Abstract
Histocompatibility, the ability of an organism to distinguish between its own cells and tissue from those of another, is a universal phenomenon in the Metazoa. In vertebrates, histocompatibility is a function of the immune system controlled by a highly polymorphic Major Histocompatibility Complex (MHC) that encodes proteins which target foreign molecules for immune cell recognition. The association of the MHC and immune function suggests an evolutionary relationship between metazoan histocompatibility and the origins of vertebrate immunity. However, the MHC is the only functionally characterized histocompatibility system; mechanisms underlying this process in non-vertebrates are unknown. A primitive chordate, the ascidian Botryllus schlosseri, also undergoes a histocompatibility reaction controlled by a highly polymorphic locus. We have isolated a candidate gene encoding an immunoglobulin superfamily member that by itself predicts the outcome of histocompatibility reactions. This is the first non-vertebrate histocompatibility gene described, and may provide insights into the evolution of vertebrate adaptive immunity.
Peptide presentation by the MHC is the foundation of the vertebrate adaptive immune system. Characterized by the presence of immunoglobulins, clonal B and T cells, somatic recombination, memory, and a number of other highly specialized processes, adaptive immunity has an incredibly complex, interrelated organization that is similar in form and function in all vertebrates, from sharks to humans1. However, as is clear by its name, the MHC was discovered because of its role in histocompatibility, and based on this dual function, it has been long suggested that there is an evolutionary connection between histocompatibility and vertebrate immunity. Ironically, MHC-based histocompatibility is thought to be an artifact of the MHC molecules role in the adaptive immune response coupled to their extraordinary polymorphism. Furthermore, there is no evidence of MHC, or direct homologs of any adaptive immune components in more primitive extant organisms, even those as closely related as the jawless fish1,2. Thus the mechanisms underlying histocompatibility in the non-vertebrates, the origins of any components of the vertebrate adaptive immune system, and the ultimate relationship of immunity and histocompatibility are completely unknown.
We are studying histocompatibility in the ascidian, Botryllus schlosseri. A member of the chordate subphylum, Urochordata, Botryllus begins life as a tadpole larva with many chordate traits, including a notochord, dorsal, hollow nerve tube, and gill slits. After a 24 hour motile phase, these chordate structures are lost when the tadpole settles and metamorphs into a sessile, invertebrate body plan, called an oozooid. In addition, Botryllus schlosseri is a colonial organism, and this initial metamorphosis is followed by a recurring, highly coordinated budding process which eventually gives rise to a large colony of asexually derived, genetically identical individuals, called zooids, united by a common vascular network (Figure 1a, panel 1).
Figure 1.
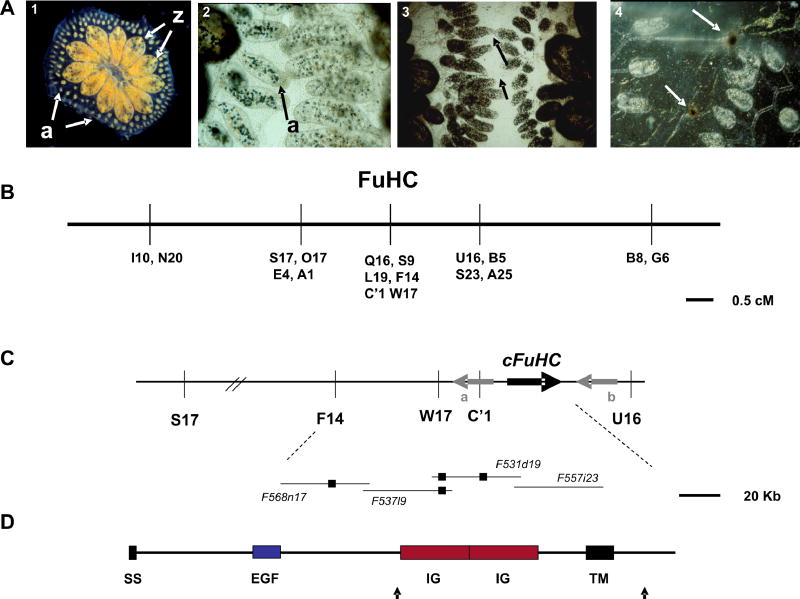
Histocompatibility and positional cloning of the FuHC locus in Botryllus schlosseri. A. An adult B. schlosseri colony (panel 1) consists of asexually-derived individuals called zooids (z) united by a common blood supply (colors are caused by pigment cells in the blood). The vasculature stops at the periphery of the colony in small protrusions called ampullae (a), where the histocompatibility reaction takes place. When two colonies grow close together the ampullae reach out and interact (panel 2), and this will result in one of two outcomes. Either the two ampullae will fuse (panel 3), allowing the circulation of the two colonies to interconnect, or they will reject each other (panel 4). Rejection is a localized inflammatory reaction where blood cells leak from the ampullae and begin killing each other, resulting in dark spots called points of rejection (bottom panel, arrows) where some melanization occurs during the reaction. Histocompatibility takes ca. 24 hrs to occur and is controlled by a single, highly polymorphic locus called the FuHC (for fusion/hisftocompatibility). Colonies will fuse if they share one or both alleles, and will reject if no alleles are shared. B. Genetic map of the FuHC region from one backcross population. Genetic markers (AFLPs) are represented by a letter and number. In one F2 cross (Table 1, F2 cross AY x AY), C’1, was found to physically lie within a small 18Kb double crossover in one individual8, this crossover did not include the cFuHC C. Physical map surrounding the candidate FuHC locus. Fosmid clones from the minimal tiling path of this region are shown on the bottom; small black boxes represent the physical location of AFLP markers, and the thick black arrow represents the location of the cFuHC locus and direction of transcription. Gray arrows show the flanking genes, a. Phospholipase A2-like, b. retinoblastoma binding protein-like D. Predicted structure of the FuHC protein, ss-signal sequence, EGF-epidermal growth factor repeat, Ig-immunoglobulin domain, TM-transmembrane domain. Black arrows indicate the location of two alternative splices, one which creates a secreted form of the protein, and another which shortens the cytoplasmic tail (see text).
