Abstract
As tools for functional genomics, expression profiling and proteomics provide correlative data, while expression cloning screens can link genes directly to biological function. However, technical limitations of gene transfer, expression, and recovery of candidate genes have limited wider application of genome-wide expression screens. Here we describe the pEYK retroviral vectors, which maintain high titers and robust gene expression while addressing the major bottleneck of expression cloning—efficient candidate gene recovery. By exploiting schemes for enhanced PCR rescue or strategies for direct isolation of proviral DNA as plasmids in bacterial hosts, the pEYK vectors facilitate cDNA isolation from selected cells and enable rapid iteration of screens and genetic reversion analyses to validate gene candidates. These vectors have proven useful to identify genes linked to cell proliferation, senescence and apoptosis.
INTRODUCTION
While new techniques for global analysis of gene and protein expression provide an enormous body of correlative data, techniques for linking gene expression directly to gene function are lacking. Expression cloning strategies are alluring because of their power to assign a phenotypic consequence—gene function—in an immediate and definitive manner. Genomic DNA transfection and plasmid library transfection have been used to clone the first cellular oncogene, Ras (1,2), the immunoglobulin recombinase gene, Rag-1 (3), and numerous lymphocyte antigens (4). But expression cloning can be tedious because of inefficient gene transfer and the formidable challenge of recovery and validation of candidate genes.
Retroviral vectors enable efficient and stable gene transfer into a variety of cell types, but the difficulty in recovering the integrated provirus has hindered their use in expression cloning. The pSVX vector was the first retroviral expression vector to employ a ‘shuttle’ strategy, incorporating antibiotic resistance markers and a bacterial origin of replication so that integrated provirus could be recovered from infected cells and transformed directly into a bacterial host for amplification as plasmid DNA (5). The pSVX vector, however, was plagued by low titers and prone to recombination during the recovery procedure. Moreover, expression cloning applications were extremely laborious until the advent of library packaging strategies that employed transient transfection for production of viral supernatant (6,7).
In the pEYK retroviral vectors described here, we utilize the same principles of proviral recovery, but have tested a variety of different vector structures that sustain high viral titer and gene expression while conferring enhanced vector versatility. In this report we provide a comparison of titers, gene expression, and cDNA recovery efficiencies for three novel retroviral vectors. Such quantitative parameters define the realistic advantages and limitations of these vectors for functional screens. These vectors have proven useful to identify genes linked to cell proliferation, senescence and apoptosis (8–11; E.Koh, T.Chen and G.Q.Daley, manuscript in preparation).
MATERIALS AND METHODS
Cell culture
Culture of the murine interleukin-3 (IL-3)-dependent BaF/3 cells has been previously described (12).
Plasmid construction
For pEYK1, the supF gene was PCR amplified from pCDNA1.1 (Invitrogen), using the primers SupF.5 (5′-GCGCGTCGACATCGACGATGGTCTTTCTCAACG-3′) and SupF.3 (5′-GCGCCTCGAGATTATCGATCTTTCGGAC-3′), and cloned into the SalI site of pMX (13). pEYK1 incorporates a mutated supF gene.
For pEYK2, site-directed mutagenesis was performed on pMX using the QuikChange kit (Stratagene) with primers 1072.F (5′-CTGTATTTGTCTGAAAAAAGGGCCCGGGCAG-3′) and 1072.R (5′-CTGGCCCGGGCCCTTTTTTCAGACAAATACAG-3′) and 1847.F (5′-CTTCCCTGACCC TGACAAGACAAGAGTTACTAAC-3′) and 1847.R (5′-GTTAGTAACTCTTGTCTTGTCAGGGTCAGGGAAG-3′).
For the 959 long terminal repeat (LTR), a dsDNA oligonucleotide that contained the NotI, loxP and AscI sites (959.UP 5′-CTAGCGCGGCCGCATAACTTCGTATAGCATACATTATACGAAGTTATTTAATTAAGGCGCGCCT-3′ and 959.DOWN 5′-CTAGAGGCGCGCCTTAATTAAATAACTTCGTATAATGTATGCTATACGAA-3′) was placed in the NheI site of the U3 region of the LTR, generating the intermediate plasmid pEYK7.
For pEYK2.1, the ble gene was PCR amplified from the pEM7/Zeo vector (Invitrogen), using the primers EM7.X (5′-GCGCCTCGAGTGTTGACAATC-3′) and ZE0.X (5′-GCGCCTCGAGTCAGTCCTGCTC-3′). The PCR product was placed in the SalI site of pEYK2. The proviral fragment was placed into pEYK7 at the NheI site to generate pEYK2.1.
