Abstract
The relationship of trehalose metabolism to fungal virulence was explored in the rice blast fungus Magnaporthe grisea. To determine the role of trehalose synthesis in pathogenesis, we identified and deleted TPS1, encoding trehalose-6-phosphate synthase. A Δtps1 mutant failed to synthesize trehalose, sporulated poorly and was greatly attenuated in pathogenicity. Appressoria produced by Δtps1 did not develop full turgor or elaborate penetration hyphae efficiently. To determine the role of subsequent trehalose breakdown, we deleted NTH1, which encodes a neutral trehalase. Nth1 mutants infected plants normally, but showed attenuated pathogenicity due to a decreased ability to colonize plant tissue. A second trehalase was also identified, required both for growth on trehalose and mobilization of intracellular trehalose during infection-related development. TRE1 encodes a cell wall-localized enzyme with characteristics of both neutral and acidic trehalases, but is dispensable for pathogenicity. Our results indicate that trehalose synthesis, but not its subsequent breakdown, is required for primary plant infection by M.grisea, while trehalose degradation is important for efficient development of the fungus in plant tissue following initial infection.
Keywords: Magnaporthe grisea/metabolism/trehalase/virulence
Introduction
Trehalose (α-d-glucopyranosyl-α-d-glucopyranoside) is a non-reducing disaccharide that is commonly found as a storage carbohydrate in eukaryotic cells (Arguelles, 2000) and implicated in the response to various environmental stresses (Thevelein, 1984, 1996). In Saccharomyces cerevisiae, trehalose has been shown to be required by cells to contend with diverse stresses such as heat shock, starvation and desiccation (De Virgilio et al., 1994). Trehalose is also known to protect enzymes from thermal stress in vitro, providing a potential means for cells to survive during adverse conditions (Hottiger et al., 1994).
Investigations to determine the biological function of trehalose in eukaryotes have been limited predominantly to the study of free-living species. Consequently, little is known about trehalose metabolism in pathogenic microorganisms. Many virulence-associated functions, including infection-related development and colonization of host tissue, require pathogens to respond to rapid changes in external environment and to mobilize storage carbohydrates. In this study, we set out to determine the role of trehalose metabolism in a plant pathogenic fungus, Magnaporthe grisea. This pathogen is the causal agent of rice blast disease, the most serious disease of cultivated rice (Talbot and Foster, 2001).
Trehalose metabolism in eukaryotic cells has been studied in most detail in S.cerevisiae. Here, trehalose is synthesized by a multienzyme complex containing trehalose-6-phosphate synthase (T6PS) encoded by TPS1, a trehalose-6-phosphatase encoded by TPS2, and two regulatory subunits encoded by the TSL1 and TPS3 genes (Vuorio et al., 1993; Bell et al., 1998). Trehalose-6-phosphate (T6P) is synthesized using UDP-glucose and glucose-6-phosphate as substrates, and then directly converted to trehalose. Three distinct trehalases are present in S.cerevisiae. Acidic trehalase, encoded by ATH1, is required for utilization of trehalose as a carbon source and is a vacuolar enzyme with optimum activity at low pH (Londesborough and Varimo, 1984; Mittenbühler and Holzer, 1988). Mobilization of intracellular trehalose is catalysed by cytoplasmic neutral trehalase, encoded by NTH1 and NTH2 (Nwaka and Holzer, 1998). In filamentous fungi such as Aspergillus nidulans, trehalose synthesis involves T6PS encoded by the tpsA gene (Fillinger et al., 2001), and trehalose metabolism involves an acidic, cell wall-localized trehalase required for utilization of exogenous trehalose (d’Enfert and Fontaine, 1997), and a cAMP-regulated neutral trehalase (Nth1) required for trehalose mobilization during spore germination (d’Enfert et al., 1999).
We reasoned that several virulence-associated functions in M.grisea might involve trehalose mobilization, e.g. germination of conidia and the development of infected cells on the leaf surface, and subsequent plant tissue colonization. Here we report the isolation and characterization of genes encoding biosynthetic and degradative enzymes involved in trehalose metabolism in M.grisea. We show that trehalose synthesis in M.grisea is mediated by a T6PS-encoding gene, TPS1, which is required for production of functional infection structures and primary plant infection. We also demonstrate that trehalose breakdown involves two trehalases; a neutral trehalase, encoded by a gene NTH1, which is important for invasive growth in plant cells, and a novel trehalase encoded by TRE1 which is required for trehalose mobilization during spore germination, but dispensable for pathogenicity. Taken together, our results indicate that trehalose synthesis is required for appressorium-mediated plant infection by a plant pathogenic fungus, while mobilization of stored trehalose is significant only after cuticle penetration.
Results
Trehalose synthesis is required for pathogenicity of M.grisea
To determine the role of trehalose synthesis in pathogenesis of M.grisea, we identified a gene encoding T6PS by designing degenerate primers to regions conserved among T6PS genes. A 650 bp product was amplified and used to identify a cDNA and genomic clone. This revealed the presence of a 1587 bp open reading frame (ORF) capable of encoding a 529 amino acid protein of 58.4 kDa. We named the gene TPS1, and the putative gene product showed 76% identity to tpsA from A.niger and 65% identity to TPS1 from S.cerevisiae (DDBJ/EMBL/GenBank accession No. AY148093; see Supplementary data available at The EMBO Journal Online). A gene replacement of TPS1 was carried out. For this, an 8 kb Pst I fragment was isolated and a 2.0 kb NcoI fragment containing part of the TPS1 promoter and most of the ORF was removed and replaced with the Hph cassette. The resulting vector was introduced into a wild-type rice pathogenic strain of M.grisea, Guy11, and 40 transformants were selected. In DNA gel blots, one transformant, T-5-13, had undergone the Δtps1 gene replacement (see Supplementary data). The putative Δtps1 mutant T-5-13 was selected and mycelial cultures prepared for measurement of intracellular trehalose. We found that trehalose was almost completely absent from the Δtps1 mutant, compared with levels observed in Guy11 and either Δnth1 or tre1::Hph mutants (Figure 1).
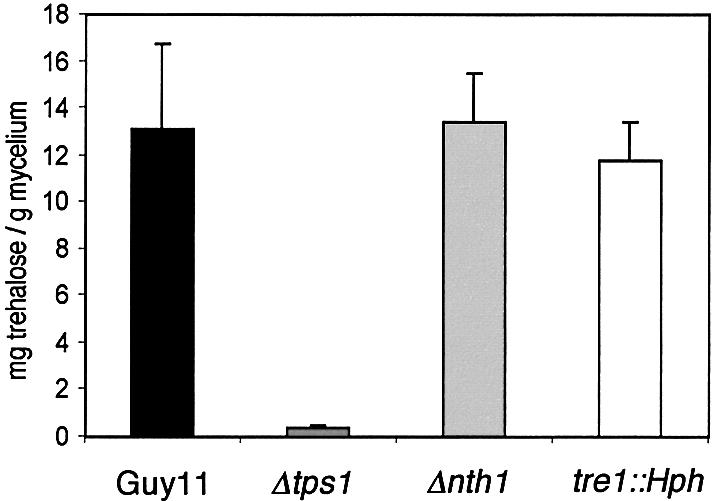
Fig. 1. Bar chart of trehalose levels in mycelium of M.grisea strains Guy11, Δtps1 mutant T-5-13, Δnth1 mutant T-2-8 and tre1::Hph mutant T-8-45. Data points represent the mean value from three experiments. The error bar represents the standard deviation.
