Abstract
Translation initiation factor 1A (eIF1A) is predicted to bind in the decoding site of the 40S ribosome and has been implicated in recruitment of the eIF2–GTP–Met-tRNAiMet ternary complex (TC) and ribosomal scanning. We show that the unstructured C-terminus of eIF1A interacts with the C-terminus of eIF5B, a factor that stimulates 40S–60S subunit joining, and removal of this domain of eIF1A diminishes translation initiation in vivo. These findings support the idea that eIF1A–eIF5B association is instrumental in releasing eIF1A from the ribosome after subunit joining. A larger C-terminal truncation that removes a 310 helix in eIF1A deregulates GCN4 translation in a manner suppressed by overexpressing TC, implicating eIF1A in TC binding to 40S ribosomes in vivo. The unstructured N-terminus of eIF1A interacts with eIF2 and eIF3 and is required at low temperatures for a step following TC recruitment. We propose a modular organization for eIF1A wherein a core ribosome-binding domain is flanked by flexible segments that mediate interactions with other factors involved in recruitment of TC and release of eIF1A at subunit joining.
Keywords: eIF1A/eIF2/eIF3/GCN4/ribosomes/translation
Introduction
The initiation of translation in eukaryoties commences with binding of charged initiator tRNAMet (Met-tRNAiMet) to the 40S ribosomal subunit. This reaction is catalyzed by the eukaryotic initiation factor 2 (eIF2) in the form of a ternary complex (TC) with Met-tRNAiMet and GTP. The TC can bind to purified 40S subunits in vitro, producing a 43S pre-initiation complex, and this reaction is enhanced by eIF1A and the multisubunit complex eIF3. The formation of a stable 48S pre-initiation complex, with Met-tRNAiMet bound to the P (peptidyl-tRNA)-site and base-paired to the AUG start codon in mRNA, also requires eIF1 and the mRNA-binding factors eIF4F, eIF4A and eIF4B. Upon AUG recognition, the GTPase-activating protein eIF5 stimulates hydrolysis of GTP in the TC, with release of eIF2-GDP and other factors from the ribosome. The resulting 40S complex joins with the 60S subunit to form the 80S initiation complex in a reaction catalyzed by eIF5B. The recycling of eIF2-GDP to eIF2-GTP, required to regenerate TC for the next round of initiation, is catalyzed by the five-subunit guanine nucleotide exchange factor eIF2B (reviewed in Hinnebusch, 2000). The functions of these factors have been defined primarily by in vitro analysis of the partial reactions of the initiation pathway using purified mammalian components. While the corresponding yeast factors have critical functions in translation initiation in vivo (Hershey and Merrick, 2000; Hinnebusch, 2000), it is often unclear whether the biochemical activities ascribed to them by in vitro studies correspond to their essential functions in living cells.
Studies on the translational control of GCN4 expression in yeast have provided strong confirmation that eIF2 is crucial for delivery of Met-tRNAiMet to 40S ribosomes, and that eIF2B is required to maintain high levels of the TC in vivo. Phosphorylation of the α-subunit of eIF2 impairs the conversion of eIF2-GDP to eIF2-GTP by eIF2B that is required for TC formation. Phosphorylation of eIF2α by protein kinase GCN2 induces GCN4 translation in amino acid-starved cells, dependent on four short open reading frames (uORFs) in the mRNA leader. After translating the first uORF (uORF1), ribosomes resume scanning downstream in both starved and non-starved cells. In non-starved cells, where TC levels are abundant, ribosomes rapidly rebind the TC and reinitiate at uORF4, preventing them from reaching the GCN4 start codon. When TC levels are reduced in starved cells by eIF2 phosphorylation, a fraction of ribosomes fail to rebind the TC until scanning past uORF4, allowing them to reinitiate at GCN4 instead. Consistently, the bypass of uORF4 and induction of GCN4 translation in starved cells is suppressed by overproducing all three subunits of eIF2, or all four essential subunits of eIF2B, both conditions that should elevate TC levels. Morever, GCN4 translation is constitutively derepressed (Gcd– phenotype) in mutants in which TC levels are lowered by defects in eIF2 or eIF2B subunits or by reduced amounts of Met-tRNAiMet (Hinnebusch, 1996). Mutations in yeast eIF3 or eIF1A that impair TC binding would be expected to allow 40S subunits to scan past uORF4 and reinitiate at GCN4 even when the TC is abundant; however, no such Gcd– mutations have been identified in an eIF3 subunit or eIF1A. Thus, it is uncertain whether eIF3 and eIF1A are critically required for TC binding to 40S ribosomes in vivo.
The central domain of eIF1A is similar in sequence and tertiary structure to bacterial initiation factor IF1 (Battiste et al., 2000). IF1 binds to the A-site (Moazed et al., 1995; Carter et al., 2001), has been cross-linked to IF2 and stabilizes IF2 binding to the 30S subunit (Gualerzi and Pon, 1990; Palacios Moreno et al., 1999). IF2 promotes binding of fMet-tRNAiMet to the P-site (Gualerzi and Pon, 1990; La Teana et al., 1996), and its release from the ribosome following 30S–50S subunit joining is dependent on the IF2 GTPase activity (Luchin et al., 1999) and also on IF1 (Benne et al., 1973). The eIF5B is an ortholog of IF2 (Lee et al., 1999) and has been implicated in Met-tRNAiMet binding to the P-site in yeast (Choi et al., 1998). As indicated above, mammalian eIF5B is required for 40S–60S subunit joining in vitro, and GTP hydrolysis triggers its release from 80S ribosomes (Pestova et al., 2000). In several respects, therefore, eIF1A and eIF5B are functional homologs of bacterial IF1 and IF2.
In accordance with evidence that IF1 and IF2 interact on the 30S ribosome, we showed previously that purified eIF5B and eIF1A from yeast interact directly and are associated in cell extracts. We mapped the eIF1A-binding domain to the C-terminal portion of eIF5B, a region critically required for eIF5B function. Interestingly, eIF1A overexpression exacerbated the growth defect of fun12 mutants lacking eIF5B or containing a C-terminally truncated form of the factor. We suggested that eIF1A is partially dependent on eIF5B for release from the 80S initiation complex, such that eIF1A overexpression in a fun12 mutant would prolong eIF1A binding to the ribosome and impede entry of the first eEF1A-GTP–aminoacyl-tRNA to the A-site (Choi et al., 2000).
Here we have mapped the binding site for eIF5B to the unstructured C-terminus of eIF1A and shown that removal of this segment impairs translation initiation in vivo. A more extensive C-terminal truncation of eIF1A produced a Gcd– phenotype that was suppressed by overproducing the TC, providing the first evidence that eIF1A promotes 40S binding of TC in vivo. We also found that the unstructured N-terminus of eIF1A interacts with eIF2 and eIF3 and is required for optimal translation in vivo. Hence, we propose a modular structure for eIF1A in which the IF1-related domain mediates ribosome binding and is flanked by C-terminal segments involved in TC binding and interaction with eIF5B, plus an N-terminal domain (NTD) that contacts other initiation factors on the ribosome.
Results
The C-terminus of eIF1A is required for interaction with N-terminally truncated eIF5B in vivo
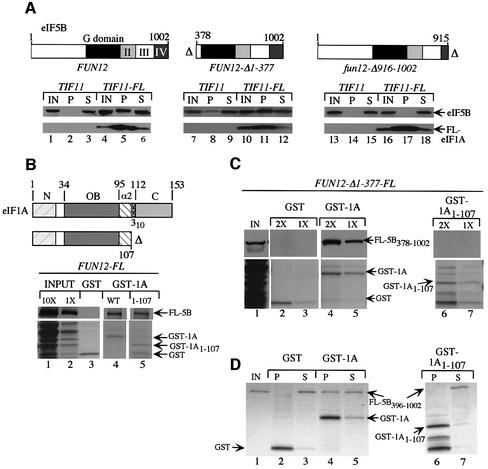
We previously reported that the C-terminal domain (CTD) of eIF5B is required for direct interaction with eIF1A in vitro in the context of a truncated version of eIF5B lacking the N-terminal 377 residues (eIF5B378–1002) (Choi et al., 2000). To confirm that the eIF5B CTD is required for eIF1A binding in vivo, we carried out immunoprecipitation experiments using strains expressing native eIF1A or FLAG epitope-tagged eIF1A (FL-eIF1A) from single copy (sc) plasmids and the eIF5B proteins shown in Figure 1A expressed from low-copy (lc) plasmids. The wild-type and FLAG-tagged alleles of TIF11 were functionally equivalent as judged by the indistinguishable growth rates of tif11Δ transformants containing each allele. Full-length eIF5B1–1002 and the N-terminally truncated protein eIF5B378–1002, but not C-terminally truncated eIF5B1–915, specifically co-immunoprecipitated with FL-eIF1A from whole-cell extracts (WCEs) (Figure 1A, lanes 5 and 11 versus 17). Thus, the eIF5B CTD is required for interaction with eIF1A in vivo, in the presence or absence of the eIF5B NTD. The fun12-Δ916–1002 allele (encoding eIF5B1–915) is almost completely defective for complementation of the Slg– phenotype of a fun12Δ mutant (Choi et al., 2000). Thus, the eIF1A-binding domain at the C-terminus of eIF5B is important for eIF5B function in vivo.