At the periphery of the colony, the vasculature stops in small protrusions called ampullae (Fig. 1a, panel 1). The ampullae are the site of a naturally occurring histocompatibility reaction initiated when two colonies asexually expand into close proximity. Once juxtaposed, ampullae of each colony will reach out and begin an interaction (Fig. 1a, panel 2). This will result in either fusion of the two ampullae (forming a single chimeric colony sharing a common blood supply, panel 3), or a rejection reaction during which the interacting ampullae are destroyed, thus preventing vascular fusion (panel 4). Fusion or rejection is governed by a single, highly polymorphic locus called the FuHC 3–5. When two colonies share one or both FuHC alleles, they will fuse, whereas rejection occurs if no alleles are in common. The FuHC is highly polymorphic, with most populations containing tens to hundreds of alleles3–5. Thus this primitive chordate undergoes a histocompatibility reaction analogous to the vertebrate MHC-based reaction, and we wished to uncover the molecular mechanisms underlying this process.
Positional Cloning of the FuHC locus
We have created conditions to raise and breed B. schlosseri in captivity and developed partially inbred lines homozygous for different FuHC alleles, allowing us to take a forward genetic approach to identify the FuHC locus6–8. As described previously8, mapping populations were created using FuHC defined individuals as parents, the FuHC locus was mapped using a combination of AFLPs and bulk segregant analysis (Figure 1b), and a genomic walk was initiated from the linked markers using both bacterial artificial chromosome (BAC) and Fosmid genomic libraries. The physical map now consists of 3 contigs spanning 1.3 Mbp, with one chromosomal breakpoint crossed (not shown). Over 1 Mbp of the minimal tiling path have been sequenced.
The strategy for identifying candidate FuHC gene(s) is based on the extraordinary polymorphism of the locus. Predicted genes from genomic sequence are analyzed for expression using RT-PCR, then compared to those from the genomic clones. The genomic (BAC and Fosmid) and cDNA libraries are made from FuHC defined, but non-histocompatible individuals, thus we can concurrently survey for both expression and polymorphism of these cDNAs. The FuHC gene(s) should have polymorphisms with the following three characteristics: 1) absolute correlation with defined histocompatibility alleles in a fusion assay; 2) co-segregation with the same alleles in a defined cross and; 3) extraordinary polymorphisms in colonies isolated from the wild. Finally, expression patterns should correlate spatially with the functional aspects of histocompatibility.
Isolation and characterization of a candidate FuHC (cFuHC) gene
A Genscan9 analysis of a sequenced contig consisting of two overlapping fosmid clones (531d19, 557i23; both segregated without recombination with the FuHC, Figure 1c) predicted a gene model encoding a transmembrane protein with an extracellular immunoglobulin (Ig) domain. A full-length cDNA was isolated via RACE and was 3.2 kb in length, predicting an open reading frame of 1007 residues, and was highly polymorphic. The cFuHC is a type I transmembrane protein with the majority of the protein (852 residues) extracellular, followed by a transmembrane domain and an intracellular tail of 128 residues.
