Abstract
Termination of translation in eukaryotes has focused recently on functional anatomy of polypeptide chain release factor, eRF1, by using a variety of different approaches. The tight correlation between the domain structure and different functions of eRF1 has been revealed. Independently, the role of prokaryotic RF1/2 in GTPase activity of RF3 has been deciphered, as well as RF3 function itself.
Keywords: polypeptide release factors/ribosomes/RNA–protein recognition/translation
Introduction
Several overviews of termination in protein synthesis have been published recently (Kisselev and Buckingham, 2000; Poole and Tate, 2000; Bertram et al., 2001; Wilson et al., 2002). In this mini-review, we will focus on some important conceptual aspects of translational termination and discuss a few novel and unexpected experimental findings.
Termination of translation exemplifies in a striking way how informational and chemical aspects of biology can be integrated fundamentally. Termination signals of polypeptide synthesis are encoded in the genome and transcribed to mRNA in the form of three different base triplets referred to as termination, stop or nonsense codons. When a stop codon has been translocated into the ribosomal A-site by the action of elongation factor EF-G or eEF2, it is decoded at the small ribosomal subunit. However, the chemical reaction that is triggered by a stop signal, the cleavage of the ester bond between the peptidyl and tRNA moieties of the peptidyl-tRNA, occurs within the large ribosomal subunit at the peptidyl transferase centre (PTC) of the ribosome. How a stop signal can be transduced from the small to the large ribosomal subunit and trigger hydrolysis of peptidyl-tRNA remains unknown.
Class-1 polypeptide release factors decode stop signals in mRNA
Decoding of the stop signals in mRNAs could, in principle, be implemented by (i) the ribosome itself; (ii) external factors that associate with the ribosome in response to a stop signal and then dissociate after completed action; and (iii) the combined action of the ribosome and external factors. It was suggested that either the small or the large rRNA can interact directly with stop codons in mRNA (see Arkov and Murgola, 1999). This version of the first hypothesis is supported by data from several experiments. First, mutations in the small and large rRNAs strongly affect translational termination (see Green and Noller, 1997; Velichutina et al., 2001; and references therein). Secondly, the stop codon is in a close contact with rRNA within the ribosome (see Poole and Tate, 2000; Chavatte et al., 2001, 2002). Thirdly, two segments of prokaryotic 23S rRNA (helices 69 and 89) can be folded into a tRNA-like shape with anticodon-like loops complementary to stop codons (Ivanov et al., 2001). However, this in silico-derived hypothesis lacks support from experiments.
It is known that one or several external factors, called class-1 polypeptide release factors (RFs), associate(s) transiently with the ribosomal A-site. In bacteria, there are two RFs, the proteins RF1 and RF2, which recognize UAA/UAG and UAA/UGA, respectively. In eukaryotes, archaea and mitochondria, in contrast, only a single protein (eRF1, aRF1 and mtRF1, respectively) exists. In Arabidopsis thaliana and in the ciliate Euplotes, eRF1 is present as two homologous molecular species (see Liang et al., 2001). It remains unknown whether these eRF1 homologues respond to the same or different stop codons. The fact that external factors exist and terminate protein synthesis in a codon-specific way does not in itself tell us whether they recognize stop codons through direct interactions (second hypothesis) or indirectly, via rRNA (third hypothesis).
There are strong arguments in favour of a direct interaction between stop codons and class-1 RFs (Moffat and Tate, 1994; Nakamura et al., 1996). First, class-1 RFs cross-link specifically with stop signals in mRNA, although with rather low yield (see Poole and Tate, 2000; Chavatte et al., 2001, 2002; Bulygin et al., 2002). Secondly, mutations in bacterial RFs (see Nakamura and Ito, 1998), yeast (Bertram et al., 2000) and human (Frolova et al., 2002; Seit-Nebi et al., 2002) eRF1s affect stop codon recognition in ways that depend on both the position and the nature of the substituting amino acid. For bacterial RFs, it was found with genetic approaches that two tripeptides at structurally homologous positions, Pro-Ala-Thr in RF1 and Ser-Pro-Phe in RF2, determine their stop codon identity (Ito et al., 2000; Nakamura and Ito, 2002). However, the most important question is how the first U of the stop codons is discriminated from C, A and G (Freistroffer et al., 2000).
For eukaryotes, where all three stop codons are decoded by eRF1, a different approach is necessary to identify putative peptide sequences determining the stop codon identity of eRF1. One possibility was offered by using lower eukaryotes with variant genetic codes (see Knight et al., 2001; Lozupone et al., 2001), where one or two out of three stop codons are reassigned to sense codons. For example, in Euplotes, UGA encodes cysteine while UAA and UAG remain as stop codons. Purified, recombinant eRF1 from this organism terminates at animal ribosomes programmed with UAA or UAG, but not with UGA (Kervestin et al., 2001). Since protein synthesis carried out in vivo with these ribosomes is terminated at any one of the canonical stop codons, it must be eRF1, rather than a ribosomal component, that is responsible for the specific decoding of stop signals. Furthermore, in vitro experiments with molecular chimeras between Tetrahymena eRF1 (terminating only at UGA with the other two stop codons reassigned to glutamine) and yeast eRF1 (recognizing all three stop codons) showed that the hybrid bearing the N domain from Tetrahymena eRF1 can terminate only at UGA on mammalian ribosomes (Ito et al., 2002). This confirmed that eRF1, rather than the ribosome, renders stop codon specificity to termination of translation in eukaryotes and also showed that recognition is associated with the N domain of eRF1 (see below).
