Abstract
CBP/p300 recruitment to enhancer-bound complexes is a key determinant in promoter activation by many transcription factors. We present a novel mechanism of activating such complexes and show that pre-assembled Elk-1–p300 complexes become activated following Elk-1 phosphorylation by changes in Elk-1–p300 interactions rather than recruitment. It is known that Elk-1 binds to promoter in the absence of stimuli. However, it is unclear how activation of Elk-1 by mitogen-acivated protein kinase (MAPK)-mediated phosphorylation leads to targeted gene transactivation. We show that Elk-1 can interact with p300 in vitro and in vivo in the absence of a stimulus through the Elk-1 C-terminus and the p300 N-terminus. Phosphorylation on Ser383 and Ser389 of Elk-1 by MAPK enhances this basal binding but, most importantly, Elk-1 exhibits new interactions with p300. These interaction changes render a strong histone acetyltransferase activity in the Elk-1-associated complex that could play a critical role in chromatin remodeling and gene activation. The pre-assembly mechanism may greatly accelerate transcription activation, which is important in regulation of expression of immediate-early response genes, in particular those involved in stress responses.
Keywords: chemokine/co-activators/Elk-1/p300/transcription activation
Introduction
Elk-1 (Ets-like protein-1), a member of the Ets family of transcription activators, shares a similar DNA-binding domain with Ets-1/2 (Rao et al., 1989). It is an essential component in the serum response ternary complex and has been shown to regulate expression of immediate-early response genes such as Zif268, MKP-1 (Sgambato et al., 1998) and chemokine cIL-8 (cCXCL8/cCAF; Li et al., 1999) that are not directly related to cell division, making it also a primary regulator of gene transcription involved in responses to stress. Furthermore, in some cases, Elk-1 does not require serum response factor to bind to DNA (Li et al., 1999, 2000), and it can be bound in the absence of a stimulus (Sharrocks, 1995; Li et al., 1999, 2000). In the case of c-fos, the Elk-1-binding site also is constitutively occupied in vivo (Herrera et al., 1989).
The functional domains of Elk-1 are well defined (Yang et al., 1998b). Activation of Elk-1 depends on phosphorylation of Ser383 and Ser389 in the transcriptional activation domain by mitogen-activated protein kinase (MAPK) (Janknecht et al., 1993; Gille et al., 1995), which causes a conformational change that enhances DNA binding affinity (Gille et al., 1995; Sharrocks, 1995; Yang et al., 1999; Li et al., 2000).
Most ‘activation’ domains of transcription factors are adaptors for recruitment of transcription co-activators. p300 and CBP are well-known coactivators that function as downstream scaffold and integrators for signal transduction. They contain multiple modules for protein– protein interactions and can serve as adaptors for transcription machinery assembly and recruitment by bridging the transcription activator with general transcription factors such as TATA box-binding protein (TBP) or TFIIB. Importantly, they contain intrinsic or associated extrinsic histone acetyltransferase (HAT) activity that acetylates core histones and releases repression of transcription initiation (Vo and Goodman, 2001).
Although p300 and CBP are highly homologous and share similar biochemical characteristics, they are not redundant in many of their functions, as shown by the different phenotypes exhibited by heterozygous knockout mice for either co-activator and by the fact that homozygous mutations in CBP or p300 are lethal (Yao et al., 1998; Kung et al., 2000). Furthermore, transcription factor pathways show differences in function between the two co-activators (e.g. Vo and Goodman, 2001). Therefore, critical questions remain concerning the identification of cofactors that cooperate with Elk-1 and how their cooperation leads to specific gene expression.
For many genes, inducible interactions between transcription factors and p300/CBP are the primary mechanism that turns on gene transcription (Xu et al., 1999). However, this apparently is not the case for genes under the control of Elk-1. The evidence indicates a pre-assembled Elk-1–CBP co-activator complex associated with target enhancer elements (Janknecht and Nordheim, 1996; Nissen et al., 2001). Elk-1 controls primarily immediate-early response genes, all of which have low levels of basal transcription but can reach very high levels <6 h after stimulation. In the case of cIL-8, elevation of mRNA occurs <7 min after thrombin stimulation (Li et al., 2000). Considering that there are a multitude of gene activation processes driven by the limited concentrations of p300/CBP inside cells, the pre-assembly mechanism may greatly accelerate transcription activation.
Several pieces of evidence suggest Elk-1-mediated gene activation mechanisms in these complexes. For example, phosphorylation of the Elk-1 activation domain leads to a conformational change and to enhancement of DNA binding in the N-terminus of Elk-1 (Yang et al., 1999; Li et al., 2000), but it is not yet known how this bound factor activates gene transcription. It also has been suggested that activated MAPK is able to phosphorylate the C-terminus of CBP, contributing to gene activation (Janknecht and Nordheim, 1996). However, this alone does not explain the prerequisite for Elk-1 phosphorylation in controlling gene activation. Given that pre-assembled complexes exist, it is critical to determine the switching mechanisms associated with the upstream signal transduction pathways. Thrombin activates cIL-8 via a G-protein-coupled receptor and transactivation of the epidermal growth factor receptor (EGFR) tyrosine kinase (Vaingankar and Martins-Green, 1998), followed by activation of the MAPK cascade and Elk-1 (Li et al., 2000). Using this system, here we elucidate the mechanisms of Elk-1 phosphorylation upon thrombin-stimulated transcription activation of the cIL-8 gene. We show that p300 is important in Elk-1 activation of transcription and that the interactions between Elk-1 and p300 change following Elk-1 phosphorylation, leading to a novel model for Elk-1-driven gene transcription.
Results
Thrombin and p300, but not CBP, enhance Elk-1-mediated transcription activation
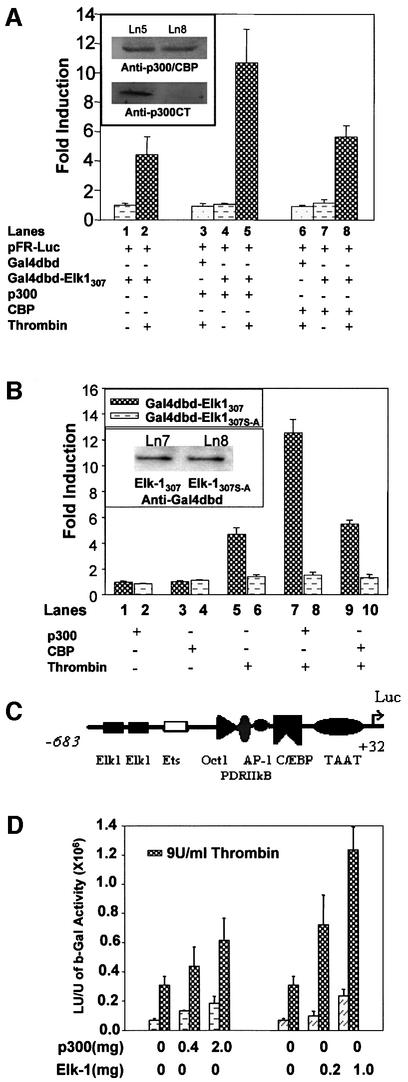
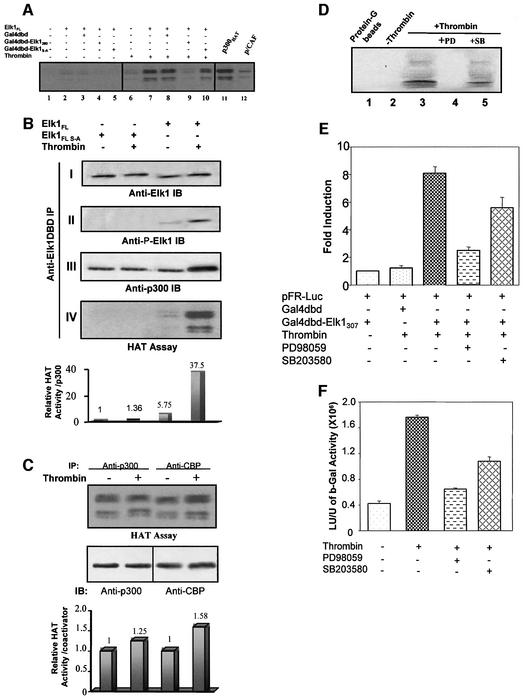
To examine whether CBP cooperates with the Elk-1 transcription factor to enhance transcription of stress response genes upon stimulation of cells by thrombin, we co-transfected primary fibroblasts with several plasmid constructs singly or in combinations. We used a luciferase reporter construct containing five Gal4 tandem elements (pFR-Luc) and two expression vectors, one containing a fusion protein with the Gal4 DNA-binding domain and Elk-1 activation domain (Gal4dbd–Elk-1307), and the other containing the co-activator CBP. pCH110 β-gal was used as internal control. To ensure that β-gal expression was not affected by co-activator transfection, we also normalized the luciferase activity to protein concentration. Exogenous expression of the Gal4dbd– Elk-1307 protein and CBP in the presence of thrombin treatment did not result in a significant increase in Elk-1-dependent gene transcription (Figure 1A, lanes 7 and 8). These observations led us to test whether thrombin-driven Elk-1 function is dependent on p300, a functional homolog of CBP. Overexpression of p300 in the presence of the Gal4dbd–Elk-1307 followed by thrombin treatment induced a synergistic effect of Elk-1-dependent transactivation (Figure 1A; compare lanes 2 and 5). Using an antibody that recognizes p300 and CBP and another that recognizes only p300, we verified that p300 and CBP are expressed at the same level in the cells (Figure 1A, insert), excluding the possibility that the lack of synergistic effect of CBP in the co-transfection experiments is due to lack of CBP expression. The same results were obtained in HEK293 cells (data not shown).
