Abstract
Generally, the presence of a consensus sequence in the promoter of a gene is taken as indication for regulation by the transcription factor that binds to this sequence. In light of the recent developments in genome research, we were interested to what extent this supposition is valid. We examined the relationship between the presence of a binding site for ComK, the competence transcription factor of Bacillus subtilis, and actual transcriptional activation by ComK. Bacillus subtilis contains 1062 putative ComK-binding sites (K-boxes) in its genome. We employed DNA macroarrays to identify ComK-activated genes, and found that the presence of a K-box is an unreliable predictor for regulation. Only ∼8% of the genes containing a K-box in the putative promoter region are regulated by ComK. The predictive value of a K-box could be improved by taking into consideration the degree of deviation from the K-box consensus sequence, the presence of extra ComK-binding motifs and the positions of RNA polymerase-binding sites. Finally, many of the ComK-activated genes show no apparent function related to the competence process. Based on our findings, we propose that the ComK-dependent activation of several genes might serve no biological purpose and can be considered ‘evolutionary noise’.
INTRODUCTION
The recent advances in bioinformatics, which, for example, made it possible to identify open reading frames (ORFs) in a genome with great accuracy, have spurred the interest in computer-based prediction of gene regulation. This field of research, however, is still in its infancy. Prediction of gene regulation on a genome-wide scale can be a powerful tool to identify proteins involved in certain cellular processes. In addition, such analyses may be helpful to elucidate the role of proteins to which no function could be attributed based on sequence comparisons. Generally, it is assumed that the location of a binding site for a certain transcription factor in a promoter region is an indication that the promoter is under control of this transcription factor. However, binding to a promoter is not necessarily proof that the transcription factor regulates the promoter, although the large number of such deductions in literature suggests otherwise. So far, the reliability of consensus transcription factor binding sites as predictors for gene regulation has not been examined in a systematic way. Studies of this kind can, for statistical reasons, not be restricted to a limited number of genes, but should cover a substantial part or, even better, all genes present on the genome to assure a reliable outcome. DNA-array technology makes it possible to determine gene expression on a genome-wide scale and in this study we use this to examine the reliability of the DNA recognition sequence of the transcription factor ComK as a predictor for ComK-dependent gene expression.
Bacillus subtilis is a Gram-positive bacterium which can differentiate into cells competent for genetic transformation by synthesizing a complex DNA-binding and -uptake system, and by activating recombination genes. Competence is a starvation-induced differentiation process (for review see 1). The various environmental signals are interpreted by a complex signal transduction cascade, and ultimately lead to the activation of comK, which encodes the competence transcription factor (2,3). ComK activates the expression of the DNA-binding and -uptake system, DNA-recombination genes, and its own expression (2,4,5). ComK binds to the promoter regions of these genes, and footprinting studies established a conserved AT-rich palindromic sequence as the ComK-recognition sequence, or K-box (6). The presence of a K-box in the promoter region of a gene suggests that the gene is regulated by ComK and encodes a protein that is likely to be involved in competence. In theory, by screening the B.subtilis genome for promoter regions that have a putative K-box, we can quickly identify the set of proteins that are involved in establishing competence in B.subtilis.
Here we have assessed the reliability of a K-box as a predictor for ComK-dependent gene expression. To identify ComK-activated genes on a genome wide scale, we have compared the transcription profiles of a wild type B.subtilis strain and a strain containing a disrupted comK gene, and correlated this data to the presence of K-boxes in promoter regions. We discuss the shortcomings of the method and describe ways to improve gene regulation predictions based on the presence of transcription factor binding sites.
MATERIALS AND METHODS
All molecular cloning and PCR procedures were carried out using standard techniques (7,8). Media for growth of Escherichia coli and B.subtilis have been described by Sambrook et al. (8) and Venema et al. (9). Bacillus subtilis strain 8G5 chromosomal DNA used as template for PCR was purified as described by Venema et al. (9).
Strain construction
A disruption of comK was obtained by a double-crossover integration of a spectinomycin resistance (Spc) marker into the comK gene of B.subtilis strain 8G5 (a derivative of B.subtilis strain 168) (10). The resulting comK mutant was labeled B.subtilis strain BV2004. The spectinomycin resistance marker was isolated by PCR using the primer pair Sp1 (5′-CGG GAT CCG CCG AAG GGG CAT CGA TTT TCG TTC GTG AAT-3′) and Sp2 (5′-CGG GAT CCG CCA AGA TGG CAT ATG CAA GGG TTT ATT-3′), and plasmid pDG1726 as template (11). The Spc marker was inserted into the unique HindII site (blunt-end ligation) of comK, and has the same transcriptional direction as comK. As control, B.subtilis strain BV2012 was constructed which contains an integrated Spc marker such that no gene or operon was disturbed. The pks locus was chosen as a ‘neutral’ site for integration of the Spc marker. The pks gene cluster encodes a polyketide synthetase implicated in synthesis of the antibiotic difficidin, yet production of difficidin by B.subtilis 168 has not been reported, and mutations in this locus showed no apparent phenotype (12,13). The Spc marker was inserted into the Eco47III site (blunt-end ligation) located between pksR and pksS by means of a double-crossover integration, and has the same transcriptional direction as pksS. In this case, the Spc marker was isolated by PCR using the primer pair Sp1 and Sp4 (5′-GCT GAG AAC ATA TGC AAG GGT TTA TT-3′), and plasmid pDG1726 as template.
Growth conditions and RNA isolation
To obtain a high percentage of competent cells we applied a two-step growth protocol (14). An aliquot of 10 ml of minimal medium supplemented with 6 mM magnesium sulphate was inoculated with 0.5 ml overnight culture and incubated at 37°C under vigorous shaking. After 3 h of growth, 10 ml of prewarmed starvation medium was added and incubation continued for another 30 min. At this stage, ∼15 min before the culture reached maximum competence, cells were harvested for isolation of RNA. Competence was tested by transformation with chromosomal DNA from a Trp+ strain, and generally reached a level of ∼0.5–1% transformants, when selecting for tryptophane-positive colonies. We experienced that inoculation of overnight cultures with colonies from plate (no older than a week), instead of inoculation from frozen stocks, resulted in better development of competence.
RNA was isolated by spinning down 2 ml of culture (30 s Eppendorf centrifuge, 14 000 r.p.m., 4°C) and resuspending the pellet in 0.3 ml ice-cold growth medium (final volume 0.4 ml). The cell suspension was added to a screw cap Eppendorf tube containing 1.5 g glass beads (75–150 µm), 0.5 ml phenol:chloroform:isoamylalcohol (12:12:1), 50 µl 10% SDS and 50 µl 3 M sodium acetate (pH 5.2). All solutions were prepared with diethylpyrocarbonate (DEPC)-treated water. After vortexing, the tube was frozen in liquid nitrogen and stored at –80°C. Cells were broken by shaking for 8 min in a shake-it-baby (15). After 5 min centrifugation (Eppendorf centrifuge, 10 000 r.p.m., 4°C) the water phase (0.4 ml) was transferred to a clean tube containing 0.4 ml chloroform. After vortexing and centrifugation (2 min, Eppendorf centrifuge, 14 000 r.p.m., 4°C), the water phase was transferred to a clean tube and the RNA was isolated with a High Pure RNA Isolation Kit (Roche). RNA was eluted in 50 µl elution buffer and quantified with GeneQuant (Amersham).
cDNA preparation and hybridization
Reverse transcription was carried out in a total volume of 50 µl. Aliquots of 1 pmol of B.subtilis ORF-specific primers (Eurogentec) were mixed with 4 µg isolated RNA, incubated for 10 min at 70°C and cooled on ice. Subsequently, 10 µl of 5× concentrated First strand buffer (Gibco BRL), 5 µl 100 mM DTT, 0.5 µl RNasin (Roche, 40 U/µl), 2.5 µl dNTP mixture (5 mM dATP, 5 mM dTTP, 5 mM dGTP, 0.1 mM dCTP) were added, and the total volume was adjusted to 42.5 µl with DEPC-treated water, and kept on ice. After addition of 5 µl [α-33P]dCTP, the mixture was incubated for 10 min at 25°C, before the addition of 2.5 µl SuperscriptII (Gibco BRL). Reverse transcription reaction was carried out for 2 h at 42°C, 15 min at 70°C, and was stopped by adding 2 µl 0.5 M EDTA, 2 µl 10% SDS, 6 µl 3 M NaOH and incubating for 30 min at 68°C. After neutralization with 6 µl 3 M HCl, the labeled cDNA was purified on a Sephadex G-25 column (Roche). Incorporation of label was checked by scintillation counting.
