Abstract
Telomerase contains two essential components: an RNA molecule that templates telomeric repeat synthesis and a catalytic protein component. Human telomerase is processive, while the mouse enzyme has much lower processivity. We have identified nucleotide determinants in the telomerase RNA that are responsible for this difference in processivity. Mutations adjacent to the template region of human and mouse telomerase RNA significantly altered telomerase processivity both in vitro and in vivo. We also identified functionally important nucleotides in the pseudoknot domain of telomerase RNA that potentially mediate the incompatibility between human TERT and mouse telomerase RNA. These experiments identify essential residues of the telomerase RNA that regulate telomerase activity and processivity.
Keywords: processivity/telomerase/telomerase RNA/telomeres/translocation
Introduction
Telomerase is a unique reverse transcriptase that is essential for telomere maintenance in nearly all eukaryotic cells. Telomeres consist of simple GT-rich repeated sequences (TTAGGG in vertebrates) and telomere-associated proteins, and are required for chromosome stability. Telomerase maintains telomeres by adding telomeric repeats to chromosome ends to counterbalance the natural shortening that occurs during DNA replication (Greider and Blackburn, 1985).
Telomerase consists of two essential core components, the catalytic protein component telomerase reverse transcriptase (TERT) and the telomerase RNA (Greider and Blackburn, 1989; Lingner et al., 1997). The RNA component of telomerase contains a short template element that specifies the sequence of telomere repeats added onto the chromosome ends (Greider and Blackburn, 1989; Yu et al., 1990; Singer and Gottschling, 1994). During repeat synthesis, the 3′-end of telomeric DNA anneals to the telomerase RNA template and TERT catalytically adds nucleotides specified by the template until the end of the template is reached. At the end of the template, the DNA product is either released or repositioned for another round of elongation. In ciliates, a specific region in the telomerase RNA template was aligned with the telomere DNA substrate (Autexier and Greider, 1994). Alteration in the template region affects both activity and processivity of ciliate telomerase (Autexier and Greider, 1995; Gilley et al., 1995; Gilley and Blackburn, 1996). The processivity of a DNA polymerase refers to the number of nucleotides that are synthesized before the enzyme dissociates from the template. For telomerase, processivity is usually defined as the number of telomere repeats synthesized before the enzyme dissociates (Greider, 1991; Hammond and Cech, 1997). For a processive telomerase, the telomere product translocates at the 5′-end of the template and realigns at the 3′-end of the template repeatedly. A non-processive enzyme synthesizes one repeat and then dissociates. Thus, the efficiency of product translocation defines telomerase processivity.
Telomerase enzymes from many organisms including human and Tetrahymena are processive (Morin, 1989; Greider, 1991). However, in some species such as mouse and Chinese hamster, telomerase displays significantly lower processivity (Prowse et al., 1993). The low processivity of the mouse and hamster telomerases was hypothesized to be due to the presence of only eight telomere-complementary bases in the template region compared with the eleven complementary nucleotides in the template region of human telomerase (Blasco et al., 1995; Maine et al., 1999).
The size and sequence of telomerase RNA vary dramatically among mammals (382–559 nt), yeast (∼1300 nt) and ciliates (148–209 nt). Ciliate telomerase RNAs share a conserved secondary structure (Romero and Blackburn, 1991; Lingner et al., 1994). The evolutionary conservation of ciliate telomerase RNA is further supported by the fact that some ciliate telomerase RNAs are functionally interchangeable in vivo (Bhattacharyya and Blackburn, 1997). A common secondary structure was also proposed for vertebrate telomerase RNAs based on a phylogenetic comparison and mutagenesis analysis (Chen et al., 2000, 2002). Although the secondary structure of vertebrate telomerase RNA is conserved, the mouse telomerase RNA (mTR) will not function with the human TERT (hTERT) in vitro (Beattie et al., 1998) and the ectopic expression of the hTERT in mouse cells inhibits endogenous mouse telomerase activity (Boklan et al., 2002). In contrast, human telomerase RNA (hTR) can form a functional telomerase complex with mouse TERT (mTERT) in vivo (Martin-Rivera et al., 1998).
To understand the structural basis for the differences in processivity and of cross-species incompatibility in mammalian telomerase, we carried out detailed activity assays of telomerase RNA mutants in an in vitro reconstitution system. We show that nucleotide residues in the pseudoknot domain of mTR mediate its low-processivity and different residues in the same domain mediate the incompatibility with hTERT.
Results
The pseudoknot domain of mTR is functionally incompatible with hTERT protein
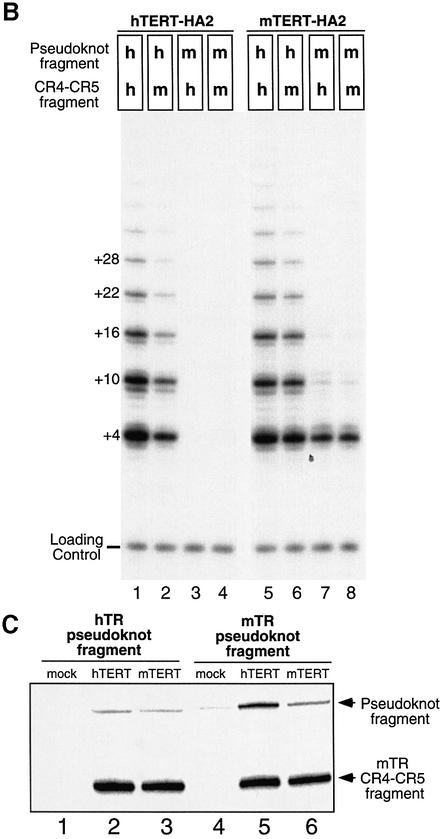
mTR was unable to reconstitute telomerase activity with hTERT protein in vitro, while hTR reconstituted activity with the mTERT protein (data not shown). To determine which RNA elements were responsible for the incompatibility between hTERT and mTR, we tested two discrete RNA fragments, the pseudoknot and CR4–CR5 fragments, of telomerase RNA for their ability to reconstitute telomerase activity in vitro with human or mouse TERT (Figure 1A). The pseudoknot fragment (nt 44–184 of hTR; nt 1–145 of mTR) contains the pseudoknot structural domain and the template region for synthesis of telomere repeats, while the CR4–CR5 fragment (nt 242–328 of hTR; nt 200–280 of mTR) contains the conserved stem–loop structural elements of the CR4–CR5 domain. These two RNA fragments can independently bind to the TERT protein and reconstitute telomerase activity in vitro (Tesmer et al., 1999; Mitchell and Collins, 2000; Chen et al., 2002). We assembled both RNA fragments of either hTR or mTR with in vitro expressed human or mouse hemagglutinin (HA)-tagged TERT protein. The assembled RNP complex was immunoprecipitated and telomerase activity was assayed directly by examining radioactive nucleotide incorporation onto telomere-primers (see Materials and methods).
