Abstract
Non-homologous insertion (NHI) of DNA fragments into genomic DNA is a method widely used in insertional mutagenesis screens. In the yeast Saccharomyces cerevisiae, the efficiency of NHI is very low. Here we report that its efficiency can be increased by γ-irradiation of recipient cells at the time of transformation. Radiation-assisted NHI depends on YKU70, but its efficiency is not improved by inactivation of RAD5 or RAD52. In a pilot study, we generated 102 transformant clones expressing a lacZ reporter gene under standard conditions (30°C, rich medium). The site of insertion was determined in a subset of eight clones in which lacZ expression was altered by UV-irradiation. A comparison with published data revealed that three of the eight genes identified in our screen have not been targeted by large-scale transposon-based insertion screens. This suggests that radiation-assisted NHI offers a more homogeneous coverage of the genome than methods relying on transposons or retroviral elements.
INTRODUCTION
Random insertional mutagenesis is a powerful tool in the analysis of gene function that is amenable to genome-wide analyses and that has been applied in a variety of organisms (1–4). In addition to studying the phenotype associated with disruption of the target sequence, by insertion of cassettes containing appropriate elements it is also possible to conduct gene expression analyses and to insert tag sequences that can be used for protein isolation or localisation studies. Insertion cassettes are often derived from mobile DNA elements, such as retroviruses or transposons, and efficient insertion into genomic DNA is then mediated by the respective mobilising apparatus (e.g. integrases or transposases). As most mobile elements exhibit a certain degree of target site specificity, uniform coverage of all genes in the genome may be difficult to achieve with this approach. For example, by insertional mutagenesis using Tn3-derived mini-transposons a large collection of Saccharomyces cerevisiae mutant clones has been established (5,6). Although, until the end of the year 2001, the site of insertion has been characterised in more than 22 000 insertion clones, less than two-thirds of the about 6200 yeast genes are represented in this collection (7). In addition to gene-size dependent biases in targeting efficiency, non-random insertion of Tn3-derived transposons (8) and unequal representation of genes in the yeast library mutagenised may account for this effect.
To achieve complete coverage of the genome, the introduction of complementary approaches is necessary. One such approach is non-homologous insertion (NHI; 9) of DNA cassettes not derived from mobile elements, a process which appears to exploit the cellular machinery that normally repairs DNA double-strand breaks (DSB) by non-homologous end joining (NHEJ; 10). Successful application of NHI strategies in large-scale insertional mutagenesis projects has, for example, been described in mammalian cells, plants and Schizosaccharomyces pombe (11–13). Similar approaches in S.cerevisiae have been precluded by the low efficiency of NHI in this organism (14).
We sought to establish an efficient NHI-based method for random insertion of a promoter-trap cassette containing a lacZ reporter gene in S.cerevisiae. Previous work demonstrated that NHI efficiency increases upon co-transformation of restriction endonucleases (14–16). Restriction-enzyme-mediated integration (REMI) was subsequently used for insertion mutagenesis in a variety of organisms (17). To avoid a sequence bias towards genomic recognition sites of the co-transformed endonuclease, we here investigate whether γ-irradiation is suitable for facilitating NHI. Within the yeast genome, DSB induced by sparsely ionising radiation appear to be distributed randomly (18), although small-scale variations due to radical shielding by DNA-associated proteins cannot be excluded. Irradiation should therefore result in a largely random distribution of insertion sites. Here, we report that γ-irradiation shortly before or after the heat-shock step of transformation enhances the rate of NHI sufficiently to allow its application in a promoter-trap scheme. To demonstrate proof-of-principle, we analysed 2000 NHI transformants for lacZ expression. Among the 102 lacZ expressing clones eight transformants were isolated that exhibited altered lacZ expression after UV-irradiation. Interestingly, three of the eight insertion sites were located in genes that so far have not been found hit in the more than 22 000 clones obtained in the large-scale transposon-based mutagenesis screen (5–7). Thus, radiation-assisted NHI of linearised plasmids may be suitable as a complementary approach in order to allow full coverage of the S.cerevisiae genome.