Labeled cDNA was hybridized to B.subtilis Panorama™ Arrays (Sigma-Genosys), as described by the manufacturer. After hybridization and washing, Cyclone phospho-imager screens (Packard) were exposed for ∼2 days. The Cyclone readouts were analyzed with Array-Pro Analyzer 4.0 (Media Cybernetics). After background subtraction, duplicate spots were averaged, and the signal was normalized against the total signal of all spots.
Data analysis and visualization
The normalized array data was subjected to a statistical analysis using Cyber-T, a program based on t-test variant combined with a Bayesian statistical framework (16). Cyber-T is available for online use at the genomics website at the University of California at Irvine (http://genomics.biochem.uci.edu/genex/cybert/). The following parameters were used in Cyber-T: the ‘minimum non-zero replicates’ was set to 2, a ‘sliding window’ of 101 was used, and the recommended ‘confidence value’ of 10 was chosen. The raw and normalized data for the complete gene sets can be downloaded from http://molgen.biol.rug.nl/publication/comk_data/. Spots were associated with gene names by using the spreadsheet B.subtilis Array information.xls, provided by Sigma-Genosys.
K-box positions were calculated using Genome-2D, an in-house-developed software package for the construction of comprehensive bacterial genome atlases (17). The B.subtilis genome sequence and gene annotation files were obtained from GenBank at NCBI (ftp://ftp.ncbi.nih.gov/genomes). Distances of K-boxes to genes, AT-boxes or RNA polymerase-binding sites, were calculated by determining the number of base pairs between the end of a K-box and the start of an ORF, start of an AT-box, or –35/–10 promoter sequence. Genome-2D contains a simple algorithm to predict putative operon organization by considering all successive genes that are transcribed in the same direction as part of an operon until a rho-independent terminator or a gene that is transcribed in the opposite direction is encountered.
The relative position of K-boxes, AT-boxes and –35 promoter sequences on the face of the DNA helix are presented in spiral curves, which are calculated as follows. First, the base pair distance (D) between a K-box and AT-box, or K-box and –35 promoter sequence was determined, and divided by 10.5 bp (C), corresponding to the length of one complete DNA helix turn (18). The cosine and sine of the resulting coefficient indicate the coordinates on the x-axis (X) and y-axis (Y), respectively. To show the number of DNA helix turns that separate the K-boxes from AT-boxes or –35 promoter sequences also, the cosine or sine and the coefficient C were added up. To prevent overlap of data points when the distances of different K-box/AT-box or K-box/–35 combinations are the same, a random number between 0 and 0.3 (rnd) was added. The complete functions used to calculate the position of the different boxes on the face of the DNA helix are: (i) C = D / 10.5, (ii) X = cos(360 × C) + C + (0.15 – rnd), (iii) Y = sin(360 × C) + C + (0.15 – rnd).
RESULTS
Distribution of K-boxes in the B.subtilis genome
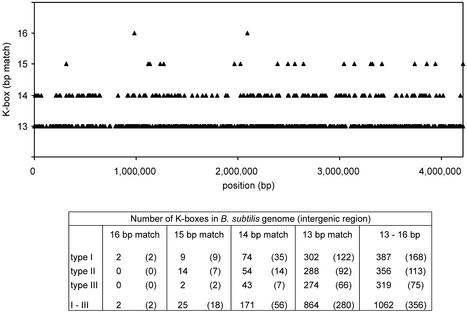
First we addressed the question how many K-boxes the B.subtilis genome contains. It has been shown that ComK functions as a tetramer composed of two dimers, each recognizing the sequence A4N5T4 or AT-box. The distance between the AT-boxes may vary between one, two and even three DNA helical turns (6). Based on this variation, three different types of K-box were distinguished, summarized in Table 1. As indicated in this table, none of the K-boxes of the known ComK-activated genes perfectly match the consensus sequence, and the deviation can amount to 3 bp. No more than 2 bp deviations per AT-box were observed, and this latter restriction was taken as a cut-off when screening the B.subtilis genome for K-boxes. As shown in Figure 1, a total of 1062 sequences corresponded to our K-box definition, roughly one K-box per 4000 bp. In a random 4.1 Mb DNA sequence with the same AT content as the B.subtilis genome only about 175 K-boxes were found. Apparently, B.subtilis contains many more ComK-binding sites than can statistically be expected.
Table 1. Known ComK-activated genes and operons arranged according to the type of ComK-binding site (K-box) present in their promoters.
| Type | K-box | Gene | Function | Deviation (bp) |
|---|---|---|---|---|
| I | (AT-box)-N8-(AT-box) | addAB | DNA recombination | 1 |
| recA | DNA recombination | 3 | ||
| uvrB | DNA recombination | 2 | ||
| II | (AT-box)-N18-(AT-box) | comC | DNA uptake | 2 |
| comE | DNA uptake | 2 | ||
| comF | DNA uptake | 3 | ||
| comG | DNA uptake | 1 | ||
| III | (AT-box)-N31-(AT-box) | comK | Regulation | 1 |
The number of base pair (bp) deviation from the consensus sequence is shown. N indicates the number of base pairs between the two ComK dimer-binding sequences (AT-box).
Figure 1.
Distribution of K-boxes in the B.subtilis genome. In the graph a distinction between K-boxes is made on the basis of the base pair deviations from the consensus sequence. The consensus sequence contains 16 defined base pairs. Only two K-boxes perfectly matched the consensus sequence (16 bp match). The table shows the number of different K-boxes present in the B.subtilis genome. The number of K-boxes in intergenic regions is placed between brackets.
It is likely that a K-box is only functional when located in the promoter region of a gene, which is generally located in an intergenic region. Figure 1 shows that about one-third of all K-boxes are located in intergenic regions. Since intergenic regions cover only 12% of the genome, these areas are substantially enriched with K-boxes. Although the AT content of intergenic regions is higher than that of coding sequences, 61.4 and 55.7%, respectively, these percentages do not explain the relatively high number of K-boxes in intergenic regions. According to Figure 1, type I is the more abundant type of K-box. This bias is primarily caused by the limited length of the intergenic regions. The average length of intergenic regions is only 130 bp; therefore the shorter the K-box, the higher the chance it matches sequences in an intergenic region.
The data above shows that the genome of B.subtilis harbors many K-boxes, which may serve as operator sites. To evaluate the validity of a K-box as predictor for regulation of gene expression by ComK, it will be necessary to link this data to the ComK-regulon.
Identification of the ComK-activated genes of B.subtilis
In order to identify genes that are regulated by ComK, we compared the expression profile of a strain in which the comK gene is disrupted by a spectinomycin resistance marker (BV2004) with the control strain BV2012. Strain BV2012 also contains a spectinomycin (Spc) marker, but inserted in such a way that neither a gene nor an operon was disturbed, to rule out any effect on transcription that may be caused by the Spc marker itself. The latter was confirmed by comparing the expression profiles with that of a wild type strain (see Supplementary Material for details). Transcription profiles of the different strains were obtained by using commercial DNA macro-arrays containing 4107 PCR-amplified ORFs representing all putative protein-encoding genes of B.subtilis. Strains were grown in competence medium (without spectinomycin selection) and cells were harvested for RNA isolation when the competence stage was reached. Competence was checked by transforming aliquots of the cultures with chromosomal DNA. RNAs isolated from simultaneously grown cultures of the two strains were successively hybridized to the same DNA-filter, and the entire procedure from growth to hybridization was performed in triplicate, on three separate filters.
In the analysis of the macroarray data the question arises how the ComK regulon is best defined. Previously, Long et al. (19) showed that a simple selection based on differences in expression is disputable. In this study we chose to combine a statistical approach with prior knowledge of ComK activation. The program Cyber-T, developed by Baldi and Long (20), incorporates a Bayesian prior to improve the t-test for DNA-array measurements. We used this program to sort our data (a more detailed comparison of three different methods of analysis—differences in expression, t-test and CyberT—is provided as Supplementary Material). Liu and Zuber (21) showed that read-through from the comF operon led to increased expression of the downstream-located flgM gene. As a consequence flgM is moderately activated by ComK. We chose the parameters of this gene (p = 0.017 and 1.36-fold up-regulation) as a low-end border to discriminate ComK-activated genes after analysis with Cyber-T. A total of 105 genes met these criteria, and were considered to be ComK activated. The genes are listed in Table 2, ordered on increasing p-values. Genes of putative operons are grouped and sorted according to their position in the operon.
