Abstract
A comprehensive investigation of ribosomal genes in complete genomes from 66 different species allows us to address the distribution of r-proteins between and within the three primary domains. Thirty-four r-protein families are represented in all domains but 33 families are specific to Archaea and Eucarya, providing evidence for specialisation at an early stage of evolution between the bacterial lineage and the lineage leading to Archaea and Eukaryotes. With only one specific r-protein, the archaeal ribosome appears to be a small-scale model of the eukaryotic one in terms of protein composition. However, the mechanism of evolution of the protein component of the ribosome appears dramatically different in Archaea. In Bacteria and Eucarya, a restricted number of ribosomal genes can be lost with a bias toward losses in intracellular pathogens. In Archaea, losses implicate 15% of the ribosomal genes revealing an unexpected plasticity of the translation apparatus and the pattern of gene losses indicates a progressive elimination of ribosomal genes in the course of archaeal evolution. This first documented case of reductive evolution at the domain scale provides a new framework for discussing the shape of the universal tree of life and the selective forces directing the evolution of prokaryotes.
INTRODUCTION
The ribosome is at the core of the translation machinery of all organisms and assures two key functions: the decoding of the genetic information contained in messenger RNA and the formation of peptide bonds. It is a ribonucleoprotein particle of 70S in prokaryotes and 80S in eukaryotes composed of two subunits (30S and 50S subunits in prokaryotes, 40S and 60S in eukaryotes). As the ribosome is universal and submitted to strong selection pressure, ribosomal RNA (rRNA) has been extensively used as a reference molecule in phylogeny and is at the origin of the division of living organisms into three domains: Bacteria, Eucarya and Archaea (1). Recent advances in structural biology have enabled the observation at high resolution of the 30S subunit of the thermophilic bacteria Thermus thermophilus (2,3), the 50S subunit of the bacteria Deinococcus radiodurans (4) and of the halophilic archaeon Haloarcula marismortui (5). The complete structure of the 70S ribosome of T.thermophilus has been determined at 5.5 Å, in the presence of a transcript molecule and cognate transfer RNA (tRNA) bound to aminoacyl, peptidyl and exit sites (6). Together, these crystal structures provide a considerable amount of information on the global architecture and protein–RNA interactions as well as details on ribosome interaction with mRNA and tRNA (reviewed in 7). They also confirm that the functional regions for peptide bond formation in the large prokaryotic subunit (3,8) and the decoding center in the small prokaryotic subunit (9) consist entirely of rRNA. Information on the eukaryotic ribosome structure is not so abundant but a recent cryo-electron microscopy reconstruction of the yeast 80S ribosome (10) confirms that the fundamental mechanism of protein synthesis is highly conserved throughout the three domains.
In parallel to the spectacular progress achieved in understanding the structure of ribosome, the development of genomics has enabled the examination of ribosomal protein (r-protein) genes in both prokaryotic and eukaryotic genomes. These analyses confirm that most of the bacterial r-protein genes are clustered in a few operons allowing coordinated regulation (11–15) and are rarely duplicated (16). In contrast, eukaryotic r-protein genes appear widely scattered across the chromosomes and show numerous duplications (17–23). Wool et al. (24) were the first to compare the rat r-proteins to the available sequence data from human, yeast, archaea and Escherichia coli. This pioneering work establishes that the rat r-proteins can be divided into three groups: (i) proteins with counterparts in both archaeal and bacterial domains; (ii) proteins with orthologs in the archaeal domain; and (iii) proteins exclusively found in Eucarya. However, no recent systematic comparison of the r-protein component of ribosome from the three primary domains is available. Here, we propose a comparative analysis of the r-proteins from 66 different species that includes r-proteins not previously reported in genome annotations. The wide range of genomes examined allows us to establish the phylogenetic distribution of r-proteins within and between each of the three primary domains, providing new insights into the emergence and evolution of the protein component of ribosomes.
MATERIALS AND METHODS
An initial set of r-proteins classified into 102 families was obtained at http://www.expasy.ch/cgi-bin/lists?ribosomp.txt. For each family, representatives of various lineages across Bacteria, Archaea and Eucarya were used as probes and systematically compared to a non-redundant protein database consisting of SwissProt, SpTrEMBL and SpTrEMBLNEW using the BlastP program (25) with a cut-off of E < 0.001. The results of the BlastP comparison were cross-validated by a TBlastN search against a complete genome database including the bacterial species Aquifex aeolicus, Thermotoga maritima, D.radiodurans, Chlamydia muridarum, Chlamydia trachomatis, Chlamydia pneumoniae, Synechocystis sp., Anabaena sp., Mycobacterium leprae, Mycobacterium tuberculosis, Bacillus halodurans, Bacillus subtilis, Listeria innocua, Listeria monocytogenes, Staphylococcus aureus, Clostridium acetobutylicum, Mycoplasma genitalium, Mycoplasma pneumoniae, Ureaplasma urealyticum, Lactococcus lactis, Streptococcus pneumoniae, Streptococcus pyogenes, Caulobacter crescentus, Brucella melitensis, Agrobacterium tumefaciens, Mesorhizobium loti, Sinorhizobium meliloti, Rickettsia conorii, Rickettsia prowasekii, Neisseria meningitidis, Ralstonia solanacearum, Campylobacter jejuni, Helicobacter pylori, Buchnera aphidicola, E.coli, Salmonella typhimurium, Salmonella enterica, Yersinia pestis, Haemophilus influenzae, Pasteurella multocida, Pseudomonas aeruginosa, Vibrio cholerae, Xylella fastidiosa, Borrelia burgdorferi, Treponema pallidum; the archaea Aeropyrum pernix, Sulfolobus solfataricus, Sulfolobus tokodaii, Pyrobaculum aerophilum, Pyrococcus abyssi, Pyrococcus horikoshii, Pyrococcus furiosus, Methanopyrus kandleri, Methanococcus jannaschii, Methanobacterium thermoautotrophicum, Archaeoglobus fulgidus, Halo bacterium sp., Thermoplasma acidophilum, Thermoplasma volcanium; and the Eucarya Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae, Schizosaccharomyces pombe and Encephalitozoon cuniculi.