MATERIALS AND METHODS
For optimising radiation-assisted NHI, plasmids pM150 and pM151 (16) were used. These pUC18 derivatives contain the URA3 gene and differ in the restriction sites present in the MCS. For constructing plasmid pFA6-kanMX6-lacZ, a 3.1 kb BamHI/SalI fragment containing the lacZ gene was isolated from plasmid pCM159 (19) and inserted in BamHI/SalI-digested plasmid pFA6-kanMX6 (20). Plasmid MKM20 was generated by PCR amplification of the lacZ gene, using pFA6-kanMX6-lacZ as a template, with primers lacZ-C-BamHI (5′-CGA ATT CGG ATC CAG CTG AAG CTT CGT ACG-3′) and lacZ-N-BglII (5′-CGA ATT CAG ATC TAC TGG CCG TCG TTT TAC-3′), and insertion of the BamHI/BglII-digested product into the BglII site of pM151. The lacZ allele thus obtained lacks the first 20 nucleotides, including the start codon, and lacZ expression is obtained only upon in-frame insertion of the BglII-linearised plasmid MKM20 into expressed genes.
NHI experiments were performed in haploid strain RSY12 (MATa leu2-3,112 his3-11,15 ura3Δ::HIS3), in which the entire URA3 open reading frame was replaced by the HIS3 gene (14). Strain RSY12 rad5 was generated by transformation of a rad5Δ::kanMX-lacZ deletion cassette generated by PCR, using primers rad5-kanMX-C (5′-CTA TTC AAA CAG CAT CTG GAT TTC TTC AAT TCT CCT TTT TCT GGG TCA CCC GGC CAG CG-3′) and rad5-lacZ-N (5′-ATG AGT CAT ATT GAA GAA AGG AAG TTT TTT AAC GAT CCC GTC GTT TTA CAA CG-3′) on template pAF6-kanMX6-lacZ. Generation of strains RSY12 yku70 and RSY12 rad52 has been described earlier (15,21).
Yeast cells were transformed using a high efficiency method (22). For transformation, plasmids pM151 or MKM20 were digested to completion with BglII, extracted with phenol, precipitated and dissolved in sterile water. Control transformations, using the same batch of cells, were performed with circular plasmid YEplac195 (23). For radiation-assisted NHI, transformation reaction mixes were γ-irradiated at room temperature, using a 60Co-γ-source (Atomic Energy of Canada, Ltd) at a dose rate of 10 Gy min–1. If not stated otherwise, irradiation took place immediately after the heat-shock step of the transformation procedure. Relative transformation frequencies were determined by normalising the rate of transformation of the linearised plasmid (per microgram of DNA) relative to the rate of transformation with the control plasmid.
Integrated plasmids and adjacent genomic sequences were recovered by plasmid rescue as described (21), except that several restriction enzymes that do not cut within the plasmid sequence (BamHI, BspEI, NheI and XhoI) were used in parallel digestions of each DNA sample, in order to enhance the efficiency of plasmid rescue. Plasmids with flanking chromosomal DNA, as identified by migration behaviour in agarose gels, were sequenced by Medigenomix GmbH (Martinsried, Germany) using sequencing primers P1 (5′-AGC GGA TAA CAA TTT CAC ACA GGA-3′) and P2 (5′-ACT CCA GCC AGC TTT CC-3′). Sequencing data were analysed by a BLASTN search on the Saccharomyces Genome Database (SGD; http://genome-www.stanford.edu/Saccharomyces/).
