RNases HII and HIII (43). RNase HII is found in most eubacteria and all 10 of the completed archaeal sequences. It is missing from the Mycoplasma group, from the Spirochaete Treponema pallidum and the γ-Proteobacterium Buchnera sp. There are different families of RNase HII proteins; those found in the low G+C Gram-positives have a ∼60 amino acid N-terminal extension relative to RNase HII from most other species, for example, while those found in the high G+C Gram positives have short C-terminal extensions.
Homologues of RNase HIII are confined to the low G+C Gram-positive bacteria, the Chlamydiae and to A.aeolicus. Since RNase H activity is essential to both E.coli and B.subtilis, inactivation of one or other of the rnh genes, but not both, is tolerated in these two organisms (43). Proteins similar to RNase HI and HII enzymes can be found in eukaryotes and, in the retroviruses, an RNase H domain is associated with reverse transcriptase (42).
Ribonucleases I, Bsn and Barnase
RNases I, Bsn and Barnase are broad-specificity extracellular nucleases originally identified in E.coli and the Bacilli. Their role is presumably to allow scavenging of nucleotides from RNAs found in the environment. RNase I is a non-specific 27 kDa endonuclease found in the periplasmic space in E.coli. It belongs to the T2 family of RNases and generates 3′ phosphoryl-,5′ hydroxl-terminated products in the presence of EDTA. A multiply mutant form of RNase I, known as RNase M, has also been identified in some E.coli strains (45). Both are products of the same gene, rna. RNase I is found in the α- and γ-subdivisions of the Proteobacteria exclusively, with S.meliloti and some Salmonella species appearing to have two copies of the gene.
RNase Bsn is an extracellular nuclease, also with no apparent sequence specificity. It can cleave RNAs endonucleolytically to yield 5′ phosphorylated oligonucleotides (46). A 51–53 amino acid N-terminal peptide is removed upon secretion of the enzyme. RNase Bsn is found in some members of the low G+C Gram-positives. Low levels of homology to proteins from the γ-Proteobacteria are provided by the related DNase, End A. In Bacillus intermedius, RNase Bsn has been termed binase II (47), to distinguish it from the better known binase I or Barnase (below).
Barnase is a guanyl-specific extracellular RNase. Although found in many of the Bacilli, it is actually not present in B.subtilis. Orthologues of Bacillus amyloliquifaciens Barnase, and its inhibitor Barstar, are also found in Clostridium acetobutylicum and the Gram-negative Yersinia pestis. Interestingly, a sequence potentially encoding the C-terminal half of Barnase, followed immediately on the chromosome by a gene encoding Barstar, can also be found in Neisseria meningitidis. The same N-terminal truncation is found in an independently sequenced N.meningitidis serogroup, suggesting this is not a simple sequencing error and that the protein may not be fully functional. The occurrence of Barstar independently of Barnase has also been reported in B.subtilis (48). Thus, it appears some organisms have lost their copy of the Barnase gene, because it was no longer required for a selective advantage. Alternatively, they acquired the resistance gene because other organisms sharing the same niche produced Barnase.
THE EXORIBONUCLEASES
Although exonucleolytic degradation of RNA can occur in both 5′→3′ and the reverse orientation in eukaryotes, only 3′→5′ degradative activity has so far been identified in bacteria. Eight 3′→5′ exoribonucleases have been described in E.coli, and their architecture and phylogenetic distribution have been extensively analysed in a recent survey by Zuo and Deutscher (3). Thus, they will only be superficially treated here and the reader is referred to this paper for more detail. PNPase, RNase II and oligoribonuclease are thought to be the primary enzymes of messenger RNA degradation (recently reviewed in 49). The remaining nucleases are mainly involved in stable RNA maturation and decay. RNases PH, BN, D and T play redundant roles in maturation of the 3′ ends of tRNAs (50–53), with RNase T also being involved in the removal of the last few nucleotides from the 3′ end of both 5S and 23S rRNA precursors (54,55). The remaining exonuclease of E.coli, RNase R, is thought to be primarily involved in ribosomal RNA degradation, aided by its greater processivity than its close relative RNase II, particularly on structured RNA (56). The phylogenetic distribution of these eight exonucleases is shown in Figure 1.