The domain structure of the cFuHC is shown in Figure 1d. The N-terminus begins with a signal sequence, followed by an extracellular EGF repeat, then two tandem Ig domains, followed by the transmembrane domain and an intracellular tail. BLAST searches show that the EGF repeat has homology to notch and tenascin at E values of 5e-05; the region encompassing the two Ig domains is homologous to Immunoglobulin Superfamily Member 4D/nectin-like 3 from a variety of vertebrate species (E = 7e-10), the highest homology is to chicken. No direct homolog was identified in other sequenced ascidian genomes. 3D modeling on the PSSM fold recognition server suggested that the Ig domains have the highest homology to the poliovirus receptor, CD155. Conserved domain searches suggest that the first Ig domain is potentially a variable or more ancient intermediate type domain, and the second is closest to a C2-type, but is divergent and may not be easily classifiable (outlined in red, Supplementary Figure 1). These analyses depend on the presence of conserved residues throughout the domain and spacing between these residues10, however, without structural data the true configuration remains unknown. The genomic structure of each Ig domain is also different: the first domain is encoded in three exons, while two exons code the second domain. The intracellular tail has several potential phosphorylation sites, but it is unknown whether these are functional.
The cFuHC gene spans 33Kb, consisting of 31 exons (Supplementary Figure 1) In addition to the transmembrane form of the protein, which consists of 27 exons; there are two alternative splice variants of the cFuHC gene (Supplementary Fig. 1, bottom). First, a short, secreted form is created by an alternative splice which adds three new exons after exon 14 (exons 15–17). These exons, physically located between exons 14 and 18, encode another putative low-homology EGF domain, followed by stop codons in all three reading frames and ca. 300 base-pairs of 3′ untranslated sequence (UTR). The predicted secreted form does not include the Ig domains, and as discussed below, is expressed in both the tadpole and adult colonies. Secondly, there is a rare splice variant in the cytoplasmic domain which has been observed in 5% of the sequences covering the region. This is due to the presence of an extra 113 bp exon which is physically located between exons 29 and 31 of the full-length clone (exon 30; Figure 2), but is normally spliced out. When present, this exon encodes a new, shorter cytoplasmic domain, and the exon ends in a stop codon. Exon 31 is still present in this alternatively spliced form, but is now 3′ UTR. The shorter cytoplasmic domain adds no known motifs, and removes the predicted phosphorylation sites encoded in exon 29. This form has also been identified in both the tadpole and adult. Finally, there are also a three splice variants with combinations of exons 15 and 16 that contain the rest of the transmembrane form of the gene (exons 18–31). However, these variants do not form open reading frames due to frameshifts between exon boundaries when these extra exons are present in any combination. These may be non-functional transcripts made during splicing of the secreted form of the protein.
Figure 2.
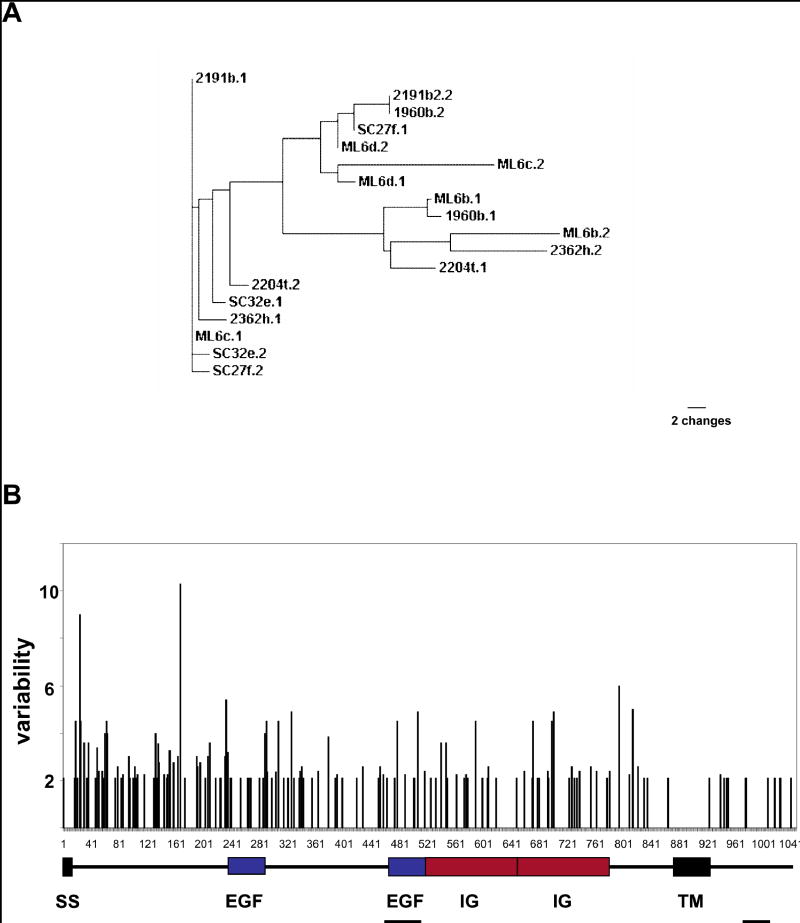
Graphic representation of the overall polymorphism of the cFuHC A A phylogram of 18 cFuHC alleles isolated from wild-type animals collected in Monterey (Numbers), Santa Cruz (SC) and Moss Landing (ML) marinas (see methods) B. the domain structure of the protein (below) is overlayed with a graphic representation of amino acid variability along the protein. The same 18 wild type alleles used in A were aligned, and the variability is defined as the ratio of the number of different amino acids to the frequency of the most common amino acid at each position. The secreted exons and new COOH tail exons are also shown (underlined). There are no hypervariable regions, and the majority of the variability (i.e., the shortest vertical line) consists of single amino acid changes spread throughout the ectodomain.
