Abstract
The U5 snRNA loop 1 is characterized by the conserved sequence G1C2C3U4U5U6Y7A8Y9 and is essential for the alignment of exons during the second step of pre-mRNA splicing in Saccharo myces cerevisiae. Despite this sequence conservation the size, rather than sequence, of loop 1 is critical for exon alignment in vitro. To determine the in vivo requirements for U5 loop 1 a library of loop 1 sequences was transformed into a yeast strain where the endogenous U5 gene was deleted. Comparison of viable mutations in loop 1 revealed that position 6 was invariant and positions 5 and 7 displayed some sequence conservation. These data indicate positions 5, 6 and 7 in loop 1 are important for U5 function in vivo. A screen for mutations that suppress the temperature-sensitive phenotype of three loop 1 mutants produced eight intragenic suppressors all containing alterations in loop 1. Further analysis of these temperature-sensitive mutants revealed that each displayed distinct cell cycle arrest phenotypes and pre-mRNA splicing inhibition patterns. The cell cycle arrest is likely attributed to inefficient splicing of α-tubulin pre-mRNA in one mutant and actin pre-mRNA in another. These results suggest that various mutations in loop 1 may affect the splicing of different pre-mRNAs in vivo.
INTRODUCTION
The removal of intron regions from pre-mRNA is catalyzed by the spliceosome, a large ribonucleoprotein complex composed of five snRNPs (U1, U2, U4/U6 and U5) and numerous non-snRNP proteins. The accepted model for assembly of the active spliceosome is the stepwise addition of snRNPs onto the pre-mRNA (reviewed in 1,2). Recently, a new model of spliceosome formation has been proposed where a pre-assembled penta-snRNP composed of all five snRNPs interacts with pre-mRNA directly to form the active spliceosome (3). Once the snRNPs are assembled with the pre-mRNA to form the spliceosome, a dynamic network of RNA–RNA interactions occurs resulting in the two transesterification reactions required for intron removal and exon ligation. These RNA–RNA interactions are certainly facilitated by the protein components of the spliceosome and involve snRNA–pre-mRNA together with snRNA–snRNA contacts (4,5).
The snRNAs perform a critical role in the recognition of conserved sequences within the introns, the tethering of spliceosomal intermediates and most likely the catalysis of splicing. For instance, the 5′ splice site is initially recognized by direct base pairing with the 5′ end of the U1 snRNA (6–8). The branchpoint is recognized by direct base pairing with the U2 snRNA resulting in a bulged adenosine required for attack of the 5′ splice site during the first step of splicing (9–12). Prior to activation of the spliceosome the U4 and U6 snRNAs are extensively base paired to each other. Upon spliceosome activation the U4/U6 base pairing is disrupted and the U6 snRNA replaces U1 at the 5′ splice site in addition to forming a complex base pairing interaction with U2 still bound at the branchpoint (5). Phosphorothioate substitution experiments have indicated that certain phosphate linkages in U6 contribute to the active site during the first catalytic step of splicing (13). In addition, the ability of U2 and U6 snRNAs in the presence of a branchpoint oligonucleotide to form a branched RNA molecule further supports a catalytic role for these RNAs (14). Finally, the U5 snRNA is known to interact with exon sequences at the 5′ and 3′ splice sites keeping the exons in the correct orientation for the second step of splicing (15).
Of all the snRNPs, U5 is the only snRNP conserved between U2 and U12 spliceosomes and therefore plays a key role in splicing (16,17). At the sequence level the U5 snRNAs from different organisms are overall variable in sequence. However, they all have in common a stem–loop that contains a conserved 9 nt sequence (G1C2C3U4U5U6Y7A8Y9) within the loop, termed loop 1 in yeast (18). In addition to sequence conservation, loop 1 includes modified nucleotides common between organisms at positions 1, 4, 6 and 9 (19).
The first clue as to the precise function of the U5 snRNA loop 1 came through genetic analysis in yeast. Mutation of the invariant G1 to A at the 5′ splice site allows accurate 5′ splice site cleavage but prevents further processing of the first step intermediates. It was found that mutations in the conserved loop 1 sequence could overcome the effects of the G1 to A mutation by activation of aberrant 5′ splice sites near the authentic site or by onward processing of dead-end intermediates produced from the authentic site (20,21). These results provided evidence that the U5 loop 1 interacted with the 5′ and 3′ exons during pre-mRNA splicing. In a mammalian system mutations in U5 loop 1 also activated aberrant 5′ splice sites with G1 to A or U2 to A mutations at the 5′ splice site (22). A direct interaction of U5 loop 1 with the 5′ and 3′ exons in mammalian and yeast wild-type pre-mRNA was demonstrated using cross-linking groups within the 5′ and 3′ exons (23–25). Recently, incorporation of cross-linking groups within the yeast and mammalian U5 loop 1 has revealed the loop 1 interactions with pre-mRNA and splicing intermediates during splicing in vitro. In yeast, replacement of loop 1 positions 4, 5, 6 or 7 with the cross-linking group 4-thio-uridine confirms 5′ exon interactions of loop 1 before and after the first step found by pre-mRNA based cross-linking (26). Cross-linking in yeast from loop 1 also identified a novel interaction of loop 1 with the conserved GU at the 5′ splice site (26). In the mammalian system cross-linking with an azidophenacyl group incorporated between loop 1 positions 5 and 6 or 6 and 7 produced cross-links only to the U of the conserved intronic GU at the 5′ splice site in the pre-mRNA and the lariat-intron intermediate (27). None of the previously discovered cross-links of exon sequences to the U5 loop 1 were found using this technique. In sum, the above data is consistent with a model where U5 loop 1 interacts intimately with sequences near the splice sites during the two steps of splicing.
Both mammalian and yeast in vitro systems have been valuable in dissecting the role of U5 loop 1 in splicing. For example, in the mammalian system loop 1 is not required for either step of splicing and it is thought that the 220 kDa protein (Prp8 in yeast) may compensate for the loss of loop 1 (28). In the yeast system, loop 1 is not required for the first catalytic step of splicing but is critical for aligning the exons for the second step of splicing (29,30). It was found that insertions and deletions in loop 1 blocked the second step of splicing in yeast indicating that the size of loop 1, not the sequence, was important for aligning exons for the second step of splicing in vitro (30). Nevertheless, even though mutations that maintain the wild-type size of loop 1 sequence do not significantly affect splicing in vitro, a number of these mutants are in fact lethal or temperature-sensitive in vivo (29,31). It was hypothesized that this was due to the inability of some U5 loop 1 mutants to generate an adequate level of mRNA from one or more of the essential intron-containing genes in the yeast genome (29).
To investigate the requirements for U5 loop 1 during pre-mRNA splicing in vivo a library of U5 loop 1 sequences was constructed. This library was employed to uncover functional U5 loop 1 sequences in a U5 deletion strain. A number of viable and temperature-sensitive variants in U5 loop 1 were discovered. All viable variants contained a U at position 6 in loop 1 in addition to some conservation in sequence at positions 5 and 7. Intragenic suppressors of temperature-sensitive variants all contained changes within loop 1. These results suggest that positions 5, 6 and 7 in loop 1 are important for U5 function in vivo. Analysis of three temperature-sensitive mutants in loop 1 revealed that each exhibited distinct growth phenotypes and pre-mRNA splicing patterns. Investigations into the pre-mRNA splicing of specific essential genes in the temperature-sensitive mutants revealed that the inefficient splicing of the α-tubulin gene could explain the cell cycle defect of one mutant and the inefficient splicing of the actin gene could explain the defect of another mutant. These results suggest that mutations in U5 loop 1 affect the splicing of different pre-mRNAs in vivo.