The putative new gene sequences detected by the TBlastN searches were examined in the light of their genomic context to eliminate false-positives ‘hits’ and further compared to the RefSeq database (26). For each r-protein family, the likely r-protein sequences obtained by the BlastP and TBlastN searches were included in a multiple alignment constructed by MAFFT (27). All alignments were refined by RASCAL (J. D. Thompson, J. C. Thierry and O. Poch, manuscript submitted) and their quality assessed by NorMD (28). These alignments were manually examined to remove false-positives observed in some r-protein families, in particular those containing ubiquitous RNA-binding domains. All the alignments are available at http://www-igbmc.u-strasbg.fr/BioInfo/Rproteins.
RESULTS
Gene detection
The distribution of 102 families of ribosomal prokaryotic and/or eukaryotic cytoplasmic proteins has been analyzed in complete genome sequences from 66 different species, including 45 bacteria, 14 archaea and 7 eukaryotes. In the case of ribosomal genes which often exhibit a biased composition and a small size, the results of a database search can greatly depend on the choice of the query sequence. Thus, for each protein family, we performed multiple searches in the non-redundant protein database using query sequences from phylogenetically distant organisms. All these sequences were also compared to complete genomic sequences, allowing the localization of the corresponding genes on the genomes. This cross-validation appears to be a prerequisite to obtain a correct picture of the phylogenetic distribution of ribosomal genes since a small but not negligible number of short genes escape annotation, as revealed by re-annotations of complete genomes (29,30). Using this approach, we detected 24 potential genes (Supplementary Material, Table S1) in the complete genomes investigated that were overlooked during the gene prediction process but are likely to encode r-proteins. Among them, 12 have already been created as provisional records of the RefSeq database (26) and integrated in the COG database (31,32). The convergence of two independent processes substantiates our method and confirms these open reading frames as functional genes.
Inter-domain distribution
The overall phylogenetic distribution of prokaryotic and/or eukaryotic cytoplasmic r-proteins is summarized in Figure 1 and a more detailed description, including nomenclatural correspondence between protein names, is provided as Supplementary Material (Table S2). We detected 57 different r-protein families in Bacteria, 68 in Archaea and 78 in Eucarya, which underlines the protein enrichment of the ribosome from Bacteria to Eucarya. Among the 102 r-protein families, 34 (15 in the SSU, 19 in the LSU) are represented in all three domains of life (the BAE set in Fig. 1) but two of them are absent in some bacteria (see below), leading to a total of 32 families conserved in all the complete genomes studied (Table 1). In E.coli, 22 of the 32 universal proteins belong to the S10-spc-alpha operons which constitute the longest array of genes conserved in bacterial genomes (11–13). Many of the universal proteins have been shown to be crucial for the ribosome assembly such as the early assembling proteins S4p, S7p, S8p, S15p, S17p, L2p, L3p, L4p, L5p, L15p, L18p (33,34) and the proteins implicated in the bridges between the two subunits (S13p, S15p, S19p, L2p, L5p, L14p) (6). Others are in contact with the tRNA (S7p, S9p, S12p, S13p, L1p and L5p) or surround the polypeptide exit channel (L22p, L24p and L29p) (6). It is noteworthy that the proportion of universal r-proteins is higher in the SSU than in the LSU: among the 22 r-protein types experimentally identified in E.coli ribosomal small subunit (35,36), more than two-thirds are conserved in all studied genomes, whereas only half of the 33 r-protein types from the E.coli ribosome large subunit are universal. The greater evolutionary stability of the SSU protein component may be linked to the higher conservation of the SSU rRNA compared to the LSU rRNA despite striking differences reported between E.coli and eukaryotic small subunit morphology (37).
Figure 1.
Venn diagram showing the general distribution of r-protein families between the three domains: Bacteria (B), Archaea (A), Eucarya (E). The number of families is indicated for each set. The two numbers enclosed by parentheses refer to r-protein families found in the small and large ribosome subunits respectively.
Table 1. Distribution of r-protein families in Bacteria (B), Archaea (A) and Eucarya (E).
| Small subunit | Large subunit | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Families | B | A | E | Families | B | A | E | Families | B | A | E | Families | B | A | E |
| S1p | x | S3ae | X | X | L1p | X | X | X | L6e | X | |||||
| S2p | X | X | X | S4e | X | X | L2p | X | X | X | L7ae | x | X | X | |
| S3p | X | X | X | S6e | X | X | L3p | X | X | X | L10e | X | X | ||
| S4p | X | X | X | S7e | X | L4p/L4e | X | X | X | L13e | x | X | |||
| S5p | X | X | X | S8e | X | X | L5p | X | X | X | L14e | x | x | ||
| S6p | X | S10e | X | L6p | X | X | X | L15e | X | X | |||||
| S7p | X | X | X | S12e | X | L9p | X | L18ae | X | ||||||
| S8p | X | X | X | S17e | X | X | L10p | X | X | X | L18e | X | X | ||
| S9p | X | X | X | S19e | X | X | L11p | X | X | X | L19e | X | X | ||
| S10p | X | X | X | S21e | x | L12p | X | X | X | L21e | X | X | |||
| S11p | X | X | X | S24e | X | X | L13p | X | X | X | L22e | X | |||
| S12p | X | X | X | S25e | x | X | L14p | X | X | X | L24e | X | X | ||
| S13p | X | X | X | S26e | x | X | L15p | X | X | X | L27e | X | |||
| S14p | X | X | X | S27ae | X | X | L16p | X | L28e | x | |||||
| S15p | X | X | X | S27e | X | X | L17p | X | L29e | X | |||||
| S16p | X | S28e | X | X | L18p | X | X | X | L30e | x | X | ||||
| S17p | X | X | X | S30e | x | X | L19p | X | L31e | X | X | ||||
| S18p | X | S31e | x | L20p | X | L32e | X | X | |||||||
| S19p | X | X | X | L21p | X | L34e | x | X | |||||||
| S20p | X | L22p | X | X | X | L35ae | x | X | |||||||
| S21p | x | L23p | X | X | X | L36e | X | ||||||||
| S22p | x | L24p | X | X | X | L37ae | X | X | |||||||
| L25p | x | L37e | X | X | |||||||||||
| L27p | X | L38e | x | x | |||||||||||
| L28p | X | L39e | X | X | |||||||||||
| L29p | X | X | X | L40e | X | X | |||||||||
| L30p | x | X | X | L41ea | X | X | |||||||||
| L31p | X | L44e | X | X | |||||||||||
| L32p | X | LXa | x | ||||||||||||
| L33p | X | ||||||||||||||
| L34p | X | ||||||||||||||
| L35p | X | ||||||||||||||
| L36p | X | ||||||||||||||
The conservation of the protein family in all investigated genomes of a primary domain is denoted by X whereas the presence of a protein family in some, but not all, representatives of a domain is indicated by x.
aUncertain distribution.