For pEYK3.1, the single LTR provirus was generated from pDOL by digesting with XbaI, eliminating the backbone, and then self-ligating to generate pDSL. For the ble-ColE1 fusion, the ble gene was PCR amplified from the pEM7/Zeo vector, using the primers EM7.X (5′-GCGCCTCGAGTGTTGACAATC-3′) and ZE0.M (5′-GCGCACGCGTTCAGTCCTGCTC-3′). The ColE1 fragment was PCR amplified from pEYK2 using the primers COLF.M (5′-GCGCACGCGTGCGTAATCTGCTGCTTGC-3′) and COLR.C (5′-GCGCATCGATGCGTTGCTGGCGTTTTTCC-3′). The ble-ColE1 fusion was created by ligating the fragments at the MluI site and PCR amplifying using the primers EM7.X and COLR.C. The resulting PCR fragment was cloned at the SalI and ClaI sites of pDSL. The 959 LTR from pEYK7 was cloned into the NheI and KpnI sites, resulting in the pEYK3 vector. The mutagenized 1 kb gag region of pEYK2 was placed into pEYK3 via the KpnI and BamHI sites, yielding pEYK3.1.
Retroviral generation and infections
293T cells (5 × 105) were plated onto 6 cm plates the day before transfection. The cells were transfected with FuGENE6 (Roche) according to the supplied protocol with 1:1 ratio of the retroviral construct to the packaging construct PCL-eco (7). The medium was changed 16 h after transfection. At 30 h after transfection, supernatants were isolated and filtered through 0.45 μm filter (Acrodisc). Using centrifugation (2500 g for 1.5 h at 33°C in a Sorvall RT 6000 tabletop centrifuge), cells were infected with the retroviral supernatant supplemented with 8 μg/ml of polybrene (Sigma) and IL-3.
PCR rescue
PCR primers for pEYK1 were designed using the Primer 3 program (Whitehead Center for Genome Research; www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi). The primers 1759 (5′-AAAGGACCTTACACAGTCCTGCTGA-3′) and 3289 (5′-CCACAGGTAATGCTTTTACTGGCCT-3′) were utilized with the Expand High Fidelity PCR kit (Roche).
Restriction enzyme and Cre rescue
For pEYK2.1, 100 ng of NotI or AscI digested genomic DNA containing single-copy provirus was ligated to 1 μg of the correspondingly digested pEYK7 plasmid. For pEYK3.1, 1 μg of NotI or AscI digested genomic DNA containing single-copy provirus was self-ligated in dilute conditions (100 μl per 1 μg of DNA). The ligation mixture was phenol/chloroform extracted and ethanol precipitated before transformation into Electromax DH10B (Life Technologies). The bacteria were selected on LB plates containing zeocin (50 μg/ml, Invitrogen) alone or together with ampicillin (100 μg/ml, Sigma). For the cre-mediated rescue of pEYK3.1, 1 μg genomic was treated with cre enzyme (Invitrogen) according to the protocol supplied by the manufacturer.
Reversion analysis
The BCR-ABL oncogene was subcloned into pEYK3.1 at the EcoRI sites, generating the pEYK3.1-B/A vector. BaF/3 cells were infected and deprived of IL-3 2 days post-infection. The factor-independent population was then infected with MIG-cre in media containing IL-3. Two days later, half the population was deprived of IL-3, while the other half continued to receive IL-3. Two days later, fluorescence activated cell sorter (FACS) analyses were performed to determine the percentage of GFP positive viable cells. Immunoblot assays and densitometric analyses were performed as previously described (12).
RESULTS
pEYK1: enhanced PCR recovery
In attempting PCR amplification of a candidate cDNA from an integrated provirus, the length and GC content are two critical but unknown factors, making PCR recovery essentially a blinded process. Our initial efforts to perform genetic screens of cDNA expression libraries using the pMX retroviral vector (13) were confounded by inefficient recovery of cDNA inserts by PCR. In the pEYK1 vector, PCR-based recovery was greatly facilitated by virtue of a unique 3′ primer binding site inserted into the vector.
The design of the pEYK1 vector was a fortuitous product of efforts to incorporate the bacterial amber stop codon tRNA suppressor gene supF into the pMX vector to enable selection in bacterial hosts (14). In vectors carrying supF, bacterial transformation efficiency decreased by >10-fold and bacterial colonies varied dramatically in size. This resulted in an undesirable reduction in library complexity and introduced unequal representation of clone frequencies in the library. Sequencing of the supF gene in several large colonies revealed inactivating mutations, reflecting selective pressures on the toxic supF gene. Though non-functional, the inactive version of supF (termed subF) served as unique sequence for primer binding and southern hybridization. Efforts to incorporate unique primer binding sequences 5′ of the cDNA insertion site led to unacceptable loss of titer and cDNA expression and were therefore abandoned.