To identify lacZ-expressing transformants, patches of cells on selection plates were covered with ∼10 ml chloroform and incubated for 5 min to permeabilize cells. After removing the chloroform, plates were air-dried for 5 min, before they were covered with X-Gal suspension (1 mg/ml X-Gal in 1% LMP-agarose solution, dissolved at 60°C). After solidification of the agarose, plates were incubated at 30°C up to 24 h to allow for colour development. To quantify UV-induced lacZ expression, cells were grown to logarithmic phase (5 × 106 cells/ml), harvested by centrifugation and re-suspended at a titre of 1 × 108 cells/ml in liquid holding recovery (LHR) buffer (67 mM K2HPO4/KH2PO4, 100 mM glucose, pH 5). Aliquots of 2 ml were UV-irradiated (254 nm) in the dark in plastic Petri dishes under constant stirring. Post-irradiation incubation of irradiated suspensions was performed in the dark at 30°C under constant shaking. At given time points, 1 ml aliquots were removed, and after centrifugation cells were re-suspended in 0.5 ml Z-buffer (75 mM Na2HPO4, 50 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) and immediately frozen at –80°C, where they were kept until detection. For detection, all samples of an experimental series were thawed simultaneously and each 50 µl of the suspensions were used to determine the OD600. The remaining cells were incubated for 1 h at 37°C with 100 µl zymolyase solution (zymolyase T100, Seikagaku Corp., Japan; 0.5 mg in 1 ml Z-buffer). After completion of lysis, 100 µl of CPRG solution (4 mg/ml Chloro-Phenol-Red-Galactopyranosid in Z-buffer) were added and incubated at room temperature. Incubation times (tinc) varied between 1 and 180 min, according to the different levels of lacZ expression. Reactions were stopped by adding 200 µl 1 M Na2CO3. After 15 min centrifugation at 15 000 r.p.m., the optical density of the supernatant was measured at 574 nm and 634 nm. The β-galactosidase activity in Miller units was calculated as: [MU] = [(OD574 – OD634) × 1000] / (OD600 × tinc). Semi-quantitative screening of lacZ expression was performed similarly, except that cell titres at the time of irradiation were not determined.
RESULTS
The frequency of NHI events increases upon irradiation
We investigated the influence of irradiation on the frequency of NHI of BglII-linearised plasmid pM151 (16), which contains the yeast URA3 gene, into the genome of strain RSY12, in which the URA3 locus had been deleted (14). The frequency of transformation with plasmid pM151 was normalised with respect to parallel control transformations with a circular episomal plasmid. γ-Irradiation of cells immediately after the heat-shock step of the transformation procedure caused an increase in NHI frequency of up to ∼15-fold (Fig. 1A). Thus, about 150 transformants can be obtained in one transformation experiment, using 7 µg of linearised plasmid. We refer to this method as radiation-assisted NHI. Comparable effects were seen when cells were irradiated immediately before the heat shock, while the effect declined with increasing duration of the period between irradiation and transformation, until, with periods exceeding 4 h, no positive effect of irradiation could be seen anymore (data not shown). Additional experiments using plasmids linearised with other enzymes showed that radiation-assisted NHI is more pronounced with fragments terminating in single-stranded overhangs than with plasmids terminating in blunt ends (data not shown).
Figure 1.
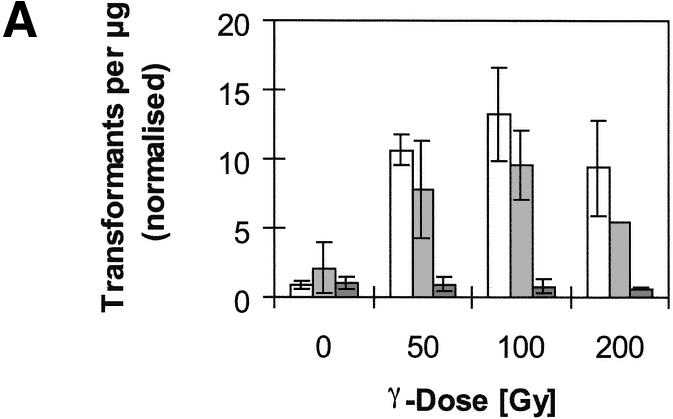
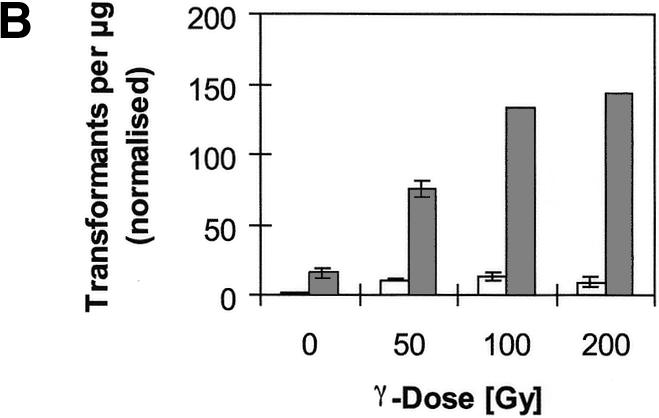
Frequency of transformation with linearised plasmid pM151 when cells were treated with γ-irradiation immediately after the heat-shock step of the transformation procedure. The number of transformants per microgram plasmid DNA was normalised with respect to parallel control transformations with circular plasmid YEplac195. (A) Comparison of wild-type strain RSY12 (white) and its rad5 (light grey) and yku70 (dark grey) mutant derivatives. Indicated are means and standard deviations from three independent experiments (except RSY12 rad5, 200 Gy: one experiment). (B) Comparison of RSY12 [white; same data as in (A)] and its rad52 (grey) mutant derivative. Note that the scale of the y-axis has changed. Data for RSY12 rad52 are from two experiments (0 and 50 Gy) or one experiment (100 and 200 Gy).