At each protein elongation step on the bacterial ribosome, there is a transient, strong interaction between the A-site codon–anticodon duplex and the nucleotides of 16S rRNA (particularly A1492 and A1493) within the small ribosomal subunit (see Ramakrishnan, 2002). Similar conclusions have been reached for 18S rRNA (Demeshkina et al., 2000). Since a functionally active ‘core’ of eRF1 (Frolova et al., 2000) mimics the shape of a tRNA (Song et al., 2000), it is conceivable that a ternary complex, consisting of a stop codon, small rRNA and a class-1 RF, can be formed at the translation termination step. This would mean a unification of the stop codon–rRNA model and the stop codon–class-1 RF model discussed above in a mechanism where each type of interaction is important for termination. Accordingly, the basic specificity of decoding would be dictated by class-1 RFs, while the strength and stereochemistry, which could enhance further the specificity of the interaction, would be strongly affected by rRNA sequences. The analogy between RFs and tRNAs with respect to decoding should not, however, be taken literally. This is evidenced by the fact that the antibiotic neomycin, which binds to conserved sequences of 16S rRNA (Moazed and Noller, 1987) and induces high levels of error in translation of sense codons by tRNAs, does not make bacterial RFs more error prone (Freistroffer et al., 2000).
Functional anatomy of eRF1
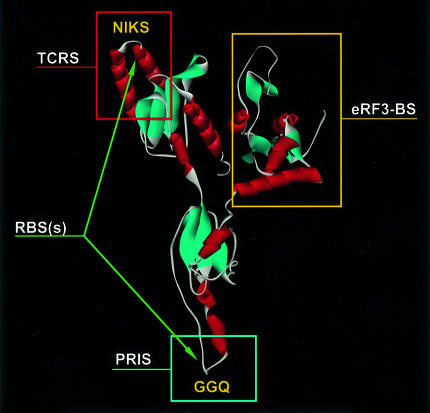
The function of class-1 RFs suggests that they should have four distinct sites (Frolova et al., 1999). There should be a ribosome-binding site (RBS), a termination codon recognition site (TCRS), a peptidyl-tRNA interaction site (PRIS) and an RF3- or eRF3-binding site [(e)RF3-BS]. The three-dimensional structure of human eRF1 (Song et al., 2000) can be used for a tentative assignment of these sites to different regions of the molecule (Figure 1).
Fig. 1. Functional anatomy of eRF1. Proposed locations of the eRF1 functional sites.
The C domain (domain 3) of eRF1 is unlikely to be a RBS for a number of reasons. First, removal of this domain from human eRF1 enhances, rather than reduces, the termination activity in vitro (Frolova et al., 2000), meaning that this region cannot be essential for ribosome binding. Secondly, the C domain is the most rapidly evolving part of eRF1 (Inagaki and Doolittle, 2001), while the ribosome structure is highly conserved. Thirdly, although there is substantial sequence divergence between the C domains of eRF1 and aRF1 (Inagaki and Doolittle, 2001), aRF1 is still able to terminate at animal ribosomes (Dontsova et al., 2000).
Presumably, the binding of eRF1 to the A-site is stabilized by interactions with both ribosomal subunits. This is supported by the observation that mutations in the GGQ and NIKS minidomains of eRF1, most probably located at the large and small ribosomal subunits, respectively (see below), reduce the binding of eRF1 to the ribosome (Seit-Nebi et al., 2001; Frolova et al., 2002). Furthermore, truncation of the N (N-terminal, or domain 1) or the M (middle, or domain 2) domain causes a gradual loss of RF activity in vitro (Frolova et al., 2000), probably due to distortions of RBSs.
The ribosomal functional sites are composed mainly of rRNAs (see Ramakrishnan, 2002), suggesting that RBSs of class-1 RFs bind primarily to rRNA sequences rather than to ribosomal proteins. This notion is consistent with the charge distribution along the consensus polypeptide chains of eRF1 and aRF1. There are two clusters of positively charged amino acid residues around positions 180 and 60–70, where the GGQ and NIKS motifs are mapped (Kisselev et al., 2000). This suggests that these two regions comprise two RBSs, which interact with rRNA in either of the two ribosomal subunits.
The existence of the RF1 RBS toward the PTC of the prokaryotic ribosome is manifested by the isolation of short RNA sequences (aptamers) containing 5′-ACCU-3′ and 5′-GAAAGC-3′ sequences identical to the 23S rRNA consensus sequences present in the PTC. These aptamers bind to RF1 and inhibit RF1 activity (Szkaradkiewicz et al., 2002).
The TCRS of class-1 eRFs is located at the N domain, as follows from: (i) in vivo genetic data with yeast eRF1 (Bertram et al., 2000) and in vitro biochemical data with human eRF1 (Frolova et al., 2002; Seit-Nebi et al., 2002), in which site-directed mutagenesis of some positions at the N domain causes profound alterations of the stop codon recognition profile for mutant eRFs; (ii) stop codon specificity of the hybrid eRF1 mentioned above (Ito et al., 2002); and (iii) data revealing cross-linking between the first U of the stop codons and the N domain of human eRF1 (Chavatte et al., 2002). However, the exact sequence and structure of TCRS in eukaryotes remain obscure.