MATERIALS AND METHODS
Yeast strain construction
To construct a Saccharomyces cerevisiae strain where the coding sequence of the U5 snRNA gene, SNR7, was replaced by the kanMX6 marker, a PCR fragment was generated by using plasmid pFA6a-kanMX6 (32) as template and two primers (U5KOF, 5′-TATTTTAAAATACTTTTCTTTCTTTTTGTTTTAAAACCTGCGTACGCTGCAGGTCGAC and U5KOR, 5′-CATGAATCAAATTTGTAGAAAAATAAAATAGAAAAGATAAATCGATGAATTCGAGCTCG). The primers have homology to the pFA6a-kanMX6 plasmid (underlined) and homology to the regions immediately upstream and downstream of the U5 snRNA coding sequence. This PCR fragment was used to disrupt the SNR7 coding sequence in the diploid S.cerevisiae strain FY1679 (MATa/MATα; ura3-52/ura3-52; trp1Δ63/TRP1; leu2Δ1/LEU2; his3Δ200/HIS3; GAL2/GAL2) by transformation using a lithium acetate procedure (33). G418-resistant cells were selected following transformation by re-suspending cells in 200 µl of YPD and incubating at 30°C with agitation for 1–2 h, then spreading on YPD plates containing 200 mg/l G418. Genomic DNA was then prepared from G418-resistant transformants and correct targeting of the kanMX6 module into the SNR7 gene was verified by PCR with two primer pairs (U5F, 5′-CGGGGTACCCGATGACAAAGGGATAATGGG and KanB, 5′-CTGCAGCGAGGAGCCGTAAT; U5R, 5′-CCGCCTCGAGGCATCTAAAGTAGGGGAAGC and KanC, 5′-TGATTTTGATGACGAGCGTAAT). A resulting strain YROK1 (MATa/MATα; ura3-52/ura3-52; trp1Δ63/TRP1; leu2Δ1/LEU2; his3Δ200/HIS3; GAL2/GAL2; snr7::kanMX6/SNR7) was transformed with plasmid pRS416-U5. The pRS416-U5 plasmid contains the SNR7 gene cloned, by PCR amplification of genomic DNA using primers U5F and U5R, into the KpnI and XhoI sites of pRS416 (CEN, URA3) (34). Viable G418-resistant and Ura+ transformants were sporulated and tetrads dissected. The haploid progeny were analyzed to identify the strain YROK2 (MATa; ura3-52; trp1Δ63; leu2Δ1; his3Δ200; GAL2; snr7::kanMX6; pRS416-U5).
Construction of U5 snRNA library containing randomized residues
A library of mutants in the S.cerevisiae U5 snRNA loop 1 positions 94–100 was created by in vitro mutagenesis of the CEN, TRP1 plasmid m571 (29). Two oligonucleotide primers (U5Ran1, 5′-CAACACCCGGATGGTTCTGGNNNNNNNCAAGAACCATGTTCGTTATAAG and U5Ran2, 5′-CTTATAACGAACATGGTTCTTGNNNNNNNCCAGAACCATCCGGGTGTTG), each complementary to opposite strands of U5 loop 1 and containing randomized nucleotides at positions 94–100, were used with the QuikChange site-directed mutagenesis method (Stratagene). Reaction products were introduced into Escherichia coli strain XL1-Blue MRF′ (Stratagene), spread on LB-ampicillin plates and plasmid DNA from ∼1 × 106 transformants recovered to form the U5 loop 1 library. This library was sequenced to confirm a random distribution of residues at each position.
In vivo selection experiments
Strain YROK2 was transformed with the U5 loop 1 library and transformants were selected on SD-URA-TRP plates at 30°C for 3 days. These were replica-plated to 5-fluoroorotic acid (5-FOA) plates and incubated at 30°C for 3 days. Transformants that grew on 5-FOA were incubated overnight in 5 ml YPD and cells recovered by centrifugation at 1800 g for 5 min. Cells were re-suspended in 0.2 ml of lysis buffer [10 mM Tris–HCl pH 8, 1 mM EDTA, 100 mM NaCl, 1% (w/v) SDS, 2% (v/v) Triton X-100] and transferred to a 1.5 ml microcentrifuge tube. To the cell suspension, 0.2 ml of acid-washed glass beads (Sigma) and 0.2 ml of phenol– chloroform–isoamyl alcohol (25:24:1, pH 8) were added before vigorous vortexing for 6 min. Following a brief centrifugation, 0.25 ml of Buffer P2 (Qiagen) was added and the tube inverted to mix. Then, 0.35 ml of Buffer N3 (Qiagen) was added and the tube inverted to mix, followed by centrifugation for 10 min. Plasmid DNA was purified from the resulting supernatant with a QIAprep spin column (Qiagen) according to the manufacturer’s instructions and eluted in a 20 µl volume. This volume of plasmid DNA was used to transform competent XL1-Blue MRF′ cells. Plasmid DNA was isolated from the resulting transformants and the U5 loop 1 region sequenced to identify functional U5 loop 1 variants. Functional variants were re-transformed into strain YROK2, then grown on 5-FOA at 16, 30 and 37°C to verify their ability to act as the sole source of U5, along with assessing their cold- and temperature-sensitivity.
Primer extension and RT–PCR
Total RNA was prepared by the hot phenol method (35) from U5 loop 1 variants following incubation for 12 h at 30 or 37°C. For primer extension, 2 µg of RNA was hybridized in RT buffer (50 mM Tris–HCl pH 8.3, 100 mM KCl, 4 mM DTT, 10 mM MgCl2) to a 32P end-labeled oligonucleotide complementary to exon 2 of the U3 snoRNA or the U5 snRNA (U3RT, 5′-CCAAGTTGGATTCAGTGGCTC; U5RT, AAA AATATGGCAGGCCTACAGTAACGG). Following hybridization, primers were extended for 30 min at 41°C in a reaction containing RT buffer, 0.5 mM each dNTP, 0.1 U/µl RNasin, and 0.2 U/µl Super RT (HT Biotech). The reaction was heated to 100°C for 1 min, then placed on ice. RNase A (10 µg) was added and incubated at 37°C for 15 min. A 0.2 ml volume of stop solution (0.3 M sodium acetate pH 5.3, 1 mM EDTA, 0.1% SDS, 25 µg/ml E.coli tRNA) was added and extension products were precipitated with 2.5 volumes of ethanol. Precipitated extension products were re-suspended in formamide gel-loading buffer and run on an 8% acrylamide 7 M urea sequencing gel. The gel was fixed, dried and exposed to X-ray film with an intensifying screen at –80°C.
For RT–PCR, poly(A)+ mRNA was isolated from 200 µg of total RNA prepared as above from cells grown at 37°C for 12 h using the GenElute mRNA miniprep kit (Sigma). Poly(A)+ mRNA was then treated with RQ1 DNase (Promega), the DNase inactivated, then phenol extracted and ethanol precipitated. Poly(A)+ mRNA was reverse transcribed using Super RT with oligo(dT) as a primer. One-twelfth of the cDNA product was used in a PCR (50 µl total) using Taq DNA polymerase (Promega) according to the manufacturer’s protocol. Reactions were subjected to denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 56°C for 1 min and 72°C for 1 min, followed by incubation at 72°C for 10 min after the final cycle. One-fifth of the PCR product was analyzed on a 1.5% agarose gel. Primers were designed to be complementary to the two exons allowing the amplification of the unspliced pre-mRNA and spliced mRNA.