cFuHC Genetics
To analyze segregation of the cFuHC, we used a fragment amplified between the two Ig domains containing parts of exons 18 and 19 and a small (250bp) intron. As described in the methods, this fragment was highly polymorphic, and included multiple substitutions and insertion/deletions. These polymorphisms segregated absolutely with histocompatibility (as assayed by fusion/rejection or genetic mapping) for all individuals in our mapping crosses (Table 1). In addition, we correlated cFuHC polymorphisms with a number of individuals from multiple generations of our main FuHC AB x AB partially inbred lines from the last 16 years. In all cases, correlation of cFuHC polymorphisms with phenotypic and/or genetic FuHC typing was absolute.
Table 1. FuHC Segregation in Defined Cross Progeny.
FuHC genotypes were compared between results obtained from genetic mapping versus results from direct typing of cFuHC alleles on an individual by individual basis. The small number of FuHC homozygotes is discussed elsewhere.12
| Cross/Genotype | n | FuHC Segregation (genetic mapping) | FuHC Segregation (cFuHC allele) | Correlation |
|---|---|---|---|---|
| AY x AY F2 | 72 | 2:64:6 | 2:64:6 | 100% |
| AD x AD F2 | 22 | 1:19:3 | 1:19:3 | 100% |
| AC x AC F2 | 60 | 4:49:8 | 4:49:8 | 100% |
| AY x CD OC | 78 | 16:21:19:22 | 16:21:19:22 | 100% |
| AA | 10 | 10 | 10 | 100% |
| BB | 6 | 6 | 6 | 100% |
| AB | 3 | 3 | 3 | 100% |
To correlate cFuHC polymorphism with functional histocompatibility, wild-type colonies were grown in the lab and assayed for phenotypic fusion/rejection. We identified five robust pairs of fusing and five pairs of rejecting genotypes, including one pair of siblings which rejected each other, as well as two pairs of fusing colonies from different geographic locations. Full-length or extracellular polymorphic regions of the cFuHC were cloned from naïve subclones of each individual and compared among genotypes. In all cases, the alleles identified by sequencing correctly predicted the outcome of histocompatibility reactions (Table 2). Thus far, we have not identified fusing genotypes which do not share an identical allele, or rejecting genotypes which do share an allele. The cFuHC polymorphisms predict the outcome of histocompatibility reactions by themselves.
Table 2. Comparison of phenotypic fusion/rejection with FuHC genotype.
Individuals were placed together and fusion and rejection visibly observed. FuHC alleles were then genotyped from naïve subclones and compared (described in methods). Zygosity at the cFuHC locus is also shown (heterozygous or homozygous)
| Colony 1 | Colony 2 | Relationship | Histocompatibility Outcome | cFuHC shared |
|---|---|---|---|---|
| 1960b (het) | 1960c (het) | sibs | Fuse | 1 |
| 2204t (het) | 2204p (het) | sibs | Fuse | 1 |
| 2190b (het) | 2190c (het) | sibs | Fuse | 1 |
| SC27f (het) | HM9aY (het) | none | Fuse | 1 |
| SC32e (het) | HM9aY (het) | none | Fuse | 1 |
| ML6c (het) | 2191b (het) | none | Reject | 0 |
| 2191b (het) | Ml6b (het) | none | Reject | 0 |
| SC27f (het) | SC32e (het) | none | Reject | 0 |
| HM9aY (het) | 2362h (het) | none | Reject | 0 |
| ML6c (het) | ML6d (het) | sibs | Reject | 0 |
These experiments also confirmed that the cFuHC is a single locus. Segregation analysis done in Table 1 revealed no aberrant banding patterns, and experiments done in Table 2 on wild-type individuals never revealed more than two alleles/genotype. In addition, screening of the entire 13X genomic library revealed only known alleles, which was confirmed by southern blotting (not shown).