Fig. 1. Synergistic effects of thrombin and p300 on Elk-1 activation of transcription. A 1 µg aliquot of Gal4dbd–Elk-1307 plasmid, 1 µg of the Gal4-controlled luciferase reporter (pFRLuc) and 2 µg of the β-gal control plasmid pCH110 were co-transfected into fibroblasts. Luciferase activity was normalized to β-gal activity and to protein concentration. (A) Cells transfected with pFRLuc and treated with thrombin (9 U/ml) showed an ∼4-fold increase in transcription (lane 2). With p300 or CBP co-transfection alone (1 µg of plasmid), there was no difference from control (lanes 3 and 6). When co-transfected cells were treated with thrombin, a synergistic response was observed for p300 (lane 5) but was not significant for CBP (compare lanes 2 and 8). Gal4dbd served as negative control. Insert: immunoblots of extracts from lanes 5 and 8 to detect p300 and CBP levels using an antibody that recognizes both proteins (upper panel) and an antibody specific to the C-terminus of p300 (α-p300CT; lower panel). (B) p300–thrombin synergistic effects require phosphorylation of Ser383 and Ser389 on Elk-1. Odd lanes confirm the results in (A). Even lanes: cells transfected with Gal4dbd–Elk-1307 S-A (which carries S383A and S389A mutations) showing that loss of phosphorylation on these serine residues eliminates the thrombin response and p300 synergy. Insert: immunoblots of protein levels of exogenous Gal4 fusion proteins in lanes 7 and 8. (C) A schematic representation of a region of the cIL-8 promoter (p683; Li et al., 2000) depicts the Elk-1 DNA-binding elements. (D) To determine whether p300-mediated activation of the cIL-8 gene is direct and specific, Elk-1 and p300 expression vectors were transfected individually with the p683 reporter of cIL-8. Increasing amounts of p300 gave a linear response in luciferase activity upon thrombin stimulation, but increasing Elk-1 showed synergy.
To confirm that this synergistic effect is specific, we performed the same experiments using Gal4dbd–Elk-1307 with double mutation of Ser383 and Ser389 (Figure 1B), two serine residues critical for phosphorylation of Elk-1 by MAPK which leads to transcription activation (Janknecht et al., 1993; Gille et al., 1995). Mutation of these two serines completely eliminated the thrombin effects and thrombin–p300 synergy, even though the wild-type and mutant fusion proteins were expressed at the same level (Figure 1B, insert), showing that the synergistic effects in Figure 1A depend on Elk-1 Ser383 and Ser389 phosphorylation.
To test for this synergy in a native eukaryotic promoter, we performed similar experiments using a luciferase reporter previously developed by us for cIL-8 (p683, Figure 1C). This reporter contains –683 to +32 bp of the cIL-8 promoter, responds to thrombin treatment, and the response is controlled by Elk-1 (Li et al., 2000). p683 was co-transfected with Elk-1- and/or p300-expressing vectors (Figure 1D). Exogenous expression of either Elk-1 or p300 alone affected the constitutive basal levels of transcription and enhanced cIL-8 gene transcription in response to thrombin. In the case of Elk-1, there was a clear dose-dependent synergistic effect, whereas, in the case of p300, the ratio of gene transcription in response to thrombin relative to the quiescent state was independent of concentration.
Interactions between Elk-1 and p300 in vitro
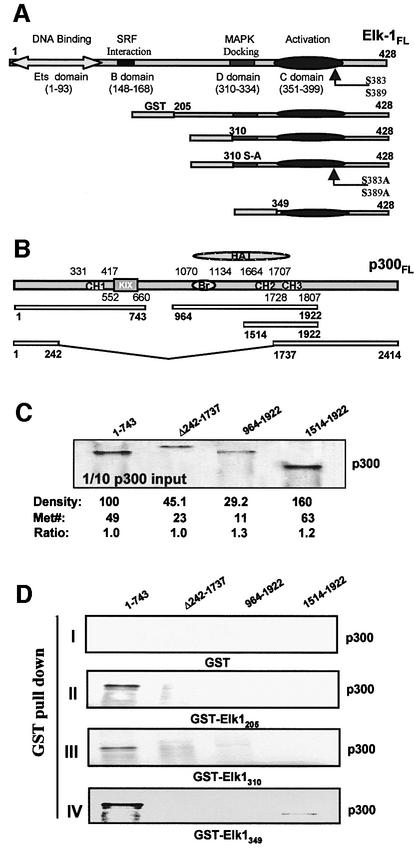
To determine whether Elk-1 and p300 physically interact, we used GST pull-down assays (Figure 2). Serial N-terminal deletions of Elk-1 cDNA were used to produce several C-terminal Elk-1 proteins that were fused to GST at their N-termini (Yang et al.,1998b). The inputs of GST–Elk-1 fusion proteins to be used in the pull-down assays were evaluated by western blot analysis and densitometry. Similarly, constructs containing various domains of p300 were expressed in vitro (Avantaggiati et al., 1997). All four constructs were used as templates for in vitro transcription and translation to obtain the various radiolabeled truncated proteins that were then run on gels for quantitation (Figure 2C). This procedure allowed us to use the same relative molar amounts for the various p300 truncated proteins, enabling detection of interactions between Elk-1 and p300 as well as changes in those interactions after exposure to activated ERK2 (Figure 3).
Fig. 2. GST pull-down assays to map Elk-1 and p300 ‘basal’ interactions in vitro. GST–Elk-1 fusion proteins on beads and 35S-labeled p300 truncated proteins, prepared by in vitro transcription and translation, were used. (A) A schematic diagram of full-length Elk-1 and several GST–Elk-1 fusion proteins. (B) A schematic diagram of the p300 truncated proteins. (C) p300 proteins used in GST pull-down binding assays. One-tenth of p300 inputs in the binding assays were examined by SDS–PAGE and autoradiography. (D) Molecular interactions between Elk-1 and p300 truncated proteins. GST beads alone were used as negative control (panel I). All three Elk-1–GST fusion proteins tested contain the activation domain (C domain) and bind to the N-terminus (1–743) of p300 (panels II–IV). However, the Elk-1 protein that contains only the activation domain (panel IV) binds with a higher affinity to p3001–743 and also binds, albeit more weakly, to p3001514–1922.