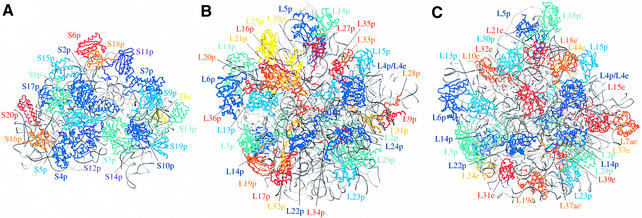
The r-protein families common to Eucarya and Archaea but absent in Bacteria constitute an extended pool (called AE in Fig. 1) of 33 members (13 in the SSU, 20 in the LSU). The high number of r-proteins specific to Eucarya and Archaea reflects the deep resemblance between the informational proteins of the two domains previously noted in comparative genomic studies (38–40) and is in agreement with most phylogenetic studies on rRNA. However, this number is substantially higher than expected from previous comparison of r-proteins (12,24). Another large set (set B in Fig. 1) corresponds to the 23 r-proteins (8 in the SSU, 15 in the LSU) exclusively found in Bacteria. The major split between Bacteria on the one hand and Archaea and Eucarya on the other hand is further supported by the absolute absence of proteins specific to bacterial and archaeal domains or to bacterial and eukaryotic domains despite the wide phylogenetic range of genomes studied here. When proteins are positioned in the available 3D structures of bacterial and archaeal ribosomal subunits (Fig. 2), no clear correlation arises between the phylogenetic and the spatial distributions of the r-proteins, except for the bacteria-specific proteins in the small ribosomal subunit which are mainly found at the periphery of the ribosome. This could be in agreement with the observation of Spahn et al. (10) who note the presence of additional proteins with no counterpart in Bacteria as well as expansion segments of rRNA at the solvent exposed surface of the yeast ribosome. The two remaining sets of r-proteins correspond to the archaeal-specific r-protein LXa (set A in Fig. 1) and to the 11 r-proteins (4 in the SSU, 7 in the LSU) exclusively found in Eucarya (set E in Fig. 1). Thus, with the exception of LXa, all the r-proteins found in Archaea are also found in Eucarya and the archaeal ribosome, which is close to the bacterial one in terms of size, appears to be a small-scale model of the eukaryotic ribosome in term of r-protein composition.
Figure 2.
(A) Overview of the 30S ribosomal subunit of T.thermophilus (2) and (B) of the 50S ribosomal subunit of D.radiodurans (4) and (C) H.marismortui (5). The 30S subunit is presented from the back side (as defined in 2) and the large subunits are presented in the crown view rotated by 180° about a vertical axis. The rRNA molecules are shown in grey and protein backbones are colored according to their phylogenetic distribution (proteins conserved in the three domains, in different shades of blue; proteins conserved in Bacteria or in Archaea/Eucarya, in different shades of red and orange). When dispensable, the bacterial r-proteins are coloured in yellow. Figures were generated using SETOR (79).
Phylogenetic distribution of r-proteins in Bacteria
The r-protein families ranging from S2p to S20p as well as the r-protein families L1p-L6p, L9p-L24p, L27p-L29p and L31p-L36p are encoded by all bacterial genomes investigated (Table 1), defining a stable pool of 50 r-proteins (19 in the SSU and 31 in the LSU) in Bacteria. Only four well established bacterial r-proteins exhibit a disparate distribution: the bacterial-specific S1p, S21p, L25p proteins and the L30p found in all three domains of life.
The pattern of absence of the S1p and S21p families is very puzzling since they are lacking in only a few lineages widely dispersed in the phylogenetic tree of Bacteria, ranging from early divergent free-living bacteria such as D.radiodurans and T.maritima to intracellular pathogens (Fig. 3). Although the S1p and S21p are adjacent in the ribosome small subunit of E.coli (41), there is no strict correlation between the absences of the two proteins; the pattern of absence seems, on the contrary, indicative of erratic and independent gene losses. The haphazard character of the phylogenetic distribution of the S1p protein is further illustrated by the discrepancies observed between closely related species since this protein which is absent in the complete genomes of M.genitalium and M.pneumoniae has been identified in Mycoplasma pulmonis under GenBank accession number Q98R80. It is noteworthy that we detect a DNA region in the T.maritima genome similar to the S1p gene but including a frameshift that could reflect a recent decay of the S1 gene in T.maritima.
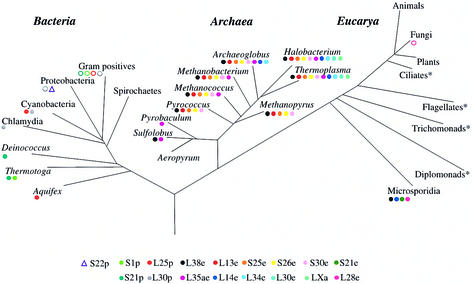
Figure 3.
Schematic representation of the universal tree of life (adapted from 49). R-proteins exhibiting a heterogeneous distribution within a primary domain are symbolized by circles or a triangle. Full circles indicate proteins absent in all complete genomes investigated in the indicated taxon. Empty circles stand for proteins absent in some complete genomes of the indicated taxon: S1p is absent in the Gram positives M.genitalium, M.pneumoniae and U. urealyticum; S21p is absent in the Gram positives M.leprae, M.tuberculosis; L25p is absent in the Gram positives C.acetobutylicum, M.genitalium, M.pneumoniae, U.urealyticum, L.lactis, S.pyogenes, S.pneumoniae; L30p is absent in the Gram positives M.genitalium, M.pneumoniae, U.urealyticum and in the Proteobacteria C.jejuni and H.pylori. The empty triangle symbolises the S22p protein exclusively found in the Proteobacteria E.coli, S.typhimurium and S.enterica. The four eukaryotic lineages marked by an asterisk are positioned in the tree for comparison purposes but have not been investigated in this study.