One additional exoribonuclease has been discovered in bacteria since the Zuo and Deutscher paper (3), the YhaM protein of B.subtilis (57). This protein was predicted to be a nuclease by virtue of the HD (His Asp) domain and OB-fold found in certain other RNases (58). YhaM has a molecular mass of 36 kDa and its role in B.subtilis is not yet known. Although primarily in the low G+C Gram-positive bacteria, homologues of YhaM can be found in Thermotoga maritima and in some members of the sulphur reducing δ-Proteo bacteria, namely Desulfovibrio vulgaris and Geobacter sulfurreducens, whose genome sequences are not yet complete. The YhaM orthologue of Staphylococcus aureus is known as CBF1, a protein that binds the replication enhancer, cmp, of plasmid pT181 (59). It has been confirmed that S.aureus CBF1 has YhaM exoribonuclease activity, in addition to its role in DNA replication. Interestingly, two YhaM/CBF1 homologues can be found in both B.halodurans and C.acetobutylicum.
CONCLUSIONS
Here we have examined the phylogenetic distribution of 22 ribonuclease proteins among 50 completely sequenced eubacteria and Archaea. Most of these proteins were identified in either E.coli or B.subtilis, and only six confirmed RNases are shared by these two species. It is no surprise that these six RNases, RNase III, RNase P, RNase HII, RNase R, PNPase and RNase PH, are also the most conserved throughout the eubacterial kingdom. RNase P is the most widespread enzyme. It is found in eukaryotes and in all the Archaea examined, and is present in all but one of the eubacteria, being curiously absent from A.aeolicus. This is the closest example of a ‘universal’ RNase. RNase III was found in all but one of the eubacterial species examined. RNase R, PNPase and RNase HII are the next most widely distributed, although each is missing from one or more eubacterial subsets.
It is interesting to compare the selection of RNases available to the different organisms for tasks relating to RNA metabolism. At first glance, the Proteobacteria would seem to have the greatest number of RNases, with more than half a dozen each of endo- and exoribonucleases in their repertoire. It is difficult to guess at this stage whether there is a genuine difference with other bacterial species or whether it simply reflects the depth of study of E.coli pathways of RNA processing and decay compared to other organisms. In the case of the Mycoplasma, which have only four endoribonucleases, three of these with highly specialised tasks (RNases P, M5 and H) and one exoribonuclease, RNase R, this presumably reflects the fact that their genomes are only about half the size of that of E.coli. As obligate intracellular parasites, the Mycoplasma can afford to have much smaller genomes than free-living organisms, since the host can presumably provide many of their functions. The list of E.coli RNases is believed to be essentially exhaustive, with very few RNase cleavages described that do not yet have an associated enzyme. One of these is the ribosome-associated cleavage of the fimbrial daa mRNA (60); others are the enzymes responsible for the maturation of the 5′ ends of 23S and 5S rRNA and the 3′ end of 16S rRNA. In B.subtilis, on the other hand, a large number of cleavages and exonucleolytic activities are known whose enzymes have not yet been identified (61–63), although it is likely that at least some of these reactions are catalysed by the same enzymes.
Whether or not a gene is essential is not an indication that it is universally, or even highly, conserved. Although this is practically the case for the gene encoding RNase P, it is not so for RNase E or oligoribonuclease, both essential genes in E.coli, but confined to the Proteobacteria. While RNase III is universally conserved in eubacteria, and is essential in B.subtilis, E.coli rnc mutants are viable. Neither RNase R nor PNPase, the most widespread of the exonucleases, is essential in either B.subtilis or E.coli. Clearly, these enzymes give a selective advantage to the organisms that carry them, without providing a critical function, at least under normal laboratory growth conditions.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by funds from the CNRS (UPR 9073), Université Paris VII and PRFMMIP from the Ministère de l’Education Nationale.