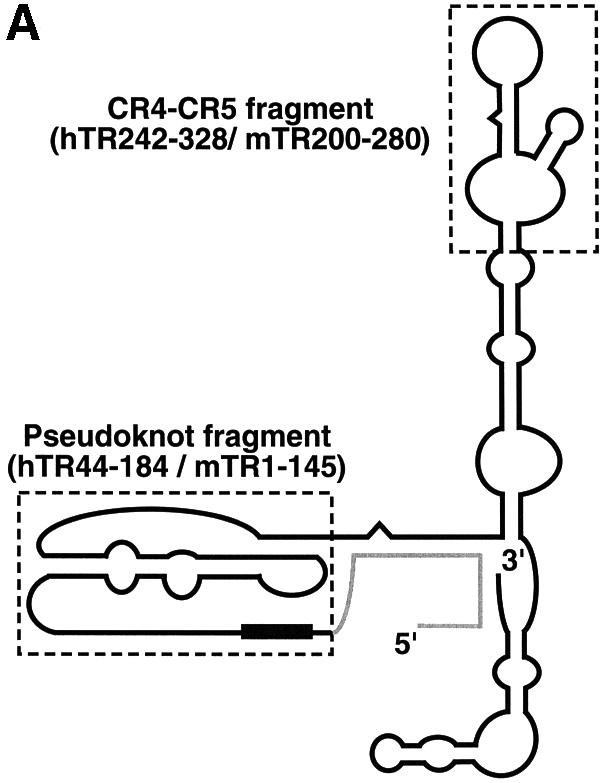
Fig. 1. Functional interspecies swapping of telomerase RNA domains and TERT proteins. (A) An outline of mammalian telomerase RNA secondary structure (Chen et al., 2000) is shown. The two fragments, pseudoknot and CR4–CR5 fragment, that are used for interspecies swapping are highlighted with dashed boxes. Nucleotide position of the 5′- and 3′-ends of both hTR and mTR fragments are indicated. (B) The pseudoknot and CR4–CR5 RNA fragments of human (h) and mouse (m) RNA were assembled with either human or mouse TERT-HA2 recombinant protein in different combinations as indicated. After immunoprecipitation, each assembled telomerase complex was assayed for telomerase activity with an 18-nucleotide telomere-primer (TTAGGG)3 using the conventional telomerase assay protocol (see Material and methods). A 32P end-labeled 12-mer oligonucleotide, (TTAGGG)2, was added as loading control before the purification and precipitation of telomeric products. The numbers on the left (+4, +10, +16, +22, +28, etc.) indicate the number of nucleotides added to the telomere-primer for each major band seen. (C) Northern analysis of cross-species RNA–protein interactions between human and mouse telomerase components. The pseudoknot RNA fragments of hTR and mTR were assembled and co-immunoprecipitated with either hTERT-HA2 (hTERT, lanes 2 and 5) or mTERT-HA2 (mTERT, lanes 3 and 6) protein, and detected by northern blotting as previously described (Chen et al., 2002). A reconstitution reaction without either protein expressed was carried out as a negative control (mock, lanes 1 and 4). The mTR CR4–CR5 RNA fragment was also added in the assembly reactions as a positive control for the immunoprecipitation of an RNA component.
Both the human and mouse CR4–CR5 fragments reconstituted telomerase activity when combined with the human pseudoknot RNA fragment and the hTERT-HA2 protein (Figure 1B, lanes 1 and 2). However, when the mouse pseudoknot fragment was used with the hTERT-HA2 protein, no activity was detected (Figure 1B, lanes 3 and 4). Thus, sequence variations in the mouse pseudoknot fragment, not the CR4–CR5 fragment, mediate the incompatibility of mTR and hTERT-HA2. The sequence variation in mTR, however, did not impair the RNA–protein interaction between the mTR pseudoknot RNA fragment and the hTERT protein (Figure 1C, lane 5).
When the same experiment was carried out with the mTERT-HA2 protein, different results were obtained. The mTERT-HA2 protein generated active enzymes at a similar level when assembled with either human or mouse pseudoknot RNA fragment. Similarly, both human and mouse CR4–CR5 fragments functioned equally with the mTERT-HA2 protein. This result suggests that mTERT can tolerate sequence differences between human and mouse RNA in the pseudoknot domain while hTERT cannot.
Interestingly, the activity of telomerase reconstituted from mTERT using the human pseudoknot fragment had significantly increased processivity than the enzyme reconstituted using the mouse pseudoknot fragment. The increase in processivity is evident from the increase in intensity of multiple-repeat products at positions +10, +16, +22 and +28 relative to the first-repeat product at position +4 (Figure 1B). This difference in processivity of in vitro reconstituted human and mouse telomerases is consistent with a previous report that mouse telomerase purified from cell extracts is significantly less processive than human telomerase (Prowse et al., 1993; Maine et al., 1999). Our data indicate that the element responsible for the low processivity of mouse telomerase is located in the pseudoknot region of mTR.
Identification of regions in the pseudoknot fragment that mediate low processivity and cross-species incompatibility
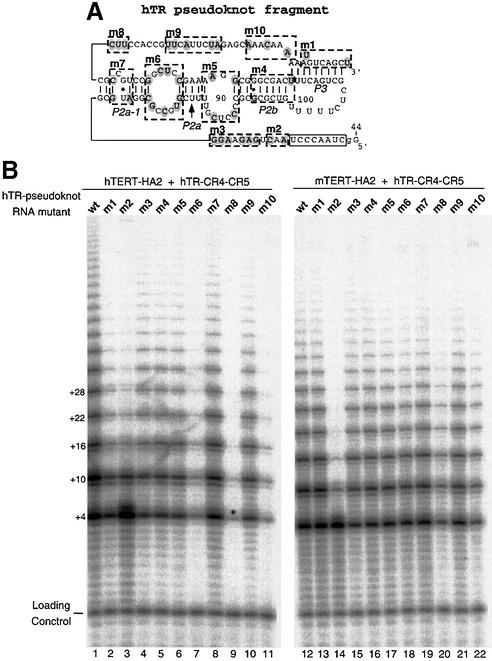
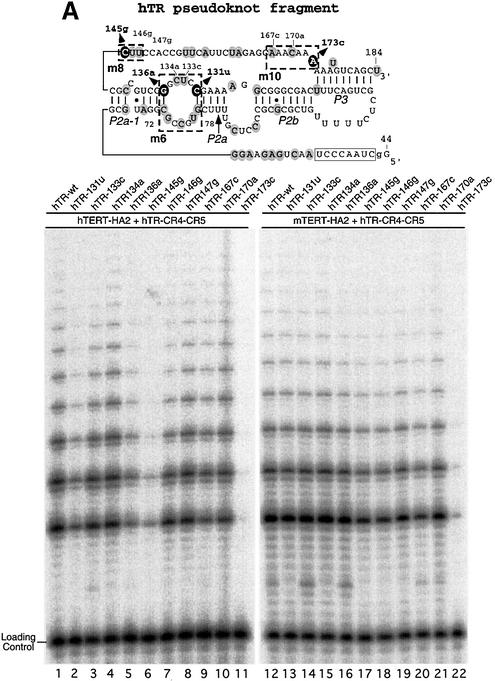
The pseudoknot fragments of hTR and mTR differ at 39 positions. To identify the nucleotides in the mouse pseudoknot domain that mediate the inability to reconstitute telomerase activity with hTERT-HA2, we systematically substituted regions of sequence in the human pseudoknot fragment with the mouse sequence. Ten different pseudoknot RNA mutants (hTR-m1 to hTR-m10) were generated (Figure 2A). Each one consists of multiple point mutations that substituted the human nucleotides with their mouse homologs in the designated regions. Each mutant pseudoknot RNA fragment was assembled with either human or mouse TERT and the human CR4–CR5 domain, and telomerase activity of each reconstituted enzyme was analyzed. Strikingly, among all RNA mutants, hTR-m2 with the nucleotides 54 and 56 changed to mouse sequence showed low processivity similar to that reconstituted with the mouse pseudoknot RNA (Figure 2B, lanes 3 and 14). This result demonstrates that the region adjacent to the template plays a major role in the processivity of telomerase, as suggested previously by others (Blasco et al., 1995; Maine et al., 1999).
Fig. 2. Effect of region-specific sequence substitution in the pseudoknot fragment of human telomerase RNA on telomerase activity. (A) The location of ten regions of the secondary structure of human pseudoknot RNA fragment in which sequence substitutions were made is indicated. Each region is indicated by dash-lined box and labeled from m1 to m10. The nucleotides that differ between human and mouse RNA are shaded with gray circles. In each case the nucleotides were changed from the human to the mouse residues at the corresponding location in the secondary structure. (B) Direct telomerase activity assay of telomerase reconstituted with each mutant RNA fragment. Each mutant pseudoknot human RNA fragment was assembled in vitro with hTERT-HA2 protein (lanes 1–11) or mTERT-HA2 (lanes 12–22) and human CR4–CR5 RNA fragment. The assembled telomerase complexes were assayed for activity after immunoprecipitation as described in Materials and methods.