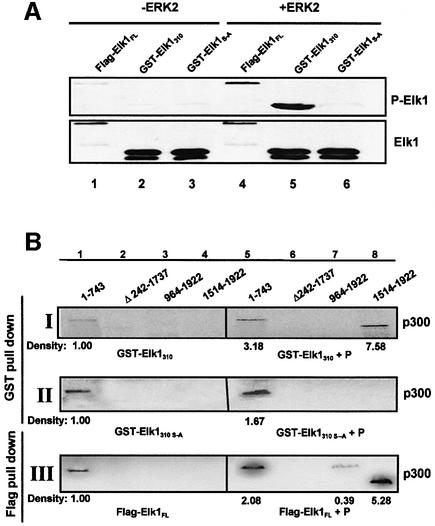
Fig. 3. Phosphorylation of Elk-1 changes its interactions with p300. (A) For the GST fusion proteins, flag-tagged full-length Elk-1 (flag-Elk-1FL), GST–Elk-1310 and GST–Elk-1310 S-A fusion proteins were phosphorylated on beads by activated ERK2. Immunoblot analysis was performed with an antibody against phospho Ser383. The same membrane stripped and reprobed with an antibody against Elk-1 shows equal loading of proteins. (B) The unphosphorylated and phosphorylated Elk-1 proteins were subjected to GST or anti-Flag pull-down assays with radiolabeled p300 truncated proteins as described in Figure 2.
The GST pull-down experiments show that the various Elk-1 truncated proteins bind to p3001–743 without requiring phosphorylation (Figure 2D, panels II and III). This was also true for the full-length Elk-1 protein (not shown in Figure 2, but see Figure 3B, panel III). No binding of Elk-1 to p300964–1922 was observed. However, weak but repeatable binding of GST–Elk349 to p3001514–1922 was seen. Interestingly, the truncated GST–Elk-1 proteins do not bind to p300Δ242–1737, indicating that the sequence of amino acids between residues 243 and 743 is necessary for the interaction between these proteins and p300. In addition, our results show that the MAPK docking domain of Elk-1 (D-domain) does not participate in the interaction of the Elk-1 proteins with p3001–743 because the GST–Elk-1349 protein (that does not contain the D-domain) binds to this region of p300, and does so with a slightly higher efficiency than the D-domain-containing proteins (Figure 2D, panel IV). Moreover, when the D-domain is absent (Elk-1349), there is an additional weak affinity binding to the region of p300 that contains CH2 and CH3 alone (p3001514–1922; Figure 2D, panel IV), suggesting that the presence of the D-domain interferes with binding of Elk-1 to the HAT activity domain of p300 under quiescent conditions. These results taken together show that Elk-1 interacts with the p300 N-terminal region (amino acids 242–743) through the transcription activation domain and without requiring phosphorylation; hence, we refer to this interaction of Elk-1 with p300 as ‘basal’ binding.
Elk-1 phosphorylation leads to changes in interactions with p300
Elk-1 constitutively binds to immediate-early response gene enhancer elements without requiring activation. As shown above, Elk-1 also can bind to p300 in vitro without requiring phosphorylation/activation. However, phosphorylation of Ser383 and Ser389 is a pre-requisite for Elk-1 function in activation of gene transcription in vivo (Janknecht et al., 1993); it causes a conformational change in Elk-1, which is accompanied by an increase in DNA-binding affinity of the Elk-1 N-terminus (Yang et al., 1999). Therefore, we tested whether phosphorylation of Elk-1 also affects its interactions with p300 and is important for activation of this co-activator. GST–Elk-1310 fusion protein contains both the MAPK docking motif and the activation domain of Elk-1, hence we used it and the Elk-1 full-length protein (Elk-1FL) in the presence of activated ERK2 to investigate the role of Elk-1 phosphorylation in co-activator binding. Figure 3A shows that recombinantly produced GST–Elk-1310 and flag-Elk-1FL become phosphorylated on Ser383 in the presence of activated ERK2. As expected, ERK2 does not phosphorylate the GST–Elk-1310 S-A protein and this latter protein cannot be detected with a specific antibody against phosphorylated Ser383. Figure 3B shows that phosphorylated and unphosphorylated forms of Elk-1 interact differently with p300. After phosphorylation by ERK2, GST–Elk-1310 not only has a higher affinity for the N-terminus of p300 (Figure 3B, panel I, compare lanes 1 and 5), but the most significant change occurs in the pairing of GST–Elk-1310 and the region of p300 that contains CH2 and CH3 (Figure 3B, panel I, compare lanes 4 and 8) which is associated with the HAT activity of this co-activator. This latter interaction is Elk-1 serine phosphorylation specific because such changes do not occur in GST–Elk-1310 S-A (Figure 3B, panel II, compare lanes 4 and 8). The small increase in binding of GST–Elk-1310 S-A to p3001–743 after exposure to activated ERK2 could be the result of phosphorylation of other serine or threonine residues present in the Elk-1 C-terminus. Interestingly, phosphorylated GST–Elk-1310 did not interact with p300964–1922 which also contains the CH2 and CH3 domains (Figure 3B, panel I, lane 7). This may be because the extra sequences present N-terminally of CH2, when not in the context of the whole molecule, interfere with the binding of GST–Elk-1310 to p3001514–1922. For example, p3001514–1922 may be an independently folding unit, but p300964–1922 fails to fold correctly. To further these studies, we performed similar experiments with flag-tagged Elk-1FL (Figure 3B, panel III). Much like Elk-1310, flag- Elk-1FL also interacts with p3001–743 and with p3001514–1922. However, in addition, it interacts with p300964–1922 which not only contains the CH2 and CH3 domains but also has the bromo domain, suggesting N-terminal involvement of full-length Elk-1 in binding to this region of p300. Taken together, these results show a novel role for Ser383 and Ser389 phosphorylation in Elk-1 function. Not only does this change enhance the N-terminal DNA-binding activity of Elk-1, but it also alters the interactions and enhances the affinity of the C-terminus of Elk-1 for the p300 co-activator, which potentially can have important effects in downstream gene activation.
Interactions between Elk-1 and p300 in vivo
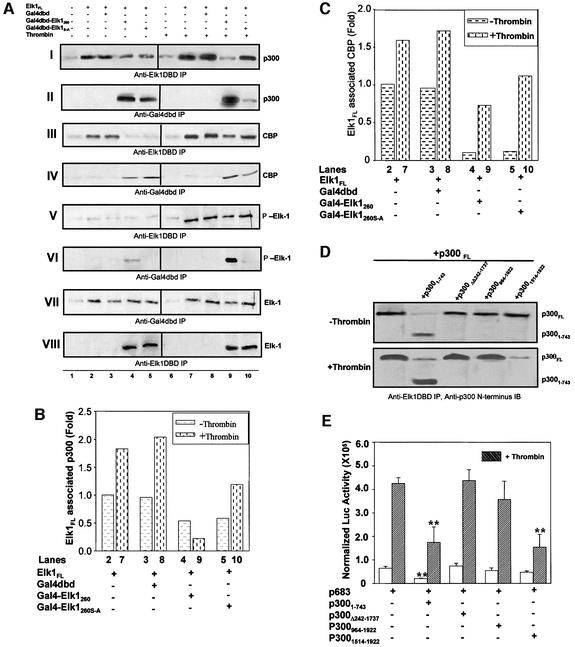
Using confocal microscopy, we determined that Elk-1 is transiently activated by thrombin in primary fibroblasts; the peak of activation occurs ∼15 min after stimulation, at which time Elk-1 and p300 co-localize in the nucleus. The nuclear Ser383 and Ser389 phosphorylation signal was attenuated after 30 min (data not shown). To address further the phosphorylation effects on Elk-1–p300 interaction in vivo, we performed competition experiments using the Gal4 fusion protein and analyzed them by co-immunoprecipitation. The fibroblasts were co-transfected with pCMV-Elk-1FL, Gal4dbd, Gal4dbd–Elk-1260 or Gal4dbd–Elk-1260 S-A. The three latter proteins were transfected in excess (twice the amount of full-length Elk-1) to facilitate detection of competition. Nuclear extracts were prepared after 15 min in the presence or absence of thrombin treatment, and subjected to two types of immunoprecipitation. One used an antibody against the Elk-1 DNA-binding domain (anti-Elk-1DBD) that does not interfere with the Elk-1–p300 binding region (which occurs through the C-terminus of Elk-1) to pull-down endogenous and overexpressed Elk-1FL. The other used an antibody to the Gal4 DNA-binding domain (anti-Gal4dbd) to pull-down Gal4–Elk-1 fusion proteins. The precipitates from both antibodies were examined for (i) p300 or CBP by immunoblotting with antibodies that specifically recognize these two co-activators (anti-p300 and anti-CBP); (ii) Elk-1 activation with an antibody that recognizes phosphorylated Elk-1 (α-P-Elk-1); and (iii) expression levels with an antibody that recognizes the Elk-1 protein (α-Elk-1).