REFERENCES
- 1.Pace N.R., Olsen,G.J. and Woese,C.R. (1986) Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell, 45, 325–326. [DOI] [PubMed] [Google Scholar]
- 2.Woese C.R. (1994) There must be a prokaryote somewhere: microbiology’s search for itself. Microbiol. Rev., 58, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuo Y. and Deutscher,M.P. (2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res., 29, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schweisguth D.C., Chelladurai,B.S., Nicholson,A.W. and Moore,P.B. (1994) Structural characterization of a ribonuclease III processing signal. Nucleic Acids Res., 22, 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelladurai B., Li,H., Zhang,K. and Nicholson,A.W. (1993) Mutational analysis of a ribonuclease III processing signal. Biochemistry, 32, 7549–7558. [DOI] [PubMed] [Google Scholar]
- 6.Nikolaev N., Schlessinger,D. and Wellauer,P.K. (1974) 30S pre-ribosomal RNA of Escherichia coli and products of cleavage by ribonuclease III: length and molecular weight. J. Mol. Biol., 86, 741–747. [DOI] [PubMed] [Google Scholar]
- 7.Regnier P. and Grunberg-Manago,M. (1990) RNase III cleavages in non-coding leaders of Escherichia coli transcripts control mRNA stability and genetic expression. Biochimie, 72, 825–834. [DOI] [PubMed] [Google Scholar]
- 8.Regnier P. and Grunberg-Manago,M. (1989) Cleavage by RNase III in the transcripts of the metY-nusA-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J. Mol. Biol., 210, 293–302. [DOI] [PubMed] [Google Scholar]
- 9.Portier C., Dondon,L., Grunberg-Manago,M. and Regnier,P. (1987) The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5′ end. EMBO J., 6, 2165–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bardwell J.C., Regnier,P., Chen,S.M., Nakamura,Y., Grunberg-Manago,M. and Court,D.L. (1989) Autoregulation of RNase III operon by mRNA processing. EMBO J., 8, 3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsunaga J., Dyer,M., Simons,E.L. and Simons,R.W. (1996) Expression and regulation of the rnc and pdxJ operons of Escherichia coli. Mol. Microbiol., 22, 977–989. [DOI] [PubMed] [Google Scholar]
- 12.Ketting R.F., Fischer,S.E., Bernstein,E., Sijen,T., Hannon,G.J. and Plasterk,R.H. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev., 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 14.Herskowitz M.A. and Bechhofer,D.H. (2000) Endoribonuclease RNase III is essential in Bacillus subtilis. Mol. Microbiol., 38, 1027–1033. [DOI] [PubMed] [Google Scholar]
- 15.Li Z., Pandit,S. and Deutscher,M.P. (1999) RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J., 18, 2878–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schedl P. and Primakoff,P. (1973) Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl Acad. Sci. USA, 70, 2091–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakano H., Yamada,S., Ikemura,T., Shimura,Y. and Ozeki,H. (1974) Temperature sensitive mutants of Escherichia coli for tRNA synthesis. Nucleic Acids Res., 1, 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reich C., Olsen,G.J., Pace,B. and Pace,N.R. (1988) Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science, 239, 178–181. [DOI] [PubMed] [Google Scholar]
- 19.Guerrier-Takada C., Gardiner,K., Marsh,T., Pace,N. and Altman,S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 20.Brown J.W. (1999) The Ribonuclease P Database. Nucleic Acids Res., 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willkomm D.K., Feltens,R. and Hartmann,R.K. (2002) tRNA maturation in Aquifex aeolicus. Biochimie, 84, 713–722. [DOI] [PubMed] [Google Scholar]