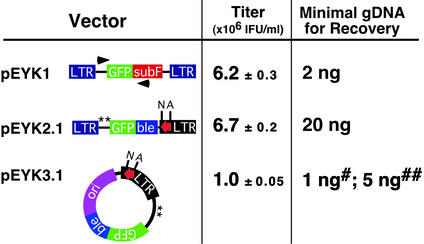
The resulting pEYK1 vector had comparable retroviral titers and only modestly decreased GFP expression levels relative to pMX (Figs 1 and 2). To select optimal primers for PCR recovery from pEYK1, we designed 15 primer pairs using unique subF sequences as a 3′ anchor, and tested each for amplification efficiency and fidelity. Using a single copy proviral template containing GFP as the cDNA insert, we reproducibly recovered the target sequence (∼700 bp) from a minimum of 2 ng of genomic DNA with the optimal pair of primers (1759 and 3289; Fig. 1). The efficiency of PCR amplification, however, depends on the size of the cDNA insert. We were unable to recover the BCR/ABL cDNA (∼7 kb) from integrated pEYK1 provirus containing this oncogene. Thus, isolation of long cDNAs remains a limitation of PCR recovery.
Figure 1.
Schematic representation and characterization of the pEYK retroviral vectors. All vectors contain the green fluorescent protein (GFP) gene. The pEYK1 vector incorporates subF, a mutated version of the suppressor tRNA gene supF, to serve as unique primer binding or probe sequence. The ble gene confers resistance to the bleomycin/phleomycin family of antibiotics. All pEYK vectors include ∼1.0 kb of retroviral gag gene sequence for enhanced retroviral packaging (19). The two ATGs within the extended gag region capable of generating gag-cDNA fusion proteins by alternative translation initiation were eliminated by site-directed mutagenesis (denoted by **). The 959 LTR (red arrow) consists of unique restriction enzyme sites (N-NotI and A-AscI) and a loxP site. Retroviral titers were determined as averages of triplicate infections with standard deviations. Titers are shown as 106 infectious units per ml. The minimal quantity of genomic DNA needed to recover the GFP insert or the intact provirus from cells containing a single-copy provirus was determined for each vector. For pEYK3.1, amount of genomic DNA required for recovery by restriction digestion and intramolecular ligation (#) or cre-mediated excision and intramolecular ligation (##).
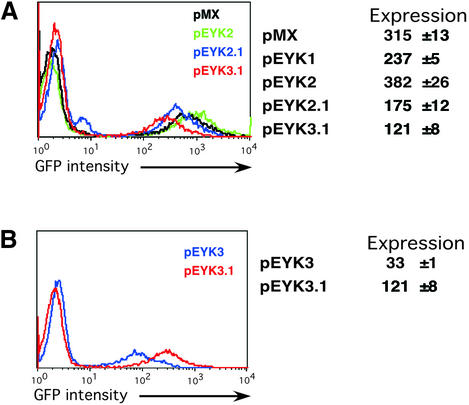
Figure 2.
Comparative levels of expression for pEYK retroviral vectors. (A) FACS plots of BaF/3 cells infected with pMX, pEYK2, pEYK2.1 and pEYK3.1 retroviral vectors demonstrate differences in GFP fluorescence (pEYK1 was not included in the overlay to allow better visualization of profiles for other vectors). Expression levels were calculated as fold-increase in geometric mean fluorescence above background fluorescence, and represent averages of triplicate infections with standard deviations. Mutagenesis of the two internal ATGs (**) within the gag region of pMX yielded pEYK2, which showed modest but consistent increases in GFP expression over pMX. This enhanced packaging region was incorporated into the pEYK2.1 and pEYK3.1 vectors. (B) FACS plots of BaF/3 cells infected with pEYK3 and pEYK3.1 demonstrate a 4-fold increase in expression levels with the incorporation of the mutagenized gag sequences into pEYK3.1. The pEYK3 vector lacks the mutagenized gag sequence. For both plots, GFP expression is measured on a logarithmic scale.
Mobilization of integrated provirus by Moloney murine leukemia virus
Recovery by superinfection with the replication competent Moloney murine leukemia virus is an indirect method for isolating integrated provirus from selected murine cells. However, the resulting retroviral supernatants typically possess a low titer of recombinant virus (102–103 IFU/ml), and the presence of multiple viral templates within infected cells can confound PCR amplification (data not shown). Applying this rescue strategy in non-murine cells necessitates use of replication competent virus whose host range includes the target cell, like amphotropic helper virus (15). Nevertheless, this technique can be useful in cases where PCR rescue fails.
pEYK2.1 and pEYK3.1: direct proviral recovery
To obviate PCR-based cDNA isolation altogether, we created two retroviral vectors that enable recovery of an intact provirus from genomic DNA in the form of a plasmid vector that can be propagated in bacterial hosts. This required engineering the LTR to facilitate excision and circularization of the provirus, and incorporating appropriate selectable markers and bacterial replicons within the retroviral sequences.