Using this approach, we compared the interaction between p300/CBP and Elk-1 in vivo (Figure 4): p300 was co-immunoprecipitated with both the endogenous Elk-1 (Figure 4A, panel I, lane 1) and overexpressed Elk-1 (panel I, lane 2), and this interaction was enhanced significantly by thrombin treatment (panel I, compare lanes 1 and 2 with lanes 6 and 7). Furthermore, Gal4dbd–Elk-1260 competed with Elk-1FL for p300 binding under both conditions (panel I, lanes 4 and 9), whereas the mutated form Gal4dbd–Elk-1260 S-A could not compete out phosphorylated Elk-1 binding to p300 (panel I, lanes 5 and 10), strongly suggesting that Elk-1 phosphorylation enhances Elk-1–p300 interactions in vivo. Indeed, the Ser383 and Ser389 phosphorylated form of Elk-1 (Figure 4A, panel II, lane 9) had a higher affinity for p300 than the S-A mutant, which cannot be phosphorylated (panel II, lane 10), even though the amount of S-A mutant is in excess. This competition is specific for Elk-1 because the control fusion protein Gal4dbd did not compete out p300 binding to full-length Elk-1 (Figure 4A, panel I, lanes 3 and 8). These results are summarized in Figure 4B. All of these affinity changes correspond to the in vitro GST pull-down data (Figure 3). These results indicate that in vivo, thrombin can control the affinity of Elk-1–p300 interactions by phosphorylation of Elk-1 on Ser383 and Ser389. Similar results were obtained when the recombinant GST–Elk-1310 was used as competitor in the co-immunoprecipitation assays (data not shown).
Fig. 4. In vivo competition assays to examine Elk-1–p300 interaction. (A) Cells overexpressing p300/CBP (2 µg of plasmid) and full-length Elk-1 (0.2 µg of plasmid) in the presence or absence of Gal4–Elk-1260 or Gal4–Elk-1260 S-A (0.4 µg of plasmid each) were treated with thrombin (9 U/ml) for 15 min and nuclear extracts prepared. Differential co-immunoprecipitation experiments were performed with antibody against the DNA-binding domain of Elk-1 (α-Elk-1DBD) or with antibody against the Gal4 DNA-binding domain (α-Gal4dbd). Panels I and II: p300 binding to Elk-1FL and Gal4dbd–Elk-1260. Panels III and IV: CBP binding to Elk-1FL and Gal4dbd–Elk-1260. Panels V and VI: immunoblots on the same membrane as panels I and II verify the phosphorylation status on Ser383 in each sample. The presence of more activated Elk-1 in lanes 4 and 9 of panel VI than that of panel V corresponds to higher levels of p300 binding to Gal4dbd–Elk-1260 in panel II, indicating that the competition shown in panels I/II and III/IV is specific. Panels VII and VIII: the same blots as V and VI reprobed with antibody against the C-terminus of Elk-1. (B and C) Summary of competition experiments. The p300/CBP band density in panels I and III was normalized to the Elk-1FL band density in VII. (D) Cells overexpressing Elk-1FL and p300FL were co-transfected with expression vectors for various p300 truncated proteins; the nuclear extracts were immunoprecipitated with an antibody to Elk-1 DBD and immunoblotted with an antibody to the N-terminus of p300. Without thrombin treatment, full-length p300 remained associated with Elk-1 in the presence of various p300 truncated proteins, except for p3001–743, and this competitor was detected by the antibody shown as the band below the full-length p300. Experiments were repeated with samples after thrombin treatment for 15 min. Competition was detected not only with p3001–743, but also with p3001514–1922. p3001514–1922 is not visible on the blot because the α-p300 antibody only recognizes the N-terminus of p300. (E) A 4 µg aliquot of p683 reporter, 1 µg of pCH110 and the same molar amount of expression vectors for the p300 truncated proteins were co-transfected into cells, and samples collected 6 h after thrombin treatment and analyzed for cIL-8 promoter activation. Overexpressed p300 truncated proteins bind to endogenous Elk-1 protein if they have affinity and titrate out the endogenous p300, impairing reporter gene activation. This effect was observed with p3001–743 in the absence and presence of thrombin, but detected significantly with p3001514–1922 only after thrombin treatment. **P < 0.01.
Similar experiments to examine Elk-1–CBP interaction patterns showed that thrombin treatment enhances CBP interaction with Elk-1. However, this increase is much less pronounced and is not as dependent on Ser383 and Ser389 phosphorylation (Figure 4A; compare lanes 9 and 10 in panels I and II, and lanes 9 and 10 in panels III and IV). This is consistent with the lack of sygnergism observed when thrombin is applied to cells overexpressing CBP with Gal4–Elk1 fusion protein (Figure 1A).
To confirm the changes in binding interactions between Elk-1 and p300 with and without thrombin treatment and to examine details of binding in vivo, we performed competition experiments with co-transfection of expression vectors for full-length p300 and various deletion fragments of p300. Nuclear extracts were collected and precipitated with an antibody specific to the Elk-1 DNA-binding domain followed by immunoblot analysis with antibody specific to the p300 N-terminus (Figure 4D). In the absence of thrombin, only p3001–743 competed with full-length p300 for Elk-1 binding, whereas in the presence of thrombin, p3001514–1922 showed affinity for Elk-1, confirming the in vitro GST pull-down assay (Figure 3). As a functional consequence, p3001–743 competes with the endogenous Elk-1–p300 interaction and results in a decrease of luciferase production in the cIL-8 p683 reporter assay in the presence or absence of thrombin, whereas p3001514–1922 only competes for binding to Elk-1 in the presence of thrombin (Figure 4E). Taken together, the in vitro and in vivo results strongly suggest that Elk-1 phosphorylation increases its affinity for p300 and that this increase involves a new interaction domain on p300.
Changes in interaction of Elk-1 and p300 are accompanied by changes in p300-associated HAT activity
p300 is a co-activator with intrinsic and associated acetyltransferase activity that normally functions as an important component of histone-modifying complexes. As shown in Figure 4, Elk-1 and p300 interact with each other and that interaction is enhanced after thrombin treatment. To determine whether this interaction leads to HAT function, standard HAT assays were performed with the same immunoprecipitates analyzed in Figure 4A. The HAT activity of Elk-1-associated complex displayed a correlation to Elk-1 phosphorylation. As shown in Figure 4A, overexpressed Elk-1 can bind significant amounts of p300. However, in the absence of thrombin treatment, Elk-1 Ser383 phosphorylation is low and the HAT activity of the associated p300 is barely detectable (Figure 5A lanes 2–5). The phosphorylation of Ser383 not only increases the amount of p300 binding but, more importantly, it dramatically raises its ability to transfer acetyl groups to the histones (Figure 5A, lane 7). This HAT activity is associated specifically with Elk-1 phosphorylation because phosphorylated Gal4–Elk-1260 competed out p300, and hence HAT activity, from full-length Elk-1, whereas Gal4–Elk-1S-A could not (Figure 5A, lanes 9 and 10; Figure 5B, panel IV). These results strongly suggest that phosphorylated Elk-1 not only changes its interactions with p300 and enhances the recruitment of this co-activator to the enhancer elements, but also induces allosteric changes in p300, causing an increase in its enzymatic function.