Fig. 1. The CTD of eIF1A is required for binding to N-terminally truncated eIF5B in vitro. (A) fun12Δ::hisG tif11Δ::hisG strains harboring TIF11 (lanes 1–3, 7–9 and 13–15) or TIF11-FL (lanes 4–6, 10–12 and 16–18) on sc plasmids p3412 and p3499, respectively, were produced by plasmid shuffling from H2971 and transformed with lc or hc plasmids containing FUN12 (pC479; lc) (lanes 1–6), FUN12-Δ1–377 (p3572; lc) (lanes 7–12) or fun12-Δ916–1002 (p3571; hc) (lanes 13–18). A 2 mg aliquot of WCEs was immunoprecipitated with anti-FLAG resin, resolved by SDS–PAGE and subjected to immunoblot analysis using antibodies against eIF5B (upper panels) or FLAG epitope (lower panels). Schematics of FUN12 alleles are presented above the relevant immunoblots, depicting locations of the G (GTP-binding) domain and domains II–IV. Lanes labeled IN, P and S contained, respectively, 5% of the input WCEs, 100% of the immunoprecipitates and 5% of the supernatants. (B) Full-length GST–eIF1A (WT GST-1A, lane 4), GST–eIF1A containing residues 1–107 (GST-1A1–107, lane 5) or GST alone (lane 3) were purified on glutathione–Sepharose from bacterial transformants harboring p3415, p3466 or pGEX-4T-1, respectively, and incubated with WCE from a transformant of strain J111 expressing FL-eIF5B (FL-5B) encoded on plasmid pC1005. Precipitated proteins were resolved by SDS–PAGE and subjected to immunoblot analysis with FLAG antibodies (upper panels). Lower panels show the Ponceau S-stained gels. Lanes 1 and 2 contain 10 and 1%, respectively, of the input WCE. The schematics depict the eIF1A segments in the GST fusions and locations of N-terminal (N), OB-fold (OB), α-helix 2 (α2), 310 helix and C-terminal domains of eIF1A. (C) The same as (B) except that WCE was obtained from a J111 transformant containing FUN12-Δ1–377-FL on plasmid pC1043, and two different amounts (1× and 2×) of GST fusion proteins were used in separate assays. IN contains 10% of the input WCE. (D) The same immobilized GST proteins as described in (B) were incubated with purified eIF5B396–1002 and the precipitated proteins were visualized by Coomassie Blue staining. IN, P and S lanes contained 10, 100 and 10% of the input, pellet and supernatant fractions, respectively.
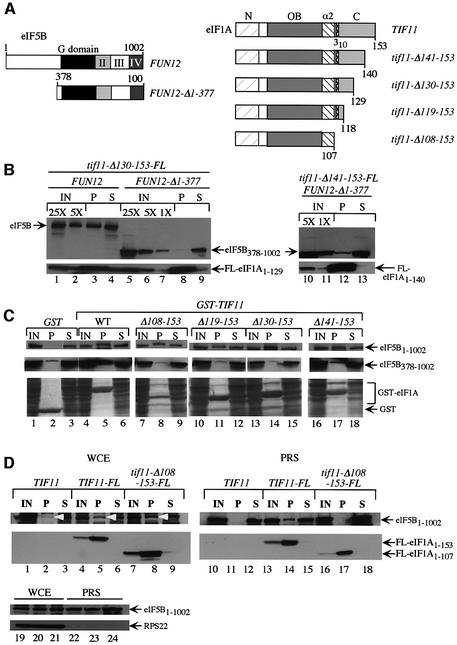
We next sought to identify the domain in eIF1A required for eIF5B interaction. First, GST–eIF1A fusions expressed in Escherichia coli were used in pull-down assays with WCEs prepared from fun12Δ transformants expressing wild-type eIF5B or N-terminally truncated eIF5B378–1002. Whereas full-length GST–eIF1A1–153 bound to both forms of eIF5B (Figure 1B, lane 4; Figure 1C, lanes 4 and 5), the GST–eIF1A1–107 fusion interacted with full-length eIF5B (Figure 1B, lane 5) but not with eIF5B378–1002 (Figure 1C, lanes 6 and 7). Note that GST–eIF1A1–107 lacks the entire unstructured C-terminus and predicted 310 helix (Figure 1B). The inability of GST–eIF1A1–107 to interact with N-terminally truncated eIF5B396–1002 was also observed using purified proteins (Figure 1D, lanes 4 and 6). We confirmed these results in vivo by showing that FL-eIF1A1–129, lacking the last 24 residues of the protein, co-immunoprecipitated with wild-type eIF5B but not with eIF5B378–1002 (Figure 2B, lanes 3 and 8). Thus, the eIF1A CTD contains a binding site for eIF5B that is required for association between these proteins in WCEs when the N-terminus of eIF5B is missing.
Fig. 2. Mapping residues in the C-terminal domain of eIF1A required for binding to eIF5B. (A) Schematics of the relevant eIF5B and eIF1A proteins. (B) fun12Δ::hisG tif11Δ::hisG strains harboring tif11-Δ130–153-FL (lanes 1–9) or tif11-Δ141–153-FL (lanes 10–13) on sc plasmids p3505 and p3503, respectively, were produced by plasmid shuffling from H2971 and transformed with plasmids containing FUN12 (pC479) (lanes 1–4) or FUN12-Δ1–377 (lanes 5–9 and 10–13). Co-immunoprecipitation of FL-eIF1A1–129 or FL-eIF1A1–140 with eIF5B or eIF5B378–1002 from WCEs using FLAG antibodies was analyzed as in Figure 1A. Lanes 1 and 2 contain 5 and 1%, respectively, lanes 5–7 contain 5, 1 and 0.2%, respectively, and lanes 10 and 11 contain 1 and 0.2%, respectively, of the relevant input WCEs. Lanes 3, 8 and 12 contain 100% of the pellets, and lanes 4, 9 and 13 contain 5% of the supernatant fractions. (C) TIF11 FUN12 strain H1895 (eIF5B1–1002, top panel) or strain J111 harboring FUN12-Δ1–377 on plasmid pC1000 (eIF5B378–1002, middle panel) were transformed with the following hc plasmids encoding GST fusions to eIF1A (WT) or C-teminally truncated eIF1A proteins lacking the indicated residues, or GST alone, all under the GAL promoter: pEG(KT) (lanes 1–3), p3559 (lanes 4–6), p3566 (lanes 7–9), p3563 (lanes 10–12), p3564 (lanes 13–15) and p3565 (lanes 14–16). WCE extracts were prepared and GST proteins purified on glutathione–Sepharose, separated by SDS–PAGE and subjected to immunoblotting with antibodies against eIF5B (upper two panels). Bottom panels show Ponceau S-stained gels corresponding to the upper panel immunoblot. IN, P and S lanes contained 5, 100 and 5% of the input, pellet and supernatant fractions, respectively. (D) WCEs (lanes 1–9) or PRSs (lanes 10–18) from FUN12 strains H2809 (TIF11), H2974 (TIF11-FL) and H3002 (hc tif11-Δ108–153-FL) were immuno precipitated with anti-FLAG resin, resolved by SDS–PAGE and subjected to immunoblot analysis using antibodies against the indicated proteins. IN, P and S lanes contained 10, 100 and 10% of the input, pellet and supernatant fractions, respectively. Lanes 19–24 contained the input samples probed by western analysis for 40S subunit protein RPS22.