The mutations in hTR-m6, -m8 and -m10 resulted in more significant reduction of telomerase activity when assembled with hTERT (Figure 2B, lanes 7, 9 and 11), than with mTERT (Figure 2B, lanes 18, 20 and 22). Mutations in the m1 region also caused a marginally reduced activity with hTERT (Figure 2B, lanes 2 and 13). Mutants in other regions resulted in little or no decrease in telomerase activity. The mutations in hTR-m6 and -m8 are located in the single-stranded regions flanking the P2a-1 helix, while the hTR-m10 mutations are located near the P3 pseudoknot helix. Both P2a-1 and P3 helical structures are highly conserved among all known mammalian telomerase RNAs (Chen et al., 2000), suggesting a functional role for residues in both regions. This result again suggests that mTERT can tolerate sequence variation in these regions of the pseudoknot fragment, while hTERT cannot.
Sequence of the template-adjacent region affects the processivity of human and mouse telomerase
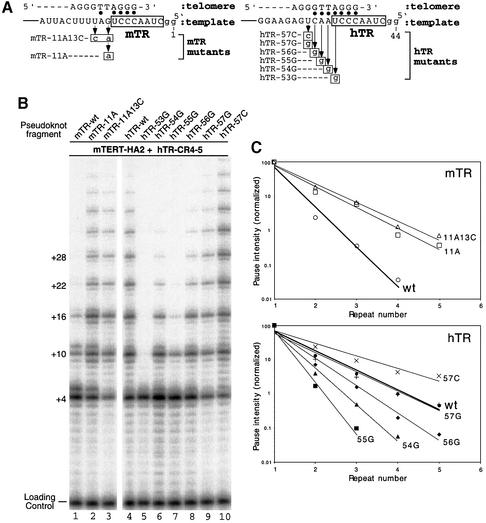
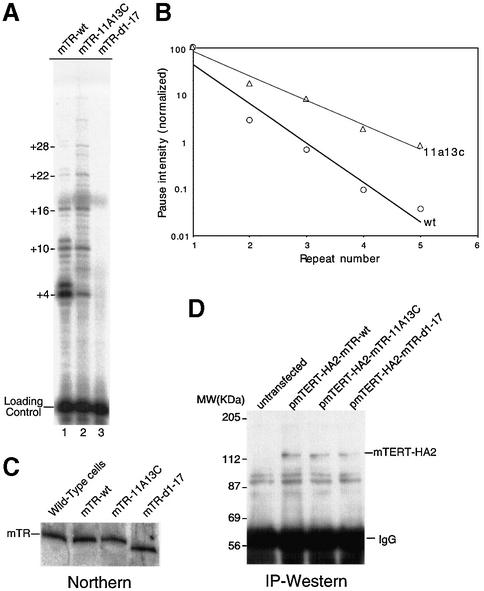
The region adjacent to the template has been termed the primer-alignment region for its role in positioning the telomere substrate (Greider and Blackburn, 1989; Autexier and Greider, 1995). The length of this primer-alignment region is variable in different species and the short length in the mouse template was proposed to account for the low processivity of mouse and hamster telomerase (Blasco et al., 1995; Maine et al., 1999). To experimentally examine the role of nucleotides in the primer-alignment region on enzyme processivity, we generated mutations in the template-adjacent region of the mouse pseudoknot RNA fragment and analyzed the processivity of the assembled mutant enzymes (Figure 3A). Alteration of the two residues at position 11 and 13 in mTR to the corresponding human residues presumably results in an increased number of base pairs between the telomere-primer and RNA template. Indeed, both mutant mouse RNAs, mTR-11A and mTR-11A13C, produced enzymes with significantly increased processivity compared with wild-type mouse RNA (Figure 3B, lanes 1–3). To quantitatively measure telomerase processivity, the intensity of each prominent band was measured and normalized as previously described (Bryan et al., 2000). The relative intensities were then plotted for each repeat. In this analysis, the processivity of each mutant is inversely related to the slope of the line. This quantitation clearly demonstrated that increasing the base pairing between the telomere-primer and the primer-alignment region in mTR results in greater processivity (Figure 3C).
Fig. 3. Effect of sequence substitution in the template region on telomerase processivity. (A) Schematic diagram of the sequence substitutions in the template region of the mouse and human pseudoknot fragments. The sequence of telomere-primer is shown above the RNA template with potential Watson–Crick base pairings indicated by closed circles. The residues substituted in each mutant RNA are indicated below the sequence of wild-type RNA. (B) Direct telomerase assay of telomerase complex reconstituted with each RNA mutant. Each pseudoknot RNA fragment was assembled with the in vitro expressed mTERT-HA2 protein and human CR4–CR5 RNA fragment. (C) Telomerase processivity quantitation of each reconstituted telomerase mutant. The intensity of each major band (+4, +10, +16, +22 and +28; also shown as repeat 1–5) from the telomerase assay in (B) was quantitated by phosphorimager analysis and analyzed as described in Materials and methods.
To further test the role of primer–RNA base pairing on processivity, we generated six mutants of the human pseudoknot RNA that reduced the base-pairing ability of the primer-alignment region with the telomere-primer. Alteration of single residues from nucleotide 53 to 56 to a guanosine residue, that would not base pair with the primer, significantly reduced processivity of the human enzyme (Figure 3B, lanes 5–8 and C). The mutation of residue 53 generated a completely non-processive enzyme (Figure 3B, lane 5 and C), while the 55G mutation reduced processivity dramatically and the mutations 54G and 56G only slightly decreased processivity (Figure 3B, lanes 6–8 and C). It appears that the effect of template mutation on the processivity correlated with the number of remaining possible base pairs in each mutant. For example, mutation at residue 56 disrupted only one base pairing and showed a minor reduction in telomerase processivity, while the mutation at residue 55 probably disrupted two base pairings (55 and 56) and thus resulted in a more significant reduction (Figure 3A). Consistently, the mismatch in the 54G mutant would be likely to destabilize its flanking base pairing and thus had greater effect in reducing the processivity than that in the 56G mutant (Figure 3C).
Strikingly, the mutation of a U to a C at position 57, which extended the possible base pairing with the telomere primer, resulted in a telomerase processivity even higher than the wild-type enzyme (Figure 3A and C). The alteration from a U to a G at the same position did not change the processivity of the enzyme (Figure 3A and C). This therefore suggests that the U residue at position 57 in wild-type RNA does not form a G:U wobble base pair with telomeric DNA. A double mutation U57C/G58C that further extended the base pairing did not further increase processivity over that seen with the U57C single mutation (data not shown). Therefore, the 12-nucleotide region from 46–57 in mammalian telomerase RNA represents the maximal template usable for telomere synthesis.