Table 2. Genes activated by ComK.
| Gene | Operon | Description | Fold | Involved in competence | Found in other study | Distance to K-box | K-box type |
|---|---|---|---|---|---|---|---|
| comGA | comG | Involved in DNA binding | 10.5 | yesa | B1,2, O | 63 | II-15 |
| comGB | comG | Involved in DNA binding | 37.1 | yesa | B1,2, O | 1186 | II-15 |
| comGC | comG | Involved in DNA binding | 193.9 | yesa | B1,2, O | 2171 | II-15 |
| comGD | comG | Involved in DNA binding | 23.5 | yesa | B1,2, O | 2457 | II-15 |
| comGE | comG | Involved in DNA binding | 25.8 | yesa | B1,2, O | 2872 | II-15 |
| comGF | comG | Involved in DNA binding | 29.9 | yesa | B1,2, O | 3245 | II-15 |
| comGG | comG | Involved in DNA binding | 26.7 | yesa | B1,2, O | 3629 | II-15 |
| yqzE | comG | No similarity to other proteins | 2.3 | B2 | 4074 | II-15 | |
| ywfM | Similar to hypothetical proteins | 11.5 | B1,2, O | 3151 | II-13 | ||
| ywpH | ywpH | Similar to single-strand DNA-binding protein | 24.5 | yesb | B1,2, O | 121 | II-15 |
| glcR | ywpH | Transcriptional regulator (DeoR family) | 2.4 | O | 71 | II-14 | |
| ywpJ | ywpH | Similar to hypothetical proteins | 2.3 | noc | B1,2 | 853 | II-14 |
| ycbP | Similar to hypothetical proteins from B.subtilis | 7.2 | B2 | 1467 | II-13 | ||
| comK | Competence transcription factor (CTF) | 13.8 | Yes | B1,2, O | 89 | III-15 | |
| comER | comE | Counter transcript in comE operon | 9.8 | noa | B1,2, O | 4314 | II-13 |
| comEA | comE | Involved in DNA binding and translocation | 22.1 | yesa | B1,2, O | 945 | II-14 |
| comEB | comE | Similar to dCMP deaminase | 4.3 | noa | B1,2, O | 1629 | II-14 |
| comEC | comE | Involved in DNA binding and translocation | 3.8 | yesa | B1,2, O | 2202 | II-14 |
| rapH | Response regulator aspartate phosphatase | 5.5 | noc | B2, O | 39 | II-13 | |
| yckB | yckB | Similar to amino acid ABC transporter (binding protein) | 4.6 | B1,2 | 3805 | II-13 | |
| yckA | yckB | Similar to amino acid ABC transporter (permease) | 3.6 | B1,2 | 4678 | II-13 | |
| nucA | nucA | Nuclease | 6.6 | yesd | B1,2, O | 198 | II-13 |
| nin | nucA | Inhibitor of NucA | 7.6 | yesd | B1,2, O | 536 | II-13 |
| yojB | No similarity to other proteins | 50.3 | 3940 | II-13 | |||
| yckC | yckC | Similar to hypothetical proteins from B.subtilis | 3.4 | B1,2 | 2612 | II-13 | |
| yckD | yckC | No similarity to other proteins | 6.4 | B1,2 | 3149 | II-13 | |
| yckE | yckC | Similar to beta-glucosidase | 9.6 | O | 3635 | II-13 | |
| comFA | comF | Involved in DNA translocation | 15.0 | yese | B1,2, O | 59 | II-13 |
| comFB | comF | No similarity to other proteins | 4.6 | noe | B1,2, O | 1510 | II-13 |
| comFC | comF | Involved in DNA translocation | 4.6 | yese | B1,2, O | 1803 | II-13 |
| yvyF | comF | Similar to flagellar protein | 1.6 | yesc | B2, O | 2566 | II-13 |
| flgM | comF | Anti-sigma factor (repressor sigma-D-dependent gene transcription) | 1.4 | B2 | 3066 | II-13 | |
| yvyG | comF | Similar to flagellar protein | 1.4 | yesc | B2 | 3348 | II-13 |
| ybdK | ybdK | Similar to two-component sensor histidine kinase (YbdJ) | 5.4 | B2, O | 172 | III-14 | |
| ybdL | ybdK | No similarity to other proteins | 1.9 | B2 | 1204 | III-14 | |
| comC | Involved in DNA binding | 10.2 | yesa | B1,2, O | 74 | II-14 | |
| maf | maf | Septum formation | 3.0 | B1,2, O | 2105 | II-14 | |
| radC | maf | Similar to DNA repair protein | 3.2 | B1,2, O | 2711 | II-14 | |
| yyaF | Similar to GTP-binding protein | 3.8 | B1,2, O | 96 | II-13 | ||
| smf | smf | DNA processing (Smf protein homolog) | 3.3 | B1,2, O | 1779 | III-13 | |
| topA | smf | DNA topoisomerase I | 1.5 | B2 | 427 | I-13 | |
| hxlR | Positive regulator hxlAB (ribulose monophosphate pathway) | 8.0 | B1,2 | 3302 | I-13 | ||
| cwlJ | Cell wall hydrolase (sporulation) | 6.1 | B1,2, O | 3713 | I-13 | ||
| yneA | yneA | No similarity to other proteins | 3.5 | O | 2522 | III-13 | |
| yneB | yneA | Similar to resolvase | 2.4 | O | 105 | II-13 | |
| sacX | sacX | Negative regulatory protein of SacY | 3.0 | B2, O | 2435 | II-13 | |
| sacY | sacX | Transcriptional antiterminator (levansucrase and sucrase synthesis) | 3.8 | O | 3868 | II-13 | |
| tagC | (dinC) Possibly involved in teichoic acid biosynthesis | 2.6 | 198 | I-13 | |||
| spoIIB | Dissolution of septal peptidoglycan during engulfment | 6.0 | 954 | II-14 | |||
| ywfK | ywfK | Similar to transcriptional regulator (LysR family) | 2.2 | B1,2 | 1190 | II-13 | |
| ywfL | ywfK | No similarity to other proteins | 1.9 | B1,2 | 2138 | II-13 | |
| hxlA | hxlA | Ribulose monophosphate pathway | 4.2 | B1,2 | 689 | II-13 | |
| hxlB | hxlA | Ribulose monophosphate pathway | 3.3 | B1,2 | 1327 | II-13 | |
| yhxD | Similar to alcohol dehydrogenase | 2.8 | nof | 2270 | III-13 | ||
| tlpC | Methyl-accepting chemotaxis protein | 2.1 | O | 1995 | II-13 | ||
| yjbF | No similarity to other proteins | 2.1 | yesc | B2 | 135 | I-14 | |
| ycbR | Similar to toxic cation resistance protein | 2.7 | 4247 | I-13 | |||
| ywfI | Similar to hypothetical proteins | 2.0 | noc | B1 | 154 | II-13 | |
| recA | Multifunctional SOS repair regulator | 2.4 | yesg | B2, O | 111 | I-13 | |
| dinB | Nuclease inhibitor (DNA-damage inducible) | 1.8 | B2 | 25488 | III-14 | ||
| yqeN | Similar to hypothetical proteins | 1.8 | B1,2 | 4936 | II-14 | ||
| yvrP | yvrP | Similar to ABC transporter | 1.7 | noc | B2, O | 114 | III-13 |
| yvrN | yvrP | Similar to ABC transporter (ATP-binding protein) | 1.6 | noc | B1,2 | 1953 | III-13 |
| yvrM | yvrP | Similar to ABC transporter (ATP-binding protein) | 1.4 | noc | B2 | 2441 | III-13 |
| bmrU | Multidrug resistance protein | 3.6 | 3764 | I-14 | |||
| yrhL | Similar to acyltransferase | 1.6 | noc | 392 | I-13 | ||
| yhzC | No similarity to other proteins | 2.0 | noh | B2, O | 147 | III-15 | |
| ykoH | Similar to two-component sensor histidine kinase (YkoG) | 1.8 | 718 | I-13 | |||
| med | med | Positive regulator of comK | 1.6 | yesi | B2 | 130 | I-14 |
| comZ | med | Positive regulator of comG | 1.5 | yesj | B2 | 1080 | I-14 |
| guaD | Guanine deaminase | 2.8 | noc | 2537 | I-13 | ||
| ydeD | Similar to hypothetical proteins | 2.2 | 1477 | II-13 | |||
| groES | groES | Molecular chaperonin (class I heat-shock protein) | 1.5 | B2 | 622 | III-13 | |
| ydiM | groES | No similarity to other proteins | 2.4 | 4193 | III-13 | ||
| ydiN | groES | No similarity to other proteins | 2.5 | 5740 | III-13 | ||
| yhcF | yhcF | Similar to transcriptional regulator (GntR family) | 1.8 | O | 4721 | I-14 | |
| yhcH | yhcF | Similar to ABC transporter (ATP-binding protein) | 1.4 | noc | O | 5803 | I-14 |
| yhcI | yhcF | No similarity to other proteins | 1.5 | noc | B2 | 6713 | I-14 |
| gsiB | General stress protein | 2.0 | 9246 | I-14 | |||
| yfhL | No similarity to other proteins | 1.7 | 5177 | III-13 | |||
| ywaF | ywaF | No similarity to other proteins | 2.2 | 5634 | I-14 | ||
| gspA | ywaF | General stress protein | 2.0 | 6457 | I-14 | ||
| manP | Putative PTS mannose-specific enzyme (IIBCA component) | 3.8 | noc | 22 | III-13 | ||
| ybyB | No similarity to other proteins | 2.1 | 2570 | III-13 | |||
| pstC | pstC | Phosphate ABC transporter (permease) | 2.2 | 7847 | I-13 | ||
| pstBA | pstC | Phosphate ABC transporter (ATP-binding protein) | 2.5 | 9681 | I-13 | ||
