Abstract
Amplification of DNA from soil is often inhibited by co-purified contaminants. A rapid, inexpensive, large-scale DNA extraction method involving minimal purification has been developed that is applicable to various soil types (1). DNA is also suitable for PCR amplification using various DNA targets. DNA was extracted from 100g of soil using direct lysis with glass beads and SDS followed by potassium acetate precipitation, polyethylene glycol precipitation, phenol extraction and isopropanol precipitation. This method was compared to other DNA extraction methods with regard to DNA purity and size.
Keywords: polymerase chain reaction; soil; DNA, Bacterial
Introduction
The inability to culture most microorganisms from environmental samples is a fundamental obstacle to understanding microbial ecology and diversity (2). The use of DNA-based techniques can overcome this limitation by allowing the fate of particular genes or organisms to be monitored directly in environmental samples. Techniques to extract DNA from soil and sediment initially used large samples of 100g (3, 4). These extracts were usually contaminated with humic acids which interfered with subsequent molecular biological manipulations. Extensive purification steps were then required to successfully amplify a PCR product, including CsCl-ethidium bromide density gradient centrifugation (4-6), or the use of commercial reagents (7-11). These steps increase both the complexity and the cost of the technique. This paper describes in detail a method for extracting DNA from soil which involves minimal purification prior to PCR amplification (1). The method is compared to other commonly used DNA extraction methods. A PCR product was obtained rapidly and inexpensively from large amounts of soil, even when contaminated with heavy metals.
Materials and Methods
Soils
Soil (loamy sand) was collected on campus at Macquarie University to compare various DNA extraction methods. Soil samples were also collected from Western Sydney, Macquarie University, Ku-Ring-Gai Chase National Park and Balmain Power Station in central Sydney, to validate the bead beating technique (1) using a variety of soils. The Ku-Ring-Gai Chase National Park and the Balmain Power Station samples represent the extremes of pristine vs polluted soils and were compared by further soil testing (Biological and Chemical Research Institute, Sydney) (Table 1).
Table 1.
Analysis of soil samples
| SOIL SAMPLE | ||
| Ku-Ring-Gai Chase | Balmain Power Station | |
| pH | 3.90 | 6.93 |
| Organic matter (%) | 5.09 | 16.3 |
| Field capacity 0.33 Bar | 7.05 | 14.9 |
| CEC (cmol (+)/kg) * | 1.1 | 18.7 |
| As (mg/kg) # | < 3 | 6.9 |
| Hg (mg/kg) # | < 0.7 | 2.1 |
| Zn (mg/kg) # | 5 | 1818 |
| Cr (mg/kg) # | 3.3 | 30.4 |
| Cd (mg/kg) # | < 0.4 | 11.4 |
| Ni (mg/kg) # | 1.7 | 98.3 |
| Pb (mg/kg) # | 15 | 520 |
| Cu (mg/kg) # | 9.5 | 268 |
| Mn (mg/kg) # | 13 | 518 |
* CEC : cation exchange capacity
# Total elements determined by acid digestion and ICPAES
pH was determined using a 1:5 w/v soil suspension in 0.01 M CaCl2 at 25° C. Electrical conductivity (EC) was determined using a 1:5 w/v soil suspension in water at 25°C.
DNA extraction using bead beating (1).
Extraction buffer (100 ml of 100 mM Tris-HCl [pH 8.0], 100 mM sodium EDTA [pH 8.0], 1.5 M NaCl) was mixed with 100g (wet weight) of soil. Glass beads (100g, Bio-Spec Products, Bartesville,U.S.) were added and the sample blended in a Bead-Beater (Bio-Spec Products) for 2 minutes. Sodium dodecyl sulphate (SDS) was added (10 ml; 20 %) and blending continued for a further 5 sec. The sample was incubated at 65°C for 1 hr, transferred to centrifuge bottles (250 ml) and centrifuged at 6000g for 10 min. The supernatant was collected, and the soil pellet re-extracted with further extraction buffer (100 ml), incubation at 65°C for 10 minutes and centrifugation as above. Supernatants were transferred to centrifuge tubes (50 ml) containing a half-volume of polyethylene glycol (30%)/sodium chloride (1.6 M), and incubated at room temperature for 2 hr. Samples were centrifuged (10,000g for 20 min) and the partially purified nucleic acid pellet resuspended in 20 ml of TE (10 mM Tris-HCl, 1 mM sodium EDTA, pH 8.0). Potassium acetate (7.5 M) was added to a final concentration of 0.5 M. Samples were transferred to ice for 5 min then centrifuged (16,000 g, 30 min) at 4°C to precipitate proteins and polysaccharides. The aqueous phase was extracted with phenol/chloroform and chloroform/isoamyl alcohol (12) and DNA was precipitated by adding 0.6 volume isopropanol. After 2 hrs at room temperature, DNA was pelleted by centrifugation (16,000g for 30 min) and resuspended in TE (1 ml).