Suppressor screen
Plasmids containing three U5 loop 1 variants (U5-1, U5-2, U5-3) identified as viable at 30°C but temperature-sensitive at 37°C were digested with KpnI and BamHI. The resulting DNA fragments containing the variant U5 genes were then cloned into KpnI and BamHI digested pRS416. These resulting plasmids were then transformed separately into the diploid strain YROK1. Viable G418 resistant and Ura+ transformants were sporulated and tetrads dissected. The haploid progeny were analyzed to identify three strains, YROK1-1, YROK1-2 and YROK1-3 (MATa; ura3-52; trp1Δ63; leu2Δ1; his3Δ200; GAL2; snr7::kanMX6; pRS416-U5 mutant). These strains exhibited the same growth characteristics as the original U5 mutant isolates. Cells from these strains were spread onto YPD plates at high density (∼5 × 106 cells/plate) and incubated at 37°C. Suppressor colonies that appeared within 3–4 days were transformed with the corresponding CEN, TRP based U5 variant plasmids. Trp+ and Ura+ transformants were plated onto 5-FOA to determine if the suppressors were intragenic or extragenic. The cells that grew on 5-FOA were then tested for growth on YPD plates at 37°C. Strains that failed to form colonies at 37°C were intragenic suppressor mutations that were linked to the original CEN URA3 plasmid. This CEN URA3 plasmid was isolated from the original intragenic suppressors and the U5 snRNA sequenced to determine the nature of the intragenic mutations.
Suppressor mutations in loop 1 that relieved growth inhibition at 37°C were tested for suppression of the cold-sensitive phenotype. The U5 genes from suppressors recovered in CEN URA3 plasmids were cut from the plasmid with EcoRI and XhoI and ligated into a similarly cut CEN TRP1 plasmid (pRS414). The resulting plasmids were transformed into strain YROK2 and Trp+ transformants isolated. Single colonies were transferred to 5-FOA plates and incubated at 16, 30 and 37°C to confirm their growth phenotype.
Microscopy
U5 loop 1 mutant strains were grown at 37°C for 3 h, washed with water and fixed for 10 min in 10% ethanol. Cells were then stained with 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 5 min and mounted with mounting media (36). Cells were visualized using a Zeiss Axioskop with a 100× Plan-Neofluar objective.
RESULTS
Randomization selection of functional variants of U5 snRNA loop 1
The high conservation of sequence in U5 loop 1 between organisms suggests that mutation of loop 1 may not be tolerated in vivo. In fact, a handful of mutations in U5 loop 1 have been found to be lethal or temperature-sensitive as the sole source of U5 in yeast (29,31). To date, however, there has not been a systematic analysis of the in vivo requirements for U5 loop 1. To investigate the function of the yeast U5 loop 1 in vivo, a CEN TRP1 plasmid library of U5 genes was constructed with particular residues in loop 1 randomized by in vitro mutagenesis. Positions 2–8 of the yeast U5 loop 1 (Fig. 1) were chosen for randomization with the thought that randomizing all nine positions of the conserved sequence might not yield viable mutants. This library was used to transform a haploid yeast strain (YROK2) in which the U5 coding sequence was deleted and complemented by a CEN URA3 plasmid containing the wild-type U5 gene. Trans formants were then replica-plated to media containing 5-FOA which selects against the wild-type U5 CEN URA3 plasmid. Colonies that grow on 5-FOA should contain functional variants of U5 loop 1. Through this procedure a number of colonies were obtained that were capable of growth on 5-FOA. Plasmids were isolated from these colonies and the region encompassing U5 loop 1 was sequenced. Approximately 100 plasmids were sequenced from individual colonies that grew on 5-FOA after 3 days. Of the sequenced plasmids roughly half contained the wild-type U5 loop 1 with the remainder containing functional variants of U5 loop 1. These functional variants in loop 1 were subsequently re-transformed into YROK2, then transferred to 5-FOA plates to confirm their ability to complement the U5 deletion. In addition, each of the variants was tested for growth at 16, 30 and 37°C on 5-FOA.
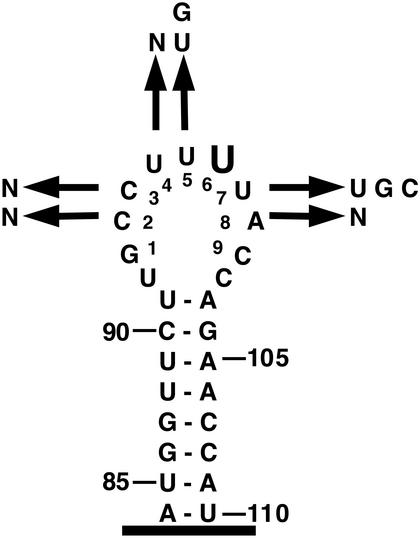
Figure 1.
Randomization of U5 snRNA loop 1. The S.cerevisiae U5 snRNA stem–loop 1 nucleotides 84–110 are depicted with the conserved U5 loop 1 sequence numbered. The nucleotides recovered at each randomized position are shown. Only variants from Table 1 that grew as wild-type at all temperatures tested are depicted. Large type indicates that only the wild-type nucleotide was recovered at that position.
In total, 47 viable mutants in U5 loop 1 were identified (Table 1). Of these mutants, four were isolated independently more than once. Three further mutants contained a single nucleotide deletion within U5 loop 1. Identification of viable, single nucleotide, deletions of loop 1 was not surprising since it has been previously demonstrated that a number of single nucleotide deletions in loop 1 are functional in vitro and viable in vivo (30; R. T. O’Keefe and A. J. Newman, unpublished results). Of the 47 functional U5 loop 1 variants, 19 grew at each temperature tested. These variants were aligned to identify the in vivo requirements for U5 loop 1 at each of the randomized positions (Table 1). In all variants that grew as wild-type, position 6 in loop 1 was invariantly U (Fig. 1). At position 5 only the wild-type U or a G residue was tolerated. At position 7 the wild-type U or a G or a C residue was tolerated. At the other positions all three possible nucleotide changes were observed. All variants also retained a run of two or more U residues. Inspection of functional sequences has not revealed any co-variation between loop 1 nucleotides. While not all U5 variants conform to this consensus for loop 1 function (see Discussion for explanation) and the screen was not saturating, these results, nevertheless, suggest that the bases at positions 5, 6 and 7 in U5 loop 1 are important for U5 function in vivo.