To enable excision of the integrated provirus, we introduced two restriction enzyme sites (NotI and AscI) and a loxP site into the U3 region of the 3′ LTR, thus generating the 959 LTR (Fig. 1). During the retroviral life cycle, the 959 U3 region is duplicated at both the 5′ and 3′ LTRs, resulting in an integrated provirus that is flanked by identical restriction enzyme and loxP sites. Insertion of various sequences into the 3′ LTR can be achieved without compromising viral function (16,17), and likewise we observed no diminution of titer or cDNA expression with the 959 LTR (data not shown).
The pEYK2.1 retroviral vector contains the 959 LTR and the bacterial marker ble, which confers bleomycin/zeocin resistance (18) (Fig. 1). When integrated in the genome, the pEYK2.1 provirus can be excised by restriction enzyme digestion of genomic DNA. (By virtue of their rare 8 bp recognition sequences, either NotI or AscI is likely to liberate full-length provirus.) Upon ligation to a plasmid backbone carrying a bacterial origin of replication and a β-lactamase gene, the recovered pEYK2.1 provirus can be directly transformed into bacterial cells and selected for dual resistance to ampicillin and zeocin. If the plasmid backbone contains the complementary LTR sequences, then half of the recovered plasmid will regenerate a functional viral vector and can be employed directly to iterate the screen. The rescue efficiency from genomic DNA carrying a single copy provirus containing the GFP cDNA was at least 100 bacterial colonies per μg. The ble gene did not diminish viral titers, but modestly decreased levels of expression compared to the parental pEYK2 vector (Fig. 2). This vector is able to tolerate larger cDNA inserts than pEYK3.1, which is described below.
The most versatile vector, pEYK3.1, incorporates a selectable marker and a bacterial origin of replication within the retrovirus itself, thereby enabling direct recovery of an excised provirus in plasmid form. Within the circular plasmid DNA, the single LTR provides both 5′ promoter and 3′ transcription termination functions. Therefore, the recovered retroviral plasmid can serve immediately as a retroviral vector (Fig. 1). Following excision of the provirus from genomic DNA by restriction digestion, recircularization by intramolecular ligation, and bacterial transformation, the rate of recovery for pEYK3.1 containing the GFP cDNA is consistently >1000 colonies per μg of genomic DNA. The structure of the recovered provirus is intact in virtually all cases (39/40, data not shown).
Alternatively, incubation of genomic DNA with the cre recombinase protein can excise the pEYK3.1 provirus by in vitro intramolecular recombination. This process yields intact, functional plasmid circles in virtually all cases (29/30, data not shown). The cre-mediated rescue, however, is five times less efficient than restriction enzyme digestion of genomic DNA followed by self-ligation (Fig. 1). In contrast to the PCR-based strategy in pEYK1, this direct recovery of the pEYK3.1 provirus is able to recover larger cDNA fragments, including BCR/ABL (∼7 kb), and the re-isolation consistently recovered structurally intact virus (25/28, data not shown). The recovery efficiency for the BCR/ABL provirus, however, was approximately three times lower than the rescue of the GFP cDNA insert (∼340 colonies per μg of genomic DNA). For both pEYK2.1 and pEYK3.1, the rescued proviral plasmids generated titers and expression levels equivalent to the parental retroviral vectors (data not shown).
Mutagenesis of gag region results in enhanced cDNA expression
Addition of genetic elements to add functionality to the retroviral vectors created a dilemma. Increasing the versatility of the virus was often at the expense of viral titers and expression levels. Therefore, we explored additional strategies to offset the loss in gene expression levels. The inclusion of the retroviral gag sequence has been proven to augment viral titers through enhanced packaging (19). In order to prevent translation of gag proteins, vectors incorporating the extended gag region carry a mutation to inactivate the initiator ATG codon (20). In order to eliminate the potential competition for translation initiation from two internal ATGs within the proviral gag region, we mutagenized these residues. Empirically, we determined that elimination of the two internal ATGs increased GFP expression levels 4-fold when incorporated into the pEYK3.1 vector (Fig. 2B). Thus overall, while the pEYK3.1 vector has modestly decreased expression levels and reduced titers (Fig. 2), this compromise is an acceptable tradeoff in order to achieve rapid, efficient proviral rescue.
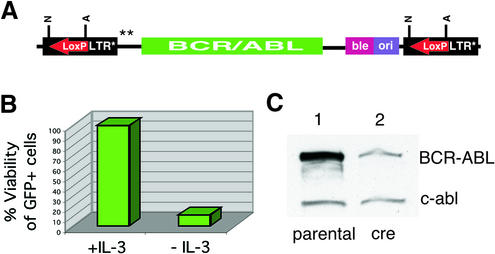
Reversion analysis of BCR-ABL transformed cells
An additional advantage of the pEYK2.1 and pEYK3.1 vectors is the capacity to perform a genetic reversion test in selected cells to confirm the link between cDNA expression and phenotype. Because the integrated provirus is flanked by loxP sites, expression of the cre recombinase gene can mediate excision in selected cells. We generated IL-3 independent BaF/3 cells transformed by the BCR/ABL oncoprotein (12) expressed from the pEYK3.1-BCR/ABL virus (Fig. 3A), and infected them with a bicistronic retrovirus expressing both the cre and GFP genes (21). Cell viability of cre-GFP positive cells decreased significantly by 48 h (Fig. 3B), accompanied by loss of BCR/ABL expression (Fig. 3C), demonstrating that in vivo excision of the BCR/ABL cDNA successfully reverts the transformed phenotype.