We mapped the eIF5B-binding domain in eIF1A more precisely by in vivo GST precipitation assays. C-terminal truncations of eIF1A (Figure 2A) were fused to GST and expressed from a galactose-inducible promoter in a TIF11 fun12Δ strain expressing wild-type or the N-terminally truncated eIF5B378–1002. As expected, wild-type eIF5B specifically co-precipitated with all of the GST–eIF1A fusions (Figure 2C, upper panels, P lanes), as the eIF1A CTD is dispensable for interaction with full-length eIF5B in WCEs. In contrast, none of the truncated GST–eIF1A fusions, except possibly GST–eIF1A1–140 (lacking residues 141–153), co-precipitated with eIF5B378–1002 at levels above the background seen with GST alone (Figure 2C, middle panels, P lanes). Consistently, a much smaller fraction of eIF5B378–1002 co-immunopre cipitated with FL-eIF1A1–140 from WCEs (Figure 2B, lanes 10–13) compared with that produced by full-length FL-eIF1A (Figure 1A, lanes 10–12). From the data in Figure 2B and C, we conclude that the last 24 residues of eIF1A are required for a strong interaction with N-terminally truncated eIF5B in vivo, and that weak interaction is retained when only the last 13 residues of eIF1A are removed.
The fact that full-length but not N-terminally truncated eIF5B interacted strongly with eIF1A lacking C-terminal residues 108–153 could indicate that the eIF5B NTD binds to eIF1A at a site N-terminal to residue 108, and that this interaction is redundant with that occurring between the CTDs of the two proteins. Alternatively, the observed association between CTD-less eIF1A and full-length eIF5B could be indirect and result from their mutual binding to the same 40S ribosomes. To distinguish between these possibilities, we asked whether co-immuno precipitation of C-terminally truncated FL-eIF1A1–107 with full-length eIF5B was retained in a post-ribosomal supernatant (PRS). As shown in Figure 2D (lanes 1–9), full-length eIF5B co-immunoprecipitated from WCE with both FL-eIF1A and C-terminally truncated FL-eIF1A1–107, in keeping with the results shown in Figure 2B. In contrast, eIF5B co-immunoprecipitated from the PRS only with full-length FL-eIF1A (compare lanes 14 and 17). We verified that 40S subunit protein S22 was nearly undetectable in the PRS (lanes 19–24). Thus, the eIF1A CTD is necessary for strong binding to full-length eIF5B when both factors are free of ribosomes.
The eIF1A CTD is sufficient for interaction with the eIF5B CTD in vivo
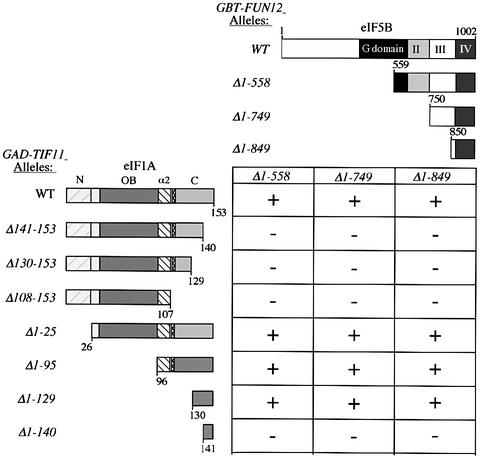
We next used yeast two-hybrid analysis to map the minimal domains sufficient for eIF1A–eIF5B interaction in vivo. Previously, we reported that the last 153 residues of eIF5B (eIF5B850–1002) showed a two-hybrid interaction with full-length eIF1A (Choi et al., 2000). Accordingly, this and two larger C-terminal fragments of eIF5B were fused to the GAL4 DNA-binding domain (GBT) and tested for two-hybrid interactions with GAL4 activation domain (GAD) fusions containing different portions of eIF1A (Figure 3). As expected, full-length GAD–eIF1A interacted with all three GBT–eIF5B fusions; however, deleting only the last 13 residues of eIF1A (eIF1A1–140) eliminated all interactions (Figure 3, rows 1 and 2). Western analysis showed that full-length GAD–eIF1A, GAD–eIF1A1–140 and GAD–eIF1A1–129 were expressed at comparable levels (data not shown). Importantly, the GAD fusion containing only the last 24 residues of eIF1A (eIF1A130–153) interacted with all three eIF5B segments, while that containing only the last 13 residues (eIF1A141–153) showed no interactions (Figure 3, last two rows). Thus, the C-terminal 24 residues of eIF1A are necessary and sufficient for strong interaction with the eIF5B CTD.
Fig. 3. The C-terminal domains of eIF1A and eIF5B are sufficient for yeast two-hybrid interaction in vivo. GAD-TIF11 alleles encoding the depicted eIF1A segments fused to the GAL4 activation domain were introduced into strain Y187 on the following hc plasmids: p3586 (WT), p3579 (Δ141–153), p3578 (Δ130–153), p3576 (Δ108–153), p3574 (Δ1-25), p3583 (Δ1–95), p3584 (Δ1–129) and p3585 (Δ1–140). GBT-FUN12 alleles encoding the depicted eIF5B segments fused to the GAL4 DNA-binding domain were introduced into strain Y190 on plasmids pC1081 (Δ1–558), pC1082 (Δ1–749) and pC1084 (Δ1–849). The Y187 and Y190 transformants were crossed, and the diploids were isolated on SC medium lacking tryptophan and leucine (SC-Trp-Leu) and tested for growth on medium lacking histidine and containing 30 mM 3-AT. Growth (+) on the latter medium indicates a two-hybrid interaction that stimulates expression of the GALUAS-HIS3 reporter present in Y190.
The C-terminus of eIF1A is critical for translation initiation and paromomycin resistance in vivo
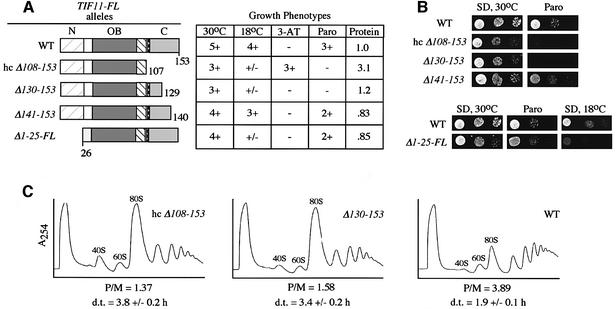
To investigate the importance of the eIF1A CTD for translation in vivo, we analyzed the phenotypes of tif11Δ yeast strains harboring T1F11-FL alleles with C-terminal truncations. Western analysis of WCEs with FLAG antibodies showed that truncated proteins lacking only 13 or 24 C-terminal residues (encoded by tif11-FL alleles Δ141–153 and Δ130–150, respectively) were expressed at nearly the same levels as wild-type FL-eIF1A, while the protein lacking 46 residues (Δ108–153) was present at only ∼15% of wild-type. When expressed from a high-copy (hc) plasmid, this last mutant protein was produced at a level ∼3-fold higher than that of wild-type FL-eIF1A from an sc plasmid (western data summarized in Figure 4A). Hence, we analyzed the phenotypes of the strain containing hc tif11-Δ108–153-FL to evaluate the functional impairment of this protein. Note that cells with wild-type TIF11-FL on either a hc or sc plasmid grew indistinguishably on all media tested.
Fig. 4. C-terminal deletions in TIF11 confer slow growth, paromomycin sensitivity and reduced rates of translation initiation in vivo. (A) The TIF11-FL alleles shown schematically were introduced into strain H2809 on sc or hc plasmids by plasmid shuffling to produce strains H2974 (sc TIF11-FL on p3499, WT), H3002 (hc tif11-Δ108–153-FL on p3604), H3001 (sc tif11-Δ130–153-FL on p3505), H3000 (sc tif11-Δ141–153-FL on p3503) and H3003 (sc tif11-Δ1–25-FL on p3501). The resulting strains were scored for growth on SD medium at 30 or 18°C, on SC-His medium containing 10 or 30 mM 3-AT and 40 mM leucine, and on SD medium containing 0 or 0.5 mM paromomycin (Paro), by spotting 10-fold serial dilutions of a saturated culture and incubating for 2–5 days. Growth was scored qualitatively based on density and colony size. The last column summarizes the western analysis of WCEs from cells grown in SC-Leu medium at 30°C using FLAG antibodies to detect FL-eIF1A proteins and antibodies against eIF2Bγ/GCD6 (analyzed as an internal control). Signals obtained with FLAG antibodies were normalized for the GCD6 signals and expressed relative to the wild-type value for TIF11-FL. (B) The results of the growth tests described in (A) on SD and SD containing 0.5 mM paromomycin. (C) WCEs were prepared from strains described in (A) after growing in yeast extract–peptone–dextrose (YPD) medium at 30°C to OD600 = 2.0 and resolved by sedimentation on 4.5–45% sucrose gradients. Fractions were collected while scanning continuously at A254. P/M is the ratio of A254 units in the combined polysome fractions to the A254 units in the 80S peak. d.t., cell doubling time in hours measured in SC medium at 30°C.