Interestingly, the L25p and the L30p proteins absent in two cyanobacteria are also the sole proteins missing in the ribosome of the spinach chloroplast compared to the bacterial one (42,43). This strong similarity between the cyanobacterial and chloroplast ribosomes is in agreement with the endosymbiotic theory in plastid evolution (44) and suggests that the gene losses occurred before the endosymbiotic event. However, the L25p and L30p gene losses are not always correlated (Fig. 3). In fact the L30p constitutes an evolutionary maverick since this protein that is lost in some bacterial lineages is conserved in Archaea and Eucarya.
Additionally, two potential small r-proteins have been identified in very few bacteria: (i) the 45 amino acids long SRA protein (stationary-phase-induced ribosome associated protein, S22p family) in E.coli (45,46) and Salmonella species; and (ii) the 26 amino acid long Thx protein in Thermus (47,48). The latter, classified in the S31e family, fits into a cavity between multiple RNA elements in the crystal structure of the 30S subunit of Thermus (2,3). Thus, in contrast to the S1p, S21p, L25p and L30p, the SRA and Thx proteins may constitute recent and specific innovation in the course of bacterial ribosome evolution. In addition to these bacterial proteins, a similarity has been detected between the L7ae family found in Archaea and Eucarya and hypothetical proteins from T.maritima and bacterial species of the Bacillus/Clostridium group but no experimental data implicates these proteins in the bacterial ribosome.
Phylogenetic distribution of r-proteins in Archaea
Among the 68 r-protein families represented in Archaea, the L41e protein exhibits such an extremely biased composition and very small size (∼20 amino acids) that no clear-cut pattern of presence/absence could be obtained. Among the 67 other archaeal r-protein families, 57 are preserved in the 14 archaeal genomes examined. Remarkably, this set of conserved archaeal r-proteins is also present in all complete genomes of Eucarya. Thus, r-protein genes stabilized in Archaea are also fixed in organisms ranging from the amitochondriate intracellular pathogen E.cuniculi to H.sapiens.
For the 10 proteins exhibiting a heterogeneous distribution within the archaeal domain (Fig. 3), the pattern of presence/absence is not patchy as observed in the case of the four dispensable bacterial r-proteins but seems to fit the tree of life with the exception of L35ae. All 10 proteins are present in at least one representative of the deeply branched kingdom of Crenarchaeota while five proteins (L38e, L13e, S25e, S26e and S30e) are missing in all representatives of the Euryarchaeota kingdom. Within this latter kingdom, the absences also seem to match the branching order established on the basis of rRNA sequences (49) and confirmed by r-protein sequences (50). For example, the early diverging Pyrococcus lineage retains five r-proteins absent in genus that emerged later such as Thermoplasma and Halobacterium. The high number of missing r-protein genes observed in this latter genome is corroborated by the crystal structure of the ribosomal LSU of H.morismortui (5), which lacks the same LSU r-proteins as its close relative, Halobacterium.
The great variation in the number of r-proteins observed within the archaeal domain is intriguing but the comparison with Bacteria and Eucarya sheds light on how the existing distribution has arisen. Excluding the archaeal-specific LXa gene, the nine dispensable genes are absent in Bacteria but are found in a wide range of Eucarya, including all complete genomes of Fungi, Plants and Animals. Moreover, all of them are detected either in the incomplete genomes of Diplomonads or Euglenozoa and are also found, with the exception of L38e and L14e (see below), in the complete genome of the amitochondriate protist E.cuniculi. The presence of these nine ribosomal genes in early divergent representatives of Eucarya and Archaea suggests their existence in the common ancestor(s) of the two domains. This makes the crenarchaeal ribosome the most ‘eukaryotic-like’ within the available archaeal ones, which is in agreement with the eukaryotic traits previously reported in Crenarchaeota in both the ribosome morphology (51) and the elongation factor EF1α (52).
The apparently dispensable nature of 10 r-proteins in Archaea raises the question of their role in the archaeal ribosome. Some of their eukaryotic counterparts have been shown to bind rRNA, tRNA, mRNA or translation factors (53–57). Unfortunately, experimental studies are lacking in Archaea, except for the L30e protein of Sulfolobus acidocaldarius, which is responsible for establishing a key bridge between the large and small subunits by specifically binding a helix–loop–helix motif (58). Nevertheless, the involvement of the 10 genes in the translation apparatus is supported by their genomic context since most of them are adjacent to other informational genes with four genes (L34e, L14e, LXa and L30e) belonging to large operonic structures that include r-proteins (Fig. 4). Interestingly, the L14e and L34e genes that share the same phylogenetic profile are located in the same cluster of genes in some archaeal genomes (see Fig. 4). This could be indicative of a physical interaction between the two proteins since co-occurrence of genes across complete genomes as well as co-localisation of genes have been frequently used to infer functional coupling (15,59).
Figure 4.
Genomic context of dispensable r-protein genes in Archaea: L34e and L14e (A), LXa (B) and L30e (C). Arrows represent genes with their relative orientation in the genomes. Contiguous arrows represent adjacent genes while broken lines indicate a loss of proximity. r-protein genes are labeled according to the r-protein families. Conserved r-protein genes are colored in orange and dispensable ones in dark orange while other genes are colored according to general functional categories: translation (light orange), protein processing (yellow), transcription (green), replication (dark green), nucleotide synthesis (blue) and unknown function (white). Abbreviations used for gene names: SECY (preprotein translocase secY subunit), ADK (adenylate kinase), CMK (cytidylate kinase), GATA [glutamyl-tRNA(Gln) amidotransferase subunit A], TRUB (tRNA pseudouridine synthase B), TRUBa (tRNA pseudouridine synthase B subunit a), TRUBb (tRNA pseudouridine synthase B subunit b), Hyp (hypothetical protein), EIF6 (translation initiation factor 6), PFDA (prefoldin alpha subunit), DPA (signal recognition particle protein), SECE (preprotein translocase secE subunit), NUSG (transcription antitermination protein nusG), ALARS (alanyl-tRNA synthetase), PDCD5 (DNA-binding protein belonging to the PDCD5 family), RFCs (replication factor C small subunit), SRP54 (signal recognition 54 kDa protein), NUSA (nusA protein homolog), EF2 (translation elongation factor 2), EF1A (elongation factor 1-alpha). RPOH, RPOB, RPOB2, RPOB1, RPOA1, RPOA2, RPOD, RPON correspond to subunits of the DNA-directed RNA polymerase. Abbreviation used for organisms: Af, A.fulgidus; Ap, A.pernix; Mj, M.janaschii; Mk, M.kandleri; Mt, M.thermoautrophicum; Pa, P.abyssi; Pf, P.furiosus; Ph, P.horikoshii; Py, P.aerophilum; St, S.tokodaii; Ss, S.solfataricus. The LXa gene of Methanococcus jannaschii is not represented since the cluster organization is disrupted for this gene.