Table 1. U5 snRNA loop 1 mutants.
| Loop 1 sequence | Growth | Loop 1 sequence | Growth | ||||
|---|---|---|---|---|---|---|---|
| 16°C | 30°C | 37°C | 16°C | 30°C | 37°C | ||
| CCUUUUA (wt) | + | + | + | CCUUUUA (wt) | + | + | + |
| UUUGUUC | + | + | + | ACUGCUC | +/– | + | +/– |
| GUUUUUG | + | + | + | GUUGCCU | – | +/– | – |
| GCGUUUC | + | + | + | UUUGUUA | +/– | + | – |
| UAUUUUCa | + | + | + | UUUUUGU (U5-2) | – | + | – |
| GCUUUUA | + | + | + | GGCUAUU | – | + | +/– |
| UCUUUCCa | + | + | + | CGUUAGC (U5-1) | + | + | – |
| GCGGUUU | + | + | + | CCCCUUC | +/– | + | + |
| UUUUUUA | + | + | + | GAUAUUG | + | + | +/– |
| UGUUUGGa | + | + | + | UGCUACC | – | + | +/– |
| GUUUUGA | + | + | + | UCUGGUC | – | + | – |
| CUGUUUA | + | + | + | ACAUUAC | + | + | – |
| CCUUUGC | + | + | + | UCCGUGC | – | + | +/– |
| GGUUUUU | + | + | + | ACAGACC | – | +/– | – |
| UUUUUUG | + | + | + | GCCGCUC | – | + | – |
| CACUUUC | + | + | + | CAGGUUU | +/– | + | + |
| AUCUUUUa | + | + | + | ACCUACC | +/– | +/– | – |
| AGUUUGC | + | + | + | GUCUGUU | – | + | +/– |
| AAAUUCA | + | + | + | GUUUAUG | + | + | +/– |
| UUUGUUC | + | + | + | CUUCCUU | – | +/– | – |
| UAUCUCG | – | +/– | – | ||||
| CAUACCU | – | +/– | – | ||||
| UUGUGU | + | + | +/– | ||||
| CGUAAU | – | +/– | – | ||||
| UUCUGU (U5-3) | – | + | – | ||||
+, growth; +/–, slow growth; –, no growth. Plates were scored after 3 days at 30 and 37°C or 5 days at 16°C.
aVariants that appeared twice.
Analysis of temperature-sensitive mutations in U5 snRNA loop 1
In an attempt to determine the defect associated with the temperature-sensitive mutations found in U5 loop 1 during this screen three mutants were chosen for further analysis. Two mutants (U5-1, CGUUAGC and U5-2, UUUUUGU) contained changes in four of the seven nucleotides in loop 1 and the third mutant (U5-3) contained changes to the sequence of loop 1 in addition to a deletion of one nucleotide (UUCUGU). These mutants exhibited either a temperature-sensitive phenotype (U5-1) or a cold- and temperature-sensitive phenotype (U5-2 and U5-3) (Fig. 2).
Figure 2.
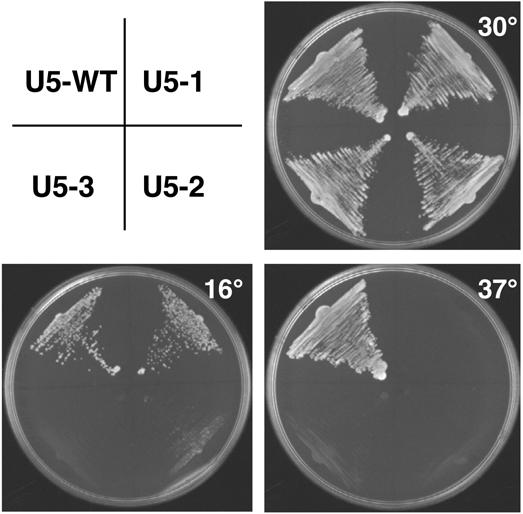
Cold- and temperature-sensitive phenotypes of U5 snRNA loop 1 mutants. Single colonies of wild-type U5 snRNA or loop 1 mutants transformed into the U5 deletion strain YROK2 were streaked onto 5-FOA plates. The plates were photographed after incubation for 2 days at 37°C, 3 days at 30°C and 5 days at 16°C.
It was presumed that these three mutants in loop 1 would either affect the stability of the U5 snRNA and/or inhibit splicing in general when grown at the restrictive temperature. Therefore, cultures of these strains were grown at 30 and 37°C for 12 h and total RNA isolated for analysis. There is a significant difference in growth of the mutant strains at the different temperatures at this time point (data not shown). The abundance and stability of the U5 snRNA was analyzed by primer extension of total RNA with a labeled primer complementary to the U5 snRNA. This analysis revealed that there is no change in the levels of U5 for any of the mutants when comparing RNA isolated following growth at 30 and 37°C (Fig. 3A). This indicated that the temperature-sensitive phenotype of the three mutants was not due to the loss of the U5 snRNA.
Figure 3.
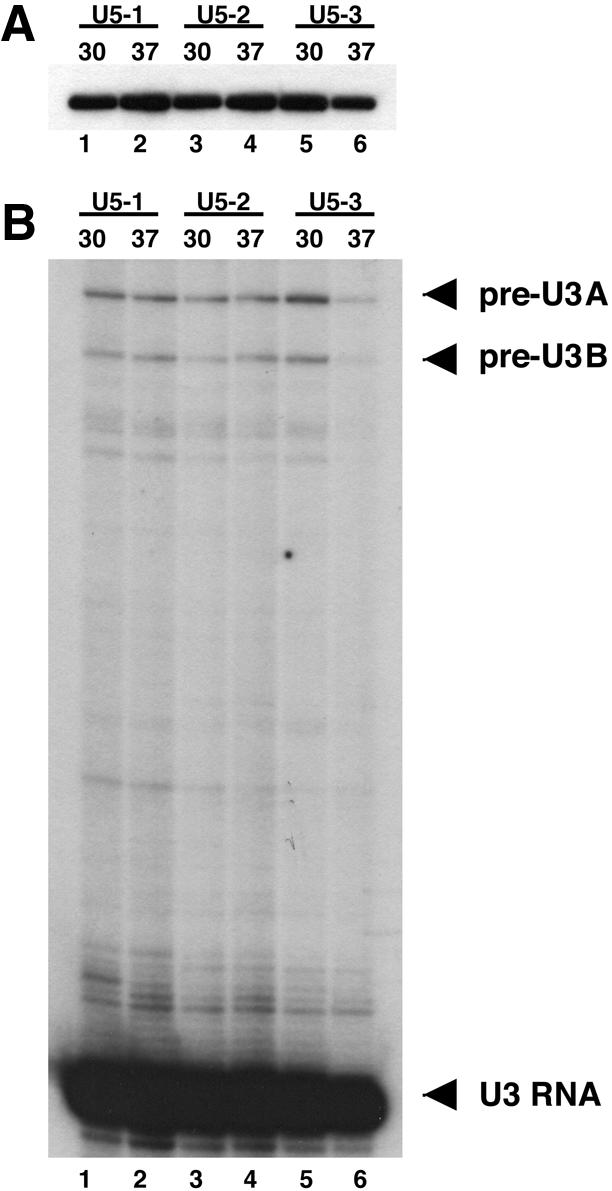
U5 snRNA levels and splicing of U3 snoRNA in U5 loop 1 temperature-sensitive mutants. Total RNA was extracted from cells grown at 30 or 37°C for 12 h and subjected to primer extension analysis. (A) Primer extension analysis to determine U5 snRNA levels. An end-labeled oligonucleotide complementary to the U5 snRNA was used for primer extension of total RNA from U5 loop 1 mutants U5-1 (lanes 1–2), U5-2 (lanes 3–4) or U5-3 (lanes 5–6) at the indicated temperatures. (B) Primer extension analysis to determine the splicing efficiency of the U3 snoRNA. An end-labeled oligonucleotide complementary to exon 2 of the U3 snoRNA was used for primer extension of total RNA from U5 loop 1 mutants U5-1 (lanes 1–2), U5-2 (lanes 3–4) or U5-3 (lanes 5–6) at the indicated temperatures. There are two genes for the U3 snoRNA (U3A and U3B) each with introns of different sizes. The U3A pre-mRNA, U3B pre-mRNA and spliced U3 RNA are indicated at the right of the panel.