Figure 3.
Reversion analysis of the pEYK3.1-B/A retroviral vector. (A) The integrated provirus contains the BCR/ABL oncogene and is flanked by loxP sites (red arrows). (B) A population of BaF/3 cells containing the pEYK3.1-B/A provirus was infected with a bicistronic virus expressing both the cre and GFP-3M genes. In the absence of IL-3, the viability of GFP positive cells was 12% after 2 days. (C) Immunoblot analysis of the BCR/ABL-transformed population with an anti-c-ABL antibody shows the loss of BCR/ABL expression after cre-medated excision (lane 2). Densitometric analyses demonstrated an ∼80% decrease in BCR/ABL protein upon cre-mediated excision, with endogenous c-abl serving as a loading control.
DISCUSSION
The advantages of retroviral vectors for stable and efficient gene transduction are widely appreciated. The vectors described here were designed to optimize retroviral titers and cDNA expression and to facilitate rapid, efficient recovery of candidate genes from complex retroviral cDNA libraries.
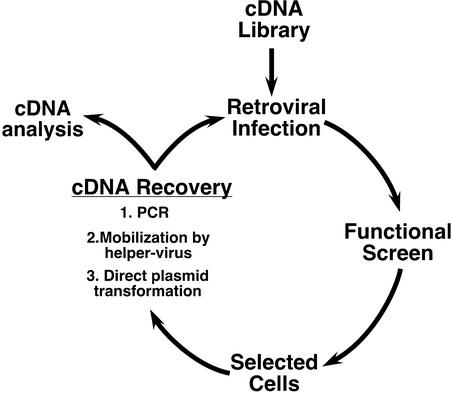
Three strategies can be employed to rescue pEYK vectors from selected cells, with particular advantages and tradeoffs for each (Fig. 4). First, cDNA candidates can be directly recovered by PCR amplification of genomic DNA from selected cells. Previously, several laboratories have developed retroviral vectors that rely upon a PCR-based recovery strategy [e.g., pCTV (14) and pMX (13)]. PCR-based recovery of cDNA inserts can clearly be successful (14,22,23), but in our hands it has been plagued by inefficiency. The reported efficiency of gene recovery in functional assays is only comparable to episomal vectors derived from Epstein Barr Virus (1 in 105) (24–26).
Figure 4.
pEYK retroviral vectors enable sequential iteration of functional genomic screens using different methods for proviral recovery.
The pEYK1 vector was designed to achieve reliable and efficient recovery of cDNAs by PCR. It tolerates the largest cDNA inserts, generates high titers, and shows improved efficiency for cDNA rescue. However, subsequent regeneration of the retroviral vector and repetition of the screen still requires laborious sub-cloning steps. Our inability to recover the BCR-ABL oncogene from the pEYK1 vector by PCR amplification demonstrates the limitations of a PCR-based strategy to recover larger cDNAs, thereby necessitating alternative strategies for recovery.
A second strategy for proviral rescue is superinfection with replication competent helper-virus (e.g., Moloney murine leukemia virus for mouse cells). This mobilizes the integrated proviruses into the cell supernatant for subsequent rounds of infection. However, this generates additional viral templates in infected cells that reduce the efficiency of PCR rescue of cDNA inserts.
The third and most expeditious strategy is to rescue the provirus in the form of a functional plasmid directly from genomic DNA of selected cells using the pEYK2.1 and pEYK3.1 vectors. This recovery scheme entails either a cre recombinase-mediated excision event or restriction enzyme digestion of LTRs followed by either ligation to an acceptor plasmid (pEYK2.1) or self-ligation (pEYK3.1) to regenerate circular provirus in plasmid form. The use of restriction enzymes to excise the provirus from genomic DNA is five times more efficient than recombinase-mediated excision, and allows an efficiency of recovery that approaches PCR-based rescue. The restriction enzyme mediated recovery of pEYK3.1 is 20-fold more efficient than for pEYK2.1, likely due to the different intrinsic efficiencies of intramolecular versus intermolecular ligation reactions.
The pEYK2.1 and pEYK3.1 vectors expedite rescreening through rapid recovery of enriched retroviral plasmid sub-libraries. These sub-libraries can be directly transfected onto packaging cells for screen iteration, critical for winnowing candidates into a few validated genes. Furthermore, recovery of the cDNA in plasmid form ultimately expedites definitive sequence analysis.