An increase of NHI frequency with irradiation is not seen in Ku70-deficient mutants [Fig. 1A; (21)], demonstrating that the insertion of transformed fragments into broken chromosomal DNA involves NHEJ. In general, NHEJ contributes little to the repair of radiation-induced chromosomal breaks in yeast (10). We hypothesised that the efficiency of radiation-induced NHI may be increased in mutant strains that exhibit more potent NHEJ. For example, the Rad5 protein, inactivation of which confers moderate radiosensitivity, has been invoked in the regulation of DSB repair pathways in S.cerevisiae. While wild-type cells repair plasmids gapped within a sequence that is also present on a chromosome almost exclusively by homologous recombination with the chromosomal donor sequence, rad5 mutants rely predominantly on NHEJ-type repair of these plasmids (24). In contrast to its influence in plasmid repair, however, the rad5 mutation does not appear to have a role in NHI, as, both with and without irradiation, the relative transformation efficiency was not significantly altered in rad5 mutants as compared with the wild-type strain (Fig.1A).
It has been proposed that NHEJ processes and homologous recombination competes for repair of DNA DSB and that the decision of which pathway is used is, at least in part, determined by the proteins initially binding to the DNA ends (25). Thus, by inactivation of Rad52, a DNA end-binding protein essential for homologous recombination, more ends may become accessible to Ku proteins and the relative frequency of NHEJ events may increase. Indeed, as compared with wild-type cells, we observe a strong increase in the relative frequency of NHI events in rad52 mutants both with and without irradiation (Fig. 1B). However, since the control transformation with circular plasmids is about 50 times less efficient in the rad52 mutant than in wild-type [(4.2 + –3.6) × 102 versus (1.9 + –0.5) × 104 transformants/µg plasmid DNA in rad52 mutants and wild-type, respectively], the absolute number of transformants is reduced in the mutant. Reduced transformation with circular plasmids in rad52 mutant strains has been observed previously (15). In addition, the mutant is much more sensitive to the cell killing effects of radiation (survival of 14 versus 45% at 100 Gy), further reducing the number of transformants obtainable.
Application of radiation-assisted NHI in a promoter-trapping scheme
The bacterial β-galactosidase gene lacZ is a widely used reporter gene, the expression of which can easily be monitored qualitatively and quantitatively (26). Insertion of a BglII/BamHI-digested PCR fragment encompassing the lacZ gene into the BglII site of pM151 created plasmid MKM20. The lacZ allele used in our studies lacks the first 25 nucleotides including the start codon. By targeted in-frame insertion into the chromosomal RAD54 gene, a gene well known for its UV inducibility (27), we verified the suitability of this allele to serve as a reporter gene in analysis of UV-induced gene induction (data not shown).
To use for promoter trapping, plasmid MKM20 was digested with BglII and transformed into strain RSY12 using a high efficiency transformation protocol. Immediately after the heat-shock step, the cells were irradiated with 50 Gy. A dose of 50 Gy was chosen to achieve both sufficient cell survival (∼60%) and high efficiency of NHI. At this dose, on average 2 DSB/cell are induced (18). Transformation efficiency with plasmid MKM20 was comparable with that of pM151. A total of about 2000 Ura+ transformants were generated and tested by X-Gal overlay for in-frame insertion of MKM20 into yeast genes expressed under standard growth conditions (YPD, 30°C). Detectable expression of the inserted lacZ gene was found in 102 transformants (5%). To identify those transformants in which the reporter gene expression was affected by UV-irradiation, a semi-quantitative screen was conducted. Cells grown to logarithmic growth phase were re-suspended in non-growth buffer, half of the sample was UV-irradiated (60 J/m2) and the other half served as mock-treated control. The non-growth condition was chosen so that changes in gene expression are not due to any secondary effect of radiation-induced cell cycle arrest. Cells were lysed 2 h after treatment and β-galactosidase activity was determined using a colourimetric assay. Out of 102 lacZ-expressing clones, 12 candidates differing at least 2-fold in expression in treated versus untreated cells were identified and chosen for further analysis. In eight of these, the site of insertion of the promoter-trapping construct was successfully determined by plasmid rescue (see Table 1). In the remaining clones, plasmid rescue and/or sequencing were not successful (three cases) or the construct was found to be fused to a fragment from the 2 µm plasmid which obviously enabled autonomous replication (one case).