Phylogenetic distribution of r-proteins in Eucarya
In the eukaryotic domain, 78 (32 small subunit and 46 large subunit) cytoplasmic r-protein families have been identified to date. Except for the L28e absent in the budding yeast, all these families are conserved in mammals (compiled in 24), S.cerevisiae (19), S.pombe (18), A.thaliana (21) and, according to our analysis, in C.elegans and D.melanogaster. One of these families, the L12P, includes three eukaryotic subfamilies of acidic ribosomal phosphoproteins: the P1 and P2 proteins (60,61) represented in all the eukaryotic species mentioned above and the P3 protein which is plant specific (62). This leads to a total of 80 r-protein types in the Eukaryotic domain, of which 78 are strictly conserved in all complete genomes of animals, plant and fungi currently available.
As shown in Figure 3, the remarkable homogeneity in r-protein composition observed within the eukaryotic ‘crown’ group is far from being verified in the E.cuniculi genome. Discarding the L41e (see above), four proteins are absent in E.cuniculi: the strictly eukaryotic S21e and L28e proteins and the L14e and L38e found in Eucarya and in some—but not all—Archaea. It is notable that the r-proteins encoded by all archaeal genomes are also preserved within all the eukaryotic genomes while r-proteins exhibiting a heterogeneous distribution within Eucarya are absent either in all archaeal genomes (such as the S21e and L28e) or in some of them (such as the L14e and L38e). As both the L14e and L38e genes have been detected in deeply branched Archaea, it seems likely that the ancestral genes have been secondarily lost in the Encephalitozoon lineage. The case of the L28e gene (also absent in S.cerevisiae) and of the S21e gene is more controversial since, both of them being eukaryotic specific, we could not infer their presence in the common ancestor of eukaryotes. However, the absence of the L28e gene at least in S.cerevisiae may result from a gene loss since the L28e gene is present in the Euglenozoa branch that diverged earlier than Fungi.
DISCUSSION
Our investigation of the complete genomes from 66 different species allows us to gain insight into the conservation of r-proteins across the three primary domains of life and within each of them. Regarding the inter-domain distribution, 32 r-proteins are strictly conserved in all the bacterial, archaeal and eukaryotic studied genomes (BAE set) in agreement with structural comparison between prokaryotic and eukaryotic ribosomes (10) which demonstrates the preservation of the core and global shape of ribosome. The high number of r-proteins conserved in all species of the wide phylogenetic range covered confirms the prevalence of r-proteins within the universal pool that may be present in the last universal common ancestor (63,64).
The distribution of the other r-proteins shows a profound rupture in the protein component of the bacterial ribosome as opposed to the archaeal and eukaryotic ones. No r-proteins are specific to Bacteria and Eucarya (BE) or to Bacteria and Archaea (BA) while 33 are common to Archaea and Eucarya (AE) and 23 r-proteins are bacterial specific (B). Even if we cannot exclude that some of the proteins of the B and AE sets have in fact the same ancestral origin but have diverged beyond recognition, the importance of the two sets testifies to a specialization of bacterial versus archaeal/eukaryotic ribosomes. An appealing hypothesis is that the B and AE proteins are involved in the folding of lineage-specific rRNA extensions shown by comparison of rRNA sequences (65). However, some of these r-proteins could also interact with domain-specific translation factors or be implicated in extra-ribosomal functions as frequently observed for r-proteins (66).
The intra-domain distribution of r-proteins shows unforeseen differences between the three domains of life. In Bacteria, a relatively simple picture of conservation emerges since only four proteins are lost in the wide collection of bacterial species investigated in our study. Gene losses are restricted to a small number of divergent species or genera suggesting that gene disruptions occurred independently in these lineages. From a physiological point of view, there is a bias toward losses in intracellular pathogens with M.genitalium and M.pneumoniae lacking three of the four dispensable proteins. In addition to these losses, some small r-proteins are found in a restricted number of bacteria such as the Thx protein in Thermus species. It is therefore possible that additional r-proteins limited to a small phylogenetic spectrum are still unknown and could lead to a slightly more diverse picture of the bacterial ribosome than expected.
The distribution of r-proteins appears more complex in Eucarya and Archaea and reveals intricate evolutionary relationships between the two domains. In Eucarya, we observe a remarkable conservation of r-proteins in all investigated genomes except in E.cuniculi that lacks at least four proteins. The homogeneous distribution of r-proteins in representatives of the eukaryotic crown group is noteworthy since rRNA exhibit numerous taxon-specific insertions in these groups (67,68). Interpretation of gene absences in Microsporidia is complicated by both their intracellular parasitic lifestyle and their uncertain phylogenetic position but gene absences appear directly correlated to the extremely small size of the rRNA which is reduced to the universal core in this species (69). The amitochondriate Microsporidia were first considered as one of the most basal eukaryotic lineages (70,71) which diverged before the endosymbiotic event that led to mitochondria. According to this evolutionary scenario, the small size of rRNA and the absence of certain r-protein genes in E.cuniculi could be considered as primitive characters. The appearance of eukaryotic-specific proteins after the emergence of the Microsporidia would be a trace of the eukaryotic ribosome enrichment in proteins in the course of evolution. However, according to a growing number of studies (reviewed in 72), Microsporidia may be atypical fungi that secondarily lost mitochondria. If this later origin is confirmed, the reduction of rRNA size and the loss of some r-proteins would participate in the general process of genome compaction revealed by the genomic sequence (73) which is probably linked to its intracellular parasitic lifestyle.