| yorB | No similarity to other proteins | 1.5 | 2760 | III-13 | |||
| trpC | trpC | Indol-3-glycerol phosphate synthase | 2.6 | 6279 | I-13 | ||
| trpB | trpC | Tryptophan synthase (beta subunit) | 1.6 | B2 | 1109 | I-13 | |
| sucC | sucC | Succinyl-CoA synthetase (beta subunit) | 1.5 | 89 | III-13 | ||
| sucD | sucC | Succinyl-CoA synthetase (alpha subunit) | 1.4 | B2 | 816 | III-13 | |
| dppC | Dipeptide ABC transporter (permease) | 1.6 | 9293 | II-13 | |||
| yqgZ | Similar to hypothetical proteins | 2.1 | 6066 | III-13 | |||
| bdbD | bdbD | Thiol-disulfide oxidoreductase | 1.4 | yesk | B1,2 | 455 | I-14 |
| bdbC | bdbD | Thiol-disulfide oxidoreductase | 1.4 | yesk | B2 | 1128 | I-14 |
| ykzA | Similar to general stress protein | 1.7 | noc | 1094 | I-13 | ||
| ykuK | No similarity to other proteins | 1.5 | noc | B2 | 441 | III-13 | |
| yuaF | yuaF | No similarity to other proteins | 1.6 | noc | 3251 | II-13 | |
| yuaG | yuaF | Similar to flotillin 1 | 1.4 | noc | 3796 | II-13 | |
| yuaI | yuaF | No similarity to other proteins | 1.6 | noc | 5343 | II-13 | |
| exoA | 3′-exo-deoxyribonuclease (multifunctional DNA-repair enzyme) | 2.2 | B2 | 2501 | II-13 | ||
| ctsR | Transcriptional repressor of class III stress genes | 1.4 | 16791 | I-13 | |||
| yxiP | No similarity to other proteins | 1.4 | 92 | I-13 | |||
| yisQ | Similar to hypothetical proteins | 1.5 | noc | 567 | I-14 | ||
| ywmF | No similarity to other proteins | 2.5 | noc | 8960 | II-13 |
Known ComK-activated genes are indicated in bold. The expression of the selected genes showed at least a 1.4-fold difference with a p-value <0.017. Genes are ordered by increasing p-values. Genes of the same operon are clustered and ordered according to the position in the operon. The functional descriptions are based on the Subtilist Web Server (http://genolist.pasteur.fr/SubtiList/) information. The abbreviation O refers to Ogura et al. (24) and B to Berka et al. (23) respectively. The suffixes 1 and 2 for B refer to the two different setups of the experiment as described in that paper. The distance to the first upstream K-box is given in base pairs, together with type and base pair match of the K-box.
aDubnau (34).
cBSFA (Bacillus subtilis functional analyses programme). Transformation percentages <10% were considered as disturbed competence.
dProvvedi et al. (35).
eLondono-Vallejo and Dubnau (36).
fvan-Sinderen et al. (37).
gLovett et al. (38).
hK.Susanna, unpublished results.
iOgura et al. (39).
jOgura and Tanaka (40).
kMeima et al. (22).
The genes that constitute the DNA-binding and -uptake machinery (comC, -E, -F, -G, nucA operons) clearly stand out in Table 2 with high induction levels of 4- to 194-fold. The presence of yhxD in the list is disputable since this gene is located adjacent to comK, and expression of this gene is likely a consequence of read through from the Spc marker inserted in comK. Table 2 encompasses a great variety of genes. The Bacillus subtilis Functional Analysis (BSFA) programme was founded by a consortium of research groups in order to determine the function of all hypothetical proteins of B.subtilis. Within this programme it was established that both yvyF and yvyG are required for competence. The BSFA consortium also showed that the two, at that time unknown, thio-disulfide oxidoreductases BdbD and BdbC are required for competence. Recently it was demonstrated that these proteins are necessary for disulfide bond formation within ComGC (22). Several genes involved in DNA processing are activated by ComK, and likely function in the DNA-integration process. Especially, the induction of an alternative single-stranded DNA-binding protein, ywpH, is very pronounced. A knockout of this gene reduces transformation considerably [(23,24) and C. Lindner, unpublished results]. Several ComK-activated genes are related to general stress response, and a substantial number of ComK-activated genes function in general metabolic pathways. So far, none of these genes appear to be required for competence and the reason for their specific activation in competent cells is still unknown.
Presence of K-boxes upstream of ComK-activated genes
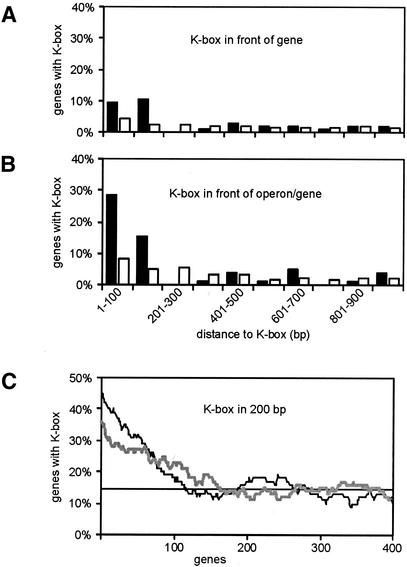
With the determination of the position of putative K-boxes and the identification of the ComK-regulon, it becomes possible to examine the upstream regions of ComK-activated genes for the presence of ComK-binding sites. In Table 2 also the distance to the nearest K-box is listed for each gene. For most of the genes this distance is rather large. We have depicted this more schematically in Figure 2. As shown in bar diagram A, ∼20% of the ComK-activated genes contain a K-box in the first 200 bp upstream, a region that generally covers the promoter. Only 7% of the non-regulated genes have a K-box in this region. Although ComK-activated genes possess significantly more K-boxes, almost 70% do not even contain an upstream located K-box within a distance of 1000 bp. This relatively high percentage of regulated genes without a box is at least partly due to the organization of genes in operons. For example, all but the first gene of the comG operon are more than 1000 bp distant from the K-box in the comG promoter (see Table 2). To take into account the effect of operons is difficult, due to the absence of distinct transcriptional start and terminator sequences. We considered contiguous genes transcribed in the same direction, and flanked by genes transcribed in the opposite direction or annotated rho-independent terminators, to form an operon (12). This definition works reasonably well, for example in case of the comG operon. However, since transcriptional terminators are poorly defined many unrealistic long operons were found, which often contain several K-boxes. Since these boxes might be used to activate downstream-located genes, we considered such K-boxes as being part of potential ComK-regulated promoters. Consequently, the presence of a K-box can determine the start (or end) of an operon, and this premise was included in the algorithm we used to define operons. When the correction for operons was taken into account the fraction of ComK-activated genes with a K-box in their putative promoter region (200 bp upstream) increased considerably to almost 45%.
Figure 2.
Presence of K-boxes in front of genes. Bar diagram (A) displays the percentage of genes containing a K-box in subsequent 100 bp upstream intervals. Only the most proximate K-box was used in the calculations. Closed bars represent ComK-activated genes, open bars represent non- regulated genes. In (B) the position of genes in putative operons is taken into account (see main text for details). Graph (C) shows a moving average analysis of genes with a K-box in their putative promoter region (K-box present at a maximum distance of 200 bp upstream gene or operon). Genes were sorted on fold difference in expression (grey line, highest fold at left end of horizontal axis), or genes were sorted on p-values (black line, lowest p-value at left end of horizontal axis). The first 400 genes are displayed. A moving average algorithm with a window size of 100 genes was used, and the average percentage is indicated by the horizontal black line.