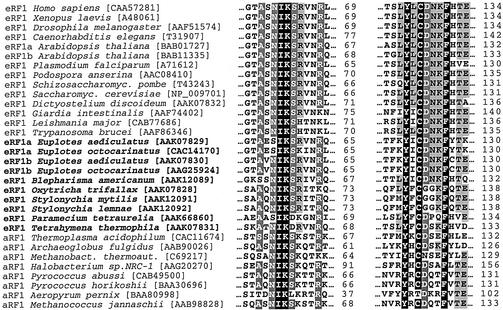
For stop codon recognition by eRF1, two types of models have been proposed, a ‘protein-anticodon’ (Nakamura et al., 2000) and a ‘cavity’ (Bertram et al., 2000; Inagaki et al., 2002) model. In the first case, a linear sequence of amino acids decodes a stop codon, while in the second case a combination of amino acid residues from different parts of the polypeptide chain clustered in space around a stop codon decodes it. Attempts to reveal a ‘protein-anticodon’ for eRF1 have failed so far. In contrast, two regions of the N domain represented by two loops containing highly conserved YxCxxxF (positions 125–131) and NIKS (positions 61–64) motifs (Figure 2) play a critical role in stop codon recognition (Seit-Nebi et al., 2002). Amino acid substitutions in these regions profoundly affect the pattern of stop codon recognition probably due to an interplay between these two loops, which are ∼15 Å apart in the crystal sructure of eRF1 (Song et al., 2000). Furthermore, in yeast, eRF1 mutations affecting stop codon recognition are scattered between positions 51 and 132 of the polypeptide chain (Bertram et al., 2000). In silico analysis (Inagaki et al., 2002) of eRF1 sequences does not support a ‘protein-anticodon’ model as well.
Fig. 2. Two minidomains of the N domain of class-1 eRF1 containing functionally essential amino acid residues in the NIKS and Y-x-C-x-x-x-F motifs. Numbering is as in human eRF1. Sequences are taken from the Swiss Prot database.
The PRIS should be located near both the peptidyl-tRNA in the ribosomal P-site and the PTC of the large ribosomal subunit. In contrast, the TCRS should interact with the decoding site of the small ribosomal subunit. From the distance between the anticodon of tRNA and its CCA end (∼75 Å), one can expect a similar distance between the PTC and the decoding site. As TCRS is assigned to part of the N domain (see above) and the C domain is not essential for the termination reaction (Frolova et al., 2000), the most probable location for PRIS is at the tip of the M domain. This suggestion gets support from a number of experimental data. All class-1 RFs, regardless of their origin and codon specificity, share the common Gly–Gly–Gln tripeptide (GGQ motif) (Frolova et al., 1999). In eRF1, it is located at the extremity of the M domain forming a highly exposed minidomain (Song et al., 2000) (Figure 1). In prokaryotes, the GGQ loop of Escherichia coli RF2 is poorly resolved in the electron density, indicating that it is mobile (Vestergaard et al., 2001). The idea that the invariant GGQ motif is located at the PTC, mimics the CCA end of tRNA and forms a part of the PRIS (Frolova et al., 1999) is supported by the fact that glycine residues of the GGQ in eukaryotic and bacterial factors are indispensable for the RF activity when tested both in vitro and in vivo (Frolova et al., 1999; Song et al., 2000; Mora et al., 2003). For example, GAQ mutants of RF1 and RF2 are between four and five orders of magnitude less efficient in the termination reaction than their wild-type counterparts, although their ability to bind to the ribosome is fully retained (Zavialov et al., 2002).
The essential role of the glycyl residues in the GGQ motif is also emphasized by observations in animal cell–virus systems. Expression of the human cytomegalovirus (HCMV) UL4 gene is inhibited by translation of a 22 codon upstream open reading frame (uORF2) (reviewed in Janzen and Geballe, 2001). The peptide product of uORF2 acts in a sequence-dependent manner to inhibit uORF2 peptidyl-tRNA cleavage. It has been shown by site-directed mutagenesis (Janzen et al., 2002) that Gly183 and Gly184 of the GGQ motif and Pro21 and Pro22 of the uORF2 (the C-terminal residues of the polypeptide) are essential for full inhibition of downstream translation. These data are consistent with the idea that the C-terminal part of the nascent polypeptide in peptidyl-tRNA is able to interact with the GGQ tripeptide at the PTC. It also suggests that this interaction potentially can obstruct translation termination via a particular structure of the C-terminus of the nascent peptidyl-tRNA.
The third amino acid residue of GGQ is also important, but some glutamine mutants retain in vitro a substantial RF activity (Seit-Nebi et al., 2000, 2001; Mora et al., 2003). This is in line with the finding (Zavialov et al. 2002) that although GGA mutants of RF1 and RF2 from E.coli are significantly impaired in the termination step, they are much more active than their GAQ counterparts. This could suggest that the role of Gln185 is in conserving the spatial structure of the GGQ minidomain (Seit-Nebi et al., 2001).
Taken together, these data are hard to reconcile with the proposal (Song et al., 2000) that the function of the glutamine is to orient a water molecule toward peptidyl-tRNA at the PTC of the ribosome. Therefore, the catalytic mechanism of the termination reaction remains unclear.
The Gln252 residue in the GGQ motif of RF2 (E.coli) was found to be N5-methylated (Dinçbas-Renqvist et al., 2000), which increases the termination efficiency of RF2, that again points to a critical role for GGQ in termination.
When the GGQ motif is located at the PTC, then the distance between the GGQ tripeptide and the TCRS should be ∼75 Å. In the crystal structure of human eRF1, the distance spanned by the NIKS motif, which cross-reacts with the first base (U) of the stop codon (Chavatte et al., 2002), and the GGQ motif is ∼100 Å (Song et al., 2000). However, it can be reduced substantially by interactions between eRF1 and the ribosome, and the YxCxxxF motif, rather than the NIKS loop, may be the major TCRS, giving a distance between PRIS and TCRS of ∼75 Å (Seit-Nebi et al., 2002).