Table 1. Insertion of the lacZ reporter relative to the trapped gene in clones displaying differential expression in a semi-quantitative screen after UV-irradiation.
| Clone ID | Gene | Insertion of lacZ N-terminus |
|---|---|---|
| MK12 | APG1 | In-frame within ORF |
| MK33 | FIR1 | In-frame within ORF |
| MK35 | MRP8 | In-frame within ORF |
| MK38 | SDS24 | In-frame within ORF |
| MK42 | UBC13 | In-frame in intron |
| MK60 | NVJ1 | In-frame within ORF |
| MK66 | BUD4 | 22 bp upstream |
| MK82 | YBR052c | In-frame within ORF |
In six of the eight events indicated in Table 1, insertion of the reporter construct occurred in-frame within a coding region. In clone 42, insertion occurred in-frame within the intron of the UBC13 gene, 46 bp upstream of the branch-point (28). In clone 66, insertion occurred 22 bp upstream of the BUD4 initiator codon. The next in-frame ATG codon is located at position –165 (as counted from the BUD4 initiator codon); since translational frameshift events occur readily in yeast (6), alternatively an out-of-frame ATG at position –36 may have been used to allow expression of the reporter gene. To investigate whether insertion events differ from events observed spontaneously, we sequenced both insertion junctions (Fig. 2). Similar to events seen in spontaneous NHI (9), in the majority (6/8) of the insertion events at least one junction apparently was facilitated by microhomology between plasmid end and genomic sequence. In five of the eight integrations, no or only one nucleotide of the genomic sequence was lost. Only one event (clone 60) was associated with a large genomic rearrangement (inversion or duplication).
Figure 2.
Junctions between plasmid MKM20 and genomic DNA, as determined by plasmid rescue and sequencing with primers P1 and P2. Remnants of the single-stranded overhangs present in the transformed plasmid after restriction digest are indicated in bold. For clarification, the first nucleotides of genomic sequence at the junction sites are labelled by shading. Regions of microhomology at the junctions are underlined; nucleotides that are inserted after plasmid integration are framed. Numbers in the chromosomal sequences refer to chromosomal positions as indicated in the SGD database. Note that in clone MK66, 16 nt of plasmid sequence were absent at the P1 junction.
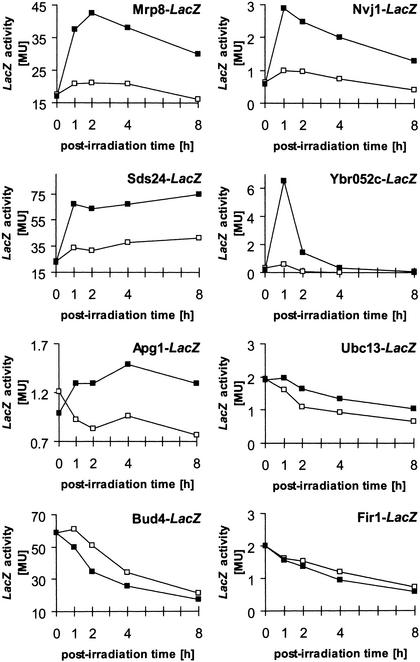
An analysis of the kinetics of expression of the lacZ-fusion proteins in response to UV irradiation with 60 J/m2 verified strong induction (>2-fold) for fusions involving MRP8, SDS24, NVJ1 and YBR052c, and 1.5-fold induction for APG1 (Fig. 3). These genes have been found by microarray analyses to be generally stress-inducible (29–31). For the fusion involving UBC13, β-galactosidase activity in UV-irradiated and mock-treated samples differed by >1.5-fold, but instead of showing a clear induction, the treated samples rather exhibited a slower decline of β-galactosidase activity during the post-treatment incubation than the mock-treated samples (Fig. 3). In microarray and northern analyses, however, increased amount of UBC13 transcripts after DNA damage have been described (29–33). We assume that the different conditions during post-treatment incubation (non-growth versus growth conditions) are responsible for the discrepancies. A slightly accelerated decline of β-galactosidase activity in treated versus untreated sample was observed during post-treatment incubation in a clone involving a fusion with BUD4, a gene which by others was described as repressed in response to DNA damage due to cell cycle arrest (30). In contrast to the results of the semi-quantitative analysis, no significant difference between treated and untreated samples was observed concerning the β-galactosidase activity of FIR1-lacZ fusions when assaying the kinetics of expression.