We investigated the role of TPS1 in growth, development and virulence of M.grisea. The Δtps1 mutant showed normal mycelial growth and did not respond differently when exposed to heat shock (not shown). Sporulation was dramatically reduced in the Δtps1 mutant. Guy11 produced a mean of 1.39 × 10 6 conidia per plate culture (n = 8) compared with 3.6 × 103 for Δtps1 strain T-5-13 (P < 0.001). Conidial germination rates and appressorium formation were not significantly affected by the Δtps1 mutation (not shown). To investigate the role of TPS1 in pathogenesis, conidial suspensions were sprayed onto seedlings of the susceptible rice cultivar CO-39. A consistent 90% reduction in disease lesion development was found in plants inoculated with Δtps1 compared with Guy11, as shown in Figure 2A (t-test, P < 0.001). The small number of disease lesions in Δtps1 infections did not expand during infection or produce sporulating hyphae. The Δtps1 mutant was also non-pathogenic on barley. The pathogenicity of Δtps1 mutants could not be restored by adding trehalose or T6P to conidia, prior to inoculation (not shown). Introduction of the TPS1 gene into Δtps1 mutant T-5-13 restored its ability to cause disease (Figure 2A). All other Δtps1 mutant phenotypes were complemented by introduction of TPS1 (not shown). We conclude that TPS1 is required for trehalose synthesis in M.grisea, and is necessary for rice blast disease.
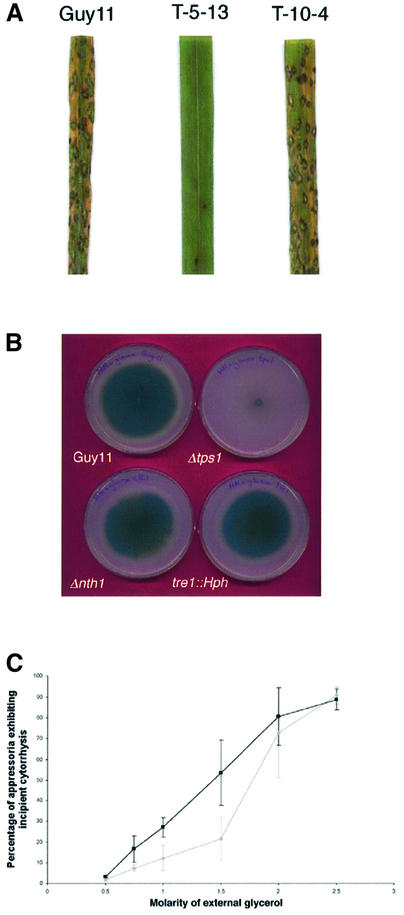
Fig. 2. Phenotypic analysis of M.grisea Δtps1 mutant T-5-13. (A) Virulence phenotype of the Δtps1 mutant. Seedlings of cv. CO-39 were inoculated with M.grisea conidial suspensions of identical concentration (2 × 104 conidia/ml) of strain Guy11 (leaf 1), Δtps1 mutant T-5-13, and a transformant (T-10-4) of Δtps1mutant T-5-13 carrying a single copy insertion of pAJF200, an 8 kb PstI fragment of TPS1. Seedlings were incubated for 4 days for development of rice blast disease. (B) Growth of M.grisea strain Guy11, Δtps1 mutant T-5-13, Δnth1 mutant T-2-8 and tre1::Hph mutant T-8-45 with glucose as sole carbon source. (C) Appressorium turgor of Guy11 (grey diamonds) and Δtps1 mutant T-5-13 (black squares) was measured by incipient cytorrhysis. The proportion of appressoria that collapsed after exposure to solutions of between 0 and 5.0 M glycerol was recorded. Each data point is the mean of three independent observations. The error bar represents the standard deviation.
TPS1 is required for appressorium-mediated cuticle penetration
To determine why Δtps1 mutants are non-pathogenic, the ability of the Δtps1 mutant to penetrate intact rice cuticles was measured. Appressoria developed normally in the Δtps1 mutant, but production of penetration hyphae and rupture of the plant cuticle were significantly decreased from 81 ± 7.2% successful penetrations by Guy11 to 26.6 ± 17.4% in Δtps1 (P < 0.001). Consistent with this, the Δtps1 mutant did not cause rice blast symptoms when spores were inoculated on intact rice leaves, but was able to produce disease symptoms when the rice cuticle was first removed by abrasion (not shown). Taken together, the experiments indicate that TPS1 is required for plant infection, but not subsequent growth of the fungus in rice tissue. Plant infection by M.grisea is brought about by the appressorium, which develops very high internal turgor to facilitate generation of mechanical force to breach the rice leaf cuticle. Appressorium turgor can be measured using an incipient cytorrhysis assay which uses hyperosmotic concentrations of a solute to collapse appressoria, thereby allowing estimation of their internal solute concentration and turgor (Howard et al., 1991; de Jong et al., 1997). To carry out this assay, appressoria were allowed to form on a hydrophobic plastic surface and then incubated in glycerol solutions of varying concentration (Figure 2C). We found that 1.5 M glycerol was sufficient to collapse 21.8 ± 10.4% of appressoria of Guy11 and 53.4 ± 15.7% of Δtps1 appressoria, indicating a significant decrease in turgor in the Δtps1 mutant (P = 0.0074). Turgor was not restored by adding trehalose or T6P to germinating conidia of Δtps1 (not shown). We conclude that TPS1 is required for appressorium-mediated cuticle penetration by M.grisea.
M.grisea TPS1 is required for growth on glucose
In S.cerevisiae, T6PS is required for regulation of glucose intake into glycolysis (Thevelein and Hohmann, 1995). As a consequence, yeast Δtps1 mutants are unable to grow on glucose. Conversely, in the filamentous fungus A.nidulans, tpsA may play a role in regulating levels of sugar-phosphates, but mutants lacking T6PS still grow on glucose (Fillinger et al., 2001). To determine whether M.grisea TPS1 is required for glycolytic regulation, we incubated the Δtps1 mutant on minimal growth medium with glucose as sole carbon source (Figure 2B). The Δtps1 mutant was unable to grow on glucose or other simple sugars including fructose, sucrose, maltose and trehalose (Table I). In addition, unlike yeast T6PS mutants, M.grisea Δtps1 failed to grow on acetate or fatty acids, indicating that gluconeogenesis is also affected by the absence of the T6PS. In rich growth media, we observed that Δtps1 mutants were able to utilize glucose, but only in the presence of a rich source of amino acids, provided either from yeast extract, peptone or casamino acids. Under these conditions, Δtps1 mutants were still unable to utilize acetate as a carbon source (Table I). We conclude that M.grisea TPS1 is required for growth on a range of rapidly fermentable carbon sources, and may be required for regulation of glycolysis and gluconeogenesis.
Table I. Growth of trehalose metabolic mutants of M.grisea on different carbon sources and in supplemented growth medium.
| Growth medium | Guy-11 | Δtps1 | Δnth1 | tre:Hph |
|---|---|---|---|---|
| Complete medium (CM)a | + + +b | + + + | + + + | + + + |
| Oatmeal agar | + + + | + | + + + | + + + |
| CM – glucose | – | – | – | – |
| CM – nitrate salts | + + + | + + + | + + + | + + + |
| CM – peptone | + + + | + + + | + + + | + + + |
| CM – yeast extract | + + + | + + + | + + + | + + + |
| CM – casamino acids | + + + | + + + | + + + | + + + |
| CM – vitamin solution | + + + | + + + | + + + | + + + |
| CM – yeast extract and casamino acids | + + + | + + + | + + + | + + + |
| CM – yeast extract and peptone | + + + | + + + | + + + | + + + |
| CM – yeast extract and glucose | – | – | – | – |
| Minimal medium (MM) + glucosec | + + + | – | + + + | + + + |
| MM + trehalose | + + + | – | + + + | – |
| MM + fructose | + + + | – | + + + | + + + |
| MM + maltose | + + + | – | + + + | + + + |
| MM + sucrose | + + + | – | + + + | + + + |
| MM + olive oil | + + | – | + + | + + |
| MM + sodium acetate | + + | – | + | + |
| MM + sodium acetate + glucose | + + + | – | + + + | + + + |
| MM + mannitol | + + | – | + | + |
| MM + mannitol + glucose | + + + | – | + + + | + + + |
| MM + galactose | – | – | – | – |
| MM + glycerol | – | – | – | – |
| MM + glycerol + glucose | + + + | – | + + + | + + + |
| MM + ethanol | – | – | – | – |
| MM + ethanol + glucose | + + + | – | + + + | + + + |
| MM – carbon source | – | – | – | – |
| MM + peptone | – | – | – | – |
| MM + peptone + glucose | + + + | + + + | + + + | + + + |
| MM + yeast extract | – | – | – | – |
| MM + yeast extract + glucose | + + + | + + + | + + + | + + + |
| MM + peptone + yeast extract + glucose | + + + | + + + | + + + | + + + |
| MM + casamino acids | – | – | – | – |
| MM + casamino acids + glucose | + + + | + + + | + + + | + + + |
| MM + casamino acids + peptone + glucose | + + + | + + + | + + + | + + + |
| MM + casamino acids + yeast extract + glucose | + + + | + + + | + + + | + + + |
| MM + casamino acids + fructose | + + + | + + + | + + + | + + + |
| MM + casamino acids + sodium acetate | + + + | – | + + + | + + + |
| MM + casamino acids + olive oil | + + | + | + + | + + |
| MM + vitamin solution + glucose | + + + | – | + + + | + + + |
| MM + vitamin solution + yeast extract + glucose | + + + | + + + | + + + | + + + |
aComplete medium is a rich growth medium containing yeast extract, peptone and casamino acids (see Materials and methods for recipe). CM normally contains glucose as the principal carbon source. The medium was supplemented (+) or depleted (–) as shown.