The signalling between the stop codon and PTC may involve conformational alterations not only of RFs themselves but also of rRNA sequences, since it is believed (see Caskey, 1980) that the catalytic reaction is carried out by PTC (which is now known to be composed of rRNA) rather than by class-1 RFs. Presumably, the GGQ minidomain opens the PTC to allow the entry of a water molecule, whereas chemical groups of certain rRNA nucleotides catalyse the cleavage reaction.
eRF1 and the class-2 RF, eRF3 (see below), interact via their C-terminal domains (see Kisselev and Buckingham, 2000). Clearly, the C domain of eRF1 is an eRF3-BS (Figure 1). It probably also binds other proteins. There are some other proteins, Upf1p, Upf2p and Upf3p (Wang et al., 2001; and references therein), Mtt1p (Czaplinski et al., 2000) and Itt1p (Urakov et al., 2001), that bind to unknown regions of eRF1. The biological significance of these interactions has been discussed elsewhere (Wang et al., 2001).
The archaean class-1 RF, aRF1, is structurally (Kisselev et al., 2000; Song et al., 2000) and functionally (Dontsova et al., 2000) similar to eRF1, but so far no aRF3 has been identified. It is, in fact, unlikely that an aRF3-BS exists in aRF1, since most aRF1s are truncated extensively from their C-termini, apparently leaving no room for such an interaction.
RF1, RF2 and mtRF1 differ profoundly in their primary structures from the eRF1/aRF1 family (Frolova et al., 1994, 1999; Kisselev et al., 2000; Song et al., 2000). Furthermore, the crystal structures of human eRF1 (Song et al., 2000) and E.coli RF2 (Vestergaard et al., 2001) are also dissimilar. Therefore, the functional anatomy of the RF1/RF2/mtRF structural family may be different from that of eRF1/aRF1. It is possible that bacterial RFs change their conformation when they bind strongly to ribosomes programmed with stop codons, an option suggested above for eRF1.
Recycling of class-1 by class-2 RFs
Class-2 RFs, RF3 in bacteria and eRF3 in eukaryotes, are G proteins not required in vitro for the peptide release reaction itself (Frolova et al., 1994; Grentzmann et al., 1994; Mikuni et al., 1994; Zhouravleva et al., 1995).
Freistroffer et al. (1997) showed that RF3 from E.coli does not accelerate the rate of binding of RF1 or RF2 to ribosomes with peptidyl-tRNA in the P-site and programmed with a stop codon in the A-site. They also demonstrated that the catalytic rate constant for ester bond hydrolysis leading to removal of the peptide from the tRNA in the ribosomal P-site is not affected by RF3. However, the rate of recycling of class-1 RFs is dramatically enhanced by RF3 in a GTP-dependent manner. From these experiments, it could be concluded that the only function of RF3 in translational termination is to remove RF1 or RF2 from ribosomes in their post-termination state, but the exact mechanism of action of RF3 remained unknown. The proper function of RF3 requires that it does not remove class-1 RFs before ester bond hydrolysis of peptidyl-tRNA. This would inhibit termination of protein synthesis, and no such deleterious effects by RF3 had been observed in experiments with well-defined release complexes carrying continuous mRNAs with a stop codon in the A-site and a peptidyl-tRNA in the P-site (Freistroffer et al., 1997). However, in other experimental systems, it had been found that RF3 and a non-hydrolysable analogue of GTP can inhibit termination (e.g. Pel et al., 1998). Furthermore, both in vivo and in vitro experiments had suggested that RF3 can abort protein synthesis by inducing premature dissociation of peptidyl-tRNA from the ribosome (‘drop-off’) (Heurgué-Hamard et al., 1998), but the mechanism for this side reaction remained unexplained.
A breakthrough in our understanding of RF3 function occurred when Zavialov et al. (2001) remeasured the binding of guanine nucleotides to this G protein. Until then, it had been assumed that free cytoplasmic RF3 is in the GTP form. This was based on the notion that the GTP/GDP ratio in the cell is very high, and on experimental data (Mortensen et al., 1995) suggesting that RF3 binds GDP only a factor of five more strongly than GTP.
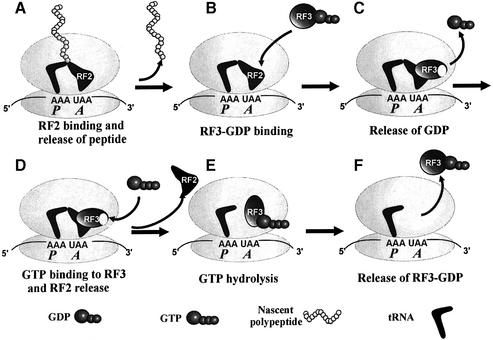
When it was found that GDP binds RF3 at least three orders of magnitude more strongly than does GTP (Zavialov et al., 2001), it meant that cytoplasmic free RF3 is in the GDP-bound rather than the GTP-bound form. Accordingly, RF3 must enter the ribosome in its GDP form, and it was found that the guanine nucleotide exchange factor (GEF) for RF3 is a ribosome with a class-1 RF bound to the ribosomal A-site programmed with the cognate stop codon for that particular RF. The mechanism of action of RF3 can now be described by the following scheme (Figure 3).
Fig. 3. The mechanism of action of RF3 in bacteria. (A) The ribosome in the pre-termination state with peptidyl-tRNA in the P-site and a class-1 release factor (RF) in the A-site. (B) The ribosome in the post-termination state with deacylated tRNA in the P-site and a RF in the A-site. (C and D) The ribosome with tRNA as in (B) and an RF in complex with RF3 in its guanine nucleotide-free state. (E) The ribosome with tRNA as in (B) in complex with RF3 in its GTP conformation. (F) The ribosome with tRNA as in (B) now free from both RF2 and RF3.