As shown in Figure 4B (panels labeled ‘SD’, for synthetic dextrose medium) and summarized in Figure 4A, all three deletion alleles conferred reduced growth rates at 30°C, but the two larger deletions (Δ130–153 and hc Δ108–153) had more severe effects than did Δ141–153. The two larger deletions also conferred strong growth defects at 18°C (Figure 4A). The immunoprecipitation results in Figure 2B showed that Δ141–153 greatly reduced binding of FL-eIF1A to eIF5B378–1002, whereas the larger deletion, Δ130–153, completely abolished the interaction. Thus, the growth phenotypes of the mutant alleles are consistent with the idea that interaction between the CTDs of eIF1A and eIF5B is required for optimum translation. To provide more direct evidence that the eIF1A CTD promotes translation initiation in vivo, we compared the polysome profiles of the wild-type, sc Δ130–153 and hc Δ108–153 TIF11-FL alleles. As shown in Figure 4C, both deletions led to a substantial decrease in polysome content and a commensurate increase in 80S monosomes. The reduction in polysome:monosome ratios for these mutants is characteristic of a reduced rate of translation initiation.
We also tested the TIF11-FL mutants for sensitivity to paromomycin. This drug binds to the A-site of prokaryotic ribosomes and decreases translational fidelity by allowing codon–anticodon mismatches at a higher frequency than normal (Schroeder et al., 2000). Bacterial IF1 binds to the A-site in a region overlapping the paromomycin-binding site (Carter et al., 2001). Given the structural similarities between eIF1A and IF1, we reasoned that eIF1A may compete with paromomycin for the A-site, and mutations that perturb A-site occupancy by eIF1A would increase sensitivity to paromomycin. Interestingly, the Δ130–153 and hc Δ108–153 tif11-FL alleles conferred strong paromomycin sensitivity (ParS phenotype) (Figure 4B; summarized in A). Thus, the unstructured C-terminus of eIF1A may contribute to proper binding of eIF1A to the A-site.
We showed above that the eIF1A CTD is required for association with eIF5B in WCEs only when the eIF5B NTD was missing. Thus, we wished to determine whether deleting the eIF1A CTD would have a more severe phenotype in cells expressing N-terminally truncated eIF5B378–1002 versus full-length eIF5B. When we compared the phenotypes of strains containing tif11-Δ130–153-FL and either wild-type FUN12 or FUN12-Δ1–377, we found no differences in the Slg– phenotypes of these two strains; however, the tif11-Δ130–153-FL FUN12-Δ1–377 double mutant was more sensitive to paromomycin (data not shown). The latter increase in paromomycin sensitivity could indicate that the eIF5B NTD contributes directly or indirectly to proper positioning of eIF1A in the A-site. The fact that truncating the N-terminus of eIF5B did not exacerbate the Slg– phenotype of tif11-Δ130–153-FL suggests that strong interaction between the CTDs of eIF1A and eIF5B is critical for a step in translation initiation, and that additionally eliminating the secondary interaction on the ribosome involving the eIF5B NTD does not further impair this step in the pathway.
If the last 24 residues of eIF1A are required only to mediate interaction with eIF5B, then deleting this segment should not affect the growth rate in a fun12Δ strain. At odds with this prediction, tif11-Δ130–153-FL fun12Δ cells grew more slowly than TIF11-FL fun12Δ cells (data not shown). Thus, the eIF1A CTD seems to perform a second function independently of eIF5B. Consistently, the results in the next section implicate the eIF1A CTD in TC binding to the 40S subunit.
Evidence that the eIF1A CTD promotes ternary complex binding in vivo
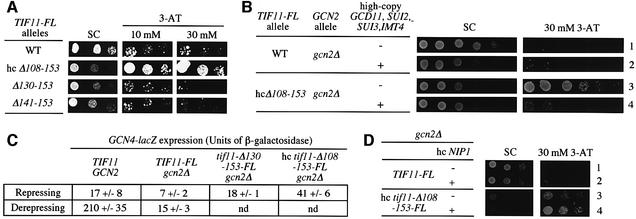
As described in the Introduction, mutations in eIF1A that impair TC binding should allow 40S subunits to scan past uORF4 and reinitiate at the GCN4 start codon. This would derepress GCN4 translation (Gcd– phenotype) even in gcn2Δ cells where TC levels cannot be lowered by eIF2 phosphorylation. To determine whether the TIF11 deletion alleles confer a Gcd– phenotype, we tested them for ability to suppress sensitivity of gcn2Δ cells to 3-aminotriazole (3-AT), an inhibitor of the histidine biosynthetic gene HIS3. Derepression of GCN4 is required for high-level HIS3 expression and resistance to 3-AT (3-ATr phenotype). Only the hc tif11-Δ108–153-FL allele had a Gcd– phenotype, conferring growth on 30 mM 3-AT plates, despite its Slg– phenotype on synthetic complete (SC) medium (Figure 5A and summarized in Figure 4A). If this Gcd– phenotype results from inefficient TC binding to 40S ribosomes, it should be suppressed by overproducing the TC. To test this prediction, we introduced hc plasmid p3000, encoding all three subunits of eIF2 and tRNAiMet, into the mutant strain. Previously, we showed that simultaneously overexpressing these factors increased the level of TC in the cell (Dever et al., 1995). Interestingly, p3000, but not the empty vector, suppressed the 3-ATR phenotype of the hc tif11-Δ108–153-FL mutant (Figure 5B, rows 3 and 4).
Fig. 5. A C-terminal deletion in TIF11 confers derepression of GCN4 expression that is suppressed by overexpressing the ternary complex. (A) The gcn2Δ strains containing the indicated TIF11-FL alleles were tested for 3-AT resistance and the results are summarized in Figure 4A. (B) The TIF11-FL and hc tif11-Δ108–153-FL strains analyzed in (A) were transformed with plasmid p3000 encoding the subunits of eIF2 and initiator tRNAMet (rows 2 and 4) or with empty vector YEp24 (rows 1 and 3) and tested for 3-AT resistance. (C) The TIF11-FL, tif11-Δ130–153-FL and hc tif11-Δ108–153-FL strains and the isogenic TIF11 GCN2 strain H1642 bearing the empty vector YCplac111, all containing a GCN4-lacZ fusion integrated at trp1, were grown in SC-leu (repressing conditions) or in SC-Leu-Ile-Val containing 1.37 µM sulfometuron methyl (derepressing conditions). WCEs were prepared from three or more independent cultures and assayed for β-galactosidase activity, defined as nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per min per mg of protein. Mean values and standard errors are provided. (D) The TIF11-FL and hc tif11-Δ108–153-FL strains were transformed with hc plasmid YEpNIP1-His-U, bearing NIP1-His, or hc empty vector YEp195, and tested for 3-AT resistance.
We quantified the Gcd– phenotype of hc tif11-Δ108–153-FL by assaying a GCN4-lacZ reporter. As shown in Figure 5C, this mutant had ∼6-fold higher GCN4-lacZ expression compared with the basal level observed in the TIF11-FL gcn2Δ cells. The tif11-Δ130–153-FL mutant also showed somewhat higher GCN4-lacZ expression, but significantly less than that seen in the hc Δ108–153 mutant, in keeping with the 3-ATS phenotype of the former (Figure 5A). Given the greater GCN4-lacZ expression seen in the GCN2 TIF11-FL strain treated with 3-AT compared with the hc tif11-Δ108–153-FL gcn2Δ strain (Figure 5C), it appears that the Δ108–153 mutation does not impair TC binding to the same extent that occurs when eIF2α is phosphorylated by GCN2 in wild-type cells.