b+++ = extensive hyphal growth and sporulation; ++ = reduced growth; + = very poor growth and sporulation; – = no growth/sparse mycelium, no sporulation. All growth tests were carried out on agar-solidified medium and monitored after 12 days incubation at 24°C.
cMinimal medium is described in Materials and methods. Supplementations were made of single or multiple carbon sources as indicated (+).
Isolation of NTH1 encoding a neutral trehalase from M.grisea
To investigate how stored trehalose in M.grisea is mobilized, we identified a gene encoding the intracellular regulated form of trehalase. A previous study to identify M.grisea genes involved in virulence using insertional mutagenesis had revealed a gene called PTH9, which encodes a protein showing similarity to neutral trehalase (Sweigard et al., 1998). We obtained the pth9 insertion mutant E508-R-6 and a PTH9 genomic clone (kindly provided by Dr J.A.Sweigard, DuPont Company, Wilmington, DE). DNA sequencing defined a 2211 bp ORF encoding a 736 amino acid protein of 84.6 kDa, showing 81% identity to the treB neutral trehalase enzyme from N.crassa and 68% identity to treB from A.nidulans (DDBJ/EMBL/GenBank accession No. AY148092; see Supplementary data). We named the gene NTH1 for neutral trehalase. The NTH1 protein sequence contains a consensus phosphorylation site for cAMP-dependent protein kinase A (PKA) and a putative Ca2+-binding site, indicating that the gene encodes a regulated form of trehalase. To investigate NTH1 expression, RNA gel blots showed that the 2.7 kb NTH1 transcript was 4-fold more abundant during conidiogenesis, than in vegetative hyphae. NTH1 was also expressed abundantly in mycelium after exposure to 0.5 M NaCl for 24 h. The same response was observed after hyperosmotic stress using sorbitol (not shown) and was independent of the presence of the osmoregulatory mitogen-activated protein (MAP) kinase gene OSM1 (Dixon et al., 1999), as shown in Figure 3A. Spatial expression of NTH1 was determined by fusing a 2.5 kb NTH1 promoter fragment upstream of the green fluorescent protein (GFP)-encoding gene sGFP (Chiu et al., 1996) and introducing the plasmid into M.grisea. Transformants containing a single copy of NTH1(p):sGFP were examined by epifluorescence microscopy (Figure 3B). NTH1(p):sGFP expression occurred during initial stages of conidiation when conidiophores develop from a foot cell and differentiate to produce a sympodial array of conidia (Figure 3B–D). Expression was also observed in invasive hyphae within plant tissue, indicating that expression of NTH1 occurs after cuticle penetration (Figure 3E–F). Sequence analysis of the NTH1 promoter revealed putative binding sites for homologues of the stuA and abaA transcriptional activators known to regulate conidiogenesis in A.nidulans (Timberlake, 1993) and three consensus stress response elements (STREs) found in genes expressed during the multistress response in yeast (Schuller et al., 1994). We conclude that NTH1 is expressed during M.grisea sporulation, plant infection and in response to environmental stress.
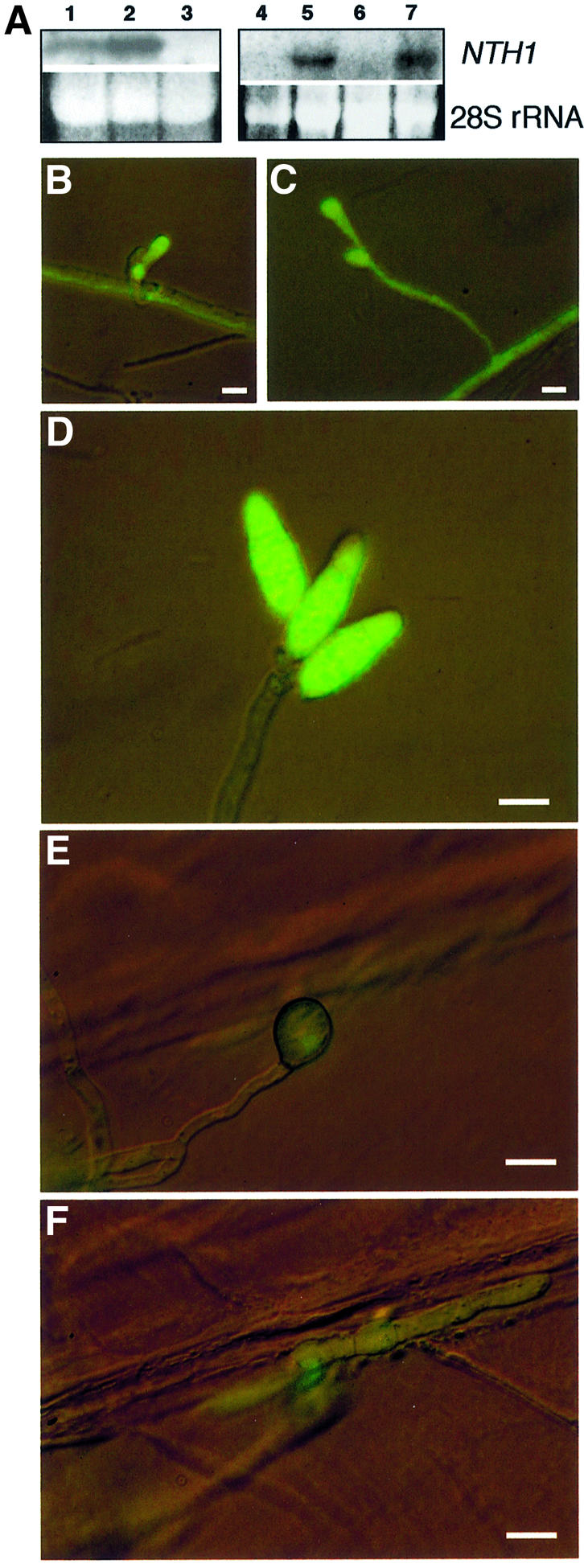
Fig. 3. Expression of M.grisea NTH1 during conidiogenesis, plant infection and in response to hyperosmotic stress. (A) RNA was extracted from a non-conidiating culture of M.grisea strain Guy11 (lane 1), a conidiating culture of Guy11 (lane 2) and a conidiating culture of nth1 mutant E508.R.6 (lane 3). RNA was also extracted from hyphal cultures of M.grisea grown in iso-osmotic media, CM (lanes 4 and 6), or subjected to acute hyperosmotic stress in CM + 0.5 M NaCl for 24 h (lanes 5 and 7). RNA was from strain Guy11 (lanes 4 and 5) or Δosm1 mutant JH73 (lanes 6 and 7). RNA gel blots were probed with NTH1. The 28S rRNA is shown as a loading standard. (B) Spatial expression of NTH1. Early conidiogenesis initiated from a foot cell in NTH1(p)::sGFP::Hph transformant T-4-11. (C) Conidial initials forming at the end of a conidiophore. (D) Mature conidiophore with three sympodially arrayed conidia. (E) Germ tube and appressorium on barley epidermis. (F) An invasive hypha entering an epidermal cell from the base of the same appressorium. Bar in all panels = 10 µm.