Polymorphism and Expression of the cFuHC
To analyze the overall polymorphism of wild-type colonies we sequenced full-length clones from 10 wild-type individuals collected from around the Monterey Bay. This resulted in the isolation of 18 cFuHC alleles; each individual was heterozygous. At the nucleotide level, any two alleles diverge by an average of 4% of the nucleotides. Amino acid polymorphisms of the cFuHC are illustrated in Figure 2. While alleles are quite divergent (Fig. 2A), polymorphism is spread throughout the protein, with no obvious highly variable regions (Fig. 2B). As shown, the majority of divergence between any two alleles is single amino acid changes located throughout the extracellular domain.
The final criteria for a candidate histocompatibility protein were the correct expression patterns. The FuHC gene should be expressed at sites of histocompatibility in the adult, and there must also be expression in the larva; histocompatibility is functional in the oozooid, thus the effector systems must be undergoing education processes prior to metamorphosis to be able to distinguish between FuHC alleles (discussed below). RT-PCR was done on cDNA isolated from different tissues (Figure 3a), and revealed that both membrane bound and secreted forms of the FuHC are expressed in the adult in both ampullae and blood, and also in the tadpole. In addition, the cFuHC does not appear to be expressed in germline tissue using these or other techniques. This corroborates with previous results suggesting that the FuHC does not appear to be involved in fertilization11,12.
Figure 3.
Expression analysis of the cFuHC. A. RT-PCR of cDNA isolated from ampullae (a), blood (b), tadpoles (t), eggs (e) and sperm (s), Top gel, 600 bp fragment including the transmembrane domain (tm); bottom gel, 350 bp fragment which includes the secreted 3′ UTR (sec). Both primer sets straddle a large intron, preventing amplification from genomic DNA B-I. Localization of cFuHC expression via in situ hybridization in eggs, tadpoles and oozooids (described in Methods). Panels B, D and G are sense controls. Eggs (Panel C) were isolated immediately prior to fertilization but show no cFuHC expression when compared to controls. In the tadpole, expression is restricted to the anterior of the larvae, and outlines the nascent ampullae (Panel E, arrows). Panel F shows a larvae in the middle of metamorphosis, with strong expression seen at the tips of the extending ampullae (arrow). Expression in the adult body is seen in the epithelia of the ampullae (Panel H). A close-up of an ampullae is shown in Panel I, with expression seen in both a subset of blood cells and the epithelium of the ampullae and blood vessel. Light blue staining is vector blue, dark purple is NBT/BCIP. The punctate, light blue staining seen in B-E is a result of vector blue precipitation that often occurs during color development.
Expression patterns were also visualized by in situ hybridization (Figure 3b). In the oozooid, strong cFuHC expression is seen in the epithelia of the ampullae, as well as a subset of blood cells. Expression in the tadpole is restricted to a region at the anterior of the larvae, outlining the nascent ampullae. Expression in isolated ripe eggs is not seen. In summary, the cFuHC is expressed strongly in tissues intimately associated with histocompatibility.
Discussion
We have isolated and characterized a highly polymorphic candidate fusion/histocompatibility (cFuHC) gene from the primitive chordate Botryllus schlosseri which encodes an immunoglobulin superfamily member. The cFuHC segregates absolutely with histocompatibility in all mapping crosses, by itself predicts the outcome of interactions between wild-type colonies, and is expressed in tissues directly associated with the natural transplantation reaction: by every criterion this candidate is directly responsible for controlling histocompatibility. Other genes with these characteristics have not been identified in the FuHC locus, and together this is strong evidence that the cFuHC is the first metazoan histocompatibility ligand ever identified outside the vertebrates.
The ability to discriminate between multiple ligands is the universal principal shared by immunity and histocompatibility, leading F.M. Burnet to hypothesize that the origins of the most complex recognition system in the metazoa, the vertebrate adaptive immune system, would be found in histocompatibility systems of more primitive organisms13.
Does identification of the cFuHC in a primitive chordate confirm this hypothesis? As a type I transmembrane protein with multiple extracellular Ig domains, the cFuHC is certainly structurally similar to the vertebrate MHC, however, it is clearly not a direct homolog; for example the Ig domains do not correspond to the C1 type found in the MHC. Alternatively, it has been suggested that vertebrate genes with the more ancient Ig domains, like human IgSF4, with which the cFuHC has high homology, may be descendents of the first antigen receptors2,30. This is also based on observations of the organization of these genes in mammalian genomes2, which may also be shared with the FuHC (not shown). Further structural and genetic analysis of the cFuHC may help elucidate the evolutionary origins of the different Ig domains, and ultimately, the molecules in adaptive immunity.