Splicing efficiency of the U3 genes SNR17A and SNR17B was next analyzed by primer extension of total RNA from each mutant strain at 30 and 37°C with a labeled oligonucleotide complementary to the common exon 2 of the two genes. Primer extension analysis revealed that none of the U5 loop 1 mutants exhibited any dramatic increase in U3A or U3B pre-mRNA at the restrictive temperature (Fig. 3B). These data indicate that U5 loop 1 mutations do not cause a global inhibition of splicing but this does not preclude the possibility that they may influence the splicing of particular individual essential pre-mRNAs.
Intragenic mutations in U5 loop 1 suppress the temperature-sensitive phenotype
To elucidate why a number of the U5 snRNA loop 1 mutants were temperature-sensitive a screen was carried out to identify mutations within the U5 snRNA that would suppress the temperature-sensitive phenotype. It was hypothesized that such mutations may define additional elements of the U5 snRNA that interact with loop 1 or mutations in loop 1 itself that would alleviate the temperature-sensitive phenotype. To this end, three yeast strains were constructed where the U5 coding sequence was deleted and complemented by a CEN URA3 plasmid containing each of the temperature-sensitive mutations in U5 loop 1 (U5-1, U5-2, U5-3). Cells from these strains were spread on YPD plates at high density and incubated at the restrictive temperature of 37°C. For each strain a number of potential spontaneous suppressors appeared following incubation for 3–4 days at 37°C.
In total, eight intragenic suppressors were identified (Table 2). All suppressors carried single nucleotide changes in the U5 loop 1 that restored growth of the mutants at 37°C. The U5-1 suppressors carried changes at position 7 from G to the original wild-type U at that position. The U5-2 suppressors contained changes from the G at position 7 in the original loop 1 mutation to C, the wild-type U or, in one instance, a deletion of the G at position 7. The two U5-3 suppressors both contained alteration of the G in the original loop 1 mutation to either C or U. There were no suppressors of mutant U5-3 that regained a nucleotide in loop 1. Owing to the one nucleotide deletion in the U5-3 mutant, the suppressors could not be assigned to a specific position but they did occupy a region of the loop that is close to position 7. These results highlight the importance of position 7 in U5 loop 1 function in vivo.
Table 2. U5 snRNA loop 1 intragenic suppressors.
| Loop 1 sequence | Growth | ||
|---|---|---|---|
| 16°C | 30°C | 37°C | |
| CGUUAGC (U5-1) | + | + | – |
| CGUUAUCa | + | + | + |
| UUUUUGU (U5-2) | – | + | – |
| UUUUUUUa | + | + | + |
| UUUUU-U | + | + | + |
| UUUUUCU | + | + | + |
| UUCUGU (U5-3) | – | + | – |
| UUCUUU | +/– | + | + |
| UUCUCU | +/– | + | + |
+, growth; +/–, slow growth; –, no growth. Plates were scored after 3 days at 30 and 37°C or 5 days at 16°C.
aVariants that appeared twice.
Two of the U5 loop 1 mutants, U5-2 and U5-3, in addition to the temperature-sensitive phenotype exhibited cold-sensitivity (Fig. 2). It was possible that the intragenic suppressors that relieved the temperature-sensitive phenotype of these mutants could also relieve the cold-sensitive phenotype. The suppressors were cloned into CEN TRP1 plasmids and tested by plasmid shuffle for complementation of the U5 deletion in YROK2. All the suppressor mutants in U5-2 formed colonies at the three temperatures tested (Table 2). In contrast, the two suppressor mutants in U5-3 could form colonies at 30 and 37°C but were very slow growing following incubation at 16°C for 5 days (Table 2). These suppressors did eventually form colonies at 16°C after 7 days incubation.
Temperature-sensitive mutants in U5 loop 1 exhibit distinct growth and pre-mRNA splicing phenotypes
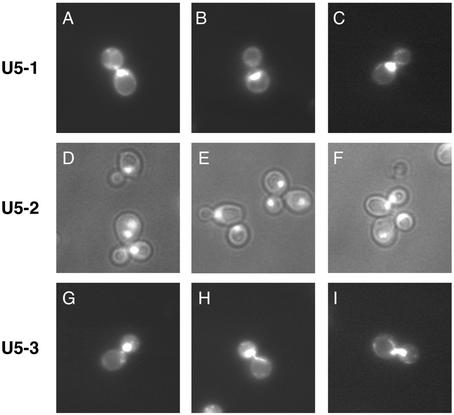
All mutations in U5 loop 1 tested to date that do not change the size of loop 1 are functional in vitro (29,30). A number of these mutants, however, do not support growth in vivo. To explain these apparently contradicting results it was hypothesized that particular mutants in U5 loop 1 may influence the splicing of individual or a subset of essential pre-mRNAs (29). If this was the case then it is possible that various U5 loop 1 mutants may display distinct phenotypes depending on the messages they influence. To determine whether the three U5 temperature-sensitive mutants (U5-1, U5-2 and U5-3) displayed any differences, the bud and nuclear morphologies of each mutant strain were visualized following switch to the restrictive temperature of 37°C. The U5-1 mutant cell population was characterized by a large proportion of cells arrested with a large bud phenotype. These cells possessed buds that were equal to or greater than half the size of the mother cell and the nucleus was wholly contained within the mother cell (Fig. 4A–C). The remaining cells were mostly unbudded cells or budded cells with the nucleus partially entering the daughter cell. In contrast, the U5-2 mutant cell population was characterized by a large proportion of cells exhibiting a multiple bud phenotype, indicative of a defect in cell separation (Fig. 4D–F). In addition, the mutant cells also displayed an increase in cell size. Finally, the U5-3 mutant cells also exhibited a distinct phenotype. A large proportion of cells in the U5-3 mutant cell population were characterized by nuclear positioning defects. These defects resembled a metaphase/anaphase arrest (Fig. 4G–I). For instance, in some cells the nucleus was stretched between the mother and daughter cells. In others the nucleus was stuck within the neck between the mother and daughter cells. Finally, in further cells the nucleus was almost completely in the daughter cell. Taken together, the distinct differences in bud and nuclear morphologies of the three U5 loop 1 mutants suggests that the mutants may affect the splicing of different pre-mRNAs.
Figure 4.
Phenotypes of U5 snRNA loop 1 temperature-sensitive mutants. Cultures of U5 snRNA loop 1 mutants were shifted to 37°C for 3 h and DNA was visualized by DAPI staining. U5-1 mutants DAPI staining (A–C). U5-2 mutants phase contrast and DAPI staining (D–F). U5-3 mutants DAPI staining (G–I).