NTH1 encodes a virulence factor for rice blast disease
The pth9 insertional mutant E508-R-6 contained a 4.0 kb plasmid insertion in the NTH1 ORF, but theoretically was capable of producing a truncated NTH1 trehalase (Figure 4A and B), although RNA gel blots suggested that the mutant did not produce an NTH1 transcript (Figure 3A). To ensure generation of a null mutant, a gene replacement was carried out by deleting 2.4 kb of NTH1 and replacing this with a hygromycin resistance cassette. DNA gel blots identified three transformants where the gene replacement had occurred from 38 selected (see Supplementary data available at The EMBO Journal Online). The three Δnth1 mutants T-2-8, T-2-30 and T-2-37 were not affected in hyphal growth, response to heat stress or the temporal dynamics of spore germination (not shown). The Δnth1 mutants did show a significant decrease in sporulation, producing ∼50% of the number of conidia in the wild-type Guy11 (t = 8.77, P < 0.001, df = 30). To test the effect of Δnth1 deletion on rice blast disease, conidial suspensions of three mutants and Guy11 (at identical concentrations) were sprayed onto seedlings of rice cultivar CO-39. The Δnth1 mutants all produced significantly fewer disease lesions than Guy11 (Figure 4). The Δnth1 mutants showed lesion densities that were 24 ± 16% of those produced by Guy11 (t-tests of lesion densities were significant at P < 0.001, n = 100 leaves). The pathogenicity of Δnth1 mutants on barley seedlings showed a similar reduction. To determine the likely cause of the pathogenicity defect, we assayed cuticle penetration and appressorium turgor generation (see Supplementary data). No significant differences were observed compared with the wild-type strain, indicating that the ability to infect plants was not compromised by the Δnth1 mutation. Instead, we observed that proliferation of invasive hyphae was not as rapid in Δnth1 mutants as in Guy11 (not shown). Re-introduction of NTH1 into a Δnth1 mutant restored pathogenicity and conidiogenesis to wild-type levels (not shown). We conclude that NTH1 is required by M.grisea to generate severe rice blast symptoms.

Fig. 4. Virulence phenotype of M.grisea Δnth1 mutants. Seedlings of rice cultivar CO-39 were inoculated with M.grisea conidial suspensions of identical concentration (2 × 104 conidia/ml) of strain Guy11 (A), and Δnth1 mutants T-2-8, T-2-30 and T-2-37 (B–D). Seedlings were incubated for 4 days for development of disease.
TRE1 encodes the major trehalase activity in germlings and hyphae of M.grisea
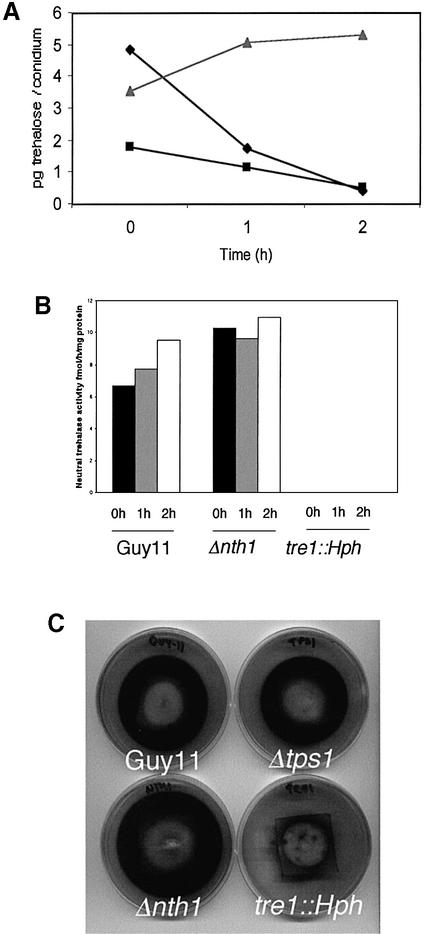
The temporal expression pattern of NTH1 and reduction in conidiation in Δnth1 mutants pointed to a potential role for NTH1 in mobilization of trehalose during spore germination. To investigate this, we measured the amount of trehalose in conidia and germlings of M.grisea at three different times during germination (Figure 5A). In Guy11, trehalose was mobilized rapidly during germination, and almost completely degraded after 2 h. A similar pattern of trehalose degradation was observed in Δnth1 mutant T-2-8, although the initial amount of trehalose in conidia was less than in Guy11. The pattern of trehalose degradation in Δnth1 mutants suggested to us that M.grisea must possess another intracellular trehalase activity, and we therefore assayed neutral trehalase activity at the same times during conidial germination. The Δnth1 mutant showed no significant difference in neutral trehalase activity during conidial germination when compared with Guy11 (Figure 5B), confirming the presence of a second trehalase in M.grisea.
Fig. 5. Trehalose mobilization and neutral trehalase activity during conidial germination of M.grisea. (A) Line graph of trehalose levels in conidia of Guy11 (diamonds), Δnth1 mutant T-2-8 (squares) and tre1::Hph mutant T-8-45 (triangles) at three intervals during germination. Data points represent the mean of three replications of the experiment. (B) Bar chart of neutral trehalase activity in conidia in the same strains at identical intervals. Each bar represents the mean of three replicate assays. (C) Neutral trehalase activity in vegetative hyphae of M.grisea assayed using an overlay test (see Materials and methods). Cultures of Guy11, Δnth1 mutant T-2-8, tre1::Hph mutant T-8-45 and Δtps1 mutant T-5-13 were grown for 6 days prior to assay. All experiments were repeated at least three times with similar results.
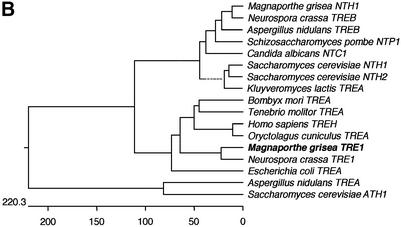
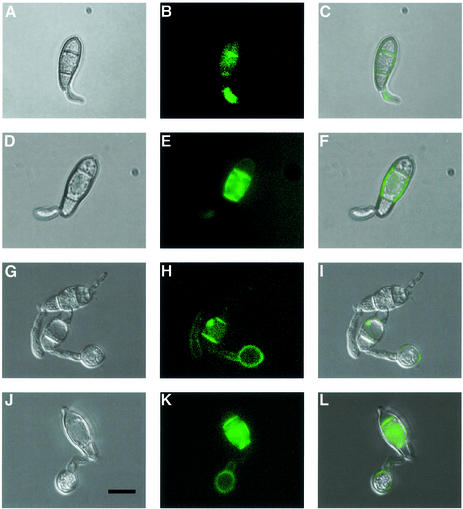
To identify the second trehalase, we used a bioinformatic approach and scanned all available DNA sequence information from M.grisea for homology to eukaryotic trehalase-encoding enzymes. We found a small fragment of a putative trehalase-encoding gene in a sequence from a bacterial artificial chromosome, used in the M.grisea genome mapping effort (Zhu et al., 1999). We amplified a corresponding 504 bp gene fragment and used this to isolate a cDNA and genomic clone spanning the locus. DNA sequencing revealed a 1914 bp ORF interrupted by four putative introns, capable of encoding a 698 amino acid protein (Figure 6A) with a putative signal peptide of 22 amino acids at the N-terminus. We named the gene TRE1 (trehalase-encoding). The putative TRE1 product showed 33% identity to the human and mouse TreA trehalases, but was distinct from both neutral and acidic trehalases previously identified from fungi (Figure 6B). Scanning the publicly available genome sequence of N.crassa revealed a gene showing 66% identity to TRE1, but no homologous sequence was observed in the Aspergillus fumigatus draft genome sequence. Thus, only closely related filamentous fungi (N.crassa and M.grisea are both pyrenomycetes) appear to possess this novel form of trehalase. To determine the cellular location of the enzyme, we produced a TRE1:GFP gene fusion by introducing the sGFP allele at the C-terminus of the TRE1 ORF. The gene fusion was transformed into Guy11, and transformants containing a single copy of the TRE1:GFP fusion were selected (Figure 7). TRE1:GFP accumulates in the septa of germinating conidia (Figure 7A–F), the germ tube apex and the walls of developing appressoria (Figure 7H and K). The temporal pattern of TRE1:GFP accumulation was consistent with RNA gel blots which showed TRE1 expression during the early stages of conidial germination (not shown). A targeted disruption of TRE1 utilized an adaptation of an in vitro transposon tagging procedure previously described (Hamer et al., 2001). We modified the Tn7 transposable element of pGPS3 (New England Biolabs), by introducing the hygromycin B resistance gene cassette. The modified transposable element was mobilized in vitro, allowing us to introduce transposon insertions throughout a cloned 8 kb genomic DNA fragment spanning the TRE1 locus. A gene disruption construct containing the 3 kb GPS-HYG transposon insertion at the 5′ end of the TRE1 coding region was introduced into M.grisea strain Guy11, and 56 hygromycin-resistant transformants were selected. Twenty-seven of these (48%) showed the presence of the gene disruption, with the 8 kb TRE1 locus replaced by the 11 kb tre1::Hph insertion allele (see Supplementary data).