In Archaea, the pattern of r-protein conservation differs dramatically from those observed in Bacteria and Eucarya. In the archaeal domain, losses include 10 r-proteins while only four proteins appear dispensable in each of the two other domains, revealing a higher than expected plasticity in the archaeal ribosome. Moreover, the losses cannot be explained by an intracellular lifestyle as in the case of eukaryotes and, to a lesser extent, bacteria, since all archaeal species considered in our study are free-living organisms. On the contrary, the pattern of gene losses indicates a progressive elimination of r-protein genes in the course of archaeal evolution, with the deeply branched Crenarchaeota exhibiting up to 10 r-proteins more than the latest divergent representatives of Euryarchaeota. This ribosome ‘striptease’ is, to our knowledge, the first tangible example of reductive evolution observed at a primary domain scale. It is all the more remarkable since informational proteins involved in a macromolecular complex are concerned. The subsequent question is why these r-protein genes have been lost. One could imagine that the loss of r-proteins is functionally and/or structurally compensated by a rRNA enlargement. The inverse mechanism has been proposed in the case of mammalian mitochondrial ribosome where the deficit of rRNA relative to the bacterial one is balanced by a protein enrichment (74). However, the situation seems more complex in Archaea since there is no indication of an rRNA shortening between the deeply branch Crenarchaeota and the later diverging Euryarchaeota. Thus, the ribosome of a Crenarchaeota, like A.pernix, may be a rich target for structural studies aimed at understanding the fundamental mechanisms underlying the reductive evolution process.
From an evolutionary perspective, our results lead to troublesome conclusions. On one hand, it seems that, with the exception of LXa, the full complement of archaeal r-proteins was present at an early stage of evolution, i.e. in the cenancestor of Archaea and Eucarya and was progressively eroded. This is in agreement with the eukaryotic-rooting tree (75,76) which proposes that prokaryotes would have evolved by simplification of an ancestral eukaryotic-like genome. On the other hand, the clear-cut opposition between bacterial and archaeal/eukaryotic r-protein complements is in agreement with the bacterial-rooting tree (77) or the symbiosis hypothesis (discussed in 78) which both explain the close relationships observed between Archaea and Eucarya. It even suggests that the ribosome specialization has been constitutive of the segregation of the bacterial lineage from the cenancestor(s) of Archaea and Eucarya, in agreement with Woese’s proposal that the translation apparatus ‘crystallizes’ first. Faced with these two opposite evolutionary scenarios, genome sequencing of early branching representatives of the three domains and comparative analyses of other macromolecular complexes will be essential in deciding whether the reductive evolution is a special trait of archaeal ribosomes or whether it constitutes a general trend in prokaryote evolution.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to acknowledge Marat Yusupov and Gula Yusupova for helpful discussions. We thank Julie Thompson, Frederic Plewniak and Claudine Mayer for a critical reading of the manuscript and their assistance during this work. This work was supported by institute funds from CNRS, INSERM, the Université Louis Pasteur de Strasbourg and the Fond de Recherche Hoechst Marion Roussel (Aventis Pharma FRHMR1-9728).
REFERENCES
- 1.Woese C.R., Kandler,O. and Wheelis,M.L. (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA, 87, 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 3.Schluenzen F., Tocilj,A., Zarivach,R., Harms,J., Gluehmann,M., Janell,D., Bashan,A., Bartels,H., Agmon,I., Franceschi,F. et al. (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell, 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 4.Harms J., Schluenzen,F., Zarivach,R., Bashan,A., Gat,S., Agmon,I., Bartels,H., Franceschi,F. and Yonath,A. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell, 107, 679–688. [DOI] [PubMed] [Google Scholar]
- 5.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 6.Yusupov M.M., Yusupova,G.Z., Baucom,A., Lieberman,K., Earnest,T.N., Cate,J.H. and Noller,H.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishnan V. (2002) Ribosome structure and the mechanism of translation. Cell, 108, 557–572. [DOI] [PubMed] [Google Scholar]
- 8.Nissen P., Hansen,J., Ban,N., Moore,P.B. and Steitz,T.A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science, 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 9.Carter A.P., Clemons,W.M., Brodersen,D.E., Morgan-Warren,R.J., Wimberly,B.T. and Ramakrishnan,V. (2000) Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature, 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 10.Spahn C.M., Beckmann,R., Eswar,N., Penczek,P.A., Sali,A., Blobel,G. and Frank,J. (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell, 107, 373–386. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe H., Mori,H., Itoh,T. and Gojobori,T. (1997) Genome plasticity as a paradigm of eubacteria evolution. J. Mol. Evol., 44 (Suppl 1), S57–S64. [DOI] [PubMed] [Google Scholar]
- 12.Fujita K., Baba,T. and Isono,K. (1998) Genomic analysis of the genes encoding ribosomal proteins in eight eubacterial species and Saccharomyces cerevisiae. Genome Inform. Ser. Workshop Genome Inform., 9, 3–12. [PubMed] [Google Scholar]
- 13.Wachtershauser G. (1998) Towards a reconstruction of ancestral genomes by gene cluster alignment. Syst. Appl. Microbiol., 21, 473–477. [Google Scholar]
- 14.Lathe W.C. III, Snel,B. and Bork,P. (2000) Gene context conservation of a higher order than operons. Trends Biochem. Sci., 25, 474–479. [DOI] [PubMed] [Google Scholar]