The percentages of genes with a K-box in Figure 2 are based on calculations using ComK-activated genes that were selected on the basis of p-value. To examine whether the alternative selection, on the basis of differences in expression levels, results in a higher percentage of ComK-activated genes containing a K-box, we calculated, using a moving average algorithm, the local percentage of genes with a K-box in their putative promoter region. As shown in Figure 2C, the fraction of ComK-activated genes that harbor a K-box in their putative promoter region is considerably higher when genes were sorted on p-values than when genes were sorted on ratio of expression differences. This outcome provides additional support for the decision to use reliability as a criterion for the selection of ComK-activated genes.
Prediction of ComK activation
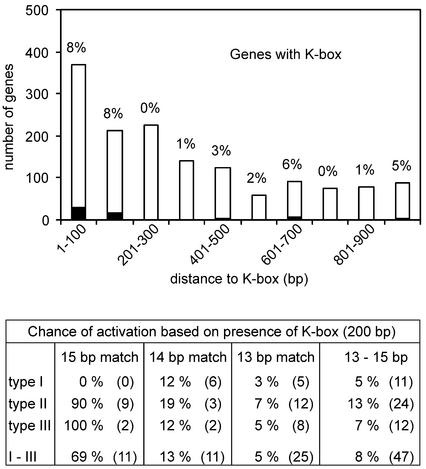
Figure 2 shows that a relative high percentage of ComK-activated genes contain a K-box in their putative promoter regions, yet, such numbers do not tell us whether the presence of a K-box is a good predictor for ComK activation. A substantial number of non-regulated genes contain a K-box in their putative promoter regions as well, and when the absolute numbers of activating and non-activating K-boxes are considered, a less appealing picture appears. Figure 3 is comparable to Figure 2, except that numbers of genes are depicted instead of percentages. The fraction of ComK-activated genes is indicated by the percentage on top of each bar. When a K-box is located at a distance of <200 bp from the start codon of a gene, the chance that this gene is activated by ComK is a mere 8%. When the maximum distance allowed is enlarged to 1000 bp this value drops to 4%. Apparently, the presence of a ComK-binding site in front of a gene is not a very reliable indication that the gene is actually activated by ComK.
Figure 3.
Chance of ComK activation based on the presence of a K-box. The bar diagram displays the number of genes which contain a K-box in subsequent 100 bp upstream intervals. Closed bars represent the number of ComK-activated genes, and open bars represent non-regulated genes. The fraction of ComK-activated genes is indicated on top of the bars. The table shows the chance of ComK activation (percentage of ComK-activated genes) related to the type and base pair match of K-boxes present at a maximum distance of 200 bp upstream gene or operon (putative promoter region). The numbers in brackets indicate the number of ComK-activated genes on which the percentages are based. In the calculations the position of genes in putative operons is taken into account.
We wondered to what extent this low reliability depends on K-box type and base pair match. The table in Figure 3 shows the chance of ComK activation related to type and base pair match of a K-box. The values for the only two perfect K-boxes (16 bp match), downstream of cspB and yocI, were omitted because neither of these genes seemed to be involved in ComK-activated gene expression. The predictive value for ComK activation is high (69%) in case the K-box matches 15 out of the 16 bp of the consensus sequence. A single additional base pair deviation strongly decreases this value, and K-boxes that match 14 or 13 bp of the consensus sequence are poor indicators for ComK activation, although these boxes have been shown to be functional. The chance of activation does not clearly depend on the spacing between the AT-boxes (type I, II, III in the table of Fig. 3). When the selection criteria for ComK-activated genes were varied (adjustment of p-values or folds expression) no clear improvement was obtained (data not shown).
In conclusion: (i) K-boxes located within the first 200 bp upstream of a gene or operon are better indicators for ComK activation than boxes that are located at a larger distance, (ii) a single base pair deviation markedly decreases the chance of ComK activation and (iii) the type of K-box does not influence its predictive value.
Improving the predictive value of K-boxes
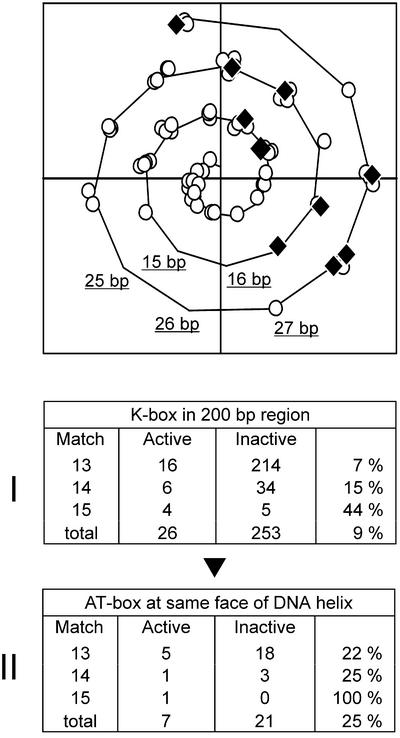
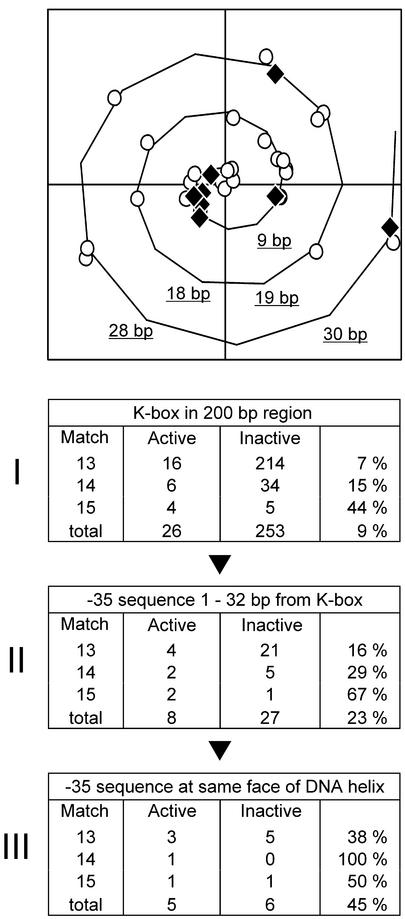
Poorly matching K-boxes appear to be poor indicators for ComK-activated transcription. Due to cooperative interactions, the presence of an additional ComK-dimer binding site (AT-box) adjacent to a K-box might enhance the affinity for ComK. This is illustrated by the recA promoter where a K-box, with a 3 bp deviation from the consensus sequence, is flanked at both sides by an extra AT-box (25). We investigated whether the chance of regulation increases when an additional AT-box is present. We focused on K-boxes located at a maximum distance of 200 bp upstream from a gene or operon, in the putative promoter regions. Of the 279 K-boxes found, 26 were located in the promoter region of activated genes and considered ‘active’ K-boxes (Fig. 4, Table 1). Any cooperative effect of an additional ComK dimer will only occur when the AT-box is in close proximity of the K-box. The greatest interval between AT-boxes can be found in type III K-boxes and amounts to 31 bp (6). This distance corresponds to three DNA helical turns, when assuming 10.5 bp per helical turn (18). Therefore, only boxes within this range were taken into account. Previous work has shown that the cooperative binding of dimers can only occur when the ComK-dimers are positioned on the same face of the DNA-helix (6). The spiral curve in Figure 4 illustrates the positions of additional AT-boxes relative to their cognate K-box on the face of the DNA helix. The reason that so many AT-boxes are depicted in the spiral curve is a consequence of the fact that a single K-box can be flanked by more than one AT-box. Only seven out of the 26 active K-boxes contain an extra AT-box, yet all of these boxes are positioned on the same face of the helix as the K-box itself (positive x-axis). Table 2 of Fig. 4 shows that when an additional AT-box with the correct phasing is taken into account, the chance that a poorly matching K-box is used as ComK operator-site increases considerably; from 7 to 22% in case of K-boxes with 3 bp deviations. In this survey only extra AT-boxes with a single base pair mismatch were allowed. The presence of extra AT-boxes with 2 bp deviations gave no improvement of the predictive value of K-boxes (data not shown).
Figure 4.