Fig. 5. HAT activity in the Elk-1–p300 complex after thrombin stimulation. (A) Elk-1FL–p300 complex-associated HAT activity was examined on protein G–Sepharose beads. Purified recombinant p300 HAT domain (lane 11) and full-length GST–p/CAF (lane 12) served as positive controls, and glutathione–agarose/protein G–Sepharose beads only as negative control. Elk-1-associated HAT activity was observed in cells stimulated by thrombin when Elk-1FL was present, except when competed out by Gal4-Elk-1260. (B) Elk-1 conformation is critical for associated p300 activity. Elk-1FL S-A (0.2 µg of plasmid) was co-transfected with p300 (2 µg of plasmid). Experiments similar to those in Figure 4 and in (A) were performed to test Elk-1-associated p300 HAT activity. Panel I: a similar level of Elk-1 proteins. Panel II: thrombin stimulated Ser383 phosphorylation of Elk-1FL protein but not of the mutant. Panel III: Elk-1–p300 interaction. Thrombin induces a 2.4-fold affinity increase upon Elk-1FL phosphorylation. Panel IV: HAT activity of the Elk-1-associated complex. Densitometry analysis of HAT activity (graph below panel IV) records the sum of densities of bands (representing H1–H4 acetylation) in each lane. The density of HAT activity bands was divided by the density of p300 bands detected by western blot in the corresponding lanes in panel III, and the results are shown in lane 1, to which we assigned a value of 1. Comparison of lanes 3 and 4 shows a 7-fold increase in HAT activity upon thrombin-stimulated phosphorylation of Elk-1FL. HAT activity from the S-A mutant was undetectable. (C) Nuclear extracts from cells transfected with 3 µg of p300 or CBP expression vectors were collected with or without 15 min thrombin treatment. α-p300 or α-CBP immunoprecipitation was performed to separate and concentrate the corresponding protein. Both co-activators show a slight increase in HAT activity, but the magnitude is small compared with changes observed when p300 is associated with Elk-1. (D) Thrombin-stimulated Elk-1-associated HAT activity can be eliminated by 25 µM of the MEK1/ERK2 inhibitor PD98509 (lane 4) and partially inhibited by 20 nM of the p38 inhibitor SB203580 (lane 5). The Gal4dbd–Elk-1307 fusion protein system (E) and the cIL-8-p683-luciferase reporter (F) were used to detect the relationship between inhibition of HAT activity and inhibition of transcriptional activity; inhibitor results are similar to those in (D).
MAPK can phosphorylate CBP in vitro, and a phosphorylation-induced activation model of CBP/p300 has been developed (Janknecht and Nordheim, 1996). According to this model, the HAT activation shown in Figure 5A could also be explained by direct phosphorylation of p300. To distinguish between these two possibilities, we performed the experiments shown in Figure 5B and C. Elk-1FL and Elk-1FL S-A were transfected individually into the cells, co-immunoprecipitated with anti-Elk-1DBD (as described in Figure 4), and the HAT activity of the precipitates was examined. The Elk-1FL S-A mutant is able to bind p300 in vivo, and thrombin-stimulated MAPK activation should have been able to phosphorylate p300, but the associated complex was unable to activate HAT activity. However, in the case of full-length Elk-1, thrombin treatment not only enhanced p300 binding (Figure 4A), but it also greatly enhanced the associated HAT activity. Furthermore, we immunoprecipitated p300 or CBP from the same nuclear extract samples and performed the HAT assay. A slight increase of p300 activity in general is detected, but it is much less than the HAT activity associated with Elk-1 (Figure 5C). These results strongly suggest that active Elk-1 is required for the function of the Elk-1–p300 complex. Whether p300 phosphorylation is necessary remains to be determined, but our results suggest that if it is required, it is not sufficient.
The requirement for Elk-1 phosphorylation for the Elk-1–p300 complex to acquire HAT activity was confirmed by signal transduction inhibitors that block thrombin pathways (Figure 5D). Treatment with PD98059, a specific inhibitor of MEK1 (that directly phosphorylates/activates ERK2) completely abolished this HAT activity. Interestingly, PD98059 treatment also greatly decreases Elk-1 activation (Figure 5E) and thrombin stimulation of cIL-8 chemokine promoter-controlled transcription (Figure 5F). A specific inhibitor for the p38 MAPK, SB203580, also inhibits thrombin-driven stimulation but to a lesser extent than PD98059 (Figure 5E and F). This smaller inhibition was paralleled by partial reduction of HAT activity (Figure 5D, lane 5). Taken together, these results show that after thrombin-stimulated phosphorylation of Elk-1, the Elk-1–p300 complex acquires HAT activity and stimulates chemokine gene activation.
To make a direct correlation of the Elk-1–p300 interaction with the cIL-8 promoter, chromatin immunoprecipitation (ChIP) was performed to detect the associated protein on Elk-1 elements (Figure 6). DNA and associated proteins were cross-linked by formaldehyde, sheared and precipitated by a variety of antibodies. The DNA fragments were then released and amplified by nested PCR to target specifically the promoter region rich in Elk-1/Ets elements. We observed that Elk-1 protein was bound to Elk-1 elements in the absence of thrombin treatment, and that thrombin stimulated an increase in this interaction (which reached a peak between 15 and 30 min). These results correlate well with those obtained by confocal microscopy. We also detected pre-assembly of p300 on Elk-1 elements and an enhancement in p300 binding only 5 min after thrombin treatment that was maintained for >60 min. On the other hand, CBP was detected on the Elk-1 elements to a much lesser extent and in a more constant manner. Even though p300 is strongly pre-assembled on the Elk-1 elements, we did not detect significant histone acetylation until 15 min after thrombin treatment, suggesting that this activity is the result of conformational changes occurring in these molecules, and/or of changes in configuration of the complex which are required for the function of p300 downstream of thrombin signaling.
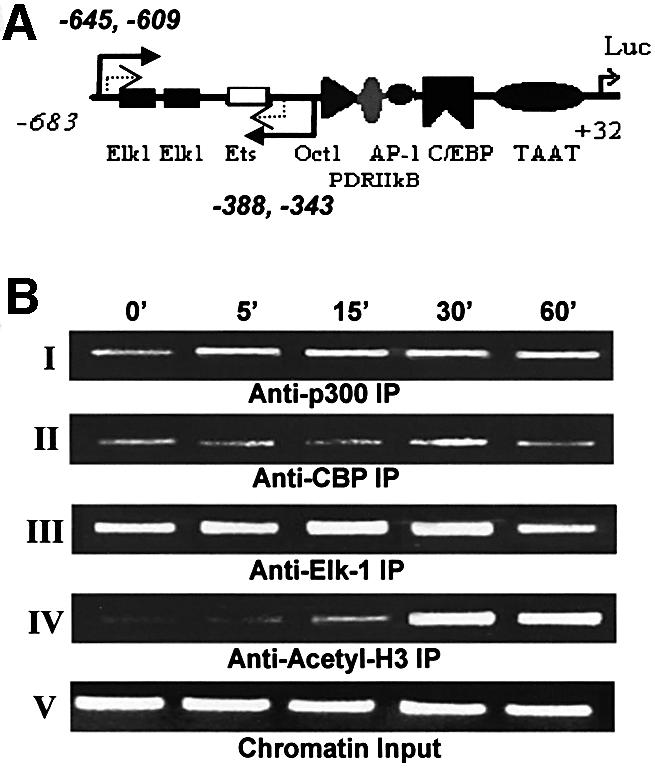
Fig. 6. Dynamics of the cIL-8 promoter-associated Elk-1 complex analyzed by ChIP. Fibroblasts were treated with thrombin over time (0–60 min), and chromatin DNA with associated proteins was cross-linked with formaldehyde. Various antibodies were used to precipitate different proteins, then DNA was released from the complex and purified to serve as a template for PCR. The cIL-8 promoter region containing Elk-1 elements was amplified specifically by two rounds of nested PCR. The amount of DNA represents the degree of occupancy of cIL-8 Elk-1 sites in the cell population. The primer choice is marked by the solid and dashed arrows in (A). The first round of PCR amplified the sequence corresponding to –645 to –343 of the cIL-8 promoter, which contains two Elk-1 elements and an Ets element, which potentially binds to Elk-1 with lower affinity. The second round of PCR was performed with the addition of nested primers specific for the –609 to –388 region to amplify the signal and avoid false priming. (B) ChIP resulted in a 222 bp band after two rounds of PCR. Panels I and II: immunoprecipitation with α-p300 and α-CBP shows the assembly of transcription activator and co-activator complex during treatment with thrombin. Panel III: immunoprecipitation with α-Elk-1 illustrates Elk-1 elements–transcription factor binding. Panel IV: immunoprecipitation with α-acetyl-H3 shows histone acetylation in the proximal promoter region of Elk-1. Panel V shows equal loading of chromatin components before precipitation.