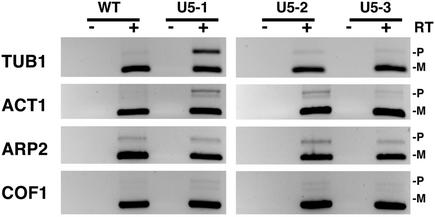
To gain further insight into the differences between U5 loop 1 mutants each mutant was tested for sensitivity to certain growth conditions and for the splicing of particular pre-mRNAs. It has recently been observed that the cell cycle phenotype exhibited by mutations in the spliceosomal proteins Cef1, Prp17 and Prp22 is in part due to the inefficient splicing of the essential gene for α-tubulin, TUB1 (37). Cells with defective microtubules are characterized by the presence of large budded cells and nuclear positioning defects. This phenotype is displayed by the U5-1 mutant. To determine if any of the three U5 mutants possessed defective microtubules each mutant was grown in the presence of the microtubule depolymerizing drug benomyl. Mutants with defective microtubules usually display hypersensitivity to benomyl. Interestingly, loop 1 mutant U5-1 displayed hypersensitivity to benomyl at 10 µg/ml, whereas mutants U5-2 and U5-3 did not exhibit any hypersensitivity (Fig. 5). In addition, splicing of the TUB1 pre-mRNA was inhibited in the U5-1 mutant but not in the U5-2 and U5-3 mutants (Fig. 6). These results suggest that defective splicing of the TUB1 pre-mRNA may explain the cell cycle defect associated with mutant U5-1.
Figure 5.
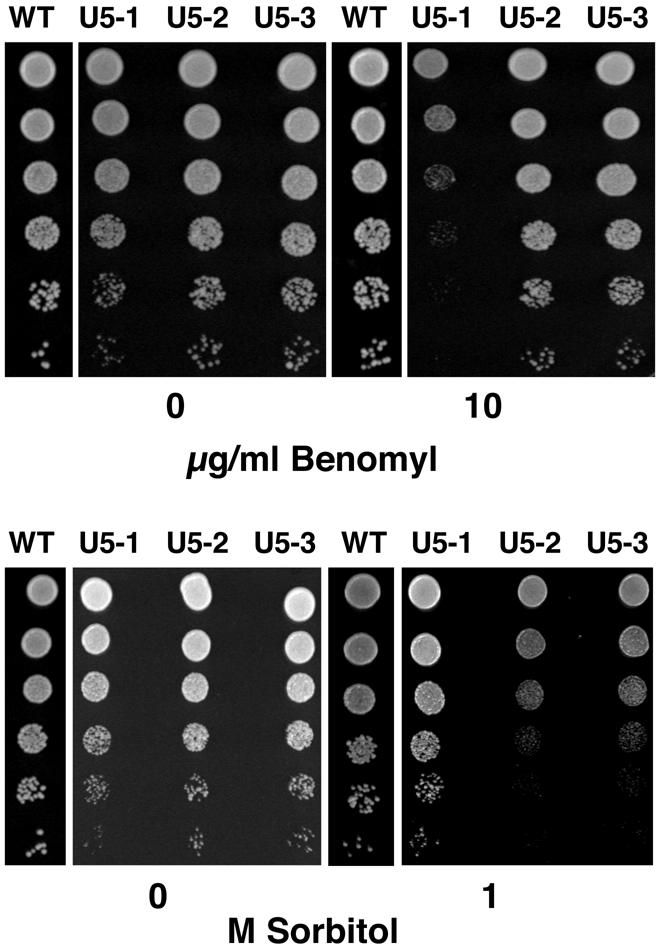
Benomyl and sorbitol sensitivity of U5 snRNA loop 1 mutants. Single colonies of U5 snRNA loop 1 mutants from 5-FOA plates were grown to an optical density of 1.0 at 600 nm then serially diluted 5-fold and spotted onto YPD plates or YPD plates containing the indicated amount of benomyl or sorbitol. Cells were grown for 3 days at 30°C.
Figure 6.
Splicing analysis by RT–PCR of specific genes in U5 snRNA loop 1 mutants. Inverted images of ethidium bromide stained agarose gels. Poly(A)+ mRNA isolated from total RNA was subjected to RT–PCR. The genes analyzed are listed to the left of each panel. The unspliced pre-mRNA (P) and spliced mRNA (M) are indicated on the right of each panel. Control reactions that did not include reverse transcription of poly(A)+ RNA are indicated above each lane with a minus (–).
The phenotype displayed by U5 loop 1 mutant U5-2, multiple buds and increase in cell size, has been observed for mutants affecting the actin cytoskeleton (38). Some mutants with actin-related defects are sensitive to high osmolarity. To determine whether any of the three U5 mutants exhibited sensitivity to high osmolarity mutants were grown on plates containing 1 M sorbitol. At this concentration of sorbitol, mutant U5-1, which was sensitive to benomyl, grew as wild-type (Fig. 5). In contrast, mutants U5-2 and U5-3 were both sensitive to 1 M sorbitol (Fig. 5). Analysis of the splicing of three genes associated with actin function, ACT1, ARP2 and COF1, revealed that mutants U5-1 and U5-2 displayed a decrease in the splicing efficiency of the ACT1 pre-mRNA, whereas U5-3 did not (Fig. 6). None of the mutants displayed any significant differences in the splicing of ARP2 and COF1 pre-mRNA compared to the wild-type strain (Fig. 6). These results suggest that the decrease in splicing efficiency of the ACT1 pre-mRNA may explain the cell cycle defect associated with mutant U5-2. In contrast, while mutant U5-1 displays decreased splicing of the ACT1 pre-mRNA, it also has a defect in TUB1 splicing which may be the major defect. It is still unclear which intron-containing genes are not spliced in the U5-3 mutant.
DISCUSSION
Randomization of U5 loop 1 in vivo has uncovered a collection of functional variants indicating a degree of flexibility in the requirements of loop 1 for pre-mRNA splicing. While loop 1 was tolerant to changes it was discovered that positions 5, 6 and 7 in loop 1 exhibited more strict requirements. A number of viable mutants in U5 loop 1 were found to be temperature-sensitive. Analysis of three temperature-sensitive mutants in U5 loop 1 revealed each displayed distinct cell cycle and pre-mRNA splicing phenotypes. These differences between U5 loop 1 mutants suggest that various sequences in loop 1 can influence the splicing of particular pre-mRNAs.
It was somewhat surprising to discover a number of viable variants in U5 loop 1 in light of the high sequence conservation of loop 1 between organisms. Examination of the U5 loop 1 variants that grew as wild-type at all temperatures tested revealed that positions 5, 6 and 7 appeared to be important for loop 1 function in vivo. In particular, position 6 was invariantly U and changes in position 7 could suppress the temperature-sensitive defect of two loop 1 mutants. It was not possible, however, to construct an exact consensus for loop 1 function in vivo as a number of the temperature-sensitive mutants conformed to the consensus found with the wild-type growers. It is apparent, therefore, that the yeast splicing machinery is somewhat flexible to alterations in U5 loop 1. It appears that certain pre-mRNAs are more sensitive to changes in loop 1 than others. Therefore, the highly conserved U5 loop 1 sequence found in most organisms may represent the optimal sequence for efficient splicing of the full complement of pre-mRNAs in a cell.
The previous in vivo and in vitro functional analysis of the yeast U5 loop 1 appeared, at the time, to be contradictory. Certain mutations in U5 loop 1 that maintained the size of loop 1 were competent for splicing of a pre-mRNA in vitro but did not support splicing in vivo (29). Two explanations could be presented to account for these earlier results. First, splicing in vitro could be less sensitive to changes in U5 loop 1 than splicing in vivo. Secondly, changes in loop 1 in vivo may only influence the splicing of a subset of essential pre-mRNAs but not all pre-mRNAs. If this second scenario is correct the use of a very limited number of efficiently spliced pre-mRNAs to analyze splicing in vitro may not accurately represent the splicing of all pre-mRNAs in vivo. The results presented here indicating that U5 loop 1 mutants affect the splicing of different pre-mRNAs supports this second hypothesis. In HeLa extracts both steps of splicing can be carried out in vitro in the absence of U5 loop 1 and the 220 kDa protein is proposed to compensate for loss of loop 1 (28). However, in light of the results presented here, it is still possible that there are pre-mRNAs that may be affected by U5 loop 1 mutations in human cells.