Both the pEYK2.1 and pEYK3.1 vectors and the MaRX retroviral vectors developed by Hannon et al. (27) embody a shuttle vector strategy with conceptual similarity to pSVX (5). However, the pEYK3.1 vector, our most versatile, incorporates several advantageous features: (i) it carries two point mutations in the gag region that enhance cDNA expression; (ii) it allows proviral recovery by the more efficient method of restriction enzyme digestion to re-isolate the integrated provirus as a circular plasmid; and (iii) it utilizes a bacterial origin that can be propagated in a broader variety of bacterial hosts.
The pEYK vectors address some of the major bottlenecks encountered in expression cloning from retroviral libraries. Our phenotypic screens with a known cDNA diluted into a plasmid library have re-isolated clones represented at a frequency of 1 in 106—an efficiency at least 10-fold greater than reported for other retroviral vectors (13,24,28). To date, the pEYK retroviral vectors have been applied in diverse screens to identify genes that overcome premature senescence in bmi-1 knock-out mouse embryonic fibroblasts (MEFs) (9), the slow-growth phenotype of fibroblasts lacking c-myc (8), ras-mediated senescence (10), senescence in MEFs conditionally transformed by SV40 large-T (11), and apoptosis in cytokine-dependent hematopoietic cells (E.Koh, T.Chen and G.Q.Daley, manuscript in preparation).
These phenotypic screens with the pEYK vector have thus far isolated cDNAs encoding a dominant gain-of-function or a dominant-negative. The identification of recessive loss-of-function phenotypes may be difficult but not impossible. With the strong retroviral promoter driving the expression of the cDNAs, generation of inhibitors that block protein function could mimic loss-of-function phenotypes. For these recessive/suppression screens, two approaches have and will become useful in identifying candidates in loss-of-function screen: antisense/genetic suppressor elements (29–33) and RNA-i (34).
Despite the difficulties in recessive phenotypic screens, several laboratories have been successful in identifying genes through the overexpression of antisense libraries. These phenotypic screens examine the role of loss of function in providing cells with a survival and/or proliferative advantage. Deiss and Kimchi (29) have identified thioredoxin as a pro-apoptotic gene, and Whitehead et al. have identified that the loss of Spi-2 gene results in 3T3 transformation (14). However, many antisense constructs have failed to cause a significant change in biological phenotypes (35,36). These failures indicate a huge barrier to using antisense strategies to cover the whole genome in recessive screens. The combination of antisense cDNA, genetic suppressor elements, and RNA-i may provide the solution, allowing saturation of loss of function at each genetic locus (34,37–39).
The pEYK vectors can be utilized to screen mutant alleles of a specific gene. The desired gene can be altered through mutagenic PCR conditions or by propagation of plasmids in mutagenic bacterial strains. Such strategies have been utilized to identify oncogenic forms of the c-mpl (thrombopoietin receptor) gene (40) and constitutively active forms of the STAT5 gene (41,42). These mutagenic screens can serve to identify neomorphic or dominant-negative alleles—both useful reagents in understanding gene function. Again, the ease in recovering the mutant alleles becomes critical in these screens; the efficient recovery in pEYK vectors allows for quick isolation and identification of mutant alleles.
Recent advances in retroviral packaging into high titer supernatants has been key to enabling efficient genetic screens with retroviral libraries. Using pCL-Eco to generate helper-free supernatants (7), the pEYK1 and pEYK2.1 vectors consistently produce titers of >5 × 106 IFU/ml. The pEYK3.1 vector generates lower titers (>106 IFU/ml), but even given conservative estimates for full-length representation in cDNA libraries, these titers are adequate to represent the estimated 30 000–40 000 genes in the human genome in a single small volume of viral supernatant, thereby enabling efficient genome-wide screens. With the completion of the genome sequences for human and mouse, it becomes possible to envision retroviral libraries comprised of a normalized set of all full-length cDNAs and splice variants. Assembling such complete clone libraries constitutes an important enabling technology for assigning gene function throughout the genome. The set of vectors described here should facilitate applications of these libraries in diverse functional genomic screens.
Acknowledgments
ACKNOWLEDGEMENTS
Drs Garry Nolan and Rene Bernards provided helpful insights on retroviral cloning technology. Critical comments on the manuscript were provided by Scott Dessain and Robert Weinberg. This work was supported by grants from the NIH (CA76418 and CA86991), the Edward Mallinckrodt, Jr Foundation, the Burroughs Wellcome Fund, and the NSF/MIT Biotechnology Process Engineering Center. G.Q.D. is the Birnbaum Scholar of the Leukemia and Lymphoma Society of America. E.Y.K. was supported by NIH training grants (GM07753-22 and CA09541).