Figure 3.
LacZ expression in samples treated with 60 J/m2 UV radiation (filled squares) and in mock-treated samples (open squares) as a function of post-irradiation incubation under non-growth conditions. LacZ activity is indicated in Miller units.
Of the genes affected by insertion, only UBC13 is known to be involved in response to DNA damage. To test whether the other genes have an as yet uncharacterised function in damage resistance, UV and γ-ray sensitivity was tested for all insertion clones. None of the clones (except for the ubc13 mutant) exhibited increased sensitivity (data not shown), confirming a recent report that only a very small proportion of the genes whose expression is altered after DNA damaging treatments actually is involved in conferring resistance to these treatments (34).
DISCUSSION
The method used to insert a mutagenising fragment into the genome may be a critical factor for successful applica tion of insertional mutagenesis screens. Use of mobile element-derived mechanisms is highly efficient, but often accompanied by a target bias that precludes uniform coverage of all genomic loci. The alternative method, random insertion of fragments without using mobilising machinery, is still poorly characterised in terms of mechanistic details. Recent evidence supports the notion that NHI is mediated by components of the NHEJ pathway of DSB repair (15,21,35; this work). Here we demonstrate that radiation-assisted NHI, similar to REMI (9), results in an increased yield of transformants. Optimal enhancement of NHI was seen when irradiation took place around the time of the heat-shock step in transformation, i.e. the time when the transformed DNA enters the cell. Similar to spontaneous and restriction enzyme-mediated NHI, radiation-assisted NHI works better on fragments terminating in single-stranded overhangs than in blunt-ended fragments. Also, the junction sites observed in radiation-assisted NHI were very similar to those observed after spontaneous or enzyme-mediated NHI in their dependence on microhomologies and generation of small deletions or insertions. Whether large genomic alterations (as seen in clone 60) occur more often in radiation-assisted NHI than in spontaneous NHI (13) remains to be tested. Our attempts to manipulate the regulatory system of DSB repair by inactivating RAD5 or RAD52 did not improve radiation-assisted NHI. In line with our results, it has recently been found that impairment of homologous recombination does not enhance NHEJ efficiency in yeast (36). Whether, as suggested by the data of these authors, the frequency of radiation-assisted NHI is increased in G1/G0 cell populations as compared with logarithmically growing populations, remains to be tested.
Expression of the lacZ reporter gene under standard growth conditions (30°C, YPD) was observed in 5% of our transformants. This is about the frequency expected, considering the proportion of coding DNA in S.cerevisiae [about 0.7; (37)], the probability of in-frame insertion in the correct orientation (0.33 × 0.5), the frequency of essential genes [0.19; (38)] and the fact that expression of some genes under standard conditions may be too low to allow detection. Among the clones expressing lacZ, ∼8% were found to increase expression upon UV-irradiation. Considering the experimental differences, this value compares well with results obtained in expression studies using microarrays (29–31,34). Our investigation differs from these studies in that we performed post-irradiation incubation of treated samples and mock-treated controls under non-growth conditions. In addition, depending on the site of insertion of the reporter construct, the amount of lacZ-fusion proteins may not only reflect promoter activity, but, at least in part, also post-transcriptional and post-translational regulation of expression. It is known that in many cases protein levels do not correlate with mRNA abundance (39).