How can altering the sequence of U5 loop 1 influence the splicing of different pre-mRNAs? Novel sequences in loop 1 could perturb the known interactions of loop 1 with the pre-mRNA and splicing intermediates. Positions 5, 6 and 7 in U5 loop 1 identified here as being important for U5 function in vivo have been previously implicated in U5 function (20–27). It is more than likely, therefore, that changes to the sequence of loop 1 at these positions may influence the interaction of U5 with particular pre-mRNAs. This, in turn, may affect the efficiency of their splicing or even inhibit splicing of some pre-mRNAs altogether.
In addition to the interactions of U5 loop 1 with the splicing substrate, there is evidence that loop 1 interacts with other RNAs and proteins within the spliceosome. For example, a cross-link between loop 1 position 7 and the U1 snRNA has been identified very early in the mammalian splicing reaction (39). In yeast, mutations in U5 loop 1 position 6 or positions 5 and 7 are synthetic lethal with mutations in the U2 snRNA residues that form helix Ia and Ib with the U6 snRNA (40). These positions in loop 1 are also synthetic lethal with the proteins Prp8, Slu7, Prp17, Slt11 and Brr2 (31,40). In fact, cross-linking has demonstrated that Prp8 directly interacts with U5 loop 1 (41,42). Changing the sequence of U5 loop 1, therefore, may also influence the interaction of loop 1 with snRNA or protein components within the spliceosome affecting the splicing efficiency of particular pre-mRNAs.
A model can be proposed to account for all in vitro and in vivo mutagenesis of the yeast U5 loop 1. Mutations that change the size of loop 1 prevent alignment of the exons for the second step of splicing and inhibit splicing in vitro and are lethal in vivo (29,30). Because these mutations influence a fundamental step in pre-mRNA splicing, exon alignment, all messages will be affected. Mutations that change the sequence of loop 1 allow splicing of the limited number of messages utilized to test splicing in vitro. These mutations in vivo can display a spectrum of growth characteristics including lethality, temperature sensitivity or wild-type growth. Loop 1 sequence changes, therefore, may only perturb slightly the function of the spliceosome and result in a decrease of splicing efficiency. In this case only certain pre-mRNAs may be affected. This model can be extended to protein-splicing factors where some mutations may affect a fundamental aspect of the splicing reaction and influence the splicing of all messages. Other mutations may only perturb the splicing efficiency of certain pre-mRNAs.
It is well documented that a number of S.cerevisiae and Schizosaccharomyces pombe protein-splicing factors manifest cell cycle phenotypes when depleted or mutated (37,43–51). However, not all splicing factors display a cell cycle phenotype when depleted or mutated. Indeed some splicing factors exhibit defects in splicing but no cell cycle phenotype (37,44). A number of hypotheses have been put forward to explain why mutation or depletion of splicing factors results in a cell cycle phenotype. The first is that inhibition of pre-mRNA splicing may change the organization of the nucleus thereby preventing cell cycle progression (43,45,46,52). Alternatively, mutations in splicing factors that disrupt splicing may elicit a checkpoint response that arrests the cell cycle (49,50). A more likely explanation is that some intron-containing genes required for cell cycle transitions may have different requirements for protein splicing factors or their splicing may be regulated (37,45,49–51). In fact, proof that mutation of a protein-splicing factor affects the splicing of a specific cell cycle gene has recently been presented for the gene CEF1 (37). The results presented here are the first evidence that mutation of an snRNA leads to cell cycle-like defects.
It is likely that U5 loop 1 mutations will affect the splicing of more than one pre-mRNA. While the current results identify two genes, TUB1 and ACT1, whose splicing is affected in two of the U5 loop 1 mutations, splicing analysis of all the intron-containing genes in S.cerevisiae is required. Splicing-specific microarrays have been recently developed to analyze mRNA processing in yeast (37,53). These would be ideally suited for the analysis of splicing in U5 loop 1 mutant strains. Analysis of all introns from the nuclear genome sequence of S.cerevisiae has revealed that there is a preference for AAAG preceding the 5′ splice site and that these nucleotides may be important for interacting with U5 loop 1 (54–56). It would be interesting to determine if the exon sequences preceding the 5′ splice site of all genes affected by U5 loop 1 mutations can be related to the changes in loop 1.
Acknowledgments
ACKNOWLEDGEMENTS
I would like to thank Colin Stirling for the FY1679 yeast strain, Riz Alvi for constructing strains YROK1, YROK2 and the U5 snRNA loop 1 library and Helen Dobbyn for designing the TUB1 and ACT1 PCR primers. I thank Andy Newman and Helen Dobbyn for critical reading of the manuscript. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosome. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Press, Cold Spring Harbor, NY, pp. 525–560.
- 2.Will C.L. and Lührmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- 3.Stevens S.W., Ryan,D.E., Ge,H.Y., Moore,R.E., Young,M.K., Lee,T.D. and Abelson,J. (2002) Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell, 9, 31–44. [DOI] [PubMed] [Google Scholar]
- 4.Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen T.W. (1998) RNA–RNA interactions in nuclear pre-mRNA splicing. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Press, Cold Spring Harbor, NY, pp. 279–307.