REFERENCES
- 1.Shih C. and Weinberg,R.A. (1982) Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell, 29, 161–169. [DOI] [PubMed] [Google Scholar]
- 2.Goldfarb M., Shimizu,K., Perucho,M. and Wigler,M. (1982) Isolation and preliminary characterization of a human transforming gene from T24 bladder carcinoma cells. Nature, 296, 404–409. [DOI] [PubMed] [Google Scholar]
- 3.Schatz D.G., Oettinger,M.A. and Baltimore,D. (1989) The V(D)J recombination activating gene, RAG-1. Cell, 59, 1035–1048. [DOI] [PubMed] [Google Scholar]
- 4.Aruffo A. and Seed,B. (1987) Molecular cloning of a CD28 cDNA by a high-efficiency COS cell expression system. Proc. Natl Acad. Sci. USA, 84, 8573–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cepko C.L., Roberts,B.E. and Mulligan,R.C. (1984) Construction and applications of a highly transmissible murine retrovirus shuttle vector. Cell, 37, 1053–1062. [DOI] [PubMed] [Google Scholar]
- 6.Pear W.S., Nolan,G.P., Scott,M.L. and Baltimore,D. (1993) Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl Acad. Sci. USA, 90, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naviaux R.K., Costanzi,E., Haas,M. and Verma,I.M. (1996) The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol., 70, 5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berns K., Hijmans,E.M., Koh,E., Daley,G.Q. and Bernards,R. (2000) A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene, 19, 3330–3334. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs J.J., Keblusek,P., Robanus-Maandag,E., Kristel,P., Lingbeek,M., Nederlof,P.M., van Welsem,T., van De Vijver,M.J., Koh,E.Y., Daley,G.Q. et al. (2000) Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19ARF) and is amplified in a subset of human breast cancers. Nature Genet., 26, 291–299. [DOI] [PubMed] [Google Scholar]
- 10.Peeper D.S., Shvarts,A., Brummelkamp,T., Douma,S., Koh,E.Y., Daley,G.Q. and Bernards,R. (2002) A functional screen identifies hDRIL1 as an oncogene that rescues RAS-induced senescence. Nature Cell Biol., 4, 148–153. [DOI] [PubMed] [Google Scholar]
- 11.Shvarts A., Brummelkamp,T., Scheeren,F., Koh,E., Daley,G.Q., Spits,H. and Bernards,R. (2002) A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19ARF-p53 signaling. Genes Dev., 16, 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klucher K.M., Lopez,D.V. and Daley,G.Q. (1998) Secondary mutation maintains the transformed state in BaF3 cells with inducible BCR/ABL expression. Blood, 91, 3927–3934. [PubMed] [Google Scholar]
- 13.Kitamura T., Onishi,M., Kinoshita,S., Shibuya,A., Miyajima,A. and Nolan,G.P. (1995) Efficient screening of retroviral cDNA expression libraries. Proc. Natl Acad. Sci. USA, 92, 9146–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehead I., Kirk,H. and Kay,R. (1995) Expression cloning of oncogenes by retroviral transfer of cDNA libraries. Mol. Cell. Biol., 15, 704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang E.H., Morgan,P.L., Lee,E.J., Pirollo,K.F., White,E.A., Patrick,D.H. and Tsichlis,P.N. (1985) Pathogenicity of retroviruses containing either the normal human c-Ha-ras1 gene or its mutated form derived from the bladder carcinoma EJ/T24 cell line. J. Exp. Pathol., 2, 177–189. [PubMed] [Google Scholar]
- 16.Reik W., Weiher,H. and Jaenisch,R. (1985) Replication-competent Moloney murine leukemia virus carrying a bacterial suppressor tRNA gene: selective cloning of proviral and flanking host sequences. Proc. Natl Acad. Sci. USA, 82, 1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergemann J., Kuhlcke,K., Fehse,B., Ratz,I., Ostertag,W. and Lother,H. (1995) Excision of specific DNA-sequences from integrated retroviral vectors via site-specific recombination. Nucleic Acids Res., 23, 4451–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatignol A., Durand,H. and Tiraby,G. (1988) Bleomycin resistance conferred by a drug-binding protein. FEBS Lett., 230, 171–175. [DOI] [PubMed] [Google Scholar]
- 19.Bender M.A., Palmer,T.D., Gelinas,R.E. and Miller,A.D. (1987) Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J. Virol., 61, 1639–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dranoff G., Jaffee,E., Lazenby,A., Golumbek,P., Levitsky,H., Brose,K., Jackson,V., Hamada,H., Pardoll,D. and Mulligan,R.C. (1993) Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific and long-lasting anti-tumor immunity. Proc. Natl Acad. Sci. USA, 90, 3539–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherry S.R., Beard,C., Jaenisch,R. and Baltimore,D. (2000) V(D)J recombination is not activated by demethylation of the kappa locus. Proc. Natl Acad. Sci. USA, 97, 8467–8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starr R., Willson,T.A., Viney,E.M., Murray,L.J., Rayner,J.R., Jenkins,B.J., Gonda,T.J., Alexander,W.S., Metcalf,D., Nicola,N.A. et al. (1997) A family of cytokine-inducible inhibitors of signalling. Nature, 387, 917–921. [DOI] [PubMed] [Google Scholar]