The most interesting aspect of this pilot study is that three of the eight trapped genes identified (FIR1, BUD4, MRP8) have not yet been targeted in a large-scale transposon-based insertional mutagenesis screen in spite of the impressive number of more than 22 000 insertion events analysed so far (5–7). We infer this from the fact that these genes are not listed in the TRIPLES database [http://ygac.med.yale.edu/triples/triples.htm; (7)], which describes all insertional mutagenesis events generated by the transposon-based approach. Another two of the genes trapped in our study (NVJ1, UBC13) have been targeted only once by transposon-based trapping. In contrast, many other genes, including APG1, SDS24 and YBR052, have by now been hit multiple times. We conclude that by radiation-assisted NHI a more complete coverage of the genome can be obtained and that this method is suitable to complement any existing large-scale mutagenesis screen.
A major concern may, however, arise from using a clastogenic agent that has a potential of introducing additional genetic and genomic alterations. At present, we cannot estimate how often the analysis of insertion-associated phenotypes will suffer from artefacts caused by radiation-induced additional alterations. It should be noted, however, that good experimental practice requires that in any insertional mutagenesis screen, including transposon-based screens, phenotypes be carefully checked for segregation with given insertion alleles.
We propose that radiation-assisted NHI is a suitable means to complement existing large-scale studies in functional genomics in S.cerevisiae and any other organisms that exhibits a low-efficiency NHEJ system. It would, of course, be desirable to use multi-purpose fragments in transformation that allow, after insertion, to simultaneously investigate a variety of endpoints.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge expert technical assistance by K. Winkler, financial support by Deutsche Forschungsge meinschaft (KI 796/1) and European Commission (FIS5-1999-00066), and travel grants by the Neuherberger Forschungsförderung.
REFERENCES
- 1.Parinov S. and Sundaresan,V. (2000) Functional genomics in Arabidospis: large-scale insertional mutagenesis complements the genome sequencing project. Curr. Opin. Biotech., 11, 157–161. [DOI] [PubMed] [Google Scholar]
- 2.Stanford W.L., Cohn,J.B. and Cordes,S.P. (2001) Gene-trap mutagenesis: past, present and beyond. Nature Rev. Genet., 2, 756–768. [DOI] [PubMed] [Google Scholar]
- 3.Vidan S. and Snyder,M. (2001) Large-scale mutagenesis: yeast genetics in the genome era. Curr. Opin. Biotech., 12, 28–34. [DOI] [PubMed] [Google Scholar]
- 4.Golling G., Amsterdam,A., Sun,Z., Antonelli,M., Maldonado,E., Chen,W., Burgess,S., Haldi,M., Artzt,K., Farrington,S. et al. (2002) Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nature Genet., 31, 135–140. [DOI] [PubMed] [Google Scholar]
- 5.Ross-Macdonald P., Sheehan,A., Roeder,G.S. and Snyder,M. (1997) A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross-Macdonald P., Coelho,P.S.R., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.-H., Sheehan,A., Symoniatis,D., Umansky,L. et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 7.Kumar A., Cheung,K.-H., Tosches,N., Masiar,P., Liu,Y., Miller,P. and Snyder,M. (2002) The TRIPLES database: a community resource for yeast molecular biology. Nucleic Acids Res., 30, 73–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies C.J. and Hutchinson,C.A. (1995) Insertion site specificity of the transposon Tn3. Nucleic Acids Res., 23, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiestl R.H., Dominska,M. and Petes,T.D. (1993) Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol. Cell. Biol., 13, 2697–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis L.K. and Resnick,M.A. (2000) Tying up loose ends: non-homologous end-joining in Saccharomyces cerevisiae. Mutat. Res., 451, 71–89. [DOI] [PubMed] [Google Scholar]
- 11.Stanford W.L., Caruana,G., Vallis,K.A., Inamdar,M., Hidaka,M., Bautch,V.L. and Bernstein,A. (1998) Expression trapping: identification of novel genes expressed in hematopoetic and endothelial lineages by gene trapping in ES cells. Blood, 92, 4622–4631. [PubMed] [Google Scholar]