Biochemical studies indicate that eIF3 also contributes to TC binding to 40S ribosomes in vitro (Hinnebusch, 2000). Interestingly, we found that overexpressing the eIF3c subunit (encoded by NIP1) exacerbated the Slg– and Gcd– phenotypes of the hc tif11-Δ108–153-FL gcn2Δ strain (Figure 5D). NIP1 mediates an interaction between eIF3 and eIFs 1, 2 and 5 in a multifactor complex (MFC) (Asano et al., 2000), and there is biochemical evidence that incorporation of TC into the MFC enhances its binding to 40S subunits (Asano et al., 2001). Overexpressing NIP1 may titrate its interacting partners in the MFC into partial complexes, reducing the concentration of intact MFC. This, in turn, may exacerbate the defect in TC binding associated with deleting the eIF1A CTD.
eIF1A interacts with initiation factors 2, 3 and 5 through its N-terminus
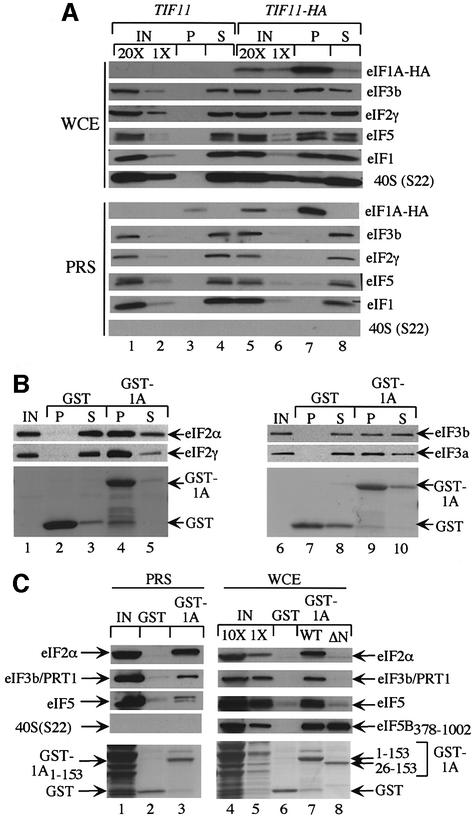
Although eIF1A is not a stable constituent of the MFC, its role in promoting TC binding to 40S ribosomes could involve physical contact with eIF2 or other MFC components. To address this possibility, we asked whether eIFs 1, 2, 3 and 5 co-immunoprecipitated with hemagglutinin (HA) epitope-tagged eIF1A expressed in yeast. The sc TIF11-HA and TIF11 alleles equally complemented the growth defect of a tif11Δ mutant (data not shown). As shown in Figure 6A (upper panel), eIFs 1, 2, 3 and 5, and 40S subunits specifically co-immunoprecipitated with HA-eIF1A from the WCEs. In contrast, no interactions of HA-eIF1A with any of these proteins were observed in the PRS (Figure 6A, lower panel), suggesting that they occur in vivo only in the context of 43S or 48S initiation complexes. As shown above, eIF5B co-immunoprecipitated with FL-eIF1A from both the WCE and PRS (Figure 2D), consistent with binding of eIF1A to eIF5B independently of the ribosome.
Fig. 6. Evidence that the eIF1A NTD mediates interactions with eIFs 2 and 3 on 40S ribosomes (A) tif11Δ::hisG strains harboring TIF11 (lanes 1–4) or TIF11-HA (lanes 5–8) on sc plasmids p3390 and p3404, respectively, were produced by plasmid shuffling from strain H2809. WCEs or post-ribosomal supernatants (PRSs) from each strain were immunoprecipitated with anti-HA antibodies, resolved by SDS–PAGE and subjected to immunoblot analysis using antibodies against the proteins indicated on the right of each panel. Lanes 1 and 5 and lanes 2 and 6 contain 20 and 1%, respectively, of the WCE or PRS employed. (B) The purified full-length GST–eIF1A fusion described in Figure 1B (GST–1A) was used in pull-down assays with 2 µg of either purified eIF2 (lanes 1–5) or purified eIF3 (lanes 6–10). Precipitated proteins were resolved by SDS–PAGE and subjected to Ponceau S staining (bottom panels) or immunoblot analysis using antibodies against the indicated factors. IN, P and S lanes contained 10, 100 and 10% of the input, pellet and supernatant fractions, respectively. (C) The full-length GST–eIF1A fusion described in (A) (GST-1A1–153) and the N-terminally truncated fusion lacking residues 1–25 (GST-1A26–153), encoded by p3465, were purified from bacteria and used in pull-down assays as described in Figure 1B with a PRS (lanes 1–3) or WCE (lanes 4–8) from a J111 transformant containing pC1043. The precipitated proteins were resolved by SDS–PAGE and subjected to Ponceau S staining (bottom panels) and to immunoblot analysis using antibodies against the indicated factors. Lane 1 contains 10% of the PRS used in the pull-down reactions in lanes 2 and 3. Lanes 4 and 5 contain 10 and 1% of the WCE used in the pull-down reactions in lanes 6–8.
We next asked whether eIF1A at high concentrations can bind to eIF2 or eIF3 independently of ribosomes. GST–eIF1A fusions expressed in E.coli were incubated with highly purified eIF2 and eIF3 prepared from yeast. As shown in Figure 6B, purified eIF2 (left panel) and eIF3 (right panel) specifically precipitated with GST–eIF1A but not with GST alone. We also detected binding of eIFs 2, 3 and 5 to GST–eIF1A when the latter was incubated with WCE or PRS from a wild-type yeast strain (Figure 6C). Thus, the non-ribosomal pool of these factors can interact with exogenous GST–eIF1A when the latter is added to cell extracts in relatively large amounts. All of the binding to eIF3 and eIF5, and much of the binding to eIF2 observed in these last assays was insensitive to micrococcal nuclease treatment (data not shown); hence, at least a large portion of the interactions between GST–eIF1A and these factors was not bridged by RNA. Deletion of residues 1–25 from GST–eIF1A abolished interaction with eIFs 2, 3 and 5, but had no effect on binding to eIF5B378–1002 (Figure 6C). In contrast, deleting residues 108–153 from the C-terminus of GST–eIF1A had no effect on the interaction with eIFs 2, 3 and 5 (data not shown), but abolished interaction with eIF5B378–1002 (Figure 1C). Thus, the NTD and CTD of eIF1A have distinct functions in binding to components of the MFC and eIF5B, respectively.
To address the importance of the eIF1A NTD in vivo, we characterized the growth phenotypes of a strain harboring tif11-Δ1–25-FL, lacking the N-terminal 25 codons. This mutant exhibited a slight Slg– phenotype but no increased sensitivity to paromomycin or resistance to 3-AT. However, it showed a strong growth defect at 18°C (Figure 4A and B). This cold-sensitive phenotype suggests that interactions between the eIF1A NTD and eIFs 2 and 3 on the 40S ribosome are required for efficient translation at low temperatures. Because a Gcd– phenotype was not observed for the tif11-Δ1–25-FL mutant, the interaction between eIF2 and the eIF1A NTD probably contributes to a step in translation following recruitment of TC.
Discussion
eIF1A and eIF5B interact primarily through their C-terminal domains
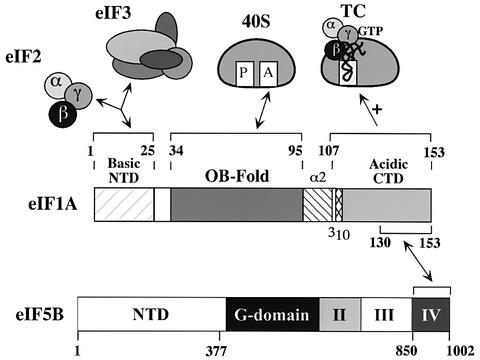
We showed previously that eIF1A and an N-terminally truncated form of eIF5B interact in vitro dependent on the CTD of eIF5B (Choi et al., 2000). Here we showed that the unstructured CTD of eIF1A is necessary and sufficient for interaction with the eIF5B CTD (Figures 1–3, summarized in Figure 7). Deleting the C-terminal 44 residues in N-terminally truncated eIF5B378–1002 (Choi et al., 2000) or the last 24 residues of full-length eIF1A (Figures 1 and 2) destroyed complex formation between these two proteins. Moreover, only the last 24 residues of eIF1A were sufficient for a two-hybrid interaction with the C-terminal 153 residues of eIF5B (Figure 3). This portion of eIF5B corresponds to domain IV in the crystal structure of archaeal eIF5B, comprising the base of the chalice-like molecule (Roll-Mecak et al., 2000). The eIF5B-binding domain in eIF1A represents ∼60% of the acidic C-terminal segment of eIF1A (Battiste et al., 2000).