RF3 enters the ribosome in the GDP form, and GDP dissociates rapidly if a class-1 RF is present on the ribosome. This reaction happens both when there is a peptidyl-tRNA and when there is a deacylated tRNA in the ribosomal P-site (Zavialov et al., 2002) and leads to a stable ribosomal complex with RF1 or RF2 and guanine nucleotide-free RF3. The existence of this stable complex explains how RF3 can promote erroneous termination at sense codons (Freistroffer et al., 2000). After, but not before, hydrolysis of the ester bond in peptidyl-tRNA, GTP can bind to RF3 on the ribosome and change its conformation (Zavialov et al., 2001, 2002). The fact that RF3 in the GTP form cannot coexist with peptidyl-tRNA on the ribosome is probably the structural basis for the abortive drop-off reaction that has been observed for RF3 (Heurgué-Hamard et al., 1998). The structural change in RF3 induced by GTP binding forces the class-1 RF out from the ribosome in a reaction that is driven forward by formation of a very stable complex between RF3 in the GTP form and the ribosome with a deacylated tRNA in the P-site. Subsequent hydrolysis of GTP leads to the GDP form of RF3 with low affinity for the ribosome, so that the factor dissociates rapidly and becomes ready for a new cycle. According to this scheme, RF3 has three functionally relevant conformations, all of which appear when the factor is in ribosomal complexes. This scenario also implies that RF3 and EF-Tu play totally opposite roles in bacterial protein synthesis. EF-Tu facilitates the entry of aminoacyl-tRNAs to the ribosomal A-site, and helps enhance the accuracy of codon recognition by making the free energy associated with hydrolysis of its GTP available for ribosomal proofreading of aminoacyl-tRNAs (Ruusala et al., 1982). The class-2 RF3, in contrast, removes RF1 or RF2 from the ribosome after termination and reduces, rather than enhances, the accuracy of codon reading by these class-1 RFs. However, in spite of their diametrically opposite functions, EF-Tu and RF3 both belong to the same class of small G proteins that require a GEF, and they have a number of functional properties in common (Zavialov et al., 2001). A challenge for future research is to clarify the mode of action of eRF3 and, in particular, to see if it has the same function as RF3, and this will be discussed next.
eRF3: distinct and common features with RF3
Common features of these two protein families are well known (see Buckingham et al., 1997; Kisselev and Buckingham, 2000; Zavialov et al., 2001). However, along with obvious similarities, there are certain distinct and essential differences. (i) eRF3 proteins are encoded by essential genes (Kushnirov et al., 1988; Wilson and Culbertson, 1988), while this is not the case for RF3 (Grentzmann et al., 1994; Mikuni et al., 1994); this genetic distinction immediately points to a potential difference in function. (ii) The eRF3 and RF3 sequences are dissimilar except for the region where the GTP-binding motifs are located. (iii) RF1/RF2 stimulate ribosome-dependent RF3 GTPase in a codon-dependent manner (Zavialov et al., 2001), while the eRF1 activates the eRF3 GTPase in a codon-independent fashion (Zhouravleva et al., 1995; Frolova et al., 1996). (iv) No significant affinity between free RF1/RF2 and RF3 has been revealed (see Nakamura et al., 1996), whereas eRF1 and eRF3 form a stable complex (for references see Kisselev and Buckingham, 2000). (v) RF1/RF2 are not required for stimulation of ribosome-dependent RF3 GTPase activity at sufficiently high RF3 concentrations (Freistroffer et al., 1997; Grentzmann et al., 1998), in contrast to the eRF3 GTPase, which is entirely eRF1 dependent (Frolova et al., 1996). (vi) In contrast to prokaryotes, where RF3 is absent or there is only one RF3, there are two class-2 RFs in eukaryotes (mouse and human) (Hoshino et al., 1998; Jacobsen et al., 2001) with primary structures that are homologous except for a part of the N domain. (vii) eRF3 binds to the poly(A)-binding protein (Hoshino et al., 1999; Cosson et al., 2002); so far, no such binding has been described for RF3.
To explain how class-1 RFs promote the GTPase activity of class-2 RFs, two assumptions, inspired by the function of other small G proteins, were made by Frolova et al. (1996). They suggested that class-1 RFs in complex with the ribosome may fulfil the role of a GTPase-activator protein (GAP) or a GEF toward class-2 RFs. The GAP hypothesis is supported by the fact that the C domain of class-1 eRF1s contains a short sequence (GILRY) that is crucial for their binding to eRF3 and for the subsequent induction of its GTPase (Merkulova et al., 1999; Frolova et al., 2000). This pentapeptide contains an invariant arginine residue flanked by highly conserved hydrophobic residues. This structural element may act as an arginine finger typical for GAPs where the arginine residues play a catalytic role (see Scheffzek et al., 1998). Eukaryotic translation initiation factor eIF5 contains a similar structural motif and acts as a classical GAP (Paulin et al., 2001). In bacteria, in contrast, a class-1 RF in complex with its stop codon-programmed ribosome acts as a GEF for RF3 (Figure 3; Zavialov et al., 2001).
At the post-termination step of bacterial mRNA translation, one more protein factor, the ribosome recycling factor (RRF), is involved. RRF and the tRNA translocation factor EF-G together recycle the ribosome back to a new round of initiation after termination (Janosi et al., 1996), presumably by splitting the ribosome into its subunits (Karimi et al., 1999). No recycling factor has been identified so far in the cytoplasm of eukaryotic cells. To explain this difference between prokaryotes and eukaryotes, it has been suggested (Buckingham et al., 1997) that the essential eRF3 can recycle both class-1 RFs (done by the non-essential RF3 in bacteria) and ribosomes (done by the essential RRF in bacteria). This hypothesis is consistent with the size of all these proteins and with the known properties of eRF3, RF3 and RRF, but has not been verified directly by experiments.