DNA extraction using sonication (modified from 13).
Extraction buffer (100 ml) was mixed with soil (50g) on ice. The mixture was sonicated using a High Intensity Ultrasonic Processor (Vibra Cell) with a standard 13mm horn solid probe for 150 seconds. The sample was cooled in ice and the sonication repeated. SDS was added (10 ml; 20%) and the sample incubated at 65°C for 1 hr. The sample was transferred to centrifuge bottles (250 ml) and centrifuged at 6000g for 10 min. The supernatant was collected, and the soil pellet re-extracted with further extraction buffer (50 ml), incubation at 65°C for 10 minutes and centrifuged as above. Extraction was then continued as per bead beating method.
DNA extraction using enzymatic lysis (modified from 11).
Extraction buffer (100 ml) containing proteinase K (5 mg) was mixed with soil (50g) in 250 ml centrifuge tubes. The sample was incubated at 37°C for 30 minutes with shaking at 180 rpm. SDS was added (10 ml; 20%) and the sample incubated at 65°C for 90 min. The supernatant was collected after centrifugation at 6000g for 10 min at room temperature. Extraction was continued as per bead beating method.
DNA extraction from bacterial cells isolated from soil (modified from 4 and 14).
The bacterial fraction of soil was separated from the inorganic or humic layer by a differential centrifugation technique (14). Bacterial cells were lysed using lysozyme and the DNA purified using ammonium acetate precipitation and ethanol precipitation (14). DNA was resuspended in TE.
Test for Co-Extraction of Contaminants
Co-extracted humic acids are the major contaminant when DNA is extracted from soil. These compounds absorb at 230 nm whereas DNA absorbs at 260 nm and protein at 280 nm. To evaluate the purity of the extracted DNA, absorbance ratios at 260 nm/230 nm (DNA / humic acids) and 260 nm/280 nm (DNA / protein) were determined (see Tables 2 and 3).
Table 2.
Comparison of DNA extraction methods using a single soil, collected from Macquarie University.
| Method * | Number of samples | A 260/230 | A 260/280 |
| Bacterial cells | 4 | 0.83 ± 0.03 | 1.10 ± 0.003 |
| Chemical lysis | 10 | 1.06 ± 0.03 | 1.31 ± 0.03 |
| Sonication | 4 | 1.20 ± 0.10 | 1.41 ± 0.07 |
| Bead beating | 6 | 1.82 ± 0.05 | 1.69 ± 0.02 |
* DNA diluted 1 : 100
Table 3.
Crude DNA ratios for different soil samples extracted using bead beating.
| Sample * | Soil type | A 260/230 | A 260/280 |
| Western Sydney | Clay loam | 1.22 | 1.42 |
| Macquarie University | Clay loam | 1.83 | 1.71 |
| Ku-Ring-Gai Chase | Loamy sand | 1.03 | 1.30 |
| Balmain Power Station | Loamy sand | 1.33 | 1.53 |
* DNA diluted 1 : 100
Polymerase Chain Reaction (PCR)
DNA (1 ml of 1:50 dilution) was mixed with 9 ml of Genereleaser™ (Bioventures Inc., Murfreesboro, Tennessee, USA) in a 0.5 ml tube and overlaid with 2 drops of sterile mineral oil. Genereleaser™ is a proprietary agent that sequesters inhibitors of PCR. Negative controls containing water only, and Genereleaser™ only, were included in each set of reactions. Reaction tubes were heated on the high setting of a 650 Watt microwave oven for 7 min (4550 W/min) in a microwave transparent rack (Bioventures Inc.). An Erlenmeyer flask containing 100 ml of water was included as a microwave sink. Tubes were incubated for at least 10 min at 80°C in an Omn-E PCR machine (Hybaid). PCR master mix (40 μl) was then added to each tube. Final concentrations of reagents were as follows: 20 mM (NH4)2SO4, 75 mM Tris-HCl (pH 9.0), 0.01% (w/v) Tween 20, 2 mM MgCl2, 0.5 mM of each primer, 0.2 mM of each deoxyribonucleotide triphosphate, and 1 U Red Hot DNA Polymerase (Advanced Biotechnologies, Surrey, UK). The following thermal cycle was performed : 94°C 3 min (1 cycle), 94°C 1 min, 55°C 1min, 72°C 2 min (35 cycles), 72°C 5 min (1 cycle).