In vitro reconstituted telomerase is processive
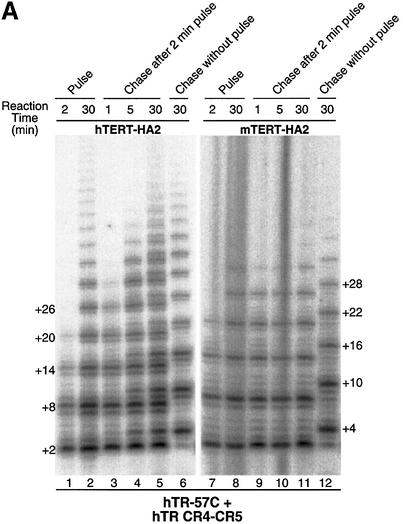
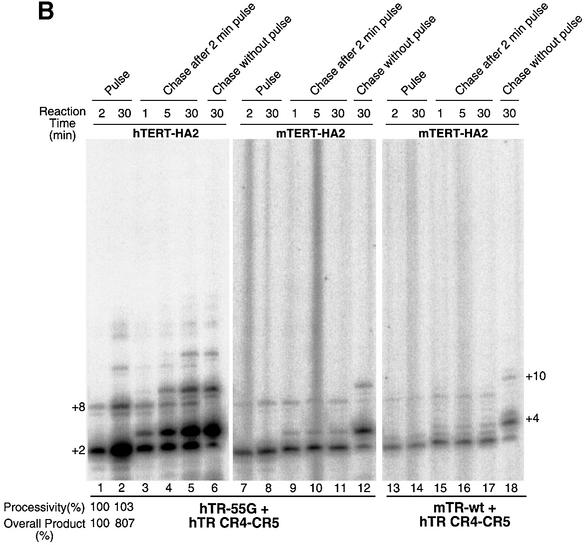
To examine whether the long telomeric products observed with the in vitro reconstitution telomerase result from a continuing elongation of a given telomeric primer by telomerase, we carried out a pulse–chase time-course assay of the both processive and non-processive enzymes. The hTR-57C pseudoknot RNA fragment was used to reconstitute processive enzymes with either human or mouse recombinant TERT protein (Figure 4A), while hTR-55G and mTR-wt pseudoknot RNA fragments were used to reconstitute non-processive enzymes (Figure 4B). Upon addition of the chase primer competitor (TTAGGG)3, the processive enzyme (hTR-57C) continued to elongate the pulse primer (AGGGTT)3 over time (Figure 4A, lanes 3–5 and 9–11). Although some elongation of the chase primer (bands at +4 and +10) was seen with the human enzyme after 5 min of incubation (Figure 4A, lanes 4 and 5), the significant elongation of the pulse primer demonstrates that the in vitro reconstituted enzyme processively adds multiple telomere repeats onto a given telomeric primer before it dissociates.
Fig. 4. Pulse–chase time course analysis of in vitro reconstituted telomerase. (A) Immunopurified human and mouse telomerase reconstituted with hTR-57C pseudoknot and hTR CR4–CR5 RNA fragments were assayed for telomerase activity, as described in Materials and methods. For the initial pulse, 1 µM of telomeric primer (AGGGTT)3 was added in the reaction mixture and the assay was carried out for either 2 (lanes 1 and 7) or 30 min (lanes 2 and 8). After 2 min of this pulse reaction, an equal volume of reaction mixture with 20 µM of the competitor primer (TTAGGG)3 was added to start the chase reaction. At the indicated time point (1, 5 and 30 min), aliquots were removed for electrophoresis analysis (lanes 3–5 and 9–11). A 30 min chase reaction without the prior 2 min pulse reaction was also carried out as a control for the ability of the chase primer to compete with the pulse primer under this reaction condition (lanes 6 and 12). The major band of elongated products of the pulse primer (AGGGTT)3 (+2, +8, +14, +20 and +26) and the chase primer (TTAGGG)3 (+4, +10, +16, +22 and +28) are labeled on the side of the gel. (B) Pulse–chase assay was carried out using telomerase enzymes reconstituted with hTR-55G (lanes 1–12) or mTR (lanes 13–18) pseudoknot fragment and hTR CR4–CR5 fragment. The processivity ratio (indicated at the bottom of lanes 1 and 2) between the 30 min reaction (lane 2) and the 2 min reaction (lane 1) was derived from the intensity of band +8 that was normalized by the intensity of band +2 and the number of radioactive nucleotides incorporated. The ratio of amount of overall products was derived from the total intensity of products accumulated in each reaction.
To determine if the low processivity of some template mutants resulted from the slow rate of product translocation, or from a slow elongation kinetics, we tested the non-processive hTR-55G mutant with both human and mouse TERT protein and the mTR-wt RNA with mouse protein using the pulse–chase experiment described above. After addition of the competitive chase primer, the product pattern of the pulse primer remained identical even after 30 min of incubation (Figure 4B, lanes 3–5, 9–11 and 15–17; note bands at +2, +8 and +14). Therefore, the low processivity of these template mutants is not due to slow elongation of the bound primer, in which an increase in the +8 and +14 bands would be seen over time.
As noted above, the hTERT, but not the mTERT, elongates some chase primer during the chase reaction (Figure 4A, lanes 3–5), which suggests a higher turnover rate of the human telomerase enzyme. The enzyme recycling of human enzyme is even more dramatic with the non-processive enzyme reconstituted from hTR55G pseudoknot RNA (Figure 4B, lanes 3–5). The intensity of the first repeat product at band +2 significantly increased over time with the human enzyme (Figure 4B, lanes 1 and 2), but not with the mouse enzyme (Figure 4B, lanes 7–11 and 13–17). In addition, there was a significant amount of elongation of the chase primer at band +4 with the hTERT (Figure 4B, lanes 3–5), but not with the mTERT (Figure 4B, lanes 9–11 and 15–17), further indicating a higher recycling for the human enzyme. Although the enzyme recycling allows an increase in overall products with the human enzyme, the processivity at 2 min and 30 min is similar (Figure 4B, lanes 1 and 2). Thus, the elongated telomeric product efficiently dissociates from the human telomerase, especially when the re-alignment of product on the template was not favorable in the non-processive enzyme. In contrast, the mouse enzyme apparently does not recycle and may not dissociate from the elongated telomeric product. This difference in enzyme turnover might result from a species-specific difference between the human and mouse TERT proteins.
Processivity of in vivo reconstituted mouse telomerase
To examine whether the processivity determinants identified in the in vitro reconstitution assay indeed mediate processivity differences in vivo, we tested the mTR mutant, mTR-11A13C, in an in vivo reconstitution system. Plasmids containing the mTERT-HA2 gene and different mTR mutant genes were co-transfected into mTR-null embryonic fibroblast cells (see Materials and methods). The wild-type and a template-deleted mTR gene, mTR-d1-17, were used as positive and negative controls respectively. Telomerase immunoprecipitated from transfected cell lysate was assayed for telomerase processivity (see Materials and methods). As seen with the in vitro reconstituted enzymes, the activity reconstituted in vivo from the telomerase RNA mutant mTR-11A13C showed an increased processivity compared with that of the wild-type telomerase (Figure 5A, lanes 1–2 and B). Telomerase reconstituted from the template-deleted mTR gene, mTR-d1-17, showed no telomerase activity as expected (Figure 5A, lane 3). Both the mTR mutant and the mTERT-HA2 genes were expressed normally in the transfected cells (Figure 5C and D). Thus, increasing the complementarity between the primer and the primer-alignment sequence increases processivity of telomerase in vivo.
Fig. 5. Processivity of in vivo reconstituted mouse telomerase. (A) Direct telomerase assay of in vivo assembled telomerase. Telomerase complex reconstituted in mouse cells transfected with the pmTERT-HA2-mTR-wt (lane 1), pmTERT-HA2-mTR11A13C (lane 2) or pmTERT-HA2-mTR-d1-17 (lane 3) was immunopurified and assayed in the presence of telomere-primer (TTAGGG)3 and [32P]dGTP as described in Materials and methods. (B) Quantitation of telomerase processivity of the in vivo reconstituted telomerase. The degree of processivity of the enzyme assayed in (A) was quantitated as described in Materials and methods. (C) Northern analysis of in vivo expressed mouse telomerase RNA mutants. Total RNA was extracted from mTR-null mouse embryonic fibroblasts (MEF) transfected with plasmids, pmTERT-HA2-mTR-wt, pmTERT-HA2-mTR-11A13C or pmTERT-HA2-mTR-d1-17. The endogenous mTR in the total RNA isolated from wild-type mouse cells was used as a size marker. (D) Western blot of immunoprecipitated mTERT-HA2 protein. The ectopically expressed mTERT-HA2 protein was extracted from transfected MEFs and immunoprecipitated using anti-HA F7 agarose beads. Protein was extracted from the beads and resolved by SDS–PAGE. After electrophoresis, the protein was transferred to membrane and detected by monoclonal anti-HA F7 antibody.