- 15.Wolf Y.I., Rogozin,I.B., Kondrashov,A.S. and Koonin,E.V. (2001) Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res., 11, 356–372. [DOI] [PubMed] [Google Scholar]
- 16.Makarova K.S., Ponomarev,V.A. and Koonin,E.V. (2001) Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol., 2, research0033.1–0033.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 546, 563–567. [DOI] [PubMed] [Google Scholar]
- 18.Gross T. and Kaufer,N.F. (1998) Cytoplasmic ribosomal protein genes of the fission yeast Schizosaccharomyces pombe display a unique promoter type: a suggestion for nomenclature of cytoplasmic ribosomal proteins in databases. Nucleic Acids Res., 26, 3319–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Planta R.J. and Mager,W.H. (1998) The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast, 14, 471–477. [DOI] [PubMed] [Google Scholar]
- 20.Kenmochi N., Kawaguchi,T., Rozen,S., Davis,E., Goodman,N., Hudson,T.J., Tanaka,T. and Page,D.C. (1998) A map of 75 human ribosomal protein genes. Genome Res., 8, 509–523. [DOI] [PubMed] [Google Scholar]
- 21.Barakat A., Szick-Miranda,K., Chang,I.F., Guyot,R., Blanc,G., Cooke,R., Delseny,M. and Bailey-Serres,J. (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol., 127, 398–415. [PMC free article] [PubMed] [Google Scholar]
- 22.Uechi T., Tanaka,T. and Kenmochi,N. (2001) A complete map of the human ribosomal protein genes: assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics, 72, 223–230. [DOI] [PubMed] [Google Scholar]
- 23.Yoshihama M., Uechi,T., Asakawa,S., Kawasaki,K., Kato,S., Higa,S., Maeda,N., Minoshima,S., Tanaka,T., Shimizu,N. et al. (2002) The human ribosomal protein genes: sequencing and comparative analysis of 73 genes. Genome Res., 12, 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wool I.G., Chan,Y.L. and Gluck,A. (1995) Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol., 73, 933–947. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K., Misawa,K., Kuma,K. and Miyata,T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res., 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson J.D., Plewniak,F., Ripp,R., Thierry,J.C. and Poch,O. (2001) Towards a reliable objective function for multiple sequence alignments. J. Mol. Biol., 314, 937–951. [DOI] [PubMed] [Google Scholar]
- 29.Natale D.A., Shankavaram,U.T., Galperin,M.Y., Wolf,Y.I., Aravind,L. and Koonin,E.V. (2000) Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs). Genome Biol., 1, research0009.1–0009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bocs S., Danchin,A. and Medigue,C. (2002) Re-annotation of genome microbial CoDing-Sequences: finding new genes and inaccurately annotated genes. BMC Bioinformatics, 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatusov R.L., Natale,D.A., Garkavtsev,I.V., Tatusova,T.A., Shankavaram,U.T., Rao,B.S., Kiryutin,B., Galperin,M.Y., Fedorova,N.D. and Koonin,E.V. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Held W.A., Ballou,B., Mizushima,S. and Nomura,M. (1974) Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem., 249, 3103–3111. [PubMed] [Google Scholar]
- 34.Rohl R. and Nierhaus,K.H. (1982) Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA, 79, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittmann H.G. (1982) Components of bacterial ribosomes. Annu. Rev. Biochem., 51, 155–183. [DOI] [PubMed] [Google Scholar]
- 36.Wittmann-Liebold B., Kopke,A.K.E., Arndt,E., Kromer,W., Hatakeyama,T. and Wittman,H.-G. (1990) Sequence comparison and evolution of ribosomal proteins and their genes. In Hill,W.E., Dahlberg,R.E., Garrett,R.E., Moore,P.B., Schlessinger,D. and Warner,J.R. (eds), The Ribosome, Structure, Function and Evolution. American Society of Microbiologists, Washington DC, pp. 598–616.
- 37.Verschoor A., Srivastava,S., Grassucci,R. and Frank,J. (1996) Native 3D structure of eukaryotic 80s ribosome: morphological homology with E. coli 70S ribosome. J. Cell Biol., 133, 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doolittle W.F. and Logsdon,J.M.,Jr (1998) Archaeal genomics: do archaea have a mixed heritage? Curr. Biol., 8, R209–R211. [DOI] [PubMed] [Google Scholar]
- 39.Makarova K.S., Aravind,L., Galperin,M.Y., Grishin,N.V., Tatusov,R.L., Wolf,Y.I. and Koonin,E.V. (1999) Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res., 9, 608–628. [PubMed] [Google Scholar]
- 40.Andrade M.A., Ouzounis,C., Sander,C., Tamames,J. and Valencia,A. (1999) Functional classes in the three domains of life. J. Mol. Evol., 49, 551–557. [DOI] [PubMed] [Google Scholar]
- 41.Sengupta J., Agrawal,R.K. and Frank,J. (2001) Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl Acad. Sci. USA, 98, 11991–11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi K. and Subramanian,A.R. (2000) The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem., 275, 28466–28482. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi K., von Knoblauch,K. and Subramanian,A.R. (2000) The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem., 275, 28455–28465. [DOI] [PubMed] [Google Scholar]
- 44.Bogorad L. (1975) Evolution of organelles and eukaryotic genomes. Science, 188, 891–898. [DOI] [PubMed] [Google Scholar]
- 45.Wada A. (1998) Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells, 3, 203–208. [DOI] [PubMed] [Google Scholar]
- 46.Arnold R.J. and Reilly,J.P. (1999) Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal. Biochem., 269, 105–112. [DOI] [PubMed] [Google Scholar]
- 47.Choli T., Franceschi,F., Yonath,A. and Wittmann-Liebold,B. (1993) Isolation and characterization of a new ribosomal protein from the thermophilic eubacteria, Thermus thermophilus, T. aquaticus and T. flavus. Biol. Chem. Hoppe Seyler, 374, 377–383. [DOI] [PubMed] [Google Scholar]
- 48.Tsiboli P., Herfurth,E. and Choli,T. (1994) Purification and characterization of the 30S ribosomal proteins from the bacterium Thermus thermophilus. Eur. J. Biochem., 226, 169–177. [DOI] [PubMed] [Google Scholar]