Concluding remarks
The genetic code is deciphered incompletely in that the mechanism of stop codon recognition remains unknown. However, the problem is now much more well defined since those regions in class-1 RFs from both prokaryotes and eukaryotes that are involved in stop codon recognition have been identified. Rather unexpectedly, it now seems that the translation machineries in prokaryotes and eukaryotes are quite different. High resolution crystal and cryo-electron microscopy structures of bacterial and eukaryotic ribosomes in complex with RFs will be necessary to elucidate specific and universal features of termination of protein synthesis.
Signal transduction from the stop codon of mRNA in the small ribosomal subunit to the PTC in the large ribosomal subunit is likely to depend not only on class-1 RFs but also on rRNAs and to involve as yet unknown specific RNA–protein interactions.
There is growing evidence that class-1 RFs recognize stop codons directly within the decoding site of the ribosome, but this attractive idea still lacks solid experimental support in both prokaryotic and eukaryotic systems. It is likely that recognition of stop codons involves an mRNA–class-1 RF–rRNA ternary complex, but its molecular basis remains to be clarified.
The chemical step of termination, i.e. cleavage of the ester bond in peptidyl-tRNA, is also a consequence of an interplay between two nucleic acids, rRNA and peptidyl-tRNA, with a class-1 RF. However, in this case also, the molecular mechanism remains to be elucidated. The post-terminal steps in protein synthesis are much better understood in prokaryotes than in eukaryotes, and to clarify ribosomal recycling in eukaryotes is a major challenge for future work.
In spite of remarkable progress in the study of translation termination, there are still large gaps in our general understanding of the process. Furthermore, the molecular basis used by nature to achieve fast and accurate termination at stop codons is virtually unknown.
Acknowledgments
Acknowledgements
The work of the L.K. laboratory has been supported during the last years by the Human Frontier Science Program, a Chaire Internationale Blaise Pascal award, the EU (via INTAS) and the Russian Foundation for Basic Research and Support for Russian Scientific Schools. M.E. has been supported by the Swedish Foundation for Strategic Research (SSF) and the Swedish Research Council.
References
- Arkov A.L. and Murgola,J. (1999) Ribosomal RNAs in translation termination: facts and hypotheses. Biochimiya (Moscow), 64, 1354–1359. [PubMed] [Google Scholar]
- Bertram G., Bell,H.A., Ritchie,D.W., Fullerton,G. and Stansfield,I. (2000) Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA, 6, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G., Innes,S., Minella,O., Richardson.J. and Stansfield,I. (2001) Endless possibilities: translation termination and stop codon recognition. Microbiology, 147, 255–269. [DOI] [PubMed] [Google Scholar]
- Buckingham R.H., Grentzmann,G. and Kisselev,L. (1997) Polypeptide chain release factors. Mol. Microbiol., 24, 449–456. [DOI] [PubMed] [Google Scholar]
- Bulygin K.N., Repkova,M.N, Ven’yaminova,A.G., Graifer,D.M., Karpova,G.G., Frolova,L.Yu. and Kisselev,L.L. (2002) Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes. FEBS Lett., 514, 96–101. [DOI] [PubMed] [Google Scholar]
- Caskey C.T. (1980) Peptide chain termination. Trends Biochem. Sci., 5, 234–237. [Google Scholar]
- Chavatte L., Frolova,L., Kisselev,L. and Favre,A. (2001) The polypeptide chain release factor eRF1 specifically contacts the s(4)UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem., 268, 2896–2904. [DOI] [PubMed] [Google Scholar]
- Chavatte L., Seit-Nebi,A., Dubovaya,V. and Favre,A. (2002) The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J., 21, 5302–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B., Couturier,A., Chabelskaya,S., Kiktev,D., Inge-Vechtomov,S., Philippe,M. and Zhouravleva,G. (2002) Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI+] propagation. Mol. Cell Biol., 22, 3301–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Majlesi,N., Banerjee,T. and Peltz,S.W. (2000) Mttl is a Upfl-like helicase that interacts with the translation termination factors and whose overexpression can modulate termination efficiency. RNA, 6, 730–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N., Repkova,V., Ven’yaminova,A., Graifer,D. and Karpova,G. (2000) Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P and E sites: a crosslinking study with mRNA analogs carrying an aryl azide group at either the uracil or the guanine residue. RNA, 6, 1727–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinçbas-Renqvist V., Engström,Å., Mora,L., Heurgué-Hamard,V., Buckingham,R.H. and Ehrenberg,M. (2000) A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J., 19, 6900–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontsova M., Frolova,L., Vassilieva,J., Piendl,W., Kisselev,L. and Garber,M. (2000) Translation termination factor aRF1 from the archaeon Methanococcus jannaschii is active with eukaryotic ribosomes. FEBS Lett., 472, 213–216. [DOI] [PubMed] [Google Scholar]
- Freistroffer D.V., Pavlov,M.Y., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J., 16, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freistroffer D.V., Kwiatkowski,M., Buckingham,R.H. and Ehrenberg,M. (2000) The accuracy of codon recognition by polypeptide release factors. Proc. Natl Acad. Sci. USA, 97, 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L. et al. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature, 372, 701–703. [DOI] [PubMed] [Google Scholar]