Nucleotide residues in the pseudoknot domain responsible for incompatibility of the mTR and the hTERT protein
To further understand the incompatibility of hTERT and mTR, we examined the three regions of nucleotide difference, that we found significantly decreased activity when changed from human to mouse sequence (Figure 2). To identify the determinants at the nucleotide level that mediate the incompatibility, we generated human pseudoknot RNA fragments with single point mutations in the three regions at positions that differ between human and mouse sequence (Figure 6A). At the homologous position, each mutant had the human residue substituted with the mouse residue. The mutants were assembled with the human CR4–CR5 RNA fragment, and with either human or mouse TERT-HA2 protein. The reconstituted telomerase complexes were immunopurified and assayed for telomerase activity.
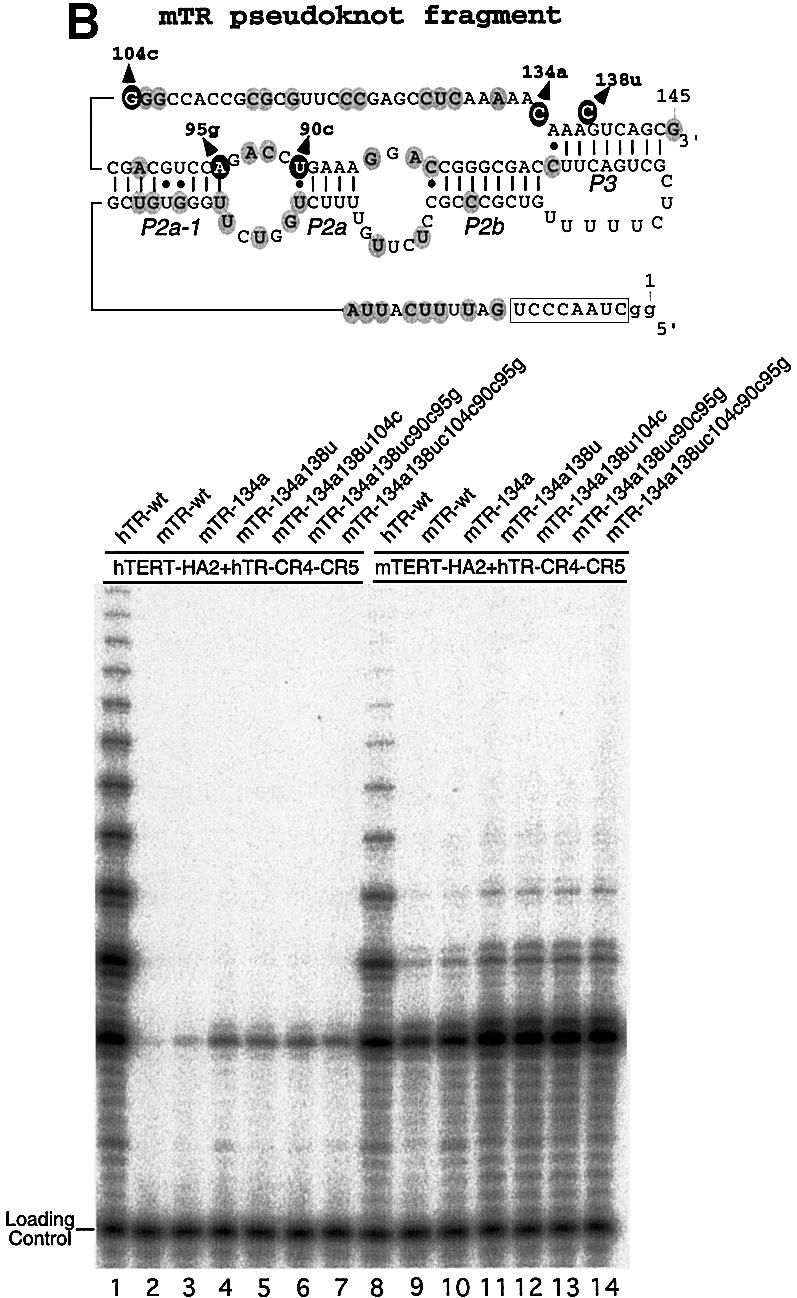
Fig. 6. Nucleotides in telomerase RNA important for compatibility with human and mouse TERTs. The positions of nucleotide substitution in each mutant are shown on the secondary structure of the human or mouse pseudoknot RNA fragment. The human CR4–CR5 RNA fragment and each pseudoknot RNA mutant were assembled with either human or mouse TERT-HA2 protein in rabbit reticulocyte lysate as described in Materials and methods. (A) Human pseudoknot RNA mutants, hTR-131u, -133c, -134a, -136a, -145g, -146g, -147g, -167c, -170a and -173c, were assembled with TERT-HA2 proteins and assayed for telomerase activity. The nucleotides that affected telomerase activity when substituted are shown in black circles. (B) Mouse pseudoknot RNA mutants, mTR-134a, -134a138u, -134a138u104c, -134a138u90c95g and -134a138u104c90c95g, were assembled with TERT-HA2 proteins and assayed for telomerase activity. The nucleotides substituted are shown in black circles.
Most of the sequence alterations had little effect on the activity or processivity of telomerase (Figure 6A). The bottom strand of region m6, 72-CGCCGUG-78, was substituted with mouse sequence, 72-UUCUGGU-78, and no difference in activity was observed (data not shown). Mutations 131U, 136A and 145G, however, showed a significant reduction in telomerase activity when reconstituted with hTERT protein (Figure 6A, lanes 2, 5 and 6), but had no effect when reconstituted with mTERT protein (Figure 6A, lanes 13, 16 and 17). Since the nucleotide alteration at these three positions had effects only when assembled with hTERT, these residues are likely to be responsible for the incompatibility between hTERT and mTR. Mutation 173C of hTR surprisingly resulted in a dramatic reduction of activity when reconstituted with either hTERT or mTERT (Figure 6A, lanes 11 and 22). Thus, the adenine residue at positions 173 might be involved in an RNA–protein or RNA–RNA interaction critical for catalysis in both human and mouse telomerase.
To examine whether these critical nucleotides have the same effect in the context of mTR, we made mutations at homologous positions in the mTR pseudoknot fragment. The C to A mutation at nucleotide 134 of mTR (the homolog of nucleotide 173 in hTR) restored a low level of activity when reconstituted with human TERT (Figure 6B, lane 3). As shown in Figure 2, mutation m1 that alters the conserved U-bulge in the P3 helix of hTR moderately reduced telomerase activity. Consistent with this, a double mutation, C134A/C138U, generated an even higher activity than the single mutation, C134A, did (Figure 6B, lane 4). Both RNA mutants also showed higher activity than wild-type mTR when reconstituted with mTERT (Figure 6B, lanes 9–11). Again, this result indicates that residues 134A and 138U play important roles in telomerase function for both human and mouse telomerase.
Although residue 145, 131 and 136 of hTR are clearly critical for reconstituting active telomerase enzyme with hTERT protein, multiple nucleotide substitutions at residue G104C, U90C and A95G in mTR did not increase telomerase activity (Figure 6B, lanes 5–7 and 12–14). Substitutions of 104G to either A or U also did not increase activity (data not shown). It is possible that these critical nucleotides are cooperatively involved in an RNA–RNA or RNA–protein interaction. Thus, substitutions at these positions in our mutant mouse RNA might not have reconstructed a functional tertiary structure or RNA motif that is specifically important for human telomerase.