Fig. 6. Predicted amino acid sequence of the M.grisea TRE1 gene product. (A) Sequences were aligned using Clustal W (Thompson et al., 1994). The TRE1 gene product is aligned with sequences of N.crassa hypothetical protein NCU00943.1 (http://www-genome.wi.mit.edu/annotation/fungi/neurospora/), rabbit TreA (P19823), Escherichia coli treA (P13482) and S.cerevisiae NTH1 (P321356). Identical amino acids are highlighted on a black background, and similar amino acids on a light grey background. Gaps are indicated by dashes. M.grisea TRE1 has DDBJ/EMBL/GenBank accession No. AF543465. (B) Dendrogram indicating the relationship of TRE1 to previously identified trehalase gene products. Genetic distance is proportional to the length of the horizontal line.
Fig. 7. Localization of TRE1 trehalase during infection-related development of M.grisea. Conidia of Guy11 TRE1:sGFP transformant T-9-14 were allowed to form appressoria on plastic surfaces. Bright-field images are shown in the left-hand panels, epifluorescence images in the centre and merged images in the right-hand panels. (A–C) A three-celled conidium undergoing germination and germ tube extension 2 h after inoculation; TRE1:GFP accumulates in septa and germ tube apex. (D–F) A germ tube extension with septal localization of TRE1:GFP, 2 h after inoculation (G–I) A germ tube undergoing hooking and appressorium formation after 6 h. The appressorium is the round structure at the end of the fungal germ tube. (J–L) Appressorium maturation with wall localization of TRE1:GFP after 7 h. Bar = 10 µm.
Trehalose mobilization was measured in germinating conidia of a tre1::Hph mutant T-8-45 (Figure 5A). Conidial trehalose levels stayed constant throughout germination, indicating that trehalose degradation does not occur in a tre1::Hph mutant (three mutants showed the same pattern). Consistent with this, we were unable to detect any trehalase activity in tre1::Hph mutants during conidial germination (Figure 5B). We also developed a plate assay for visualizing trehalase activity which results in production of a brown precipitate in the presence of trehalase. The assay showed that trehalase activity was absent from mycelium of the tre1::Hph mutant and suggested that TRE1-encoded trehalase is secreted because activity was observed as a dark halo surrounding hyphal cultures (Figure 5C). Furthermore, tre1::Hph mutants failed to grow on trehalose as a sole carbon source (Figure 8), and TRE1 showed enhanced expression in response to trehalose (not shown). TRE1 therefore encodes a trehalase with attributes of both acidic trehalase, which has been shown to be required for trehalose utilization in S.cerevisiae and A.nidulans, and neutral trehalase, which is required for intracellular trehalose mobilization in both fungi (Destruelle et al., 1995; Nwaka et al., 1995; d’Enfert and Fontaine, 1997; d’Enfert et al., 1999). Infection assays on rice showed that tre1::Hph mutants were as virulent as the wild-type Guy11 (Figure 8). TRE1-encoded trehalase activity is thus dispensable for fungal pathogenicity.
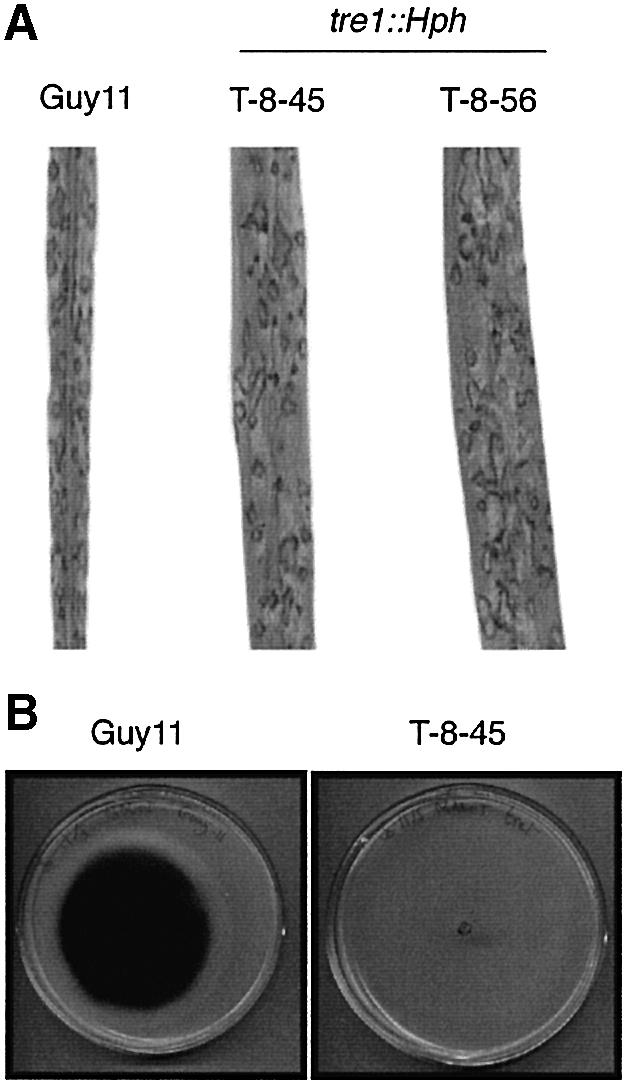
Fig. 8. Phenotypic analysis of M.grisea tre1 mutants. (A) Pathogenicity of M.grisea tre1 mutants. Seedlings of rice cultivar CO-39 were inoculated with M.grisea conidial suspensions of Guy11 and tre1::Hph mutants T-8-45 and T-8-56. Seedlings were incubated for 4 days for development of disease. (B) Growth of M.grisea strain Guy11 and tre1::Hph mutant T-8-45 with trehalose as the sole carbon source.
Discussion
Trehalose is a versatile compound that can fulfil a wide variety of functions in cells, acting as a reserve carbohydrate for survival during stress, a membrane protectant, a compatible solute, or acting to stabilize protein structures during adverse conditions (Thevelein, 1984). In this report, we have shown that the main trehalose biosynthetic enzyme T6PS, encoded by TPS1, is required for fungal virulence, affecting the ability of the fungus to breach the plant cuticle, while a trehalase-encoding gene NTH1 has an effect on subsequent stages of disease development. Taken together, these results point to a significant role for trehalose metabolism in allowing the fungus to be able to cause plant disease.