- 23.Deng H.K., Unutmaz,D., KewalRamani,V.N. and Littman,D.R. (1997) Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature, 388, 296–300. [DOI] [PubMed] [Google Scholar]
- 24.Onishi M., Kinoshita,S., Morikawa,Y., Shibuya,A., Phillips,J., Lanier,L.L., Gorman,D.M., Nolan,G.P., Miyajima,A. and Kitamura,T. (1996) Applications of retrovirus-mediated expression cloning. Exp. Hematol., 24, 324–329. [PubMed] [Google Scholar]
- 25.Wong B.Y., Chen,H., Chung,S.W. and Wong,P.M. (1994) High-efficiency identification of genes by functional analysis from a retroviral cDNA expression library. J. Virol., 68, 5523–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson C. and Legerski,R. (1991) High-frequency transformation of human repair-deficient cell lines by an Epstein-Barr virus-based cDNA expression vector. Gene, 107, 279–284. [DOI] [PubMed] [Google Scholar]
- 27.Hannon G.J., Sun,P., Carnero,A., Xie,L.Y., Maestro,R., Conklin,D.S. and Beach,D. (1999) MOLECULAR GENETICS:MaRX: an approach to genetics in mammalian cells. Science, 283, 1129–1130. [DOI] [PubMed] [Google Scholar]
- 28.Rayner J.R. and Gonda,T.J. (1994) A simple and efficient procedure for generating stable expression libraries by cDNA cloning in a retroviral vector. Mol. Cell. Biol., 14, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deiss L.P. and Kimchi,A. (1991) A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science, 252, 117–120. [DOI] [PubMed] [Google Scholar]
- 30.Levy-Strumpf N. and Kimchi,A. (1998) Death associated proteins (DAPs): from gene identification to the analysis of their apoptotic and tumor suppressive functions. Oncogene, 17, 3331–3340. [DOI] [PubMed] [Google Scholar]
- 31.Kissil J.L., Deiss,L.P., Bayewitch,M., Raveh,T., Khaspekov,G. and Kimchi,A. (1995) Isolation of DAP3, a novel mediator of interferon-gamma-induced cell death. J. Biol. Chem., 270, 27932–27936. [DOI] [PubMed] [Google Scholar]
- 32.Kimchi A. (1998) DAP genes: novel apoptotic genes isolated by a functional approach to gene cloning. Biochim. Biophys. Acta, 1377, F13–F33. [DOI] [PubMed] [Google Scholar]
- 33.Kissil J.L. and Kimchi,A. (1998) Death-associated proteins: from gene identification to the analysis of their apoptotic and tumour suppressive functions. Mol. Med. Today, 4, 268–274. [DOI] [PubMed] [Google Scholar]
- 34.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 35.Kerr S.M., Stark,G.R. and Kerr,I.M. (1988) Excess antisense RNA from infectious recombinant SV40 fails to inhibit expression of a transfected, interferon-inducible gene. Eur. J. Biochem., 175, 65–73. [DOI] [PubMed] [Google Scholar]
- 36.Leiter J.M., Krystal,M. and Palese,P. (1989) Expression of antisense RNA fails to inhibit influenza virus replication. Virus Res., 14, 141–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gudkov A.V., Kazarov,A.R., Thimmapaya,R., Axenovich,S.A., Mazo,I.A. and Roninson,I.B. (1994) Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proc. Natl Acad. Sci. USA, 91, 3744–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestov D.G., Grzeszkiewicz,T.M. and Lau,L.F. (1998) Isolation of growth suppressors from a cDNA expression library. Oncogene, 17, 3187–3197. [DOI] [PubMed] [Google Scholar]
- 39.Pestov D.G. and Lau,L.F. (1994) Genetic selection of growth-inhibitory sequences in mammalian cells. Proc. Natl Acad. Sci. USA, 91, 12549–12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onishi M., Mui,A.L., Morikawa,Y., Cho,L., Kinoshita,S., Nolan,G.P., Gorman,D.M., Miyajima,A. and Kitamura,T. (1996) Identification of an oncogenic form of the thrombopoietin receptor MPL using retrovirus-mediated gene transfer. Blood, 88, 1399–1406. [PubMed] [Google Scholar]
- 41.Ariyoshi K., Nosaka,T., Yamada,K., Onishi,M., Oka,Y., Miyajima,A. and Kitamura,T. (2000) Constitutive activation of STAT5 by a point mutation in the SH2 domain. J. Biol. Chem., 275, 24407–24413. [DOI] [PubMed] [Google Scholar]
- 42.Onishi M., Nosaka,T., Misawa,K., Mui,A.L., Gorman,D., McMahon,M., Miyajima,A. and Kitamura,T. (1998) Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol., 18, 3871–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]