Discussion
Role of telomerase RNA in telomerase processivity
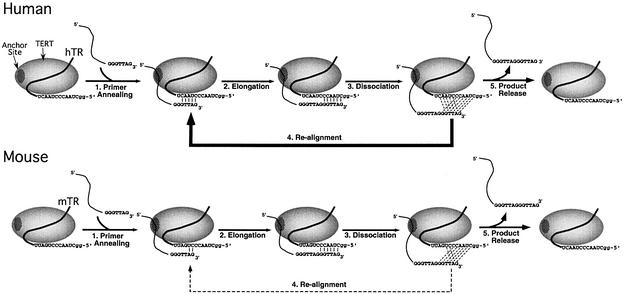
Unlike other polymerases, the processivity of telomerase depends on a translocation step rather than only on each nucleotide addition step (Figure 7). As proposed for ciliate telomerase, elongation is initiated by a proper alignment of the 3′-end of telomere-primer with the telomerase RNA template (Autexier and Greider, 1994, 1995; Gilley et al., 1995; Gilley and Blackburn, 1996) and is facilitated by the interaction between an anchor site on the TERT protein and the 5′ region of primers (Harrington and Greider, 1991; Morin, 1991; Collins and Greider, 1993). After telomere elongation, the extended product transiently dissociates from the template and translocates to the 3′-end of the RNA template for another round of telomere addition. The efficiency of initiation and nucleotide incorporation affects the overall activity, while the re-alignment of telomere product on RNA template affects the processivity.
Fig. 7. Models for human and mouse telomerase action. Step 1, the telomere-primer is recognized by the enzyme at two sites; an anchor site in the protein component that interacts with an upstream region of the primer, and the template region in the RNA component that base pairs with the 3′-end of the primer (Harrington and Greider, 1991; Morin, 1991; Collins and Greider, 1993, 1995). Step 2, the annealed primer is elongated with the sequence specified by the RNA template. Step 3, the extended telomere product dissociated from the template, but with the anchor-site interaction maintained. Step 4, the re-alignment of the telomere product with the beginning of the template is facilitated by the base-pairing interaction between the template adjacent region of the human RNA and the 3′-end of the elongated telomere product. The base-pairing interaction is depicted by dashed lines between each Watson–Crick base pair. Step 5, the telomere product is released from the enzyme. For the processive human telomerase, the more favorable product re-alignment step is illustrated by a thicker line, whereas for the less processive mouse enzyme, the less favorable product re-alignment step is illustrated by a dashed line.
We found that the amount of telomeric sequence complementarity in the RNA template plays a major role in telomerase processivity for both human and mouse enzymes. The steps in the telomerase reaction are summarized in Figure 7. First the telomere-primer binds and interacts with the template and the anchor site (step 1). Next the primer is elongated (step 2). Transient dissociation from the template (step 3) can be followed by re-alignment at the template 3′ end (step 4) or by product release (step 5). The probability of product re-alignment primarily determines the processivity of the enzyme. In human telomerase, the stronger binding (5 bp) between the telomerase template and telomere-primer presumably increases the probability of correct re-alignment of the telomere product for the next round of telomere addition. In contrast, mouse telomerase has a weaker interaction (2–3 bp) between the template and the primer, and thus has a lower probability of re-alignment. Efficient translocation involves both a transient dissociation and re-alignment of the telomeric DNA. Enzyme recycling appears to have little effect on telomerase processivity, since the human enzyme recycles much better than the mouse enzyme and yet their intrinsic processivities are similar (Figure 4). It has been suggested that a constant number of base pairs between telomere DNA and template RNA is maintained during nucleotide addition for each position during elongation (Collins and Greider, 1993; Hammond and Cech, 1998). Thus, if the amount of base pairing is the same during elongation for long and short alignment regions, a longer RNA template is likely to have a greater effect in the product re-alignment step rather than the product dissociation step.
We found that human telomerase processivity can be increased over wild-type by increasing the base pairs between the primer and template adjacent region. This remarkable increase of processivity further indicates that stable base pairing between the RNA template alignment region increases the probability of re-alignment of telomere product during translocation. Interestingly, a single amino acid substitution in Tetrahymena TERT was also shown to improve telomerase processivity (Bryan et al., 2000). Such improvement in enzyme action suggests a lack of evolutionary advantage for a highly processive telomerase in vivo. Nonetheless, it remains to be tested whether or not telomerase processivity plays any role in the telomere-length regulation.
Besides the mutations in telomerase RNA template and TERT protein, telomerase processivity in vitro can be altered by changing factors such as temperature, salt-concentration and dGTP concentration (Maine et al., 1999; Sun et al., 1999). Consistent with our model, altering the reaction conditions, such as lowering temperature and increasing salt concentration, presumably promotes the base-pairing interaction of primer–DNA and template RNA, and thus results in higher processivity. It has been suggested that dGTP might increase dissociation of telomere product and RNA template, and thus enhance processivity (Hammond and Cech, 1998). The dGTP-dependence of processivity may be due to a separate role for dGTP during translocation (Hammond and Cech, 1997, 1998; Maine et al., 1999; Hardy et al., 2001).
The potential template region for vertebrate telomerase RNA ranges from 8–11 nucleotides (Figure 8). Eight nucleotides in the template region are absolutely conserved among all known vertebrate telomerase RNAs (Figure 8). The first six conserved nucleotides of the hTR template, 46-CUAACC-51, serve as the actual template for DNA polymerization, while the other two conserved nucleotides, 52-CU-53, at the 3′-end of the template are required for primer alignment during translocation. Consistent with this sequence conservation, our data demonstrates that 52-CU-53 in the primer-alignment region are sufficient to generate basal processivity. Therefore, telomerase RNA requires a minimum template length of eight nucleotides to generate a processive enzyme for synthesis of the typical six-nucleotide telomere repeats.

Fig. 8. Sequence conservation in the template region of vertebrate telomerase RNA. The sequence alignment of the template and its adjacent region of 35 vertebrate telomerase RNA is shown (Chen et al., 2000). The 100% conserved nucleotides are highlighted in black. Nucleotides that are moderately conserved are shown in gray. The proposed function for each sequence element is indicated as (1) template for polymerization, 5′-CUAACC-3′, (2) essential for telomere alignment, 5′-CU-3′, (3) enhance telomerase processivity.
The four bases adjacent to the eight essential nucleotides are not universally conserved in all vertebrate sequences (Figure 8). Nonetheless, when these residues are complementary to the telomere sequence, they cumulatively enhance processivity (Figure 3). Therefore, the processive human telomerase possesses an 11-nucleotide template, while the less processive mouse telomerase uses a minimal 8-nucleotide template.
The increase in processivity with increased complementarity, however, has a limit. For example, telomerase enzymes from several yeasts have a very long RNA template and are not processive (Prescott and Blackburn, 1997). In Kluyveromyces lactis, the repeat synthesized is 25 nt and the template is 30 nt (Tzfati et al., 2000). Telomere synthesis from such a long template might result in a decreased ability to translocate, and therefore the enzyme will be non-processive.
Role of telomerase RNA in telomerase activity and cross-species compatibility
The telomerase RNA and TERT protein components functionally co-evolved as a ribonucleoprotein complex for maintaining telomere length. Such co-evolution within a species might result in interspecies incompatibility between enzyme components. The degree of incompatibility correlates with the evolutionary distance: for example, telomerase RNA from rabbit, a more closely related species, is functionally compatible with hTERT (Xiang et al., 2000), while the RNA from mouse is not. The elements that mediate such cross-species incompatibility thus might represent a species-specific RNA–RNA or RNA–protein interaction.
The P2a-1 helix and the adjacent regions are important for telomerase function. Alteration of three nucleotides flanking the helix P2a-1 in hTR resulted in an activity reduction only with hTERT, but not mTERT, and therefore might be involved in critical RNA–RNA or RNA–protein interactions specific for human telomerase function. Antisense oligonucleotides targeting positions 137–151 in the P2a-1 and the downstream region, inhibits telomerase activity (Pruzan et al., 2002). Thus, the P2a-1 and its flanking region may play a direct role in human telomerase enzyme function.