This study is the first report concerning trehalose metabolism in a plant pathogenic fungus, and several features appear distinct from previous analyses of S.cerevisiae and filamentous fungi such as A.nidulans. Trehalose synthesis in M.grisea requires the T6PS-encoding gene TPS1 which is necessary for efficient sporulation and growth on glucose. In this way, M.grisea TPS1 resembles the S.cerevisiae TPS1 gene which is needed for ascospore production and survival on glucose (Bell et al., 1998). In contrast, the A.nidulans ΔtpsA mutants grow normally on glucose (Fillinger et al., 2001). In yeast, three different models have been proposed to account for the role of Tps1 in glycolytic regulation (Arguelles, 2000). Trehalose synthesis may be a means of removing excess sugar-phosphates, thereby recycling phosphate for use by glyceraldehyde-3-phosphate dehydrogenase at a later stage of glycolysis. Alternatively, T6P, the product of T6PS, may inhibit hexokinase II activity. Inhibition of hexokinase occurs in vitro using physiologically relevant concentrations of T6P, and would provide a means of limiting glucose-6-phosphate entry into glycolysis (Bell et al., 1998). Finally, Tps1 may directly affect sugar uptake and phosphorylation (Thevelein and Hohmann, 1995). Interestingly, M.grisea appears to integrate trehalose synthesis with glycolytic regulation in a similar manner to S.cerevisiae, but also requires T6PS in order to utilize acetate or lipid as carbon sources. Activation of the glyoxylate cycle, or subsequent production of glucose via gluconeogenesis, may therefore be disrupted by the absence of T6PS in M.grisea. The inability of Δtps1 mutants of M.grisea to grow on glucose could be overcome by addition of amino acids to the medium, suggesting that synthesis of complex organic nitrogen compounds might also be affected by lack of T6PS. However, the presence of amino acids did not restore the ability of Δtps1 mutants to grow on acetate, indicating that gluconeogenesis is affected by loss of T6PS regardless of whether the fungus is utilizing nitrate or amino acids as a nitrogen source.
The mechanism of trehalose degradation in M.grisea differs substantially from that in S.cerevisiae and A.nidulans due to the presence of the novel TRE1-encoded trehalase. This trehalase is distinct from both neutral and acidic trehalases and is required both for growth on trehalose and mobilization of intracellular trehalose. Its localization in the cell wall is consistent with the presumed location of acidic trehalase in A.nidulans and N.crassa, but does not account for its role in intracellular trehalase activity. It may be that sufficient cytoplasmic accumulation of TRE1 occurs during spore germination, or that the localization of TRE1 in septa may be associated with this function. Although NTH1 is highly expressed during spore germination, its product does not appear to contribute to neutral trehalase activity at this time, and may be activated at a later stage of invasive growth. The presence of a consensus PKA phosphorylation motif in NTH1 and the known importance of cAMP-dependent signalling to disease progression in M.grisea (Talbot and Foster, 2001) provide a possible mechanism for post-translational activation of neutral trehalase after plant infection.
The role of trehalose metabolism in rice blast disease
Plant infection by M.grisea involves the action of a specialized infection cell, the appressorium, a structure widely utilized by pathogenic fungi to invade the tissues of their hosts (Tucker and Talbot, 2001). Turgor generation in appressoria results from accumulation of up to 3 M glycerol in the appressorium (de Jong et al., 1997) and, therefore, mobilization of storage reserves is likely to be critical for appressorium function. Our results indicate that TPS1 is required for appressorium function and cuticle penetration. The simplest explanation for this observation is that trehalose is used as a reserve carbohydrate that contributes to appressorium turgor by acting as a compatible solute itself, or being converted to the pool of intracellular glycerol in the appressorium. Trehalose could also act as an accessory compatible solute or membrane protectant to allow appressoria to withstand high turgor. Trehalose possesses physico-chemical properties including high hydrophilicity and chemical stability that might allow cellular proteins to operate effectively within mature appressoria (Arguelles, 2000). The addition of trehalose or T6P to germinating conidia of a Δtps1 mutant, however, did not restore pathogenicity or appressorium turgor. Furthermore, trehalose degradation during conidial germination, which is mediated by TRE1, is clearly not required for appressorium function because tre1 mutants are fully pathogenic. When considered together, these results suggest that although the presence of TPS1 is required for appressorium function, the generation and utilization of trehalose within the infection cell may not be as significant.
An alternative role for TPS1 in pathogenicity of M.grisea, which we favour, arises because of its role in regulation of glycolysis and gluconeogenesis. During appressorium turgor generation by M.grisea, it is likely that the major route for glycerol biosynthesis is via lipid metabolism (Thines et al., 2000). Lipid bodies move to the appressorium during maturation and are degraded by triacylglycerol lipase during turgor generation. Appressoria develop on the leaf surface in the absence of external nutrients. Under these glucose- and nitrogen-deficient conditions, the metabolism of acetyl-CoA, produced from fatty acid β-oxidation via the glyoxylate cycle and gluconeogenesis, may be essential for the fungus to carry out growth functions such as cell wall biosynthesis. The absence of TPS1 may mis-regulate the balance between glycolysis and gluconeogenesis during turgor generation. The fact that M.grisea Δtps1 mutants cannot grow on acetate or fatty acids is consistent with such a function and is distinct from the phenotype observed in S.cerevisiae tps1 mutants (Arguelles, 2000).
The role of NTH1 in pathogenicity is very different from that of TPS1. Although disease lesion number was reduced in infections by Δnth1, there was no reduction in the rate of cuticle penetration. Rather, the ability of the mutant to grow once it had entered the plant appeared to be impaired. The reduction in symptom development may arise due to inability of the fungus to proliferate effectively in plant tissue and withstand desiccation, which is a visible consequence of tissue necrosis during rice blast infections. The reduction in virulence of Δnth1 was clearly not due to a reduction in trehalose mobilization during pre-penetration stage development, because of the importance of TRE1 in these processes and its dispensability for pathogenicity.
In summary, trehalose metabolism plays a pivotal role in the ability of M.grisea to cause disease. Trehalose biosynthesis, either directly or via a regulatory effect, is necessary for initial plant infection, while trehalose mobilization is involved in virulence-associated functions that follow host colonization. Deletion of TPS1 in the human pathogenic fungus Candida albicans also impairs its ability to cause disease (Zaragoza et al., 1998), highlighting the potential widespread significance of trehalose metabolism to microbial pathogenesis.
Materials and methods
Fungal strains, growth conditions and DNA analysis
Magnaporthe grisea maintenance, media composition, DNA extraction, RNA extraction and transformation were all as described previously (Talbot et al., 1993). Minimal medium (MM) is 6 g/l NaNO3, 0.52 g/l KCl, 0152 g/l MgSO4·7H2O, 1.52 g of KH2PO4, 0.001% thiamine, 0.1% trace elements supplemented with 10 g/l glucose or an equivalent amount of alternative carbon sources used. Complete medium (CM) is MM supplemented with 2 g/l peptone, 1 g/l yeast extract and 1 g/l casamino acids. All molecular biology methods were performed using standard procedures (Sambrook et al., 1989). Sequence alignments were performed using the Clustal_W program (Thompson et al., 1994). Dendrograms were made by applying neighbour-joining methods (Saitou and Nei, 1987) to the distance matrix generated by Clustal_W in Phylip format (Felsenstein, 1989). The calculated phylogenetic tree was viewed using the program TreeView (Page, 1996).
Identification of NTH1, TPS1 and TRE1
To isolate TPS1, degenerate primers were designed based on the amino acid sequences of known fungal T6PS-encoding genes. The nucleotide sequences of the primers were as follows: PTPS1, 5′-YTNTGGCC NYTNTTYCAYTAY-3′; PTPS2, 5′-GGTNCANCAYTAYCAYYTNA TG-3′; PTPS3, 5′-NTWYTGRTAYTCYTCNACRTG-3′; and PTPS4, 5′-GNGGNAYNCCYTTDATRTARTC-3′.
The 650 bp amplicon was cloned into pGEM-T (Promega) and used as a probe to obtain the corresponding genomic and cDNA clones. To isolate NTH1, three restriction fragments were subcloned from a 14 kb EcoRV fragment in pCB1378 and sequenced (Sweigard et al., 1998). TRE1 was identified from a BAC-end sequence during the M.grisea genome mapping project (Zhu et al., 1999). Primers were designed to amplify this sequence from genomic DNA: PTREA1, 5′-CAACATCTCCAAGA ACACCGT-3′; and PTREA2, 5′-GCCTTTTTGGACTCGGTAAC-3′.