From a functional standpoint, there is another possible relationship between histocompatibility in Botryllus to the origins of vertebrate immunity. In Botryllus, individuals sharing only a single allele are compatible. It seems unlikely that the hundreds of FuHC alleles could evolve to clearly distinguish themselves via only homotypic interactions, suggesting that effector systems in Botryllus work by missing self recognition, a strategy first identified in vertebrate Natural Killer (NK) cells14. We hypothesize that a population of effector cells are educated to both FuHC alleles, and that recognition of just one allele blocks a default rejection reaction. Histocompatibility in Botryllus evolutionarily links an innate function of vertebrate NK cells, scanning for altered MHC expression, to a highly polymorphic, Ig superfamily ligand. Thus missing-self recognition and natural killing effector systems may have originated in histocompatibility systems of non-vertebrates. In fact, the genome of the related ascidian, Ciona intestinalis contains a number of genes highly-homologous to those involved in natural killing in the vertebrates33. A major goal of future research will be to identify the cells, molecules and mechanisms underlying the function and education of this effector system. Interestingly, we have already found what appears to be a putative polymorphic receptor encoded within 200Kb of the cFuHC. However, these polymorphisms play no role in histocompatibility outcomes (Nyholm, et. al., in preparation).
Vertebrate MHC-based histocompatibility is thought to be an unintended consequence of peptide presentation and polymorphism, but histocompatibility and, more importantly, extraordinary polymorphism, have a well-defined function in B. schlosseri. After compatible colonies fuse, blood-borne germline or totipotent stem cells transfer between colonies and have the ability to expand and differentiate in the newly arising, asexually derived individuals of the vascular partner. This can often result in a situation where only one of the genotypes is represented in the gametic output of the fused individuals, a situation which can remain constant for life15–17. FuHC polymorphism functions to restrict vascular fusion, and the possibility of this germline parasitism, to kin. This stem cell parasitism is thought to be widespread, and the selective force driving the evolution of polymorphic histocompatibility systems in other phyla18,19. Moreover, this movement of stem cells in Botryllus is reminiscent of the transfer of hematopoietic stem cells between fetal mammals sharing a common placenta, the basis of natural tolerance20. These same studies did not identify germline chimeras despite the fact that mammalian germline precursors are also migratory during development20,21. Thus it may be that histocompatibility is still one function of vertebrate immunity, providing both a historical origin, and contemporary selective force to explain extraordinary MHC polymorphism. Alternatively, the FuHC may represent an undiscovered Ig-based system of allorecognition which still exists in the vertebrates; for example, NK cells have recently been found to interact with a non-MHC ligand via missing-self recognition22,23 .
Furthermore, CD155, a nectin family member with which the cFuHC shares structural homology, is a ligand for the NK activating receptor CD22631. In any case, the cFuHC is the first non-vertebrate histocompatibility ligand every identified, and as an Ig superfamily member provides the first structural link between invertebrate histocompatibility and vertebrate adaptive immunity.
Methods
Mariculture, Collection, Genetic and Physical Mapping
Conditions for raising and crossing Botryllus schlosseri in the laboratory, techniques for assaying phenotypic histocompatibility, genetic and physical mapping of the FuHC locus, sequencing, DNA and RNA isolation and cDNA synthesis have been comprehensively described elsewhere6,8. Enzymes were from NEB (Beverly, Mass., USA). All chemicals were purchased from Fisher (Pittsburgh, Pa., USA). Oligonucleotides were synthesized by Operon (Chatsworth, Calif., USA). Kits were used for specific processes as described below.
For fusing or rejecting pair analysis, pregnant colonies were collected from the wild, hatched and reared in the lab. One naïve subclone was removed and DNA and RNA isolated, another subclone was used in a fusion/rejection assay3–5. Animals are named according to their collection site. Those starting with a number are from the Monterey Marina, other collection sites are Half Moon Bay, (HM), Santa Cruz (SC), and Moss Landing (ML), CA. This is followed by a lowercase letter which indicates a F1 individual, thus individuals 1960b and 1960c are progeny from the same wild-type colony.