- 49.Woese C.R. (1996) Whither microbiology? Phylogenetic trees. Curr. Biol., 6, 1060–1063. [DOI] [PubMed] [Google Scholar]
- 50.Matte-Tailliez O., Brochier,C., Forterre,P. and Philippe,H. (2002) Archaeal phylogeny based on ribosomal proteins. Mol. Biol. Evol., 19, 631–639. [DOI] [PubMed] [Google Scholar]
- 51.Lake J.A., Henderson,E., Oakes,M. and Clark,M.W. (1984) Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc. Natl Acad. Sci. USA, 81, 3786–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rivera M.C. and Lake,J.A. (1992) Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science, 257, 74–76. [DOI] [PubMed] [Google Scholar]
- 53.Perez-Gosalbez M., Vazquez,D. and Ballesta,J.P. (1978) Affinity labelling of yeast ribosomal peptidyl transferase. Mol. Gen. Genet., 163, 29–34. [DOI] [PubMed] [Google Scholar]
- 54.Synetos D., Amils,R. and Ballesta,J.P. (1986) Photolabeling of protein components in the pactamycin binding site of rat liver ribosomes. Biochim. Biophys. Acta, 868, 249–253. [DOI] [PubMed] [Google Scholar]
- 55.Stahl J. and Kobetz,N.D. (1984) Affinity labelling of rat liver ribosomal protein S26 by heptauridylate containing a 5′-terminal alkylating group. Mol. Biol. Rep., 9, 219–222. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka M., Tanaka,T., Harata,M., Suzuki,T. and Mitsui,Y. (1998) Triplet repeat-containing ribosomal protein L14 gene in immortalized human endothelial cell line (t-HUE4). Biochem. Biophys. Res. Commun., 243, 531–537. [DOI] [PubMed] [Google Scholar]
- 57.Dabeva M.D. and Warner,J.R. (1993) Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J. Biol. Chem., 268, 19669–19674. [PubMed] [Google Scholar]
- 58.Vilardell J., Yu,S.J. and Warner,J.R. (2000) Multiple functions of an evolutionarily conserved RNA binding domain. Mol. Cell, 5, 761–766. [DOI] [PubMed] [Google Scholar]
- 59.Dandekar T., Snel,B., Huynen,M. and Bork,P. (1998) Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci., 23, 324–328. [DOI] [PubMed] [Google Scholar]
- 60.Ramirez C., Shimmin,L.C., Newton,C.H., Matheson,A.T. and Dennis,P.P. (1989) Structure and evolution of the L11, L1, L10, and L12 equivalent ribosomal proteins in eubacteria, archaebacteria, and eucaryotes. Can. J. Microbiol., 35, 234–244. [DOI] [PubMed] [Google Scholar]
- 61.Shimmin L.C., Ramirez,C., Matheson,A.T. and Dennis,P.P. (1989) Sequence alignment and evolutionary comparison of the L10 equivalent and L12 equivalent ribosomal proteins from archaebacteria, eubacteria, and eucaryotes. J. Mol. Evol., 29, 448–462. [DOI] [PubMed] [Google Scholar]
- 62.Szick K., Springer,M. and Bailey-Serres,J. (1998) Evolutionary analyses of the 12-kDa acidic ribosomal P-proteins reveal a distinct protein of higher plant ribosomes. Proc. Natl Acad. Sci. USA, 95, 2378–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koonin E.V. and Mushegian,A.R. (1996) Complete genome sequences of cellular life forms: glimpses of theoretical evolutionary genomics. Curr. Opin. Genet. Dev., 6, 757–762. [DOI] [PubMed] [Google Scholar]
- 64.Kyrpides N., Overbeek,R. and Ouzounis,C. (1999) Universal protein families and the functional content of the last universal common ancestor. J. Mol. Evol., 49, 413–423. [DOI] [PubMed] [Google Scholar]
- 65.Wuyts J., Van de Peer,Y. and De Wachter,R. (2001) Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA. Nucleic Acids Res., 29, 5017–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wool I.G. (1996) Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci., 21, 164–165. [PubMed] [Google Scholar]
- 67.Schnare M.N., Damberger,S.H., Gray,M.W. and Gutell,R.R. (1996) Comprehensive comparison of structural characteristics in eukaryotic cytoplasmic large subunit (23 S-like) ribosomal RNA. J. Mol. Biol., 256, 701–719. [DOI] [PubMed] [Google Scholar]
- 68.Wuyts J., De Rijk,P., Van de Peer,Y., Winkelmans,T. and De Wachter,R. (2001) The European Large Subunit Ribosomal RNA Database. Nucleic Acids Res., 29, 175–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peyretaillade E., Biderre,C., Peyret,P., Duffieux,F., Metenier,G., Gouy,M., Michot,B. and Vivares,C.P. (1998) Microsporidian Encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a LSU rRNA reduced to the universal core. Nucleic Acids Res., 26, 3513–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vossbrinck C.R., Maddox,J.V., Friedman,S., Debrunner-Vossbrinck,B.A. and Woese,C.R. (1987) Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature, 326, 411–414. [DOI] [PubMed] [Google Scholar]
- 71.Cavalier-Smith T. (1989) Molecular phylogeny. Archaebacteria and Archezoa. Nature, 339, l00–l01. [DOI] [PubMed] [Google Scholar]
- 72.Van de Peer Y., Ben Ali,A. and Meyer,A. (2000) Microsporidia: accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene, 246, 1–8. [DOI] [PubMed] [Google Scholar]
- 73.Katinka M.D., Duprat,S., Cornillot,E., Metenier,G., Thomarat,F., Prensier,G., Barbe,V., Peyretaillade,E., Brottier,P., Wincker,P. et al. (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature, 414, 450–453. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki T., Terasaki,M., Takemoto-Hori,C., Hanada,T., Ueda,T., Wada,A. and Watanabe,K. (2001) Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem., 276, 21724–21736. [DOI] [PubMed] [Google Scholar]
- 75.Poole A.M., Jeffares,D.C. and Penny,D. (1998) The path from the RNA world. J. Mol. Evol., 46, 1–17. [DOI] [PubMed] [Google Scholar]
- 76.Brinkmann H. and Philippe,H. (1999) Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol. Biol. Evol., 16, 817–825. [DOI] [PubMed] [Google Scholar]
- 77.Iwabe N., Kuma,K., Hasegawa,M., Osawa,S. and Miyata,T. (1989) Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl Acad. Sci. USA, 86, 9355–9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lopez-Garcia P. and Moreira,D. (1999) Metabolic symbiosis at the origin of eukaryotes. Trends Biochem. Sci., 24, 88–93. [DOI] [PubMed] [Google Scholar]
- 79.Evans S.V. (1993) SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.