- 6.Zhuang Y. and Weiner,A.M. (1986) A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell, 46, 827–835. [DOI] [PubMed] [Google Scholar]
- 7.Séraphin B., Kretzner,L. and Rosbash,M. (1988) A U1 snRNA-pre-mRNA base pairing interaction is required early in spliceosome assembly but does not uniquely define the 5′ splice site. EMBO J., 7, 2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siliciano P.G. and Guthrie,C. (1988) 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev., 2, 1258–1267. [DOI] [PubMed] [Google Scholar]
- 9.Parker R., Siliciano,P.G. and Guthrie,C. (1987) Recognition of the TACTAAC box during mRNA splicing in yeast involves base-pairing to the U2-like snRNA. Cell, 49, 229–239. [DOI] [PubMed] [Google Scholar]
- 10.Wu J. and Manley,J.L. (1989) Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev., 3, 1553–1561. [DOI] [PubMed] [Google Scholar]
- 11.Zhuang Y., Goldstein,A.M. and Weiner,A.M. (1989) UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc. Natl Acad. Sci. USA, 86, 2752–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Query C.C., Moore,M.J. and Sharp,P.A. (1994) Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev., 8, 587–597. [DOI] [PubMed] [Google Scholar]
- 13.Yean S.-L., Wuenschell,G., Termini,J. and Lin,R.-J. (2000) Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature, 408, 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valadkhan S. and Manley,J.L. (2001) Splicing-related catalysis by protein-free snRNAs. Nature, 413, 701–707. [DOI] [PubMed] [Google Scholar]
- 15.Newman A.J. (1997) The role of U5 snRNP in pre-mRNA splicing. EMBO J., 16, 5797–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall S.L. and Padgett,R.A. (1996) Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science, 271, 1716–1718. [DOI] [PubMed] [Google Scholar]
- 17.Tarn W.-Y. and Steitz,J.A. (1996) A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell, 84, 801–811. [DOI] [PubMed] [Google Scholar]
- 18.Frank D.N., Roiha,H. and Guthrie,C. (1994) Architecture of the U5 small nuclear RNA. Mol. Cell. Biol., 14, 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szkukalek A., Myslinski,E., Mougin,A., Lührmann,R. and Branlant,C. (1995) Phylogenetic conservation of modified nucleotides in the terminal Loop 1 of the spliceosomal U5 snRNA. Biochimie, 77, 16–21. [DOI] [PubMed] [Google Scholar]
- 20.Newman A. and Norman,C. (1991) Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell, 65, 115–123. [DOI] [PubMed] [Google Scholar]
- 21.Newman A.J. and Norman,C. (1992) U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell, 68, 743–754. [DOI] [PubMed] [Google Scholar]
- 22.Cortes J.J., Sontheimer,E.J., Seiwert,S.D. and Steitz,J.A. (1993) Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5′ splice sites in vivo. EMBO J., 12, 5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyatt J.R., Sontheimer,E.J. and Steitz,J.A. (1992) Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev., 6, 2542–2553. [DOI] [PubMed] [Google Scholar]
- 24.Sontheimer E.J. and Steitz,J.A. (1993) The U5 and U6 small nuclear RNAs as the active site components of the spliceosome. Science, 262, 1989–1996. [DOI] [PubMed] [Google Scholar]
- 25.Newman A.J., Teigelkamp,S. and Beggs,J.D. (1995) snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA, 1, 968–980. [PMC free article] [PubMed] [Google Scholar]
- 26.Alvi R.K., Lund,M. and O’Keefe,R.T. (2001) ATP-dependent interaction of yeast U5 snRNA loop 1 with the 5′ splice site. RNA, 7, 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McConnell T.S. and Steitz,J.A. (2001) Proximity of the invariant loop of U5 snRNA to the second intron residue during pre-mRNA splicing. EMBO J., 20, 3577–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ségault V., Will,C.L., Polycarpou-Schwarz,M., Mattaj,I.W., Branlant,C. and Lührmann,R. (1999) Conserved loop I of U5 small nuclear RNA is dispensable for both catalytic steps of pre-mRNA splicing in HeLa nuclear extracts. Mol. Cell. Biol., 19, 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Keefe R.T., Norman,C. and Newman,A.J. (1996) The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell, 86, 679–689. [DOI] [PubMed] [Google Scholar]
- 30.O’Keefe R.T. and Newman,A.J. (1998) Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J., 17, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank D., Patterson,B. and Guthrie,C. (1992) Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol. Cell. Biol., 12, 5197–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wach A., Brachat,A., Alberti-Segui,C., Rebischung,C. and Philippsen,P. (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast, 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- 33.Gietz D., St. Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohrer K. and Domdey,H. (1991) Preparation of high molecular weight RNA. Methods Enzymol., 194, 398–405. [DOI] [PubMed] [Google Scholar]
- 36.Johnson G.D. and Nogueira Araujo,G.M. (1981) A simple method of reducing the fading of immunofluorescence during microscopy. J. Immunol. Methods, 43, 349–350. [DOI] [PubMed] [Google Scholar]
- 37.Burns C.G., Ohi,R., Mehta,S., O’Toole,E.T., Winey,M., Clark,T.A., Sugnet,C.W., Ares,M. and Gould,K.L. (2002) Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol., 22, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruyne D. and Bretscher,A. (2000) Polarization of cell growth in yeast II. The role of the cortical actin cytoskeleton. J. Cell Sci., 113, 571–585. [DOI] [PubMed] [Google Scholar]
- 39.Ast G. and Weiner,A.M. (1997) A novel U1/U5 interaction indicates proximity between U1 and U5 snRNAs during an early step of mRNA splicing. RNA, 3, 371–381. [PMC free article] [PubMed] [Google Scholar]
- 40.Xu D., Field,D.J., Tang,S.-J., Moris,A., Bobechko,B.P. and Friesen,J.D. (1998) Synthetic lethality of yeast slt mutations with U2 small nuclear RNA mutations suggests functional interactions between U2 and U5 snRNPs that are important for both steps of pre-mRNA splicing. Mol. Cell. Biol., 18, 2055–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dix I., Russell,C.S., O’Keefe,R.T., Newman,A.J. and Beggs,J.D. (1998) Protein–RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA, 4, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urlaub H., Hartmuth,K., Kostka,S., Grelle,G. and Lührmann,R. (2000) A general approach for identification of RNA–protein crosslinking sites within native human spliceosomal snRNPs: Analysis of RNA–protein contacts in native U1 and [U4/U6.U5] snRNPs. J. Biol. Chem., 275, 41458–41468. [DOI] [PubMed] [Google Scholar]
- 43.Lundgren K., Allan,S., Urushiyama,S., Tani,T., Ohshima,Y., Frendewey,D. and Beach,D. (1996) A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol. Biol. Cell, 7, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shea J.E., Toyn,J.H. and Johnston,L.H. (1994) The budding yeast U5 snRNP Prp8 is a highly conserved protein which links RNA splicing with cell cycle progression. Nucleic Acids Res., 22, 5555–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald W.H., Ohi,R., Smelkova,N., Frendewey,D. and Gould,K.L. (1999) Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol., 19, 5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potashkin J., Kim,D., Fons,M., Humphrey,T. and Frendewey,D. (1998) Cell division cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet., 34, 153–163. [DOI] [PubMed] [Google Scholar]
- 47.Urushiyama S., Tani,T. and Ohshima,Y. (1996) Isolation of novel pre-mRNA splicing mutants of Schizosaccharomyces pombe. Mol. Gen. Genet., 253, 118–127. [DOI] [PubMed] [Google Scholar]
- 48.Tang Z.H., Yanagida,M. and Lin,R.J. (1998) Fission yeast mitotic regulator Dsk1 is an SR protein-specific kinase. J. Biol. Chem., 273, 5963–5969. [DOI] [PubMed] [Google Scholar]
- 49.Russell C.S., Ben-Yehuda,S., Dix,I., Kupiec,M. and Beggs,J.D. (2000) Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA, 6, 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Yehuda S., Russell,C.S., Dix,I., Beggs,J.D. and Kupiec,M. (2000) Extensive genetic interactions between PRP8 and PRP17/CDC40, two yeast genes involved in pre-mRNA splicing and cell cycle progression. Genetics, 154, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ben-Yehuda S., Dix,I., Russell,C.S., McGarvey,M., Beggs,J.D. and Kupiec,M. (2000) Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics, 156, 1503–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi K., Yamada,H. and Yanagida,M. (1994) Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell, 5, 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clark T.A., Sugnet,C.W. and Ares,M. (2002) Genome wide analysis of mRNA processing in yeast using splicing-specific microarrays. Science, 296, 907–910. [DOI] [PubMed] [Google Scholar]
- 54.Long M., de Souza,S.J. and Gilbert,W. (1997) The yeast splice site revisited: new exon consensus from genomic analysis. Cell, 91, 739–740. [DOI] [PubMed] [Google Scholar]
- 55.Lopez P.J. and Seraphin,B. (1999) Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice-site recognition. RNA, 5, 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spingola M., Grate,L., Haussler,D. and Ares,M. (1999) Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA, 5, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]