The resulting 500 bp amplicon was sequenced and used to screen M.grisea cDNA and genomic libraries. An 8 kb PstI–SacI fragment containing the entire TRE1 locus was selected.
Construction of targeted gene replacement vectors
All gene replacements employed the Hph gene which encodes hygromycin phosphotransferase under the control of the A.nidulans TrpC promoter (Carroll et al., 1994). Gene replacement of TPS1 was carried out by selecting and sequencing an 8 kb PstI fragment spanning the locus. A 2.0 kb NcoI fragment containing the 5′ end and the majority of the TPS1 coding region was replaced with an AflIII-linked Hph cassette produced by PCR using primers HYGAFLIII 1, 5′-CCAGACATGTCACGACGTTGTAAAACGACGG-3′; and HYGAF LIII 2, 5′-CCAGACATGTGTCGACTCTAGAGGATCCCC-3′ using pCB1004 (Carroll et al., 1994) as a template. The resulting plasmid was digested with PstI and used to transform M.grisea. Complementation of Δtps1 was carried out by cloning the 8 kb TPS1 PstI fragment into pCB1578 and selecting sulfonylurea-resistant transformants. Trans formants containing a single integration of the TPS1 were selected. For replacement of NTH1, a 2.4 kb ApaI fragment was excised from pCB1378 and the vector re-circularized to create pAJF12. The Hph gene from pCB1003 (Carroll et al., 1994) was used a template for amplification of an ApaI-linked Hph fragment using the universal forward primer and primer HYGAPA1, 5′-AGTCAGGGGCCCAATTAACCCTCACTATAAAGG G-3′. The ApaI-linked Hph cassette was ligated to Apa I-digested pAJF12. The resulting vector pAJF14 was partially digested with NcoI to release a 9 kb fragment which was introduced into Guy11. Complementation of Δnth1 required cloning of the 14 kb EcoRV fragment from pCB1378 into pCB1578, which contains a sulfonylurea resistance selectable marker (Carroll et al., 1994). The resulting plasmid pAJF87 was introduced into Δnth1 mutant T-2-8, and transformants containing a single integrated copy of the plasmid were selected.
TRE1 was disrupted using an adaptation of the TAG-KO method (Hamer et al, 2001). The kanamycin resistance gene was excised from pGPS3 (New England Biolabs) and the vector ligated to a PCR amplicon containing the Hph gene and Cat chloramphenicol resistance gene from pCB1004. Primers used to amplify the Hph Cat PCR product were: HYGCAM1, 5′-CGCGGATCCGACGTTGATCGGCACGTAAGAGG-3′; and HYGCAM2, 5′-CGCGGATCCGAAGAACGTTTTCCAATGA TGATGGCAC-3′.
The resulting plasmid, pGPS-HYG, contains the selectable markers between Tn7-based transposon ends and therefore represents an immobilized transposable element (Craig, 1996). The element was mobilized using TnsABC Transposase complex in vitro (New England Biolabs). An 8 kb PstI–Sac I fragment of TRE1, pTRE-G, was used as the target. An insertion was found within the coding regions of TRE1, close to the 5′ end of the ORF, and confirmed by sequencing. This plasmid, pTRE1-GPS-Hyg, was digested with PstI and SacI to liberate an 11 kb linear gene disruption cassette, which was transformed into Guy11.
Measurement of intracellular trehalose and neutral trehalase activity
To assay trehalose content of conidia, a suspension of conidia was harvested from 10-day-old plate cultures of M.grisea and allowed to germinate at a concentration of 1 × 106 conidia/ml on plastic coverslips at 24°C for 1 or 2 h. Conidia were then recovered by centrifugation and re-suspended in 500 µl of water. The suspension was boiled for 5 min and sonicated before removal of cell debris by centrifugation. A 60 µl aliquot was added to 60 µl of 0.1 M sodium citrate buffer. Duplicate samples were incubated in either the presence or absence of 4 µl porcine kidney acidic trehalase (Sigma) at 37°C overnight. The concentration of glucose in each sample was assayed using a commercial kit (Roche). A plate assay for neutral trehalase was developed for M.grisea from the enzymatic overlay test of Kopp et al. (1993). Plate cultures were grown for 3 days. A filter paper disc (Whatman 3 mm) was cut to the size of a Petri dish and sterilized, before being placed on the culture and incubated for 3 days. The assay was then as described by Kopp et al. (1993) except that incubations were carried out at 24°C rather than 30°C. The colour change is due to hydrolysis of trehalose to glucose by trehalase from the fungus; the resulting glucose is hydrolysed to gluconic acid and hydrogen peroxide by glucose oxidase. A peroxidase–hydrogen peroxide complex then oxidizes the chromogen o-diansidine to cause the colour change.
To assay neutral trehalase in conidia, a suspension of conidia was re-suspended in 15 ml of extraction buffer [50 mM Tris–HCl pH 7.0 and EDTA-free protease inhibitor cocktail tablets (Roche)]. The suspension was disrupted using a Bead-Beater™ (Biospec Products). A 60 µl aliquot of supernatant was mixed with 60 µl of assay buffer (50 mM Tris pH 7.0, 10 mM CaCl2, 18 mM MgCl2, 1 mM ATP, 20 µM cAMP) in the presence or absence of 40 mM trehalose. Reaction samples were incubated at 30°C for 2 h and then boiled for 5 min. Glucose concentration was determined using a commercial kit (Roche). The concentration of protein was determined using the Bio-Rad Protein Assay Dye Reagent (Bio-Rad).
Gene expression studies
The NTH1p-sGFP plasmid was constructed by amplification of the 5′ end of NTH1 using primer NTHGFP, 5′-GAGCTTCGCCATGGTGA ATGACTT-3′, and the vector-specific T7 primer (Stratagene). The 2.6 kb promoter fragment was cloned into pGEM-T (Promega) to give pAJF18. The sGFP allele (Chiu et al., 1996) carrying the A.nidulans trpC terminator (Punt et al., 1987) was excised as an NcoI–SacI fragment from plasmid pMJK-80 (Kershaw et al., 1998) and ligated to pAJF18. The resulting plasmid, pAJF19, was linearized and ligated to ApaI-linked Hph to create pAJF20. The in-frame fusion of the NTH1 promoter with the sGFP gene was checked by DNA sequencing. Transformants containing a single integration of pAJF20 were selected. Epifluoresence microscopy was used to detect GFP using a Nikon Optiphot-2 microscope (Swindon, UK). The TRE1:sGFP gene fusion was made by amplifying a 4.8 kb fragment of TRE1 containing 2.8 kb of upstream promoter and the entire protein-coding sequence using the vector-specific T7 primer (Stratagene) and primer GFPTRE: 5′-TTTTCCATGGCACCTCCCATGACGTT CCC-3′. This primer adds an NcoI site (underlined) at the last codon of TRE1. The amplicon was digested with SacI and NcoI and cloned into SacI–NcoI-digested pAJF20 from which the NTH1 promoter sequence had been removed. This provided an in-frame fusion of TRE1, under control of its native promoter, to sGFP. The entire insert was sequenced. The TRE1:sGFP construct was transformed into Guy11, and transformants carrying a single insertion were selected. A total of four TRE1:sGFP transformants were characterized, with identical results.
Plant infections assays
Plant infections were as described previously (Talbot et al., 1993). Conidial suspensions were diluted in 0.1% gelatin to 1 × 104 conidia/ml for rice infections using the dwarf Indica rice cultivar, CO-39. Conidia were spray-inoculated using an artist’s airbrush (Badger, Franklin Park, IL) onto 14-day-old (2–3 leaf stage) plants. Penetration of onion epidermal strips was assessed by the procedure of Chida and Sisler (1987). Incipient cytorrhysis of appressoria was assessed by determining the percentage of appressoria that collapsed after exposure to a range of glycerol concentrations from 0.5 to 5 M (de Jong et al., 1997). For each concentration, three replicates of 100 conidia were examined.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors thank Mark Jones for microscopy. This work was supported by a grant to N.J.T. from the Biological Sciences and Biotechnology Research Council (P17477) and studentships to support A.J.F. and J.M.J.