- 12.Budziszewski G.J., Lewis,S.P., Glover,L.W., Reineke,J., Jones,G., Ziemnik,L.S., Lonowski,J., Nyfeler,B., Aux,G., Zhou,Q. et al. (2001) Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics, 159, 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chua G., Taricani,L., Stangle,W. and Young,P.G. (2000) Insertional mutagenesis based on illegitimate recombination in Schizosaccharomyces pombe. Nucleic Acids Res., 28, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schiestl R.H. and Petes,T.D. (1991) Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 88, 7585–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiestl R.H., Zhu,J. and Petes,T.D. (1994) Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 4493–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manivasakam P. and Schiestl,R.H. (1998) Nonhomologous end joining during restriction enzyme-mediated DNA integration in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riggle P.J. and Kumamoto,C.A. (1998) Genetic analysis in fungi using restriction-enzyme-mediated integration. Curr. Opin. Microbiol., 1, 395–399. [DOI] [PubMed] [Google Scholar]
- 18.Kraxenberger A., Friedl,A.A. and Kellerer,A.M. (1994) Computer simulation of pulsed field gel runs allows the quantitation of radiation-induced double-strand breaks in yeast. Electrophoresis, 15, 128–136. [DOI] [PubMed] [Google Scholar]
- 19.Gari E., Piedrafita,L., Aldea,M. and Herrero,E. (1997) A set of vectors with a tetracycline-regulated promoter system to modulate gene expression in Saccharomyces cerevisiae. Yeast, 13, 837–848. [DOI] [PubMed] [Google Scholar]
- 20.Wach A., Brachat,A., Alberti-Segui,C., Rebischung,C. and Philippsen,P. (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast, 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- 21.Kiechle M., Friedl,A.A., Manivasakam,P., Eckardt-Schupp,F. and Schiestl,R.H. (2000) DNA integration by Ty integrase in yku70 mutant Saccharomyces cerevisiae cells. Mol. Cell. Biol., 20, 8836–8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gietz D., St. Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz R.D. and Sugino,A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- 24.Ahne F., Jha,B. and Eckardt-Schupp,F. (1997) The RAD5 gene product is involved in the avoidance of non-homologous end-joining of DNA double strand breaks in the yeast Saccharomyces cerevisiae. Nucleic Acids Res., 25, 743–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Dyck E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- 26.Mount R.C., Jordan,B.E. and Hadfield,C. (1996) Reporter gene systems for assaying gene expression in yeast. Methods Mol. Biol., 53, 239–248. [DOI] [PubMed] [Google Scholar]
- 27.Averbeck D. and Averbeck,S. (1994) Induction of the genes RAD54 and RNR2 by various DNA damaging agents in Saccharomyces cerevisiae. Mutat. Res., 315, 123–138. [DOI] [PubMed] [Google Scholar]
- 28.Davis C.A., Grate,L., Spingola,M. and Ares,M. (2000) Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res., 28, 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasch A.P., Spellman,P.T., Kao,C.M., Carmel-Harel,O., Eisen,M.B., Storz,G., Botstein,D. and Brown,P.O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell, 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasch A.P., Huang,M., Metzner,S., Botstein,D., Elledge,S.J. and Brown,P.O. (2001) Genomic expression response to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell, 12, 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jelinsky S.A., Estep,P., Church,G.M. and Samson,L.D. (2000) Regulatory network revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol., 20, 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brusky J., Zhu,Y. and Xiao,W. (2000) UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet., 37, 168–174. [DOI] [PubMed] [Google Scholar]
- 33.Ulrich H.D. (2001) The srs2 suppressor of UV sensitivity acts specifically on the RAD5- and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res., 29, 3487–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birrell G.W., Brown,J.A., Wu,H.I., Giaever,G., Chu,A.M., Davis,R.W. and Brown,J.M. (2002) Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl Acad. Sci. USA, 99, 8778–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manivasakam P., Aubrecht,J., Sidhom,S. and Schiestl,R.H. (2001) Restriction enzymes increase efficiencies of illegitimate DNA integration but decrease homologous integration in mammalian cells. Nucleic Acids Res., 29, 4826–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karathanasis E. and Wilson,T.E. (2002) Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics, 161, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison P.M., Kumar,A., Lang,N., Snyder,M. and Gerstein,M. (2002) A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res., 30, 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giaever G., Chu,A.M., Ni,L., Connelly,C., Riles,L., Véronneau,S., Dow,S., Lucau-Danila,A., Anderson,A., André,B. et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature, 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 39.Griffin T.J., Gygi,S.P., Ideker,T., Rist.,B., Eng,J., Hood,L., Aebersold,R. (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics, 1, 323–333. [DOI] [PubMed] [Google Scholar]