- 22.Hall T.A. and Brown,J.W. (2002) Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA, 8, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiffer S., Rosch,S. and Marchfelder,A. (2002) Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J., 21, 2769–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiffer S., Helm,M., Theobald-Dietrich,A., Giege,R. and Marchfelder,A. (2001) The plant tRNA 3′ processing enzyme has a broad substrate spectrum. Biochemistry, 40, 8264–8272. [DOI] [PubMed] [Google Scholar]
- 25.Sogin M.L. and Pace,N.R. (1974) In vitro maturation of precursors of 5S ribosomal RNA from Bacillus subtilis. Nature, 252, 598–600. [DOI] [PubMed] [Google Scholar]
- 26.Stahl D.A., Pace,B., Marsh,T. and Pace,N.R. (1984) The ribonucleoprotein substrate for a ribosomal RNA-processing nuclease. J. Biol. Chem., 259, 11448–11453. [PubMed] [Google Scholar]
- 27.Condon C., Rourera,J., Brechemier-Baey,D. and Putzer,H. (2002) Ribonuclease M5 has few, if any, mRNA substrates in Bacillus subtilis. J. Bacteriol., 184, 2845–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Condon C., Brechemier-Baey,D., Beltchev,B., Grunberg-Manago,M. and Putzer,H. (2001) Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: Mature 5S rRNA is dispensable for ribosome function. RNA, 7, 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aravind L., Leipe,D.D. and Koonin,E.V. (1998) Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res., 26, 4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiGate R.J. and Marians,K.J. (1992) Escherichia coli topoisomerase III-catalyzed cleavage of RNA. J. Biol. Chem., 267, 20532–20535. [PubMed] [Google Scholar]
- 31.Sekiguchi J. and Shuman,S. (1997) Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell, 1, 89–97. [DOI] [PubMed] [Google Scholar]
- 32.Py B., Higgins,C.F., Krisch,H.M. and Carpousis,A.J. (1996) A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature, 381, 169–172. [DOI] [PubMed] [Google Scholar]
- 33.Miczak A., Kaberdin,V.R., Wei,C.-L. and Lin-Chao,S. (1996) Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA, 93, 3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanzo N.F., Li,Y.S., Py,B., Blum,E., Higgins,C.F., Raynal,L.C., Krisch,H.M. and Carpousis,A.J. (1998) Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev., 12, 2770–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghora B.K. and Apirion,D. (1978) Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell, 15, 1055–1066. [DOI] [PubMed] [Google Scholar]
- 36.Li Z. and Deutscher,M.P. (2002) RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA, 8, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ow M.C. and Kushner,S.R. (2002) Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev., 16, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tock M.R., Walsh,A.P., Carroll,G. and McDowall,K.J. (2000) The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem., 275, 8726–8732. [DOI] [PubMed] [Google Scholar]
- 39.Kaberdin V.R., Miczak,A., Jakobsen,J.S., Lin-Chao,S., McDowall,K.J. and von Gabain,A. (1998) The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl Acad. Sci. USA, 95, 11637–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa T., Pickett,G.G., Kogoma,T. and Kornberg,A. (1984) RNase H confers specificity in the DnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc. Natl Acad. Sci. USA, 81, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kogoma T., Subia,N.L. and von Meyenburg,K. (1985) Function of ribonuclease H in initiation of DNA replication in Escherichia coli K-12. Mol. Gen. Genet., 200, 103–109. [DOI] [PubMed] [Google Scholar]
- 42.Crouch R.J. (1990) Ribonuclease H: from discovery to 3D structure. New Biol., 2, 771–777. [PubMed] [Google Scholar]
- 43.Itaya M., Omori,A., Kanaya,S., Crouch,R.J., Tanaka,T. and Kondo,K. (1999) Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J. Bacteriol., 181, 2118–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtani N., Haruki,M., Morikawa,M., Crouch,R.J., Itaya,M. and Kanaya,S. (1999) Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry, 38, 605–618. [DOI] [PubMed] [Google Scholar]