Fig. 7. Summary of functional domains in eIF1A. The interactions ascribed to each domain in eIF1A are depicted schematically with double-headed arrows. The C-terminal domain of eIF1A also functions to promote ternary complex (TC) binding to the 40S ribosome (see Discussion for details).
The eIF1A CTD is required for association with N-terminally truncated but not full-length eIF5B in WCEs (Figures 1B, and 2B and C); however, the eIF1A CTD is needed for interaction with full-length eIF5B in a PRS. Based on the latter, we conclude that association between the C-termini of these proteins is responsible for their strong interaction independently of ribosomes. The eIF5B NTD could make an additional contact with the NTD or OB-fold domain of eIF1A, but this interaction would be too weak to sustain complex formation free of the ribosome. Another possibility is that the association between full-length eIF5B and CTD-less eIF1A observed in WCEs occurred through binding of both proteins to the same 40S ribosomes without direct contact between them. Tacit assumptions of this latter view are that the eIF5B NTD is required for ribosome binding in the absence of CTD interactions between eIF5B and eIF1A, and that deletion of the eIF5B CTD eliminates its interaction with both eIF1A and the ribosome.
In vivo consequences of disrupting the eIF1A–eIF5B interaction
We reported previously that deleting the eIF5B CTD (fun12-Δ916–1002) almost completely inactivated eIF5B function in vivo, conferring a growth defect only slightly less severe than that of a complete deletion of FUN12. In contrast, deleting only the eIF5B NTD (fun12-Δ1–377) had no effect on cell growth (Choi et al., 2000). These phenotypes are consistent with the idea that the interaction between eIF1A and the eIF5B CTD is required for wild-type translation in vivo, whereas that involving the eIF5B NTD, whether direct or indirect, is dispensable. We showed here that deleting the last 24 residues of eIF1A (tif11-Δ130–153-FL) decreased the rate of translation initiation in vivo, reducing the polysome content and cell growth rate (Figure 4). Because this mutation destroys the key eIF5B-binding domain in eIF1A, we propose that eIF1A–eIF5B association through their C-termini is required for an important step in the initiation pathway.
Previously, we hypothesized that eIF1A–eIF5B interaction facilitates release of eIF1A from the A-site. If so, then disrupting the CTD interactions between eIF5B and eIF1A by the Δ130–153 mutation in TIF11 would prolong eIF1A binding in the A-site, interfering with subsequent steps in translation. This hypothesis can explain our finding that overexpressing wild-type eIF1A exacerbated the growth defect of the fun12-Δ916–1002 C-terminal mutant (Choi et al., 2000). According to this model, eIF1A dissociates from the A-site very slowly in a strain expressing C-terminally truncated eIF5B, which cannot interact productively with eIF1A. When eIF1A is overexpressed, the inefficient, eIF5B-independent dissociation of eIF1A occurring in the fun12 mutant would be reduced further by mass action, leading to an intolerably high retention time in the A-site. Eliminating interaction between the CTDs of eIF1A and eIF5B could likewise impede dissociation of eIF5B from the ribosome after subunit joining, analogous to the stimulatory effect of IF1 on IF2 release in prokaryotes (Benne et al., 1973).
Considering that IF1–IF2 association mutually stabil izes the binding of these factors to the 30S ribosome (Gualerzi and Pon, 1990; Palacios Moreno et al., 1999), it is possible that interaction between the C-termini of eIF1A and eIF5B enhances their association with the 40S ribosome early in the pathway, as well as facilitating their release at the end of the process. The ParS phenotype of tif11-Δ130–153-FL, and exacerbation of this phenotype when the N-terminus of eIF5B was deleted, is consistent with this idea. In the absence of its interaction with eIF5B, eIF1A would compete less effectively with paromomycin for the A-site. It would not be surprising if different phenotypes of TIF11 and fun12 mutations would result from defective eIF1A binding to the A-site (paromomycin sensitivity) or impaired release from the ribosome (slow growth), if the two factors are interdependent for both reactions.
Evidence that the C-terminal domain of eIF1A promotes TC binding
The Δ108–153 deletion in TIF11, which removes all of the unstructured CTD and the predicted 310 helix of eIF1A, produced a Gcd– phenotype as well as the Slg– and ParS phenotypes observed for the smaller deletion (Δ130–153) discussed above. The fact that the Gcd– phenotype was suppressed by overproducing TC suggests that it reflects diminished TC binding to 40S subunits scanning the GCN4 mRNA leader after translating uORF1. The delayed rebinding of TC to these 40S subunits would allow a fraction of the latter to bypass uORFs 2–4 and reinitiate at GCN4 instead (Hinnebusch, 1996). This interpretation is consistent with biochemical evidence indicating that mammalian eIF1A facilitates TC binding to 40S subunits in vitro (for a review see Hinnebusch, 2000). Our results provide the first evidence that eIF1A is important for efficient TC binding to 40S ribosomes in vivo. They additionally imply that eIF1A is involved in the specialized reinitiation process occurring on GCN4 mRNA. The fact that overexpressing NIP1 exacerbated the Gcd– phenotype of hc tif11-Δ130–153-FL is consistent with an additive effect of eIF1A and eIF3 in promoting TC binding during reinitiation on GCN4 mRNA.
A recent study using purified eIFs 1, 1A, TC and ribosomes from yeast confirmed that yeast eIF1A is critically required for the formation of 48S complexes in vitro (Algire et al., 2002). In accordance with our in vivo findings, recombinant mutant eIF1A proteins with the Δ130–153 or Δ108–153 truncations were defective for 48S assembly (D.Maag, D.S.Olsen, A.G.Hinnebusch and J.R.Lorsch, unpublished observations). In fact, both mutants showed only a few percent of wild-type activity in this assay. Given the very weak Gcd– phenotype of the Δ130–153 mutation, and the moderate Gcd– phenotype of the Δ108–153 allele, the strong defects in TC binding conferred by these mutations in the reconstituted system must be ameliorated in living cells, perhaps by the stimulatory effects of eIF3 or eIF5 on TC binding.
Evidence that eIF1A interacts with initiation factors 2 and 3 via the N-terminus
We identified physical interactions between eIF1A and other initiation factors besides eIF5B. Bacterially expressed GST–eIF1A interacted with eIFs 2, 3 and 5 in the PRS, and it also bound to purified eIFs 2 and 3 (Figure 6B and C). By co-immunoprecipitation experiments, we found that endogenously expressed eIF1A-HA was associated with eIFs 1, 2, 3 and 5 in WCEs but not in the PRS (Figure 6A). Hence, we propose that eIF1A is an integral component of initiation complexes and can interact directly with eIF2 and eIF3 only when all of the factors are bound simultaneously to the same 40S subunit. Presumably, the interactions we observed between eIF1A and eIFs 2 and 3 free of the ribosomes were enhanced by the high concentrations of recombinant GST–eIF1A used in the binding reactions.
The NTD of eIF1A was required for its binding to eIF2 and eIF3 in vitro (Figure 6C). This region of eIF1A is extremely basic in character and was not resolved in the solution structure of human eIF1A (Battiste et al., 2000). Deletion of the N-terminal 25 residues of eIF1A had only a moderate effect on growth, and did not confer ParS or Gcd– phenotypes at 30°C. A more severe growth defect was observed for the tif11-Δ1–25-FL mutant at 18°C (Figure 4A and B), however, suggesting that interactions of the eIF1A NTD with eIF2 and eIF3 are critically required only at low growth temperatures. Kainuma and Hershey (2001) reported that deleting the N-terminal 31 residues of eIF1A had a more severe effect on growth at 30°C than that observed here for tif11-Δ1–25-FL. This may be attributed to the fact that residues 25–32 comprise one of two extended strands that, together with two C-terminal α-helices, comprise the additional structured domain that packs against the OB-fold in human eIF1A (Battiste et al., 2000).