Gel Electrophoresis
An aliquot (7 μl) of each amplification reaction was analysed on 2% w/v agarose gels cast and run in TBE buffer (pH 8.3) (12). Gels were stained with ethidium bromide and photographed using transmitted U.V. light and Polaroid film (12). A 100 base pair marker (Pharmacia, LKB) was included on every gel.
Results and Discussion
DNA extraction from soil has three requirements: extraction of high molecular weight DNA; extraction of DNA free from inhibitors for subsequent molecular biological manipulations to be performed; and representative lysis of microorganisms within the sample. In this paper, we tested a number of DNA extraction methods for their ability to fulfill these requirements.
DNA extracted using sonication was more degraded than for the other methods tested. The size of DNA extracted ranged from less than 500 bp to greater than 20 kb in size. Methods that shear DNA, such as sonication, generally result in DNA of 100-500 bp (13). Higher molecular weight DNA is desirable for PCR since the greater the size of the DNA, the less likely is the formation of chimeras during PCR (15). The bead beating method used here performed better than those previously reported which usually extract DNA of less than 10 kb in size (3). The DNA extraction methods that did not use sonication all produced DNA of greater than 20 kb.
Organic matter is the major source of inhibitors that may be co-extracted from soil with the microbial DNA. In particular, humic acids pose a considerable problem and will interfere in enzymatic manipulations of DNA (5, 14, 16). DNA polymerases have been found to be inhibited by as little as 1 μl of undiluted humic-acid-like extract, regardless of the amount of DNA present (16).
The humic materials in soil have similar size and charge characteristics to DNA resulting in their co-purification (17), evident by the extractions being brown in colour. Humic contaminants also interfere in DNA quantitation since they exhibit absorbance at both 230nm and at 260nm, the later used to quantitate DNA. This characteristic can be used to determine the level of contamination of humic material by examining absorbance ratios. A high 260/230 ratio (>2) is indicative of pure DNA, while a low ratio is indicative of humic acid contamination and a high 260/280 ratio (>1.7) is indicative of pure DNA, while a low ratio is indicative of protein contamination. When the DNA extraction methods were compared (Table 2), the bead beating method consistently extracted DNA with higher 260/230 and 260/280 ratios. This indicated that the DNA was contaminated with fewer humic acid-like compounds. Although the extracts were still brown in colour, dilution of the DNA to 1:50 from all methods was suitable to produce a PCR product. Heavy metal ions, such as are present in the Balmain soil (Table 1), also contribute to inhibitory effects (18). Here we have demonstrated that a PCR product from soil DNA contaminated with humic acids and heavy metals can be obtained without the use of expensive purification products.
To determine the diversity of microorganisms from which DNA had been extracted, different primer sets were tested (Table 4), including both multi- and single-copy genes. The multi-copy targets included the prokaryotic small subunit rRNA (19), prokaryotic rRNA intergenic spacer region (20), the eukaryotic rRNA internal transcribed spacer (ITS) region (21), the ITS region for lichen fungi (22), and the HSP70 family of proteins (23) while the low abundance targets included fungal β-tubulin (24), and nifH genes (25). With dilution of DNA from each extraction technique, successful PCR amplification was achieved with all primers tested (see Fig. 1). The only exception to this was for the DNA extracted after differential centrifugation to separate the bacterial cells. An amplification product was not found when the eukaryotic specific primers for fungal ITS were used. Due to the centrifugation step in this method, fungal cells will be underrepresented. The PCR results provide good evidence for representative lysis of organisms with all other extraction methods.
Table 4.