Isolation and Analysis of a Candidate FuHC Gene
BAC and Fosmid sequences were analyzed using BLAST24 and GENSCAN9 Putative expressed genes were analyzed for topology and functional domains using the HMMTOP transmembrane predictor 25 and ELM. Sequences which caught our interest were then isolated by RACE 27 and RT-PCR using the SMART II RACE kit as described (Clonetech). Initial Primers for the cFuHC were in two regions. STS 1 covered the Ig domain: sense: 5′-gtatgggacaacacaggaaattctac-3′; antisense: 5′-gtgacgttttagtccataggatatcag-3′. STS 2 was 5′ of this region. Sense 5′-tactattgagtgtatgaacggtgatgt-3′; antisense 5′-ttctatcgcccctatatagttttgtaa-3′. These same primers were used to isolate the entire gene by RACE. The entire gene was amplified with the following primers. The N terminus by the sense primer 5′-aacgatgaatgggttcgcgattttc-3′ which encompasses the start codon, and one of two antisense primers located near the stop codons of each form of the mRNA. The secreted form antisense primer is: 5′-tcgttatgctgtattcattcaa-3′. For the membrane bound form, the primer is antisense, 5′-aagcttctttcagagctactatcttca-3′. For the membrane bound form, we usually amplified in two overlapping ca. 1.5 Kb fragments from primers designed over the exon 14/exon 18 junction. The forward primer was 5′-tacgttgattggaactgtcgacttgaagta-3′ for amplifying a 3′ piece, the reverse primer was 5′-tacttcaagtcgacagttccaatcaacgta-3′ for amplifying the 5′ fragment, used in conjunction with the primers described above. This second strategy was done as there are a number of alternative splice variants which encode non-functional transcripts in exons 15–17. None of these variants form ORFs, due to frameshifts in the exon splice boundries (not shown). As these are near the secreted alternative splice site (after exon 16), we hypothesize that they are nonfunctional transcripts made during mRNA processing.
Segregation and Expression Analysis
The majority of segregation analysis was done using STS 1, above, as the forward and reverse primers were each on different exons and split by a small (250bp) intron. This intron was highly polymorphic, and the nature of these polymorphisms allowed a PCR-RFLP approach to distinguish between all the FuHC alleles in our mapping populations. The FuHC A, B, and Y alleles could be differentiated by EcoRI, HindIII and HincII polymorphisms, techniques have been previously described8.Segregation was confirmed by sequencing of the entire clone from recombinant individuals in each cross (not shown). To detect expression via RT-PCR we used STS 1 and 2, as well as two other primers sets tailored for the membrane bound or secreted form. The membrane bound form specific RT-PCR set was: sense Ig STS1 F (shown above), antisense 5′-gtacctcaagtaccacacgccccaat-3′. The secreted form was sense 5′-tgcagcttggaagtttttg-3′; antisense 5′-tcgttatgctgtattcattcaa-3′. For both wild-type fusion/rejection, and polymorphism analysis, pregnant colonies were brought in from the field (from marinas in Monterey, Moss Landing and Santa Cruz, CA) and tadpoles hatched and reared in the lab. Fusion/rejection was visibly assayed, and the cFuHC was isolated as described above. Protein polymorphisms and phylograms were analyzed using ClustalW32.
In situ hybridizations
In situ probes, amplified from primers described above, were subcloned into T-vector (Promega), sequenced to determine orientation, then re-amplified with T7 and sp6 specific primers. After PCR cleanup (Qiagen), in situ hybridization probes were made using T7 and sp6 RNA polymerase according to the manufacturers suggestions (Boehringer). The whole-mount in situ hybridization protocol in this study was carried out according to references 28 and 29, on lab-reared individuals. AP detection was done using either a BCIP/NBT or Vector® Blue alkaline phosphatase substrate kit (Vector Laboratories, Burlingame, CA) with levamisole. After color development, samples were post-fixed in 4% paraformaldehyde in PBS (20 min at 4 °C ), washed in PBS, mounted on slides and visualized on an Olympus BH-2 microscope.
Supplementary Material
Acknowledgments
We thank Chris Amemiya, Dan Rokhsar, Ron Davis, Audrey Southwick, Molly Miranda, David Ransom, John Cannon, Gary Litman, Robert Haire, Ayelet Voskoboynik, Jason Wallace, Erin Johnson, This study was supported by grants from the NIH (AI04588; Trans-BAC Sequencing) and the Community Sequencing Program at the DOE/JGI.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
References
- 1.Flajnik, M.F., Miller, K. & Dupasquier, L. Evolution of the immune system in: W.E. Paul, Editors, Fundamental Immunology (5th ed), Lippincott, Williams & Wilkins, Philadelphia pp. 519–570. (2003),
- 2.Dupasquier L, Zucchetti I, De Santis R. Immunoglobulin superfamily receptors in protochordates : before RAG time. Immunol Rev. 2004;198:233–248. doi: 10.1111/j.0105-2896.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- 3.Oka H, Watanabe H. Problems of colony specificity in compound ascidians. Bull Biol Stn Asamushi. 1960;10:153–155. [Google Scholar]