As shown in Figure 7, we propose that eIF1A can be divided into several functional domains. Based on its strong similarity to bacterial IF1 (Sette et al., 1997; Battiste et al., 2000), the OB-fold in eIF1A probably mediates binding to the A-site of 40S subunits. Indeed, NMR analysis has identified residues in the OB-fold and α-helical domain of mammalian eIF1A that may contact rRNA (Battiste et al., 2000). Roughly the last half of the unstructured C-terminus of eIF1A is responsible for binding to eIF5B, and we hypothesize that this interaction regulates ribosome binding and release of eIF1A. Considering the weak Gcd– phenotype of the tif11-Δ130–153-FL mutant (Figure 5C), it appears that the extreme C-terminus of eIF1A also contributes to TC binding to the 40S ribosome. A segment comprising the rest of the unstructured C-terminus and the predicted 310 helix in eIF1A clearly has a role in TC binding, as deleting the entire region C-terminal to residue 107 derepressed GCN4 translation. As deleting the CTD did not reduce binding of GST–eIF1A to eIF2 in WCEs (data not shown), we have no evidence that the eIF1A CTD interacts directly with eIF2. Perhaps it promotes TC binding indirectly by producing a conformational change in the ribosome that increases P-site affinity for TC. The unstructured NTD of eIF1A (residues 1–25) mediates direct interactions with eIFs 2 and 3 on the 40S ribosome. Because the tif11-Δ1–25-FL mutant did not have a Gcd– phenotype, the interaction between eIF2 and the eIF1A NTD probably contributes to a step in the pathway following TC recruitment.
Materials and methods
Yeast strains and plasmids
The plasmids and yeast strains employed in this work are listed in Tables I and II, respectively. Details of their construction are available on request.
Table I. Plasmids used in this study.
| Plasmid | Description | Source |
|---|---|---|
| p3498 | lc LEU2 TIF11 FUN12 in pRS315 backbone | This study |
| p3387 | int tif11Δ::hisG::URA3 in pBSII(KS-) backbone | This study |
| p3570 | sc URA3 TIF11 FUN12 in YCplac33 backbone | This study |
| p3412 | sc LEU2 TIF11 in YCplac111 backbone | This study |
| p3499 | sc LEU2 TIF11-FL in YCplac111 backbone | This study |
| pC479 | lc URA3 FUN12 in pRS306 backbone | Choi et al. (1998) |
| p3572 | lc URA3 FUN12-Δ1–377 in pRS306 backbone | This study |
| p3571 | hc URA3 fun12-Δ916–1002 in pRS426 backbone | This study |
| pGEX-4T-1 | Bacterial expression vector with tac promoter | Amersham Pharmacia-Biotech |
| pGEX-4T-2 | Bacterial expression vector with tac promoter | Amersham Pharmacia-Biotech |
| p3415 | GST-TIF11 under the tac promoter in pGEX-4T-1 backbone | Choi et al. (2000) |
| p3466 | GST-TIF11-Δ108–153 under the tac promoter in pGEX-4T-1 backbone | This study |
| p3465 | GST-TIF11-Δ1–25 under the tac promoter in pGEX-4T-1 backbone | This study |
| pC1005 | FUN12-FL in pRS316 backbone | Choi et al. (2000) |
| pC1043 | FUN12-Δ1–377-FL in pRS316 backbone | Choi et al. (2000) |
| pC484 | GST-FUN12-Δ1–395 under the tac promoter in pGEX-4T-2 backbone | Choi et al. (2000) |
| p3503 | sc LEU2 tif11-Δ141–153-FL in YCplac111 backbone | This study |
| p3505 | sc LEU2 tif11-Δ130–153-FL in YCplac111 backbone | This study |
| pEG(KT) | hc GST under the GAL promoter | Mitchell et al. (1993) |
| p3559 | hc URA3 GST-TIF11 under the GAL promoter in pEG(KT) backbone | This study |
| p3563 | hc URA3 GST-TIF11-Δ119–153 under the GAL promoter in pEG(KT) backbone | This study |
| p3564 | hc URA3 GST-TIF11-Δ130–153 under the GAL promoter in pEG(KT) backbone | This study |
| p3566 | hc URA3 GST-TIF11-Δ108–153 under the GAL promoter in pEG(KT) backbone | This study |
| p3565 | hc URA3 GST-TIF11-Δ141–153 under the GAL promoter in pEG(KT) backbone | This study |
| pC1000 | lc LEU2 FUN12-Δ1–377-FL in pRS315 backbone | This study |
| pGBT9 | hc TRP1 GAL4 DNA-BD vector with ADH1 promoter | Clontech |
| pGAD424 | hc LEU2 GAL4 AD vector with ADH1 promoter | Clontech |
| pC1081 | hc TRP1 FUN12-Δ1–558 under the ADH1 promoter, in pGBT9 backbone | Choi et al. (2000) |
| pC1082 | hc TRP1 FUN12-Δ1–749 under the ADH1 promoter, in pGBT9 backbone | Choi et al. (2000) |
| pC1084 | hc TRP1 FUN12-Δ1–849 under the ADH1 promoter, in pGBT9 backbone | Choi et al. (2000) |
| p3586 | hc LEU2 TIF11 under the ADH1 promoter, in pGAD424 backbone | This study |
| p3579 | hc LEU2 TIF11-Δ141–153 under the ADH1 promoter, in pGAD424 backbone | This study |
| p3578 | hc LEU2 TIF11-Δ130–153 under the ADH1 promoter, in pGAD424 backbone | This study |
| p3576 | hc LEU2 TIF11-Δ108–153 under the ADH1 promoter, in pGAD424 backbone | This study |
| p3574 | hc LEU2 TIF11-Δ1–25 under the ADH1 promoter, in pGAD424 backbone | This study |
| p3583 | hc LEU2 TIF11-Δ1–95 under the ADH1 promoter, in pGAD424 backbone | This study |
| p3584 | hc LEU2 TIF11-Δ1–129 under the ADH1 promoter, in pGAD424 backbone | This study |
| p3585 | hc LEU2 TIF11-Δ1–140 under the ADH1 promoter, in pGAD424 backbone | This study |
| p3604 | hc LEU2 tif11-Δ108–153-FL in YEplac181 backbone | This study |
| p3505 | sc LEU2 tif11-Δ130–153-FL in YCplac111 backbone | This study |
| p3503 | sc LEU2 tif11-Δ141–153-FL in YCplac111 backbone | This study |
| p3501 | sc LEU2 tif11-Δ1–25-FL in YCplac111 backbone | This study |
| YEpNIP1-His-U | hc URA3 NIP1-His in YEp195 backbone | L.Valášek |
| YEp195 | hc URA3 vector | |
| p3000 | hc URA3 SUI2 SUI3 GCD11 IMT4 in YEp24 backbone | Asano et al. (1999) |
| YCplac111 | sc LEU2 | Gietz and Sugino (1988) |
| p3390 | sc LEU2 TIF11 in YCplac111 backbone | Choi et al. (2000) |
| p3404 | sc LEU2 TIF11–3HA in YCplac111 backbone | This study |
Table II. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| J111 | MATa ura3-52 leu2-3,-112 fun12Δ::hisG | Choi et al. (2000) |
| H2971 | MATα ura3-52 leu2-3,-112 fun12Δ::hisG tif11Δ::hisG <p3498: TIF11 FUN12 LEU2> | This study |
| H1895 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ <p1108: GCN4-lacZ at TRP1> | Kawagishi-Kobayashi et al. (1997) |
| Y187 | MATα gal4Δ gal80Δ his3 trp1-901 ade2-101 ura3-52 leu2-3,112 met– URA3::GAL-lacZ | Harper et al. (1993) |
| Y190 | MATa gal4Δ gal80Δ his3-Δ200trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL-lacZ LYS2::GAL(UAS)-HIS3 cyhr | Harper et al. (1993) |
| H2809 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ tif11Δ::hisG <p1108: GCN4-lacZ at TRP1> <p3392: TIF11, URA3> | Choi et al. (2000) |
| H2974 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ tif11Δ::hisG <p1108: GCN4-lacZ at TRP1> <p3499: TIF11-FL, LEU2> | This study |
| H3000 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ tif11Δ::hisG <p1108: GCN4-lacZ at TRP1> <p3499: TIF11-FL-Δ141–153, LEU2> | This study |
| H3001 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ tif11Δ::hisG <p1108: GCN4-lacZ at TRP1> <p3499: TIF11-FL-Δ130–153, LEU2> | This study |
| H3002 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ tif11Δ::hisG <p1108: GCN4-lacZ at TRP1> <p3499: TIF11-FL-Δ108–153, LEU2> | This study |
| H3003 | MATa ura3-52 leu2-3 leu2-112 trp1Δ63 gcn2Δ tif11Δ::hisG <p1108: GCN4-lacZ at TRP1> <p3499: TIF11-FL-Δ1–25, LEU2> | This study |
| H1642 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 <p1108: GCN4-lacZ at TRP1> | Dever et al. (1992) |
Genetic methods
Yeast strains were constructed using standard techniques of yeast transformation (Ito et al., 1983), gene replacement (Rothstein, 1983) and plasmid shuffling (Boeke et al., 1987). Yeast two-hybrid analysis was conducted as described previously (Choi et al., 2000).