Primers from which successful amplification was achieved using microbial DNA from soil
| Target for amplification | Primer sequence | Reference |
| Prokaryotic 16S rRNA | FORB 5' AGAGTTTGATCCTGGCTCAG, REVB 5' GGTTACCTTGTTACGACTT | 19 |
| Prokaryotic rRNA intergenic spacer (IGS) region | SPRRNAF 5' GAAGTCGTAACAAGG SPRRNAR 5' CAAGGCATCCACCGT | 20 |
| Eukaryotic rRNA internal transcribed spacer (ITS) region | ITS1 5' TCCGTAGGTGAACCTGCGG ITS4 5' TCCTCCGCTTATTGATATGC | 21 |
| Fungal specific ITS |
LICHITSF 5' GCGGAAGGATCATTACTGAG LICHITSR 5' GGGTATCCCTACCTGATCCG |
22 |
| HSP70 |
HSP1 5' CA(AG)GC(I)AC(I)AA(AG)GA(CT)GC(I)GG HSP2 5' GC(I)AC(I)GC(CT)TC(AG)TC(I)GG(AG)TT |
23 |
| Fungal β -tubulin |
GEN C 5' GAGGAATTCCCAGACCGTATGATG GEN D 5' GCTGGATCCTATTCTTTGGGTCGAACAT |
24 |
| nifH gene |
NIFH1 5' CTG(CT)GA(CT)CC(ACGT)AA(AG)GC(ACGT)GA NIFH2 5' G(AGT)(ACGT)GCCATCAT(CT)TC(ACGT)CC |
25 |
Fig. 1.
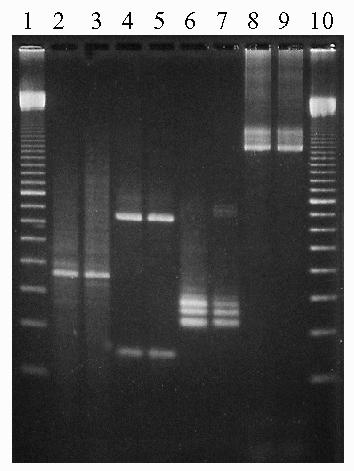
Example of PCR amplification products using various DNA targets with soil extracted by enzymatic lysis or bead beating. Lane 1: 100 bp marker; lane 2: enzymatic lysis DNA with 16S rRNA primers (19); lane 3: bead beating DNA with 16S rRNA primers (19); lane 3: enzymatic lysis DNA with fungal ITS primers (22); lane 4: bead beating DNA with fungal ITS primers (22); lane 5: enzymatic lysis DNA with hsp70 primers (23); lane 6: bead beating DNA with hsp70 primers (23); lane 7: enzymatic lysis DNA with nifH primers (25); lane 8: bead beating DNA with nifH primers (26); lane 9: 100 bp marker.
Of the DNA extraction methods tested, sonication did not produce high molecular weight DNA while the differential centrifugation method generated a DNA pool dominated by bacterial DNA and therefore not suitable for analysis of eukaryotes. Neither of these methods fulfilled all requirements for a suitable DNA extraction method. The enzymatic lysis method relies on proteinase K digestion of microbial cells to release DNA while bead beating relies on ballistic disintegration of all cells. Therefore, bead beating is more likely to result in effective lysis of all soil organisms. Due to ease of the method, the reduced co-extraction of inhibitors (Tables 2 and 3) and the greater confidence that bead beating would lyse all microbial cells in the soil, this was the method of choice and concentrated on for further analysis (see 1). Bead beating has been found to have a lysis efficiency of greater than 90% (3). The PCR results reported here provide further evidence to support this with products from both bacterial and fungal elements of the soil microbiota being obtained. The bead beating direct lysis method described here extracts between 1.5 and 2.35 mg/ml of DNA from 100g of soil or 15-23.5 μg DNA/g soil. Extraction methods using small soil samples ranging from 5g to 100 mg of soil have extracted 9-25 μg DNA/g soil (6), 12 μg/g (18), 1-100 μg/g (26), and 2.5-26.9 μg/g (11). The method described here is therefore at least as efficient as the above methods.
The focus of DNA extraction methods has moved to rapid performance of molecular techniques, avoiding extensive purification steps (7, 27). Using the bead beating DNA extraction method described here, crude microbial DNA could be extracted from a variety of soil types and dilution of this DNA was sufficient for successful PCR from both high- and low-copy number genes. The described method allows the use of large scale preparations providing greater probability of detecting genes present in low abundance in the soil environment. Because this method is applicable to even the more challenging heavily contaminated soils, molecular microbial biodiversity assessment can now be more readily applied.