Presence of extra AT-boxes. The spiral curve shows face and distance of AT-boxes on the DNA helix relative to the corresponding K-box (10.5 bp is used as one complete helix turn, some base pair distances are shown, see Materials and Methods for a description of spiral curves). An interval of 8 bp between AT-box and K-box positions both boxes at the same face of the DNA-helix, and the distances between AT-boxes and corresponding K-boxes are corrected such that an 8 bp distance coincides with the positive x-axis (6). Open circles represent AT-boxes related to K-boxes of unregulated genes or operons (‘Inactive’ in tables), and closed rhombuses represent AT-boxes related to K-boxes of ComK-activated genes or operons (‘Active’ in tables). K-boxes can be flanked by more than one AT-box as a consequence of which some circles and rhombuses belong to the same K-box. The tables indicate the number of K-boxes after subsequent selection steps. K-boxes are distributed based on homology to the consensus sequence (13, 14 or 15 bp match), and whether they relate to unregulated (Inactive) or ComK-activated (Active) genes or operons. The fraction of activated K-boxes is presented in the right column of each table. Table 1 displays the number of K-boxes present at a maximum distance of 200 bp upstream gene or operon (putative promoter region). Table 2 displays K-boxes with an extra AT-box at the same face of the DNA helix (distances allowed between K-box and AT-box; 6–10, 16–21 and 27–31 bp).
Based on the close proximity of ComK- and RNA polymerase-binding sites in several known ComK-activated promoters, it was assumed that ComK activates transcription by recruitment of RNA polymerase (6). In the comC, -E, -F and -G promoters the distance between K-boxes and –35/–10 core promoter elements is 5 or 6 bp (6). In the addAB promoter this distance is 15 bp, thus including one additional DNA helical turn. The location of RNA polymerase binding-sites could therefore be a potential tool to distinguish the relevant K-boxes. Unfortunately, the exact locations of the majority of RNA polymerase-binding sites are still unknown. All known competence genes are transcribed from promoters with –35/–10 sequences recognized by the house-keeping sigma factor, sigma-A. Recently, Jarmer et al. (26) used a Hidden Markov Model to find potential sigma-A recognition sites in the B.subtilis genome. We used their list of putative –35/–10 promoter elements to examine whether only the active K-boxes are followed by putative RNA polymerase-binding sites. In only eight out of 26 cases a K-box was followed immediately, within three helical turns (32 bp), by a –35/–10 promoter sequence. Despite the low numbers, Figure 5 shows that the presence of a RNA polymerase-binding site as a selection criterion seems to improve the prediction of active K-boxes. The spiral curve of Figure 5 indicates the position of –35/–10 promoter elements on the DNA helix relative to their cognate K-boxes. Five of the putative active K-boxes are separated from a –35/–10 promoter sequence by 4–7 bp, corresponding to the situation in the known ComK-activated promoters. Of these known promoters only comF and comG were counted for in Figure 5. When the position on the DNA helix is taken into account, and the distance between K-box and –35/–10 promoter sequences is limited to two DNA helix turns, the reliability of a K-box as indicator for ComK-activated gene expression increases further. It should be mentioned that the latter selection step is rather stringent and discards valid ComK-activated promoters such as the promoter of comK itself. The distance between the K-box and –35/–10 sequences of this promoter is 23 bp. However, the regulation of the comK promoter is very complex and concerns at least five different transcription factors [(3,27) and L. Hamoen, unpublished results]. Possibly, ComK uses a different mechanism of activation in this case.
Figure 5.
Presence of putative –35/–10 promoter elements. The spiral curve shows face and distance of putative –35/–10 promoter elements on the DNA helix relative to the corresponding K-box. Open circles represent –35/–10 promoter elements related to K-boxes of inactive genes or operons (‘Inactive’ in tables), and closed rhombuses represent –35/–10 promoter elements related to K-boxes of ComK-activated genes or operons (‘Active’ in tables). The tables indicate the number of K-boxes after subsequent selection steps. K-boxes are distributed based on homology to the consensus sequence (13, 14 or 15 bp match), and whether they relate to unregulated (Inactive) or ComK-activated (Active) genes or operons. The fraction of active K-boxes is presented in the right column of each table. Table 1 displays the number of K-boxes present at a maximum distance of 200 bp upstream gene or operon (putative promoter region). Table 2 displays the number of K-boxes containing a downstream located –35/–10 promoter elements at a maximum distance of 32 bp. Table III presents K-boxes with –35/–10 promoter elements located at the same face of the DNA helix like the known ComK-activated promoters (distances allowed between K-box and –35/–10 promoter element; 4–7 and 14–18 bp).
Although caution is in place since the percentages in Figures 4 and 5 are based on small numbers, the above results strongly suggest that substantial improvement of the predictive value of transcription factor binding sites can be obtained by consideration of structural features, such as additional binding sites and RNA polymerase-recognition sequences.
Expression differences and K-boxes
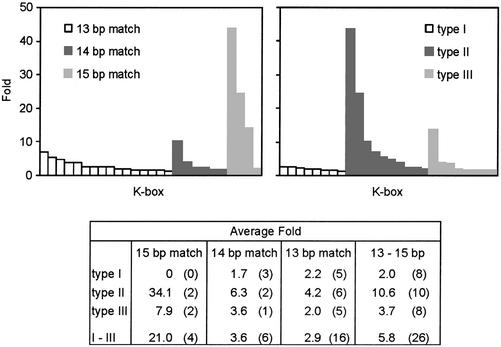
An interesting aspect not dealt with so far is whether a relationship exists between K-box type and the magnitude of expression differences. In order to investigate this, we plotted the fold expression differences against base pair match or type of K-box in the ComK-activated promoters (Fig. 6). In case of putative operons, the fold-values were averaged over the genes. The bar diagrams in Figure 6 show a large dispersion in expression differences measured. Despite this dispersion, the trends emerging from the table are also observed in the bar diagrams. Increasing deviations from the consensus sequence seem to reduce the measure of activation. This is conceivable assuming that such deviations lead to a reduced binding affinity for ComK. However, the K-box type appears to be important as well. Type II boxes, with a spacing between the ComK dimers of two DNA helix turns, show on average the highest activation. It was shown that this K-box type bends DNA about 70°, and it was speculated that such bending facilitates the wrapping of DNA around RNA polymerase so that the binding to the promoter is enhanced (6,28). Possibly, the spacing between ComK dimers in type I and type III boxes leads to non-ideal DNA bending angles for the wrapping of DNA around RNA polymerase. Further research into the mechanism of ComK activation will be required to validate this supposition.
Figure 6.
Relationship between expression levels and K-box types. The left bar diagram displays fold differences in expression when K-boxes were sorted on base pair match, and the right bar diagram displays fold differences in expression when K-boxes were sorted on type. Only putative active K-boxes present at a maximum distance of 200 bp upstream gene or operon are considered (26 in total). In the case of operons, folds were averaged over the activated genes. The table shows the average fold expression differences. The number of putative active K-boxes on which the calculations were based is placed in brackets.
DISCUSSION
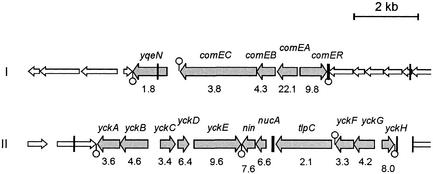
During the analysis of the transcriptome data we encountered a potentially important problem that is generally disregarded. This is best illustrated by transcription of the comE operon. In Figure 7, a schematic representation of the comE locus is shown together with the observed expression differences. The transcriptional start of the comE operon is located immediately downstream of comER, close to the K-box indicated by the vertical black line. The DNA-array experiments indicated that the expression of comER is also ComK dependent, yet this gene is transcribed in the opposite direction of the comE operon and previous studies have irrefutably shown that comER is not expressed during competence (29). This discrepancy arises from the fact that DNA-array filters are generally spotted with double-stranded DNA amplicons, due to which no discrimination can be made between sense and anti-sense RNA. This leads to false positives such as comER. It is likely that this problem is not limited to the comE operon, as is illustrated in Figure 7 by the nucA locus. ComK-dependent expression of the nucA-nin transcription unit has been documented, yet the observed ComK-dependent expression of yckC, -D and -E might be a consequence of read-through transcription from the nucA promoter (30). To what extent detection of anti-sense RNA influences the overall transcriptome data we do not know. Future use of DNA-arrays spotted with single-stranded DNA probes instead of double-stranded probes can overcome this problem.
Figure 7.
Anti-sense RNA detection in the comE and nucA loci. The organization of the comE and nucA loci in the genome are schematically depicted with ComK-activated genes in grey. Genes are drawn to scale, and the position of putative terminators and K-boxes are indicated by circles and bars, respectively. The numbers below the genes indicate the fold differences in expression.