Biochemical methods
Preparation of yeast and bacterial WCEs and yeast PRSs, GST pull-down assays using bacterially expressed GST fusion proteins, and immunoprecipitation of HA-tagged eIF1A proteins with anti-HA-protein A–Sepharose, were conducted essentially as described previously (Choi et al., 2000). Immunoprecipitation of FLAG-tagged eIF1A proteins was carried out similarly, with the following changes. M2 anti-FLAG agarose resin (Sigma) was prepared according to the vendor’s directions by washing three times in 10 vols of glycine-HCl and equilibrating with lysis buffer. A 1 mg aliquot of yeast WCE (or an equivalent volume of PRS) was added to 50 µl of a 50% slurry of M2 resin. The procedures for conducting pull-down assays with GST–eIF1A fusions expressed in yeast are provided in the Supplementary data available at The EMBO Journal Online.
The eIF5B396–1002 used in Figure 1D was purified after cleavage of GST–eIF5B396–1002 expressed in E.coli from pC484, as described previously (Choi et al., 2000). The eIF2 (Krishnamoorthy et al., 2001) and eIF3 (Phan et al., 1998) employed in Figure 6B were purified according to published protocols. Polysome analysis was carried out as described previously (Foiani et al., 1991), as was analysis of GCN4-lacZ expression (Moehle and Hinnebusch, 1991).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Leo Valášek for YEpNIP1-His-U, Tom Dever for many helpful suggestions, comments on the manuscript and gifts of antibodies, Tom Donahue and Ernie Hannig for antibodies, Jon Lorsch and Dave Maag for useful comments on the manuscript and for communicating unpublished results, and Jane Lin for help in preparing the manuscript.
References
- Algire M.A. et al. (2002) Development and characterization of a reconstituted yeast translation initiation system. RNA, 8, 382–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Krishnamoorthy,T., Phan,L., Pavitt,G.D. and Hinnebusch,A.G. (1999) Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP–GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J., 18, 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Clayton,J., Shalev,A. and Hinnebusch,A.G. (2000) A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5 and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev., 14, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Shalev,A., Phan,L., Nielsen,K., Clayton,J., Valášek,L., Donahue,T.F. and Hinnebusch,A.G. (2001) Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J., 20, 2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste J.B., Pestova,T.V., Hellen,C.U.T. and Wagner,G. (2000) The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell, 5, 109–119. [DOI] [PubMed] [Google Scholar]
- Benne R., Naaktgeboren,N., Gubbens,J. and Voorma,H.O. (1973) Recycling of initiation factors IF-1, IF-2 and IF-3. Eur. J. Biochem., 32, 372–380. [DOI] [PubMed] [Google Scholar]
- Boeke J.D., Trueheart,J., Natsoulis,G. and Fink,G.R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. [DOI] [PubMed] [Google Scholar]
- Carter A.P., Clemons,W.M.,Jr, Brodersen,D.E., Morgan-Warren,R.J., Hartsch,T., Wimberly,B.T. and Ramakrishnan,V. (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science, 291, 498–501. [DOI] [PubMed] [Google Scholar]
- Choi S.K., Lee,J.H., Zoll,W.L., Merrick,W.C. and Dever,T.E. (1998) Promotion of Met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science, 280, 1757–1760. [DOI] [PubMed] [Google Scholar]
- Choi S.K., Olsen,D.S., Roll-Mecak,A., Martung,A., Remo,K.L., Burley,S.K., Hinnebusch,A.G. and Dever,T.E. (2000) Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol., 20, 7183–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E., Feng,L., Wek,R.C., Cigan,A.M., Donahue,T.D. and Hinnebusch,A.G. (1992) Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell, 68, 585–596. [DOI] [PubMed] [Google Scholar]
- Dever T.E., Yang,W., Åström,S., Byström,A.S. and Hinnebusch,A.G. (1995) Modulation of tRNAiMet, eIF-2 and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2·GTP·Met-tRNAiMet ternary complexes. Mol. Cell. Biol., 15, 6351–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Cigan,A.M., Paddon,C.J., Harashima,S. and Hinnebusch,A.G. (1991) GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 3203–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Adami,G.R., Wei,N., Keyomarsi,K. and Elledge,S.J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B. and Merrick,W.C. (2000) Pathway and mechanism of initiation of protein synthesis. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–88.
- Hinnebusch A.G. (1996) Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 199–244.
- Hinnebusch A.G. (2000) Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp. 185–243.
- Ito H., Fukada,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainuma M. and Hershey,J.W.B. (2001) Depletion and deletion analyses of eucaryotic translation initiation factor 1A of Saccharomyces cerevisiae. Biochimie, 83, 505–514. [DOI] [PubMed] [Google Scholar]
- Kawagishi-Kobayashi M., Silverman,J.B., Ung,T.L. and Dever,T.E. (1997) Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2α. Mol. Cell. Biol., 17, 4146–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy T., Pavitt,G.D., Zhang,F., Dever,T.E. and Hinnebusch,A.G. (2001) Tight binding of the phosphorylated α subunit of initiation factor 2 (elF2α) to the regulatory subunits of guanine nucleotide exchange factor elF2B is required for inhibition of translation initiation. Mol. Cell, 21, 5018–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A., Pon,C.L. and Gualerzi,C.O. (1996) Late events in translation initiation. Adjustment of fMet-tRNA in the ribosomal P-site. J. Mol. Biol., 256, 667–675. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Choi,S.K., Roll-Mecak,A., Burley,S.K. and Dever,T.E. (1999) Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc. Natl Acad. Sci. USA, 96, 4342–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchin S., Putzer,H., Hershey,J.W., Cenatiempo,Y., Grunberg-Manago,M. and Laalami,S. (1999) In vitro study of two dominant inhibitory GTPase mutants of Escherichia coli translation initiation factor IF2. Direct evidence that GTP hydrolysis is necessary for factor recycling. J. Biol. Chem., 274, 6074–6079. [DOI] [PubMed] [Google Scholar]
- Mitchell D.A., Marshall,T.K. and Deschenes,R.J. (1993) Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast, 9, 715–722. [DOI] [PubMed] [Google Scholar]
- Moazed D., Samaha,R.R., Gualerzi,C. and Noller,H.F. (1995) Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol., 248, 207–210. [DOI] [PubMed] [Google Scholar]
- Moehle C.M. and Hinnebusch,A.G. (1991) Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PalaciosMoreno J.M., Drskjotersen,L., Kristensen,J.E., Mortensen,K.K. and Sperling-Petersen,H.U. (1999) Characterization of the domains of E.coli initiation factor IF2 responsible for recognition of the ribosome. FEBS Lett., 455, 130–134. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin,I.B., Lee,J.H., Choi,S.K., Dever,T.E. and Hellen,C.U.T. (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature, 403, 332–335. [DOI] [PubMed] [Google Scholar]
- Phan L., Zhang,X., Asano,K., Anderson,J., Vornlocher,H.P., Greenberg,J.R., Qin,J. and Hinnebusch,A.G. (1998) Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol., 18, 4935–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., Cao,C., Dever,T.E. and Burley,S.K. (2000) X-Ray structures of the universal translation initiation factor IF2/eIF5B. Conformational changes on GDP and GTP binding. Cell, 103, 781–792. [DOI] [PubMed] [Google Scholar]
- Rothstein R.J. (1983) One-step gene disruption in yeast. Methods Enzymol., 101, 202–211. [DOI] [PubMed] [Google Scholar]
- Schroeder R., Waldsich,C. and Wank,H. (2000) Modulation of RNA function by aminoglycoside antibiotics. EMBO J., 19, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette M., van Tilborg,P., Spurio,R., Kaptein,R., Paci,M., Gualerzi,C.O. and Boelens,R. (1997) The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif. EMBO J., 16, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]