Discussion
The work presented here addresses the mechanisms by which MAPK-activated signal transduction regulates Elk-1–p300 interactions to control gene transcription. Previous studies on the CREB–CBP complex (and other transcription factor–co-activator complexes) indicate that recruitment of CBP following CREB phosphorylation is the key event in triggering transcriptional activation through CBP (e.g. Janknecht and Hunter, 1996). Here we show that, rather than recruitment, the activation of complexes containing Elk-1 and the CBP-related protein, p300, can occur through changes in transcription factor– co-activator interactions following Elk-1 phosphorylation. Specifically, we show that: (i) p300 is the co-activator that participates with activated Elk-1 in chemokine gene transcription in vivo; (ii) the Elk-1 transactivation domain interacts with the N-terminus of p300 without need for Elk-1 phosphorylation in vitro and in vivo; (iii) thrombin-stimulated phosphorylation of Elk-1 induces a change in interactions between Elk-1 and p300 both in vitro and in vivo; and (iv) Elk-1 phosphorylation/activation dramatically increases HAT activity in the Elk-1–p300 complex, and this HAT activity correlates well with gene transcription.
We propose a new mechanism by which Elk-1 transactivates gene expression upon stimulation by thrombin (Figure 7). In this model, Elk-1 and p300 associate in an inactive conformation bound to the enhancer elements of the gene upstream regulatory region. Upon thrombin-induced phosphorylation of Elk-1 by MAPK, the complex changes conformation, leading to systemic allosteric effects. Under these conditions, the affinities between Elk-1 and DNA and between the Elk-1 C-terminus and p300 are significantly enhanced, and the HAT activity of the complex is activated, resulting in histone acetylation and changes in nucleosome structure. These conformational changes may also lead to interactions between p300 and the basal transcription machinery (Yuan et al., 1996), overcoming the threshold for activation of gene transcription. This activating process takes ∼15–30 min to reach its peak, measured by histone acetylation. Shortly after fully fledged activation, the co-repressor mSin3A is recruited to the N-terminus of Elk-1 and attenuates histone acetylation in 60–120 min (Yang et al., 2001). This sequential up- and down-regulatory process carried out by different domains of proteins illustrates well how signal transduction mediates transcription factor conformational changes (Yang et al., 1999) and therefore tightly controls the timing of immediate-early response gene activation.
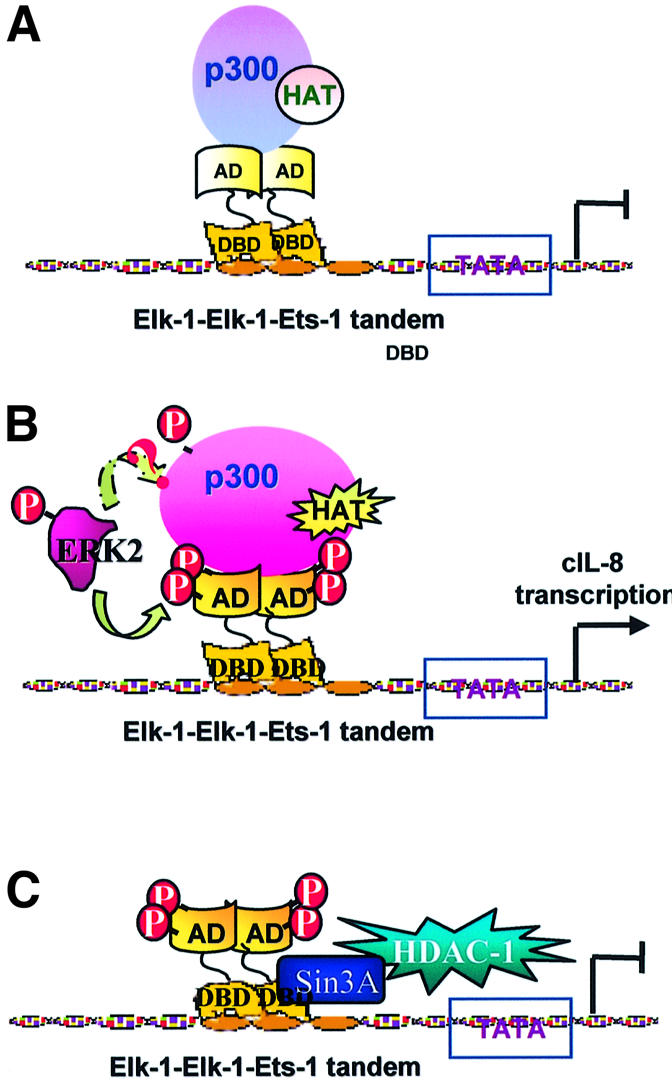
Fig. 7. Thrombin-induced Elk-1–p300 transcription activation model. (A) In the absence of thrombin, Elk-1 is hypophosphorylated, bound to the promoter, inactive and associated with p300. p300 is also in a quiescent conformation and the histones are associated with the DNA, forming nucleosomes. (B) In the presence of thrombin, Elk-1 becomes hyperphosphorylated on Ser383 and S389 (p300 also is possibly phosphorylated by MAPK). This phosphorylation results in changes in interaction between Elk-1 and p300 that render a stronger binding of Elk-1 to DNA and of p300 to Elk-1. The changes also trigger HAT activity that potentially leads to acetylation of the histones in neighboring nucleosomes, releasing them from the complex and allowing transcription to be initiated. (C) Shortly after fully fledged activation, Elk-1 recruits the co-repressor mSin3A which binds to its DNA-binding domain (N-terminus) and attenuates histone acetylation (Yang et al., 2001).
The enhanced Elk-1–p300 interaction induced through phosphorylation of Elk-1 by MAPK is not only due to binding through the pre-assembly domain, but probably involves a new region of p300 (Figures 2–4; data not shown). Phosphorylation of Elk-1 has been shown to promote complex conformational changes throughout the protein (Yang et al., 1999). Such conformational changes are likely to result in the appearance of new binding surfaces on Elk-1 that might participate in co-activator binding. These new surfaces may then be able to bind to p300 and hence provide the novel interactions (p3001515–1922) we observed. Alternatively, the phosphoacceptor motifs themselves might create this new binding surface.