References
- Arguelles J.C. (2000) Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol., 174, 217–224. [DOI] [PubMed] [Google Scholar]
- Bell W., Sun,W.N., Hohmann,S., Wera,S., Reinders,A., De Virgilio,C., Wiemken,A. and Thevelein,J.M. (1998) Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J. Biol. Chem., 273, 33311–33319. [DOI] [PubMed] [Google Scholar]
- Carroll A.M., Sweigard,J.A. and Valent,B. (1994) Improved vectors for selecting resistance to hygromycin. Fung. Genet. Newslett., 41, 22. [Google Scholar]
- Chida T. and Sisler,H.D. (1987) Restoration of appresorial penetration ability by melanin precursors in Pyricularia oryzae treated with antipenetrants and in melanin-deficient mutants. J. Pesticide Sci., 12, 49–55. [Google Scholar]
- Chiu W.L., Niwa,Y., Zeng,W., Hirano,T., Kobayashi,H. and Sheen,J. (1996) Engineered GFP as a vital reporter in plants. Curr. Biol., 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Craig N.L. (1996) Transposon Tn7. Curr. Top. Microbiol., 204, 27–48. [DOI] [PubMed] [Google Scholar]
- d’Enfert C. and Fontaine,T. (1997) Molecular characterization of the Aspergillus nidulans treA gene encoding an acid trehalase required for growth on trehalose. Mol. Microbiol., 24, 203–216. [DOI] [PubMed] [Google Scholar]
- d’Enfert C., Bonini,B.M., Zapella,P.D.A., Fontaine,T., da Silva,A.M. and Terenzi,H.F. (1999) Neutral trehalases catalyse intracellular trehalose breakdown in the filamentous fungi Aspergillus nidulans and Neurospora crassa. Mol. Microbiol., 32, 471–483. [DOI] [PubMed] [Google Scholar]
- de Jong J.C., McCormack,B.J., Smirnoff,N. and Talbot,N.J. (1997) Glycerol generates turgor in rice blast. Nature, 389, 244–245. [Google Scholar]
- Destruelle M., Holzer,H. and Klionsky,D.J. (1995) Isolation and characterization of a novel yeast gene, ATH1, that is required for vacuolar acid trehalase activity. Yeast, 11, 1015–1025. [DOI] [PubMed] [Google Scholar]
- De Virgilio C., Hottiger,T., Dominguez,J., Boller,T. and Wiemken,A. (1994) The role of trehalose synthesis for the acquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant. Eur. J. Biochem., 219, 1–2, 179–186. [DOI] [PubMed] [Google Scholar]
- Dixon K.P., Xu,J-R., Smirnoff,N. and Talbot,N.J. (1999) Independent signalling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell, 10, 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989) PHYLIP (Phylogeny Interface Package) version 3.2. Cladistics, 5, 164–166. [Google Scholar]
- Fillinger S., Chaveroche,M.K., van Dijck,P., de Vries,R., Ruijter,G., Thevelein,J. and d’Enfert,C. (2001) Trehalose is required for the acquisition of tolerance to a variety of stresses in the filamentous fungi Aspergillus nidulans. Microbiology, 147, 1851–1862. [DOI] [PubMed] [Google Scholar]
- Hamer L. et al. (2001) Gene discovery and gene function assignment in filamentous fungi. Proc. Natl Acad. Sci. USA, 98, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger T., De Virgilio,C., Hall,M.N., Boller,T. and Wiemken,A. (1994) The role of trehalose synthesis for the acquisition of thermotolerance in yeast II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur. J. Biochem., 219, 187–193. [DOI] [PubMed] [Google Scholar]
- Howard R.J., Ferrari,M.A., Roach,D.H. and Money,N.P. (1991) Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl Acad. Sci. USA, 88, 11281–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw M.J., Wakley,G. and Talbot,N.J. (1998) Complementation of the Mpg1 mutant phenotype in Magnaporthe grisea reveals functional relationships between fungal hydrophobins. EMBO J., 17, 3838–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M., Müller,H. and Holzer,H. (1993) Molecular analysis of the neutral trehalase gene from Saccharomyces cerevisiae. J. Biol. Chem., 268, 4766–4774. [PubMed] [Google Scholar]
- Londesborough J. and Varimo,K. (1984) Characterisation of two trehalases in bakers yeast. Biochem. J., 219, 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenbühler K. and Holzer,H. (1988) Purification and characterisation of acid trehalase from the yeast Suc2 mutant. J. Biol. Chem., 263, 8537–8543. [PubMed] [Google Scholar]
- Nwaka S. and Holzer,H. (1998) Molecular biology of trehalose and trehalases in the yeast Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol., 58, 197–237. [DOI] [PubMed] [Google Scholar]
- Nwaka S., Kopp,M. and Holzer,H. (1995) Expression and function of the trehalase genes NTH1 and YBR0106 in Saccharomyces cerevisiae. J. Biol. Chem., 270, 10193–10198. [DOI] [PubMed] [Google Scholar]
- Page R.D.M. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci., 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Punt P.J., Oliver,R.P., Dingemanse,M.A., Pouwels,P.H. and Van den Hondel,C.A.M.J.J. (1987) Transformation of Aspergillus based on the hygromycin-B resistance marker from Escherichia coli. Gene, 56, 117–124. [DOI] [PubMed] [Google Scholar]
- Saitou N. and Nei,M. (1987) The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schuller C., Brewster,J.L., Alexander,M.R., Gustin,M.C. and Ruis,H. (1994) The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J., 13, 4382–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard J.A., Carroll,A.M., Farrall,L., Chumley,F.G. and Valent,B. (1998) Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant Microbe Interact., 11, 404–412. [DOI] [PubMed] [Google Scholar]
- Talbot N.J., Ebbole,D.J. and Hamer,J.E., (1993) Identification and characterisation of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell, 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot N.J. and Foster,A.J. (2001) Genetics and genomics of the rice blast fungus Magnaporthe grisea: developing an experimental model for understanding fungal diseases of cereals. Adv. Bot. Res., 34, 263–287. [Google Scholar]
- Thevelein J.M. (1984) Regulation of trehalose mobilisation in fungi. Microbiol. Rev., 48, 42–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J.M. (1996) Regulation of trehalose metabolism and its relevance to cell growth and function. In Brambl,R. and Marzluf,G.A. (eds), The Mycota. III. Biochemistry and Molecular Biology. Springer Verlag, Berlin, Germany, pp. 395–420.
- Thevelein J.M. and Hohmann,S. (1995) Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem. Sci., 20, 3–10. [DOI] [PubMed] [Google Scholar]
- Thines E., Weber,R.W.S. and Talbot,N.J. (2000) MAP kinase and protein kinase A-dependent mobilisation of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell, 12, 1703–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timberlake W.E. (1993) Translational triggering and feedback fixation in the control of fungal development. Plant Cell, 5, 1453–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S.L. and Talbot,N.J. (2001) Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol., 39, 385–417. [DOI] [PubMed] [Google Scholar]
- Vuorio O.E., Kalkkinen,N. and Londesborough,J. (1993) Cloning of two related genes encoding the 56-kDa and 123-kDa subunits of trehalose synthase from the yeast Saccharomyces cerevisiae. Eur. J. Biochem., 216, 849–861. [DOI] [PubMed] [Google Scholar]
- Zaragoza O., Blasquez,M.A. and Gancedo,C. (1998) Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phosphate synthase impairs formation of hyphae and decreases infectivity. J. Bacteriol., 180, 3809–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Blackmon,B.P., Sasinowski,M. and Dean,R.A. (1999) Physical map and organisation of chromosome 7 in the rice blast fungus, Magnaporthe grisea. Genome Res., 9, 739–750. [PMC free article] [PubMed] [Google Scholar]