In this study we have focused on the up-regulated genes, and we did not discuss possible negative regulation by ComK for two reasons. So far, direct transcriptional repression by ComK has not been demonstrated. Secondly, development of competence is limited to a subpopulation of B.subtilis cells. Even when all environmental conditions are optimal only about 10% of the cells express ComK and become competent (1). Due to this heterogeneity, possible transcriptional repression by ComK will be masked by the presence of a majority of non-competent cells. In a recent microarray study Berka et al. (23) bypassed this problem, by using a B.subtilis strain containing a mecA deletion. This strain produces high levels of ComK in all cells of the population. They found that only a small number of genes was downregulated by ComK in this ComK-overproducing strain, supporting the notion that ComK is a transcriptional activator. As expected, in our experimental set-up we found no genes with a strong ComK-dependent reduction in expression. Only six genes appeared to be repressed significantly when the flgM standards were applied, however the differences in expression were no more than –1.4 to –1.7 fold (see Table S2 of Supplementary Material).
We compared our list of activated genes with two recently published DNA-array studies (23,24). Of the genes identified in our study, 68% were picked up in both or in one of the two studies (indicated in Table 2). Aside from wild type B.subtilis, Berka et al. (23) analyzed ComK-dependent expression in a mecA-knockout as well. Of the ComK-activated genes in Table 2, 63 were also activated in this ComK-overproducing strain. However, when they compared the expression profiles of wild type B.subtilis with that of a comK-knockout strain, they identified only 39 of the genes listed in Table 2. This list also shows that 38 genes are in common between our study and the ComK-regulon defined by Ogura et al. The core ComK-regulon, defined here as the genes identified in all three studies, consists of the comE-, comF-, comG- and nucA-operons, comK, comC, ywfM, ywpH, maf, radC (belonging to the putative maf operon), yyaF, smf, cwiJ, yvyF (belonging to the putative comF operon), yvrP, yhzC, ybdK, sacX, recA and rapH and represents 30% of all genes identified in our study. It should be mentioned that this core ComK-regulon does not encompass all genes required for competence. For example, the essential bdbC, -D genes were not found by Ogura et al. (24).Two transcriptional units that are known to be regulated by ComK, uvrB and addAB, are absent in all three studies.
A considerable number of genes, 34 of the 105 genes in Table 2, have not been identified by the other two studies, and also the ComK-regulons of Berka et al. (23) and the one of Ogura et al. (24), encompassing a total of 165 and 61 genes, respectively, differ substantially from each other. There are many factors that can contribute to these differences. First of all, the experimental set-ups differed considerably. Not only the growth conditions and subsequent steps varied between the three studies, but also the type of DNA-arrays used. In addition, all three groups used different derivatives of B.subtilis strain 168 for their experiments, which exhibit slight differences at a genetical level (10,23,24). Furthermore, the criteria used to define regulated genes are, by their very nature, an important cause of deviations between different array studies. As discussed before, we preferred a more statistical approach to classify our data (see also Supplementary Material). Since both Berka et al. (23) and Ogura et al. (24) used different selection criteria to distil the ComK-regulon from their expression profiles, this will account for some of the differences found.
In many transcription-profiling studies regulation of gene transcription is interrelated with the presence of a certain transcription factor-binding site. However, these studies usually do not show how many genes with a putative transcription factor-binding site are not regulated. This makes it impossible to assess whether the presence of the transcription factor-binding site can be used to predict gene regulation. We strongly advocate that this data be included in future publications. In our study we found that a substantial fraction of the ComK-activated genes contain a ComK-binding site in their putative promoter region, yet the majority of K-boxes were located upstream of unregulated genes. Therefore, prediction of ComK regulation based on the presence of a K-box seems to be rather inaccurate: only when the ComK-binding site is highly homologous to the consensus sequence, with just a single base pair deviation (15 bp match), the chance of ComK regulation is >50%. With 2 bp deviations from the consensus sequence (14 bp match) the chance of regulation drops below 15%, and with 3 bp deviations (13 bp match) the chance is a mere 5%. These results stress that prediction of regulation solely based on the presence of a transcription factor-binding site in the putative promoter region of a gene can be rather inaccurate.
The observations above lead to the question why the predictive value of K-boxes is relatively low. Apparently, the presence of a ComK-binding site is insufficient and important additional information is required. Based on the close proximity of ComK- and RNA polymerase-binding sites it was suggested that ComK stimulates binding of RNA polymerase by interacting with the alpha-subunit of RNA polymerase (31). The alpha-subunit itself displays also some sequence-specific affinity and can bind to short AT-rich regions, so called upstream activating sequences or UP elements (32,33). Possibly, active K-boxes harbor UP elements within or close to the AT-box motifs, in order to properly activate RNA polymerase. More detailed knowledge on the activation mechanism of ComK will indicate whether additional sequence motifs are involved in ComK activation. Aside from a limited knowledge of the ComK-activation mechanism, the lack of information on promoter location and operon structure may be hampering the prediction even more. We applied an algorithm, based on predicted rho-independent terminator sequences, transcription direction and K-box positions, to define an operon, and already such a simple algorithm improved the predictive value of K-boxes. However, this algorithm is far from complete. One of the difficulties in operon prediction is the exact determination of the transcriptional start site. Jarmer et al. (26) showed that a Hidden Markov algorithm can be used to identify putative –35/–10 promoter elements; yet, despite their successful approach, several known promoters (such as the comC promoter) remained undetected. To be able to perform reliable in silico analyses of gene regulation in the future, it will be essential to obtain more knowledge on such basic processes as transcriptional initiation and transcriptional termination.
Finally, the finding of many activated genes that do have a K-box, but have no established link with competence is rather puzzling. Several explanations are conceivable. First of all it might be that these genes represent processes which are in fact relevant to competence, but which have not yet been characterized. Secondly, it is possible that the expression of these genes represents a distinct physiological state of which competence is only one aspect, and that the genes are for that reason concomitantly transcribed. This hypothesis led Berka et al. (23) to rename competence to K-state. However, there is the possibility that the regulation of these genes by ComK might have no biological significance at all, but rather can be considered as ‘evolutionary noise’. The genome of B.subtilis harbors over 2300 K-boxes that match only 12 bp of the consensus sequences. A single point-mutation can upgrade these boxes to ‘valid’ sites for ComK-binding. Since a large fraction of these 12 bp match K-boxes are located in intergenic regions, it is reasonable to assume that in some cases this will influence the activity of a promoter, making it ComK regulated. As long as the increased expression of the gene driven by this mutated promoter does not substantially affect the viability of the cell, there will be no evolutionary pressure to nullify such a mutation. According to Figure 6, poorly matching K-boxes display low levels of activation, suggesting that an upgrade from a 12 to a 13 bp match K-box is likely to result in only a moderate level of activation by ComK. Especially since competence occurs only during a limited period of time, in a small fraction of the cells, and competent cells do not divide, it can be envisioned that an increased expression of certain genes will not easily impair the overall fitness. Along these lines one can assume that during evolution several (or many) ComK-activated genes have originated in B.subtilis, with no apparent relation to the competence process. This is what we seem to observe in this study. The situation for ComK can most likely be extended to many other transcription factors. Hence, we postulate that several (or many) genes identified by means of DNA-array experiments originate from ‘evolutionary noise’, i.e. gene regulation that does not serve a specific biological function, but arises from the random origination of transcription factor-binding sites in promoter regions during the process of evolution. Comparative genomics could provide a method to establish the true nature of transcription factor binding sites, since it is likely that biologically functional sites are conserved in related species. As mentioned before, the three transcriptome analyses of competent B.subtilis cells, which are now available, have been performed with three different derivatives of B.subtilis strain 168 (23,24). From Table 2 it can be deduced that the strongly activated genes are found in all three studies, but that the three independently defined ComK-regulons differ notably in the weakly activated genes. As mentioned earlier, these discrepancies might be a consequence of the different protocols used, yet it is tempting to assume that it could be a display of evolutionary noise. Comprehensive comparisons of regulons of related species will eventually indicate whether evolutionary noise is a phenomenon which should be reckoned with in the analyses of transcriptome data.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank D. Dubnau and H. Jarmer for communicating results prior to publication. We are grateful to S. Bron for help and critically reading the manuscript. This work was supported by the Groningen Biomolecular Science and Biotechnology Institute (GBB), University of Groningen.