- 45.Subbarayan P.R. and Deutscher,M.P. (2001) Escherichia coli RNase M is a multiply altered form of RNase I. RNA, 7, 1702–1707. [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura A., Koide,Y., Miyazaki,H., Kitamura,A., Masaki,H., Beppu,T. and Uozumi,T. (1992) Gene cloning and characterization of a novel extracellular ribonuclease of Bacillus subtilis. Eur. J. Biochem., 209, 121–127. [DOI] [PubMed] [Google Scholar]
- 47.Hahnen E., Znamenskaya,L., Koczan,D., Leshchinskaya,I. and Hobom,G. (2000) A novel secreted ribonuclease from Bacillus intermedius: gene structure and regulatory control. Mol. Gen. Genet., 263, 571–580. [DOI] [PubMed] [Google Scholar]
- 48.Belitsky B.R., Gustafsson,M.C., Sonenshein,A.L. and Von Wachenfeldt,C. (1997) An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol., 179, 5448–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kushner S.R. (2002) mRNA decay in Escherichia coli comes of age. J. Bacteriol., 184, 4658–4665, 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh R.K. and Deutscher,M.P. (1978) Purification of potential 3′ processing nucleases using synthetic tRNA precursors. Nucleic Acids Res., 5, 3831–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deutscher M.P., Marlor,C.W. and Zaniewski,R. (1985) RNase T is responsible for the end-turnover of tRNA in Escherichia coli. Proc. Natl Acad. Sci. USA, 82, 6427–6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J.R. and Deutscher,M.P. (1988) Transfer RNA is a substrate for RNase D in vivo. J. Biol. Chem., 263, 17909–17912. [PubMed] [Google Scholar]
- 53.Li Z. and Deutscher,M.P. (1996) Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell, 86, 503–512. [DOI] [PubMed] [Google Scholar]
- 54.Li Z. and Deutscher,M.P. (1995) The tRNA processing enzyme RNase T is essential for maturation of 5S rRNA. Proc. Natl Acad. Sci. USA, 92, 6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z., Pandit,S. and Deutscher,M.P. (1999) Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA, 5, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng Z.F. and Deutscher,M.P. (2002) Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem., 277, 21624–21629. [DOI] [PubMed] [Google Scholar]
- 57.Oussenko I.A., Sanchez,R. and Bechhofer,D.H. (2002) Bacillus subtilis YhaM, a member of a new family of 3′ to 5′ exonucleases in Gram-positive bacteria. J. Bacteriol., 184, 6250–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aravind L. and Koonin,E.V. (2001) A natural classification of ribonucleases. Methods Enzymol., 341, 3–28. [DOI] [PubMed] [Google Scholar]
- 59.Gennaro M.L. and Novick,R.P. (1986) cmp, a cis-acting plasmid locus that increases interaction between replication origin and initiator protein. J. Bacteriol., 168, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loomis W.P. and Moseley,S.L. (1998) Translational control of mRNA processing in the F1845 fimbrial operon of Escherichia coli. Mol. Microbiol., 30, 843–853. [DOI] [PubMed] [Google Scholar]
- 61.Condon C., Putzer,H., Luo,D. and Grunberg-Manago,M. (1997) Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J. Mol. Biol., 268, 235–242. [DOI] [PubMed] [Google Scholar]
- 62.Drider D., DiChiara,J.M., Wei,J., Sharp,J.S. and Bechhofer,D.H. (2002) Endonuclease cleavage of messenger RNA in Bacillus subtilis. Mol. Microbiol., 43, 1319–1329. [DOI] [PubMed] [Google Scholar]
- 63.Ludwig H., Homuth,G., Schmalisch,M., Dyka,F.M., Hecker,M. and Stulke,J. (2001) Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol., 41, 409–422. [DOI] [PubMed] [Google Scholar]
- 64.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olsen G.J., Woese,C.R. and Overbeek,R. (1994) The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol., 176, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]