Our results also show that phosphorylation of Elk-1 on Ser383 and Ser389 has important functional consequences; they result in acquisition of HAT activity (Figure 6). Two potential mechanisms may be involved in this functional switch. One possibility is that the activation of HAT is the result of allosteric or conformational changes in p300. Importantly for this enhancement of p300 function, the Elk-1 conformational changes must be necessary, if not sufficient, because the Elk-1S-A mutant is a dominant-negative in vivo whereas it does not interfere with Elk-1–p300 pre-assembly. Indeed, the importance of transcription factor on HAT activity of an associated co-activator has also been reported recently for HNF-1α (Soutoglou et al., 2001) and some b-ZIP transcription factors (Zta, NF-E2 and C/EBPα; Chen et al., 2001).
In addition to the role of Elk-1, direct phosphorylation of p300 or its associated cofactors by MAPK is another potential activating mechanism. MAPK has been shown to phosphorylate the C-terminus of CBP in vitro (Janknecht and Nordheim, 1996), but details are still unknown. One possibility is that p300/CBP phosphorylation serves to determine which HAT enzyme is selected (Xu et al., 1998). Elk-1-associated HAT can function on all histone proteins (Figure 5B); therefore, p300 probably is directly involved rather than p/CAF, which can only acetylate histones 3 and 4 (Figure 5A, lane 12). Another potential mechanism is direct activation of the co-activator upon phosphorylation. For example, CBP-induced transcription increase is uncoupled from its recruitment to CREB, and its HAT activity is calmodulin kinase IV dependent (Chawla et al., 1998; Hu et al., 1999). We examined this possibility in Figure 5C. The change in HAT activity of nuclear p300 or CBP upon thrombin stimulation was weak compared with that found with the Elk-1-associated complex, suggesting that Elk-1–p300 interactions dominate the activation process.
Recently, the importance of composition and conformational changes in regulation of co-activator function has been brought to light. The recruitment of co-activator may not be sufficient to promote gene activation, but rather conformational changes in the complexes may be the final determining factor. It has been reported that different transcription activators target different subunits of the metazoan mediator complex CRSP which then result in conformational changes of the complex. This conformation specificity contributes to enhancer-specific function (Taatjes et al., 2002). Our results strongly suggest an allosteric or conformational regulation of the p300 co-activator complex by the transcription factor Elk-1. For example, thrombin treatment is not sufficient to trigger p300 activity when the Elk-1S-A mutant is overexpressed even though, under these conditions, the potential effects of phosphorylation on p300 should not be impaired.
In conclusion, we show that, following thrombin-stimulated Elk-1 activation through phosphorylation by MAPK, a change occurs in the pattern of interaction of Elk-1 with the pre-assembled co-activator p300, leading to stimulation of HAT activity and rapid gene transcription. This ability of pre-assembled ‘ready-to-go’ complexes to undergo rapid stimulation could have broad physiological implications for responses to stress-inducing agents such as thrombin. This enzyme is a multifunctional serine protease generated at sites of injury and is important in initiating the cascade of events that lead to inflamma tion and other responses to traumatic and pathological conditions.
Materials and methods
Key reagents
Bovine thrombin, α-Flag agarose beads, glutathione–agarose beads, reduced glutathione and substrate histones were from Sigma; PD98059 and SB203580 (in dimethylsulfoxide) were from Calbiochem; and Fast-Flow protein G–Sepharose and [14C]acetyl-CoA were from Pharmacia. Primary antibodies used were: α-Elk-1307 (against amino acids 379–392); α-phospho-Ser383 Elk-1 (from Cell Signaling); α-Ets (Elk-1DBD), α-Gal4dbd, α-CBP C-terminus and α-p300 N-terminus (from Santa Cruz Biotech); α-p300/CBP (cross-reactive with both p300 and CBP), α-p300 C-terminus and α-acetyl-histone-3 (from Upstate Biotechnology); and α-cIL-8 (Martins-Green and Feugate, 1998). Secondary antibodies were goat α-rabbit or α-mouse antibodies conjugated to horseradish peroxidase for enhanced chemiluminescence (ECL; Amersham).
Plasmids
The Gal4 element-controlled luciferase (pFR-Luc) and p683 cIL-8(cCAF) reporter systems were described previously (Li et al., 2000). p300 truncated protein expression vectors were a gift from Dr Giordano (Avantaggiati et al., 1997). GST–Elk-1 vectors (Yang et al., 1998a): Gal4-Elk-1260 and Gal4-Elk-1260S-A are cytomegalovirus-driven eukaryotic expression vectors for fusion proteins containing the Gal4 DNA-binding domain and amino acids 260–428 of Elk-1 with or without S383A and S389A mutation, respectively; pAS278 is for Flag-tagged full-length wild-type Elk-1 in Escherichia coli; pAS383 is for Flag-tagged full-length wild-type Elk-1 (Elk-1FL) and pCMV-Elk-1FL S-A is for the Elk-1FL S383A and S389A mutation in eukaryotic cells.
Cell culture
Cultures of primary and secondary chicken embryo fibroblasts were prepared from 10-day-old chicken embryos as described previously (Vaingankar and Martins-Green, 1998).
Immunoblots
The procedures used to prepare and probe the immunoblots were described previously (Vaingankar and Martins-Green, 1998).
Transient transfection assay, thrombin activation and cell lysate collection
The Ca2PO4 precipitation method without glycerol shock and the luciferase assay were performed as described previously (Li et al., 2000). The specific amount of plasmid DNA to be tested for each experiment is included in the respective figure legends. For all experiments, transfected cells were incubated in the modified 199 medium for 36 h and then stimulated with thrombin for 3 h prior to lysis. Treatment with inhibitors was for 30 min before thrombin stimulation.
Luciferase and β-galactosidase assay
The procedures were described previously (Li et al., 2000). For each experiment, triplicate samples were tested and the data expressed as mean light units of luminescence per unit of β-galactosidase activity, and also normalized to per µg of protein.
Recombinant expression of GST–Elk-1 truncated proteins
pGST–Elk-1 truncated proteins were prepared in BC21 E.coli as described previously (Jiang et al., 1995). The amount of the pertinent protein for each construct was measured by BandScan (Glyko) software to adjust the GST pull-down input.
In vitro transcription and translation
The TNT Quick Coupled Transcription/Translation System (Promega) was used to prepare the labeled p300 truncated proteins. For each 50 µl volume reaction, 30 µCi of Redivue™ l-[35S]methionine (Amersham) and 1 µg of plasmid construct were used, and the reaction was incubated at 30°C for 90 min. Proteins were separated by 7.5% SDS–PAGE and identified by molecular weight after autoradiography. BandScan software was used for quantification. The molar ratio for the GST pull-down assay for the various truncated p300 proteins was adjusted by the number of methionine residues in each sequence.
GST pull-down assay
Equal molar concentrations of GST or GST–Elk-1 truncated fusion proteins were incubated with equal molar amounts of 35S-labeled p300 peptides in NETN buffer for 1 h at room temperature. The p300 proteins that interacted with Elk-1 were eluted and separated by boiling for 10 min in SDS-containing sample loading buffer, analyzed by 7.5% SDS–PAGE and quantified by BandScan software.
Chromatin immunoprecipitation (ChIP) and nested PCR
The ChIP procedure was modified from previously published procedures (Shang et al., 2000). The PCR was carried out with the first round primers 5′-GACCTAGCGTTACCATAA-3′ and 5′-CTTTGTGATAATAACT GAC-3′ (94°C, 45 s; 45°C, 45 s; 68°C, 60 s) for 20 cycles to be in the linear range of the PCR, then followed by the second round of nested PCR with addition of Advantage 2 polymerase (Clontech) and primer pair 5′-ATTAACAATACTGACACT-3′ and 5′-GTATTTATTTCTGTTG AAG-3′ (94°C, 45 s; 43°C, 45 s; 68°C, 60 s) for another 20 cycles. The final products were separated on 1.5% agarose.