- Frolova L., Le Goff,X., Zhouravleva,G., Davydova,E., Philippe,M. and Kisselev,L. (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA, 2, 334–341. [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Y., Tsivkovskii,R.Y., Sivolobova,G.F., Oparina,N.Y., Serpinsky,O.I., Blinov,V.M., Tatkov,S.I. and Kisselev,L.L. (1999) Mutations in the highly conserved GGQ motif of class-1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA, 5, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Y., Merkulova,T.I. and Kisselev,L.L. (2000) Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA, 6, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L., Seit-Nebi,A. and Kisselev,L. (2002) Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA, 8, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. and Noller,H.F. (1997) Ribosomes and translation. Annu. Rev. Biochem., 66, 679–716. [DOI] [PubMed] [Google Scholar]
- Grentzmann G., Brechemier-Baey,D., Heurgué,V., Mora,L. and Buckingham,R.H. (1994) Localization and characterization of the gene encoding release factor RF3 in Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 5848–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G., Kelly,P.J., Laalami,S., Shuda,M., Firpo,M.A., Cenatiempo,Y. and Kaji,A. (1998) Release factor RF-3 GTPase activity acts in disassembly of the ribosome termination complex. RNA, 8, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgué-Hamard V., Karimi,R., Mora,L., MacDougall,J., Leboeuf,C., Grentzmann,G., Ehrenberg,M. and Buckingham,R.H. (1998) Ribosome release factor RF4 and termination factor RF3 are involved in dissociation of peptidyl-tRNA from the ribosome. EMBO J., 17, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S., Imai,M., Mizutani,M., Kikuchi,Y., Hanaoka,F., Ui,M. and Katada,T. (1998) Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J. Biol. Chem., 273, 22254–22259. [DOI] [PubMed] [Google Scholar]
- Hoshino S., Imai,M., Kobayashi,T., Uchida,N. and Katada,T. (1999) The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem., 274, 16677–16680. [DOI] [PubMed] [Google Scholar]
- Inagaki Y. and Doolittle,W.F. (2001) Class I release factors in ciliates with variant genetic codes. Nucleic Acids Res., 29, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y., Blouin,C., Doolittle,W.F. and Roger,A.J. (2002) Con vergence and constraint in eukaryotic release factor (eRF1) domain 1: the evolution of stop codon specificity. Nucleic Acids Res., 30, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Uno,M. and Nakamura,Y. (2000) A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- Ito K., Frolova,L., Seit-Nebi,A., Karamyshev,A., Kisselev,L. and Nakamura,Y. (2002) Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within the domain 1. Proc. Natl Acad. Sci. USA, 99, 8494–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov V., Beniaminov,A., Mikheev,A. and Minyat,E. (2001) A mechanism for stop codon recognition by the ribosome: a bioinformatic approach. RNA, 7, 1683–1692. [PMC free article] [PubMed] [Google Scholar]
- Jacobsen C.G., Segaard,T.M.M., Jean-Jean,O., Frolova,L. and Justesen,J. (2001) Identification of eRF3b, a human polypeptide chain release factor with eRF3 activity in vitro and in vivo. Mol. Biol., 35, 672–681. [PubMed] [Google Scholar]
- Janosi L., Hara,H., Zhang,S. and Kaji,A. (1996) Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv. Biophys., 32, 121–201. [DOI] [PubMed] [Google Scholar]
- Janzen D.M. and Geballe,A.P. (2001) Modulation of translation termination mechanisms by cis- and trans-acting factors. Cold Spring Harbor Symp. Quant. Biol., 66, 459–467. [DOI] [PubMed] [Google Scholar]
- Janzen D.M., Frolova,L. and Geballe,A. (2002) Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol. Cell. Biol., 22, 8562–8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R., Pavlov,M.Y., Buckingham,R.H. and Ehrenberg,M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Kervestin S., Frolova,L., Kisselev,L. and Jean-Jean,O. (2001) Stop codon recognition in ciliates: Euplotes release factor does not respond to reassigned UGA codon. EMBO Rep., 2, 680–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev L.L. and Buckingham,R.H. (2000) Translation termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- Kisselev L.L., Oparina,N.Yu. and Frolova,L.Yu. (2000) Class-1 polypeptide chain release factors are structurally and functionally similar to suppressor tRNAs and comprise different structural– functional families of prokaryotic/mitochondrial and eukaryotic/archaebacterial factors. Mol. Biol., 34, 427–442. [PubMed] [Google Scholar]
- Knight R.D., Freeland,S.J. and Landweber,L.F. (2001) Rewiring the keyboard: evolvability of the genetic code. Nat. Rev. Genet., 2, 49–58. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V., Ter-Avanesyan,M.D., Telckov,M.V., Surguchov,A.P., Smirnov,V.N. and Inge-Vechtomov,S.G. (1988) Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene, 66, 45–54. [DOI] [PubMed] [Google Scholar]
- Liang A., Brunen-Nieweler,C., Muramatsu,T., Kuchino,Y., Beier,H. and Heckmann,K. (2001) The ciliate Euplotes octocarinatus expresses two polypeptide release factors of the type eRF1. Gene, 262, 161–168. [DOI] [PubMed] [Google Scholar]
- Lozupone C.A., Knight,R.D. and Landweber,L.F. (2001) The molecular basis of nuclear genetic code change in ciliates. Curr. Biol., 11, 65–74. [DOI] [PubMed] [Google Scholar]
- Merkulova T.I., Frolova,L.Y., Lazar,M., Camonis,J. and Kisselev,L.L. (1999) C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett., 443, 41–47. [DOI] [PubMed] [Google Scholar]