REFERENCES
- 1.Dubnau D. (1993) Genetic exchange and homologous recombination. In Sonenshein,A.L., Hoch,J.A. and Losick,R. (eds), Bacillus subtilis and Other Gram-Positive Bacteria. American Society for Microbiology, Washington,D.C., pp. 555–584.
- 2.van-Sinderen D., Luttinger,A., Kong,L., Dubnau,D., Venema,G. and Hamoen,L. (1995) comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol., 15, 455–462. [DOI] [PubMed] [Google Scholar]
- 3.Hahn J., Luttinger,A. and Dubnau,D. (1996) Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol. Microbiol., 21, 763–775. [DOI] [PubMed] [Google Scholar]
- 4.Haijema B.J., Hamoen,L.W., Kooistra,J., Venema,G. and van-Sinderen,D. (1995) Expression of the ATP-dependent deoxyribonuclease of Bacillus subtilis is under competence-mediated control. Mol. Microbiol., 15, 203–211. [DOI] [PubMed] [Google Scholar]
- 5.Haijema B.J., van-Sinderen,D., Winterling,K., Kooistra,J., Venema,G. and Hamoen,L.W. (1996) Regulated expression of the dinR and recA genes during competence development and SOS induction in Bacillus subtilis. Mol. Microbiol., 22, 75–85. [DOI] [PubMed] [Google Scholar]
- 6.Hamoen L.W., Van-Werkhoven,A.F., Bijlsma,J.J., Dubnau,D. and Venema,G. (1998) The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev., 12, 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidham,J.G., Smith,J.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- 8.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Venema G., Pritchard,R.H. and Venema-Schroder,T. (1965) Fate of transforming deoxyribonucleic acid in Bacillus subtilis. J. Bacteriol., 89, 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bron S. and Venema,G. (1971) Ultraviolet inactivation and excision-repair in Bacillus subtilis. I. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat. Res., 15, 1–10. [DOI] [PubMed] [Google Scholar]
- 11.Guerout-Fleury A.M., Shazand,K., Frandsen,N. and Stragier,P. (1995) Antibiotic-resistance cassettes for Bacillus subtilis. Gene, 167, 335–336. [DOI] [PubMed] [Google Scholar]
- 12.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S. et al. (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 13.Scotti C., Piatti,M., Cuzzoni,A., Perani,P., Tognoni,A., Grandi,G., Galizzi,A. and Albertini,A.M. (1993) A Bacillus subtilis large ORF coding for a polypeptide highly similar to polyketide synthases. Gene, 130, 65–71. [DOI] [PubMed] [Google Scholar]
- 14.Bron S. (1990) Plasmids. In Harwood,C.R. and Cutting,S.M. (eds), Molecular Biological Methods for Bacillus. John Wiley & Sons Ltd, Chichester, UK, pp. 75–174.
- 15.van de Guchte M., Kok,J. and Venema,G. (1991) Distance-dependent translational coupling and interference in Lactococcus lactis. Mol. Gen. Genet., 227, 65–71. [DOI] [PubMed] [Google Scholar]
- 16.Schreiber J., Enderich,J. and Wegner,M. (1998) Structural requirements for DNA binding of GCM proteins. Nucleic Acids Res., 26, 2337–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuipers O.P., de Jong,A., Baerends,R.J.S., van Hijum,S.A.F.T., Zomer,A.L., Karsens,H.A., den Hengst,C.D., Kramer,N.E., Buist,G. and Kok,J. (2002) Transcriptome analysis and related databases of Lactococcus lactis. Antonie Van Leeuwenhoek, 82, 113–122. [PubMed] [Google Scholar]
- 18.Lane D., Prentki,P. and Chandler,M. (1992) Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol. Rev., 56, 509–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long A.D., Mangalam,H.J., Chan,B.Y., Tolleri,L., Hatfield,G.W. and Baldi,P. (2001) Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem., 276, 19937–19944. [DOI] [PubMed] [Google Scholar]
- 20.Baldi P. and Long,A.D. (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics., 17, 509–519. [DOI] [PubMed] [Google Scholar]
- 21.Liu J. and Zuber,P. (1998) A molecular switch controlling competence and motility: competence regulatory factors ComS, MecA and ComK control sigmaD-dependent gene expression in Bacillus subtilis. J. Bacteriol., 180, 4243–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meima R., Eschevins,C., Fillinger,S., Bolhuis,A., Hamoen,L.W., Dorenbos,R., Quax,W.J., van Dijl,J.M., Provvedi,R., Chen,I. et al. (2002) The bdbDC operon of Bacillus subtilis encodes thiol-disulphide oxidoreductases required for competence development. J. Biol. Chem., 277, 6994–7001. [DOI] [PubMed] [Google Scholar]
- 23.Berka R.M., Hahn,J., Albano,M., Draskovic,I., Persuh,M., Cui,X., Sloma,A., Widner,W. and Dubnau,D. (2002) Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol., 43, 1331–1345. [DOI] [PubMed] [Google Scholar]
- 24.Ogura M., Yamaguchi,H., Kobayashi,K., Ogasawara,N., Fujita,Y. and Tanaka,T. (2002) Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol., 184, 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamoen L.W., Haijema,B., Bijlsma,J.J., Venema,G. and Lovett,C.M. (2001) The Bacillus subtilis competence transcription factor, ComK, overrides LexA-imposed transcriptional inhibition without physically displacing LexA. J. Biol. Chem., 276, 42901–42907. [DOI] [PubMed] [Google Scholar]
- 26.Jarmer H., Larsen,T.S., Krogh,A., Saxild,H.H., Brunak,S. and Knudsen,S. (2001) Sigma A recognition sites in the Bacillus subtilis genome. Microbiology, 147, 2417–2424. [DOI] [PubMed] [Google Scholar]
- 27.Hoa T.T., Tortosa,P., Albano,M. and Dubnau,D. (2002) Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol., 43, 15–26. [DOI] [PubMed] [Google Scholar]
- 28.Coulombe B. and Burton,Z.F. (1999) DNA bending and wrapping around RNA polymerase: a ‘revolutionary’ model describing transcriptional mechanisms. Microbiol. Mol. Biol. Rev., 63, 457–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn J., Inamine,G., Kozlov,Y. and Dubnau,D. (1993) Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA. Mol. Microbiol., 10, 99–111. [DOI] [PubMed] [Google Scholar]
- 30.van Sinderen D., Kiewiet,R. and Venema,G. (1995) Differential expression of two closely related deoxyribonuclease genes, nucA and nucB, in Bacillus subtilis. Mol. Microbiol., 15, 213–223. [DOI] [PubMed] [Google Scholar]
- 31.Ptashne M. and Gann,A. (1997) Transcriptional activation by recruitment. Nature, 386, 569–577. [DOI] [PubMed] [Google Scholar]
- 32.Estrem S.T., Ross,W., Gaal,T., Chen,Z.W., Niu,W., Ebright,R.H. and Gourse,R.L. (1999) Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev., 13, 2134–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fredrick K. and Helmann,J.D. (1997) RNA polymerase sigma factor determines start-site selection but is not required for upstream promoter element activation on heteroduplex (bubble) templates. Proc. Natl Acad. Sci. USA, 94, 4982–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubnau D. (1997) Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins—a review. Gene, 192, 191–198. [DOI] [PubMed] [Google Scholar]
- 35.Provvedi R., Chen,I. and Dubnau,D. (2001) NucA is required for DNA cleavage during transformation of Bacillus subtilis. Mol. Microbiol., 40, 634–644. [DOI] [PubMed] [Google Scholar]
- 36.Londono-Vallejo J.A. and Dubnau,D. (1993) comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases. Mol. Microbiol., 9, 119–131. [DOI] [PubMed] [Google Scholar]
- 37.van-Sinderen D., ten-Berge,A., Hayema,B.J., Hamoen,L. and Venema,G. (1994) Molecular cloning and sequence of comK, a gene required for genetic competence in Bacillus subtilis. Mol. Microbiol., 11, 695–703. [DOI] [PubMed] [Google Scholar]
- 38.Lovett C.M., Love,P.E. and Yasbin,R.E. (1989) Competence-specific induction of the Bacillus subtilis RecA protein analog: evidence for dual regulation of a recombination protein. J. Bacteriol., 171, 2318–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogura M., Ohshiro,Y., Hirao,S. and Tanaka,T. (1997) A new Bacillus subtilis gene, med, encodes a positive regulator of comK. J. Bacteriol., 179, 6244–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogura M. and Tanaka,T. (2000) Bacillus subtilis comZ (yjzA) negatively affects expression of comG but not comK. J. Bacteriol., 182, 4992–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.