A link between human disease, dyskeratosis congenita and aplastic anaemia and mutations in human telomerase RNA was recently reported (Vulliamy et al., 2001, 2002). Among the hTR mutations identified, three point mutations G58A, C72G, G107A/C108G and a deletion of 110-GACU-113 are located in the pseudoknot region of the RNA. The mutation G107A/C108G and the deletion of 110-GACU-113 would probably disrupt the highly conserved P3 helical structure and conceivably result in loss of enzyme activity. In contrast, our results indicate that two other mutations, G58A and C72G, which are implicated to confer a phenotype, have no effect on telomerase activity in vitro (Figure 2B and data not shown). Therefore, the role of these two mutations in telomerase function and telomere length regulation remains to be directly tested.
Using the in vitro reconstitution system, we can distinguish those RNA residues important for catalytic function from the nucleotides important for RNA–protein binding. Although the mTR pseudoknot fragment does not function with hTERT protein, it binds efficiently to both hTERT and mTERT proteins. Thus the human-specific residues might be involved in critical aspects of catalysis in human telomerase. Identification of the residues critical for catalysis in the human telomerase RNA will advance our understanding of the molecular mechanism of telomerase action and lead to new ways to manipulate telomerase to treat human disease.
Materials and methods
Plasmid construction and mutagenesis
For in vitro reconstitution, hTERT and mTERT genes were subcloned from pCI-neo-hEST2-HA (a gift from Dr Robert A.Weinberg) and pcDNA-mTERT (a gift from Dr Lea Harrington) into pCITE-4a (Novagen) to generate pCITE-hTERT-HA2 and pCITE-mTERT-HA2, respectively. For in vivo reconstitution, the HA-tagged mTERT gene was subcloned into the mammalian expression vector, pIRES-puro3 (ClonTech), behind the CMV promoter, to generate pIRES-mTERT-HA2. A 1.1 kb BstYI DNA fragment that contains the mTR gene and its native promoter sequence was subcloned into the BamHI site of pIRES-mTERT-HA2 plasmid to generate the pmTERT-HA2-mTR-wt plasmid. In addition to the wild-type mTR gene, two mutant mTR genes, one containing a G11A/U13C double point mutation and the other containing a 17-residue deletion of the template region, were also subcloned into the pIRES-mTERT-HA2 to generate pmTERT-HA2-mTR-11a13c and pmTERT-HA2-mTR-d1-17, respectively. All mutant RNA genes were sequenced.
DNA template for T7 transcription
The DNA templates for T7 in vitro transcription of telomerase RNA fragments were generated by PCR amplification of plasmids containing either hTR or mTR genes. For the pseudoknot-fragment, the following forward (F) and reverse (R) primers were used: hTRm0-gF, 5′-GGGTACCTAATACGACTCACTATAGGCTAACCCTAACTGAGA AGGGCGTAGGCGCCGTG-3′; mTRm0-gF, 5′-GGGTACCTAATA CGACTCACTATAGGCTAACCCTGATTTTCATTAGCTGTGGGTT CTGG-3′; hTR184R-wt, 5′-AGCTGACATTTTTTGTTTGCTC-3′, and mTR145R-wt, 5′-CGCTGACGTTTGTTTTTGAGG-3′. The 5′-ends of both human and mouse pseudoknot fragments started with two guanosine residues upstream of the template region to eliminate sequence differences at the 5′-end of these two RNAs and to increase efficiency of T7 transcription of RNA. The primers used to amplify the CR4–CR5-fragment are: hTR242-T7F, 5′-GGGGTACCTAATACGACTCA CTATAGCCCGCCTGGAGGCCGC-3′; hTR-328R, 5′-GACCCGCG GCTGACAGAGC-3′; mTR200-T7F, 5′-GGGGTACCTAATACGAC TCACTATAGACCCGCCTACAGGCCGC-3′; mTR-280R, 5′-GCCCC GCGGCTGACAGAGG-3′. To introduce specific mutations, the RNA gene was PCR amplified using oligonucleotides that were synthesized with the desired sequence changes. The PCR products were purified using MiniElute PCR purification Kit (Qiagen) and eluted in 12 µl of 10 mM Tris–HCl pH 8.5.
In vitro transcription of telomerase RNA
Telomerase RNA fragments were prepared by run-off in vitro transcription with T7 RNA polymerase using PCR DNA templates. T7 in vitro transcription was carried out in 40 µl of reaction containing 0.2 µg of PCR DNA template, 1× T7 buffer (0.4 M Tris–HCl pH 8.0, 10 mM spermidine, 50 mM DTT, 0.1% Triton X-100, 0.2 M MgCl2 and 0.5 µg/µl BSA), 4 mM rNTPs, 10 mM DTT, 5 mM MgCl2, 0.04 units yeast pyrophosphatase (USB), 16 units recombinant RNasin (Promega) and 100 units T7 RNA polymerase (Invitrogen) at 37°C for 12–16 h. In vitro transcribed RNAs were gel-purified and eluted in 0.3 M NaOAc, 0.01% SDS, 25 mM Tris–HCl pH 8.0, 1 mM EDTA and an equal volume of acid phenol/chloroform (Ambion). The aqueous phase was removed, chloroform-extracted and eluted RNA was ethanol precipitated.
In vitro transcription/translation reactions of TERT protein
Epitope-tagged human and mouse TERT proteins were expressed from phTERT-HA2 and pmTERT-HA2 by using the TnT quick coupled transcription/translation system (Promega). Each 50 µl reaction contained 40 µl TnT-quick mix, 2 µl PCR enhancer (Promega), 1 µl 1 mM methionine, 7 µl water and 200 ng plasmid DNA. After incubation at 30°C for 1 h, 200 ng of gel-purified T7 transcribed telomerase RNA fragment was added and incubated at 30°C for 1 h. Lysate was diluted 10-fold with immunoprecipitation (IP) buffer (10 mM HEPES pH 7.5, 100 mM potassium glutamate, 1 mM MgCl2, 1 mM DTT and 10% glycerol). After centrifugation at 16 000 × g for 20 min, the supernatant was transferred to a new tube, followed by immunoprecipitation.
Immunoprecipitation
Reconstituted telomerase complexes were affinity-purified using the C-terminal HA tag of recombinant TERT protein with anti-HA F7 agarose beads (Santa Cruz Biotechnology). Each lysate was diluted to 500 µl with IP buffer and 15 µl of anti-HA F7 agarose beads was added for immunoprecipitation at 4°C for 2 h or overnight. Beads were washed with IP buffer four times, transferred to a new tube, and washed two more times. The beads were resuspended in either 1× telomerase assay buffer (see below) or in SDS-gel loading buffer for western blotting.
Telomerase activity assay
Telomerase activity of immunopurified telomerase complex reconstituted either in vitro or in vivo was determined by a direct assay protocol modified from Sun et al. (1999). Briefly, the reaction mixture (20 µl) contained 1× telomerase assay buffer (50 mM Tris–HCl pH 8.0, 50 mM KCl, 1 mM MgCl2, 5 mM β-mercaptoethanol and 1 mM spermidine), 1.0 µM telomere primer, 0.5 mM dATP, 0.5 mM dTTP, 2 µM dGTP and 1.25 µM [α-32P]dGTP (800 Ci/mmol) with 6 µl immunopurified telomerase complex. The reaction was incubated at 30°C for 1 h and the products were precipitated with the addition of 100 µl 3.6 M NH4OAc, 20 µg glycogen and 450 µl ethanol. After incubation at –80°C for 1 h followed by centrifugation at 4°C for 20 min, the pellet was washed with 75% ethanol and resuspended in 1× gel-loading buffer (40% formamide, 10 mM Tris–HCl pH 8.0, 10 mM EDTA, 0.05% xylene cyanol). The heat-denatured samples were loaded onto a 10% polyacrylamide/1× TBE/8M urea denaturing gel for electrophoresis. After electrophoresis, the gel was dried and exposed to Fuji BAS1500 phosphorimager screen or X-ray film for an appropriate period of time. Telomerase processivity was quantitated as described previously (Bryan et al., 2000). In brief, the intensity of each major repeat band was measured, and normalized to both the intensity of the first band and the number of 32P-labeled nucleotides incorporated. Normalized intensities were then plotted versus the repeat number.