- Mikuni O., Ito,K., Moffat,J., Matsumura,K., McCaughan,K., Nobukuni,T., Tate,W. and Nakamura,Y. (1994) Identification of the prfC gene, which encodes peptide chain release factor 3 of Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 5798–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. and Noller,H.F. (1987) Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature, 327, 389–394. [DOI] [PubMed] [Google Scholar]
- Moffat J.G. and Tate,W.P. (1994) A single proteolytic cleavage in release factor 2 stabilizes ribosome binding and abolishes peptidyl-tRNA hydrolysis activity. J. Biol. Chem., 269, 18899–18903. [PubMed] [Google Scholar]
- Mora L., Heurgué-Hamard,V., Champ,S., Ehrenberg,M., Kisselev,L.L. and Buckingham,R.H. (2003) The essential role of the invariant GGQ motif in the function and the stability in vivo of bacterial release factors RF1 and RF2. Mol. Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- Mortensen K.K., Hansen,H.F., Grentzmann,G., Buckingham,R.H. and Sperling-Petersen,H.U. (1995) Osmo-expression and fast two-step purification of Escherichia coli translation termination factor RF-3. Eur. J. Biochem., 34, 732–736. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. and Ito,K. (1998) How protein reads the stop codon and terminates translation. Genes Cells, 3, 265–278. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. and Ito,K. (2002) A tripeptide discriminator for stop codon recognition. FEBS Lett., 514, 30–33. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito,K. and Isaksson,L.A. (1996) Emerging understanding of translation termination. Cell, 87, 147–150. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito,K. and Ehrenberg,M. (2000) Mimicry grasps reality in translation termination. Cell, 101, 349–352. [DOI] [PubMed] [Google Scholar]
- Paulin E.M., Campbell,L.E., O’Brien,K., Loughlin,J. and Proud,C.G. (2001) Eukaryotic translation initiation factor 5 (elF5) acts as a classical GTPase-activator protein. Curr. Biol., 11, 55–59. [DOI] [PubMed] [Google Scholar]
- Pel H.J., Moffat,J.G., Ito,K., Nakamura,Y. and Tate,W.P. (1998) Escherichia coli release factor 3: resolving the paradox of a typical G protein structure and atypical function with guanine nucleotides. RNA, 4, 47–54. [PMC free article] [PubMed] [Google Scholar]
- Poole E. and Tate,W. (2000) Release factors and their role as decoding proteins: specificity and fidelity for termination of protein synthesis. Biochim. Biophys. Acta, 1493, 1–11. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. (2002) Ribosome structure and the mechanism of translation. Cell, 108, 557–572. [DOI] [PubMed] [Google Scholar]
- Ruusala T., Ehrenberg,M., Caskey,C.T. and Nirenberg,M. (1982) Is there proof-reading during polypeptide synthesis? EMBO J., 1, 741–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A., Frolova,L., Ivanova,N., Poltaraus,A. and Kisselev,L. (2000) Substitutions of the glutamine residue in ubiquitous GGQ tripeptide in human eRF1 do not entirely abolish the release factor activity. Mol. Biol. (Mosk.), 34, 899–900. [PubMed] [Google Scholar]
- Seit-Nebi A., Frolova,L., Justesen,J. and Kisselev,L. (2001) Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res., 29, 3982–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A., Frolova,L. and Kisselev,L. (2002) Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep., 9, 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian,M.R. and Wittinghofer,A. (1998) GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci., 23, 257–262. [DOI] [PubMed] [Google Scholar]
- Song H., Mugnier,P., Das,A.K., Webb,H.M., Evans,D.R., Tuite,M.F., Hemmings,B.A. and Barford,D. (2000) The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- Szkaradkiewicz K., Nanninga,M., Nesper-Brock,M., Gerrits,M., Erdmann,V.A. and Sprinzl,M. (2002) RNA aptamers directed against release factor 1 from Thermus thermophilus. FEBS Lett., 514, 90–95. [DOI] [PubMed] [Google Scholar]
- Urakov V.N., Valouev,I.A., Lewitin,E.I., Paushkin,S.V., Kosorukov,V.S., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (2001) IttIp, a novel protein inhibiting translation termination in Saccharomyces cerevisiae. BMC Mol. Biol., 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina I.V., Hong,J.Y., Mesecar,A.D., Chernoff,Y.O. and Liebman,S.W. (2001) Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18S rRNA. J. Mol. Biol., 305, 715–727. [DOI] [PubMed] [Google Scholar]
- Vestergaard B., Van,L.B., Andersen,G.R., Nyborg,J., Buckingham,R.H. and Kjeldgaard,M. (2001) Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell, 8, 1375–1382. [DOI] [PubMed] [Google Scholar]
- Wang W., Czaplinski,K., Rao,Y. and Peltz,W. (2001) The role of Upf proteins in modulating the translation read-trough of nonsense-containing transcripts. EMBO J., 20, 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.N., Blaha,G., Connell,S.R., Ivanov,P.V., Jenke,H., Stelzl,U., Teraoka,Y. and Nierhaus,K.H. (2002) Protein synthesis at atomic resolution: mechanistics of translation in the light of highly resolved structures for the ribosome. Curr. Protein Peptide Sci., 3, 1–53. [DOI] [PubMed] [Google Scholar]
- Wilson P.G. and Culbertson,M.R. (1988) SUF12 suppressor protein of yeast. A fusion protein related to the EF-1 family of elongation factors. J. Mol. Biol., 199, 559–573. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Buckingham,R.H. and Ehrenberg,M. (2001) A post-termination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell, 107, 115–124. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Mora,L., Buckingham,R.H. and Ehrenberg,M. (2002) Release of peptide promoted by the GGQ-motif of class-1 release factors regulates the GTPase activity of RF3. Mol. Cell, 10, 789–798. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova,L., Le Goff,X., Le Guellec,R., Inge-Vechtomov,S., Kisselev,L. and Philippe,M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]