Elk-1 co-immunoprecipitation and p300 western blot
Nuclear extracts were prepared from cells treated in the presence or absence of thrombin, as described previously (Li et al., 2000). A 50 µg aliquot of total nuclear protein and 4 µg of the antibody to the Elk-1 DNA-binding domain (anti-Elk-1DBD) were incubated at 4°C overnight. A 50 µl aliquot of Fast-Flow protein G–Sepharose beads was added and incubated for 1 h at room temperature. The protein-bearing beads were washed three times in 150 mM NaCl RIPA and for the in vitro HAT assay they were washed three times with HAT assay buffer (see below). A 30 µl aliquot of beads was boiled for 10 min in SDS sample loading buffer and then separated by 7.5% SDS–PAGE. Protein transfer to nitrocellulose membranes was performed at 200 mA in p300 transfer buffer (20 mM Tris, 200 mM glycine, 0.1% SDS and 20% methanol) overnight at 4°C. The membranes were split in half horizontally; the upper half was probed with anti-p300 and the lower half with anti-P-Elk-1. Membranes were then stripped and probed with anti-Elk-1.
HAT activity assay
These assays were performed with the 20 µl of protein G–Sepharose beads left over from the anti-Elk-1DBD immunoprecipitations. The beads were incubated with 1.8 µg of histones H1–H4, 200 µM dithiothreitol (DTT), 100 µCi of [14C]acetyl-CoA and 1 mM phenylmethylsulfonylfluoride (PMSF) in HAT buffer (50 mM Tris pH 8.0, 0.1 mM EDTA, 50 mM KCl, 25% glycerol and 10 mM sodium butyrate) at 30°C for 30 min. The reaction was stopped with sample loading buffer and boiling for 10 min before separation by 10% SDS–PAGE, autoradiography and phosphoimaging analysis.
Acknowledgments
Acknowledgements
We thank Drs A.Giordano (Tomas Jefferson University) for the p300 constructs, E.Martinez (University of California, Riverside) for the purified p300 HAT domain-containing protein, R.Davis (University of Massachusetts Medical Center) for pCMV-Elk-1 expression vector, J.Stinson for full-length Elk-1 protein and X.Liu (University of California, Riverside) for GST control vector, the protocol for p300 western blot and helpful discussions. This work was supported in part by a Dissertation Research Grant from UC to Q.J.L., by the NIDDK to F.M.S., by the Wellcome Trust and the Lister Institute of Preventive Medicine to A.D.S. and by the NIGMS and AHA to M.M.-G.
References
- Avantaggiati M.L., Ogryzko,V., Gardner,K., Giordano,A., Levine,A.S. and Kelly,K. (1997) Recruitment of p300/CBP in p53-dependent signal pathways. Cell, 89, 1175–1184. [DOI] [PubMed] [Google Scholar]
- Chawla S., Hardingham,G.E., Quinn,D.R. and Bading,H. (1998) CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science, 281, 1505–1509. [DOI] [PubMed] [Google Scholar]
- Chen C.-J., Deng,A., Kim,A., Blobel,G. and Lieberman,P. (2001) Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol., 21, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H., Kortenjann,M., Thomae,O., Moomaw,C., Slaughter,C., Cobb,M.H. and Shaw,P.E. (1995) ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J., 14, 951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera R.E., Shaw,P.E. and Nordheim,A. (1989) Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature, 340, 68–70. [DOI] [PubMed] [Google Scholar]
- Hu S.C., Chrivia,J. and Ghosh,A. (1999) Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron, 22, 799–808. [DOI] [PubMed] [Google Scholar]
- Janknecht R. and Hunter,T. (1996) Transcription. A growing coactivator network. Nature, 383, 22–23. [DOI] [PubMed] [Google Scholar]
- Janknecht R. and Nordheim,A. (1996) MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem. Biophys. Res. Commun., 228, 831–837. [DOI] [PubMed] [Google Scholar]
- Janknecht R., Ernst,W.H., Pingoud,V. and Nordheim,A. (1993) Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J., 12, 5097–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Nepomuceno,L., Hopkins,K. and Sladek,F.M. (1995) Exclusive homodimerizaiton of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol. Cell. Biol., 15, 5131–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung A.L., Rebel,V.I., Bronson,R.T., Ch’ng,L.E., Sieff,C.A., Livingston,D.M. and Yao,T.P. (2000) Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev., 14, 272–277. [PMC free article] [PubMed] [Google Scholar]
- Li Q., Vaingankar,S.M., Green,H.M. and Martins-Green,M. (1999) Activation of the 9E3/cCAF chemokine by phorbol esters occurs via multiple signal transduction pathways that converge to MEK1/ERK2 and activate the Elk-1 transcription factor. J. Biol. Chem., 274, 15454–15465. [DOI] [PubMed] [Google Scholar]
- Li Q.J., Vaingankar,S., Sladek,F.M. and Martins-Green,M. (2000) Novel nuclear target for thrombin: activation of the Elk-1 transcription factor leads to chemokine gene expression. Blood, 96, 3696–3706. [PubMed] [Google Scholar]
- Martins-Green M. and Feugate,J. (1998) The 9E3/CEF4 gene product is a chemotactic and angiogenic factor that can initiate the wound-healing cascade in vivo. Cytokine, 10, 522–535. [DOI] [PubMed] [Google Scholar]
- Nissen L.J., Gelly,J.C. and Hipskind,R.A. (2001) Induction-independent recruitment of CREB-binding protein to the c-fos serum response element through interactions between the bromodomain and Elk-1. J. Biol. Chem., 276, 5213–5221. [DOI] [PubMed] [Google Scholar]
- Rao V.N., Huebner,K., Isobe,M., ar-Rushdi,A., Croce,C.M. and Reddy,E.S. (1989) elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science, 244, 66–70. [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Viollet,B., Vaxillaire,M., Yaniv,M., Pontoglio,M. and Talianidis,I. (2001) Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J., 20, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgambato V., Pages,C., Rogard,M., Besson,M.J. and Caboche,J. (1998) Extracellular signal-regulated kinase (ERK) controls immediate early gene induction on corticostriatal stimulation. J. Neurosci., 18, 8814–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D. (1995) ERK2/p42 MAP kinase stimulates both autonomous and SRF-dependent DNA binding by Elk-1. FEBS Lett., 368, 77–80. [DOI] [PubMed] [Google Scholar]
- Taatjes D.J., Naar,A.M., Andel,F.,3rd, Nogales,E. and Tjian,R. (2002) Structure, function and activator-induced conformations of the CRSP coactivator. Science, 295, 1058–1062. [DOI] [PubMed] [Google Scholar]
- Vaingankar S.M. and Martins-Green,M. (1998) Thrombin activation of the 9E3/CEF4 chemokine involves tyrosine kinases including c-src and the epidermal growth factor receptor. J. Biol. Chem., 273, 5226–5234. [DOI] [PubMed] [Google Scholar]
- Vo N. and Goodman,R.H. (2001) CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem., 276, 13505–13508. [DOI] [PubMed] [Google Scholar]
- Xu L. et al. (1998) Signal-specific co-activator domain requirements for Pit-1 activation. Nature, 395, 301–306. [DOI] [PubMed] [Google Scholar]
- Xu L., Glass,C.K. and Rosenfeld,M.G. (1999) Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev., 9, 140–147. [DOI] [PubMed] [Google Scholar]
- Yang S., Yates,P., Whitmarsh,A., Davis,R. and Sharrocks,A. (1998a) The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol. Cell. Biol., 18, 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Whitmarsh,A., Davis RJ, Sharrocks AD. (1998b) Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J., 17, 1740–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Shore,P., Willingham,N., Lakey,J. and Sharrocks,A. (1999) The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J., 18, 5666–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Vickers,E., Brehm,A., Kouzarides,T., Sharrocks,A. (2001) Temporal recruitment of the mSin3A–histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol., 21, 2802–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.P. et al. (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell, 93, 361–372. [DOI] [PubMed] [Google Scholar]
- Yuan W., Condorelli,G., Caruso,M., Felsani,A. and Giordano,A. (1996) Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem., 271, 9009–9013. [DOI] [PubMed] [Google Scholar]