Cell culture, transfection and cell extract preparation
Cell culture and transfection of pmTERT-HA2-mTR-wt, pmTERT-HA2-mTR-11A13C and pmTERT-HA2-mTR-d1-17 plasmids were carried out as described previously (Chen et al., 2002). Transfection efficiency was monitored by co-transfection of a pIRES-EGFP plasmid and consistently ranged between 50 and 60%. After 48 h, cells were trypsinized and centrifuged at 200 g for 2 min. For northern analysis, total RNA was isolated from cell pellets using TriZol reagent following vender’s instruction (Invitrogen). For telomerase activity assay, transfected cells harvested from each 6-well plate were washed twice in PBS and once in cell washing buffer (10 mM HEPES–KOH pH 7.5, 1.5 mM MgCl2, 10 mM KCl and 1 mM DTT). Washed cells were resuspended in 500 µl of hypotonic solution containing 1× protease inhibitor cocktail (Roche) and incubated on ice for 1 h. Extracts were centrifuged for 20 min at 4°C at 16 000 g and the supernatants were added with equal volume of IP buffer for subsequent immunoprecipitation.
Acknowledgments
Acknowledgements
We thank Drs Robert A.Weinberg and Lea Harrington for the human and mouse TERT plasmids. We thank Drs Rachel Green and Jon R.Lorsch, and members of Greider lab for reading the manuscript and helpful discussions. This research was supported by National Institutes of Health Grant (AG09383) to C.W.G. J.L.Chen is a fellow of the Leukemia & Lymphoma Society.
References
- Autexier C. and Greider,C.W. (1994) Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes Dev., 8, 563–575. [DOI] [PubMed] [Google Scholar]
- Autexier C. and Greider,C.W. (1995) Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev., 9, 2227–2239. [DOI] [PubMed] [Google Scholar]
- Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (1998) Reconstitution of human telomerase activity in vitro. Curr. Biol., 8, 177–180. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A. and Blackburn,E.H. (1997) A functional telomerase RNA swap in vivo reveals the importance of nontemplate RNA domains. Proc. Natl Acad. Sci. USA, 94, 2823–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M.A., Funk,W., Villeponteau,B. and Greider,C.W. (1995) Functional characterization and developmental regulation of mouse telomerase RNA. Science, 269, 1267–1270. [DOI] [PubMed] [Google Scholar]
- Boklan J., Nanjangud,G., MacKenzie,K.L., May,C., Sadelain,M. and Moore,M.A. (2002) Limited proliferation and telomere dysfunction following telomerase inhibition in immortal murine fibroblasts. Cancer Res., 62, 2104–2114. [PubMed] [Google Scholar]
- Bryan T.M., Goodrich,K.J. and Cech,T.R. (2000) A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem., 275, 24199–24207. [DOI] [PubMed] [Google Scholar]
- Chen J.-L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- Chen J.-L., Opperman,K.K. and Greider,C.W. (2002) A critical stem–loop structure in the CR4–CR5 domain of mammalian telomerase RNA. Nucleic Acids Res., 30, 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. and Greider,C.W. (1993) Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev., 7, 1364–1376. [DOI] [PubMed] [Google Scholar]
- Collins K. and Greider,C.W. (1995) Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J., 14, 5422–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D. and Blackburn,E.H. (1996) Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol. Cell. Biol., 16, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D., Lee,M.S. and Blackburn,E.H. (1995) Altering specific telomerase RNA template residues affects active site function. Genes Dev., 9, 2214–2226. [DOI] [PubMed] [Google Scholar]
- Greider C.W. (1991) Telomerase is processive. Mol. Cell. Biol., 11, 4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn,E.H. (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature, 337, 331–337. [DOI] [PubMed] [Google Scholar]
- Hammond P.W. and Cech,T.R. (1997) dGTP-dependent processivity and possible template switching of euplotes telomerase. Nucleic Acids Res., 25, 3698–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond P.W. and Cech,T.R. (1998) Euplotes telomerase: evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry, 37, 5162–5172. [DOI] [PubMed] [Google Scholar]
- Hardy C.D., Schultz,C.S. and Collins,K. (2001) Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem., 276, 4863–4871. [DOI] [PubMed] [Google Scholar]
- Harrington L.A. and Greider,C.W. (1991) Telomerase primer specificity and chromosome healing. Nature, 353, 451–454. [DOI] [PubMed] [Google Scholar]
- Lingner J., Hendrick,L.L. and Cech,T.R. (1994) Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev., 8, 1984–1998. [DOI] [PubMed] [Google Scholar]
- Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- Maine I.P., Chen,S.F. and Windle,B. (1999) Effect of dGTP concentration on human and CHO telomerase. Biochemistry, 38, 15325–15332. [DOI] [PubMed] [Google Scholar]
- Martin-Rivera L., Herrera,E., Albar,J.P. and Blasco,M.A. (1998) Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc. Natl Acad. Sci. USA, 95, 10471–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.R. and Collins,K. (2000) Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell, 6, 361–371. [DOI] [PubMed] [Google Scholar]
- Morin G.B. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell, 59, 521–529. [DOI] [PubMed] [Google Scholar]
- Morin G.B. (1991) Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature, 353, 454–456. [DOI] [PubMed] [Google Scholar]
- Prescott J. and Blackburn,E.H. (1997) Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev., 11, 528–540. [DOI] [PubMed] [Google Scholar]
- Prowse K.R., Avilion,A.A. and Greider,C.W. (1993) Identification of a nonprocessive telomerase activity from mouse cells. Proc. Natl Acad. Sci. USA, 90, 1493–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzan R., Pongracz,K., Gietzen,K., Wallweber,G. and Gryaznov,S. (2002) Allosteric inhibitors of telomerase: oligonucleotide N3′→P5′ phosphoramidates. Nucleic Acids Res., 30, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D.P. and Blackburn,E.H. (1991) A conserved secondary structure for telomerase RNA. Cell, 67, 343–353. [DOI] [PubMed] [Google Scholar]
- Singer M.S. and Gottschling,D.E. (1994) TLC1-template RNA component of Saccharomyces cerevisiae telomerase. Science, 266, 404–409. [DOI] [PubMed] [Google Scholar]
- Sun D., Lopez-Guajardo,C.C., Quada,J., Hurley,L.H. and Von Hoff,D.D. (1999) Regulation of catalytic activity and processivity of human telomerase. Biochemistry, 38, 4037–4044. [DOI] [PubMed] [Google Scholar]
- Tesmer V.M., Ford,L.P., Holt,S.E., Frank,B.C., Yi,X., Aisner,D.L., Ouellette,M., Shay,J.W. and Wright,W.E. (1999) Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell. Biol., 19, 6207–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y., Fulton,T.B., Roy,J. and Blackburn,E.H. (2000) Template boundary in a yeast telomerase specified by RNA structure. Science, 288, 863–867. [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Marrone,A., Goldman,F., Dearlove,A., Bessler,M., Mason,P.J. and Dokal,I. (2001) The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature, 413, 432–435. [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Marrone,A., Dokal,I. and Mason,P.J. (2002) Association between aplastic anaemia and mutations in telomerase RNA. Lancet, 359, 2168–2170. [DOI] [PubMed] [Google Scholar]
- Xiang H., Wang,J., Mao,Y.W. and Li,D.W. (2000) hTERT can function with rabbit telomerase RNA: regulation of gene expression and attenuation of apoptosis. Biochem. Biophys. Res. Commun., 278, 503–510. [DOI] [PubMed] [Google Scholar]
- Yu G.L., Bradley,J.D., Attardi,L.D. and Blackburn,E.H. (1990) In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature, 344, 126–132. [DOI] [PubMed] [Google Scholar]