Acknowledgments
Thanks are due to Mr. Bruce Dowling of Pacific Power, Sydney for supplying soil from Balmain Power Station and to Dr. Geoff Humphreys, School of Earth Sciences, Macquarie University for identifying soil types. This is publication number 259 of the Key Centre for Biodiversity and Bioresources.
References
- Yeates C, Gillings MR, Davison AD, Altavilla N, Veal DA. PCR amplification of crude microbial DNA extracted from soil. Letters in Applied Microbiology. 1997;25:303–307. doi: 10.1046/j.1472-765x.1997.00232.x. [DOI] [PubMed] [Google Scholar]
- Atlas RM. Diversity of microbial communities. Advances in Microbial Ecology. 1984;7:1–47. [Google Scholar]
- Ogram A, Sayler GS, Barkay T. The extraction and purification of microbial DNA from sediments. Journal of Microbiological Methods. 1987;7:57–66. [Google Scholar]
- Steffan RJ, Goksoyr J, Bej AK, Atlas RM. Recovery of DNA from soils and sediments. Applied and Environmental Microbiology. 1998;54:2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben WE, Jansson JK, Chelm BK, Tiedje JM. DNA probe method for the detection of specific microorganisms in the soil bacterial community. Applied and Environmental Microbiology. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteous LA, Armstrong JL. Recovery of bulk DNA from soil by a rapid, small-scale extraction method. Current Microbiology. 1991;22:345–348. [Google Scholar]
- Borneman J, Skroch PW, O'Sullivan KM., Palus JA, Rumjanek NG, Jansen JL, Nienhuis J, Triplett EW. Molecular microbial diversity of an agricultural soil in Wisconsin. Applied and Environmental Microbiology. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- More MI, Herrick JB, Silva MC, Ghiorse WC, Madsen EL. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Applied and Environmental Microbiology. 1994;(60):1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbe CC, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Applied and Environmental Microbiology. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YL, Olson BH. Rapid method for direct extraction of DNA from soil and sediments. Applied and Environmental Microbiology. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Applied and Environmental Microbiology. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning - a laboratory manual. Cold Spring Harbor, USA, Cold Spring Harbor Laboratory Press; 1987.
- Picard C, Ponsonnet C, Paget E, Nesme X, Simonet P. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Applied and Environmental Microbiology. 1992;58:2717–2722. doi: 10.1128/aem.58.9.2717-2722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan RJ, Atlas RM. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Applied and Environmental Microbiology. 1998;54:2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16Sr DNA analysis of a mixed culture of strict barophilic bacteria. Microbial Ecology. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- Tsai YL, Olson BH. Detection of low numbers of bacterial numbers in soils and sediments by polymerase chain reaction. Applied and Environmental Microbiology. 1992;58:754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben WE. Isolation and purification of bacterial DNA from soil. Methods of Soil Analysis, Part2. Microbiological and Biochemical Properties. Madison, USA, Soil Science Society of America:727-751;1994
- Tsai YL, Olson BH. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Applied and Environmental Microbiology. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Research. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Webster JA, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Applied and Environmental Microbiology. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. M.A. Innis, D.H. Gelfand, J.J. Sninsky and T.J. White, AcademicPress:315-322;1990.
- Gillings MR. Personal communication.
- Yap WH, Li X, Soong TW, Davies JE. Genetic diversity of soil microorganisms assessed by analysis of hsp70 (dnaK) sequences. Journal of Industrial Microbiology. 1996;17:179–184. [Google Scholar]
- Koenraadt H, Somerville SC, Jones AL. Characterisation of mutations in the beta-tubulingene of benomyl resistant field strains of Venturia inaequalis and other pathogenic fungi. Phytopathology. 1992;82:1348–1354. [Google Scholar]
- Zehr JP, McReynolds LA. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Applied and Environmental Microbiology. 1989;552:5222–526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteous LA, Armstrong JL, Seidler RJ, Watrud LS. An effective method to extract DNA from environmental samples for polymerase chain reaction amplification and DNA fingerprint analysis. Current Microbiology. 1994;29:301–307. doi: 10.1007/BF01577445. [DOI] [PubMed] [Google Scholar]
- Volossiouk T, Robb EJ, Nazar RN. Direct DNA extraction for PCR-mediated assays of soil organisms. Applied and Environmental Microbiology. 1995;61:3972–3976. doi: 10.1128/aem.61.11.3972-3976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