- 4.Sabbadin A. Le basi genetiche dell capacita di fusione fra colonie in Botryllus schlosseri (Ascidiacea) Atti Accad Na Lincei Rend. 1962;32:1031–1035. [Google Scholar]
- 5.Scofield VL, Schlumpberger JM, West LA, Weissman IL. Protochordate allorecognition is controlled by an MHC-like gene system. Nature. 1982;295:499–502. doi: 10.1038/295499a0. [DOI] [PubMed] [Google Scholar]
- 6.Boyd HC, Brown SK, Harp JA, Weissman IL. Growth and sexual maturation of laboratory-cultured Monterey Botryllus schlosseri. Biol Bull. 1986;170:91–109. [Google Scholar]
- 7.De Tomaso AW, Saito Y, Ishizuka KJ, Palmeri KJ, Weissman IL. Mapping the genome of a model protochordate I. A low resolution genetic map encompassing the fusion/histocompatibility (Fu/HC) locus of Botryllus schlosseri. Genetics. 1998;149:277–287. doi: 10.1093/genetics/149.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Tomaso AW, Weissman IL. Initial characterization of a protochordate histocompatibility locus. Immunogenetics. 2003;55:480–90. doi: 10.1007/s00251-003-0612-7. [DOI] [PubMed] [Google Scholar]
- 9.Burge CB, Karlin S. Finding the genes in genomic DNA. Curr Opin Struct Biol. 1998;8:346–354. doi: 10.1016/s0959-440x(98)80069-9. [DOI] [PubMed] [Google Scholar]
- 10.Williams AF, Barclay AN. The immunoglobulin superfamily-domains for surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 11.Grosberg RK, Hart MW. Mate selection and the evolution of highly polymorphic self/nonself recognition genes. Science. 2000;289:2111–4. doi: 10.1126/science.289.5487.2111. [DOI] [PubMed] [Google Scholar]
- 12.De Tomaso AW, Weissman IL. Evolution of a protochordate allorecognition locus. Science. 2004;303:277. doi: 10.1126/science.1094952. [DOI] [PubMed] [Google Scholar]
- 13.Burnet FM. Self-recognition in colonial marine forms and flowering plants in relation to the evolution of immunity. Nature. 1971;232:123–126. doi: 10.1038/232230a0. [DOI] [PubMed] [Google Scholar]
- 14.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 15.Izzard CS. Migration of germ cells through successive generations of pallial buds in Botryllus schlosseri. BiolBull. 1968;135:424–31. [Google Scholar]
- 16.Sabbadin A, Zaniolo G. Sexual differentiation and germ cell transfer in the colonial ascidian Botryllus schlosseri. J Exp Zool. 1979;207:279–301. [Google Scholar]
- 17.Stoner DS, Rinkevich B, Weissman IL. Heritable germ and somatic cell lineage competitions in chimeric colonial protochordates Proc. Nat Acad Sci USA. 1999;96:9148–9153. doi: 10.1073/pnas.96.16.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buss LW. Somatic cell parasitism and the evolution of somatic tissue Proc Natl. Acad Sci USA. 1982;79:5337–5341. doi: 10.1073/pnas.79.17.5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buss L.W. The evolution of individuality Princeton University Press, Princeton, New Jersey (1987).
- 20.Owen RD. Immunologic consequence of vascular anastomoses between twins. Science. 1945;210:400–401. doi: 10.1126/science.102.2651.400. [DOI] [PubMed] [Google Scholar]
- 21.Mintz B, Russel ES. Gene-induced embryological modifications of primordial germ cells in the mouse. J Exp Zool. 1957;134:207–237. doi: 10.1002/jez.1401340202. [DOI] [PubMed] [Google Scholar]
- 22.Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci USA. 2004;101:3527–32. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–7. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schiffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search Programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tusnády GE, Simon I. Principles Governing Amino Acid Composition of Integral Membrane Proteins: Applications to Topology Prediction". J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 26.Puntervoll P. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts; amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1987;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson B, Swalla Bj. A molecular analysis of ascidian metamorphosis reveals activation of an innate immune response. Development. 2002;129:4739–4751. doi: 10.1242/dev.129.20.4739. [DOI] [PubMed] [Google Scholar]
- 29.Bassham S, Postlethwait J. Brachyury (T) expression in embryos of a larvacean urochordate, Oikopleura dioica, and the ancestral role of T. Dev Biol. 2000;220:322–332. doi: 10.1006/dbio.2000.9647. [DOI] [PubMed] [Google Scholar]
- 30.Cannon JP, Haire RN, Rast JP, Litman GW. The phylogenetic origins of the antigen-binding receptors and somatic diversification mechanisms. Immunol Rev. 2004;200:200–212. doi: 10.1111/j.0105-2896.2004.00166.x. [DOI] [PubMed] [Google Scholar]
- 31.Bottino C, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson WR, Lipman DJ. Improved Tools for Biological Sequence Comparison. Proc Natl Acad Sci USA. 1988;1998;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azumi K, et al. Genomic analysis of immunity in a basal chordate and evolution of the vertebrate immune system: waiting for Godot. Immunogenetics. 2003;55:570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.