Abstract
The assembly of iron-sulfur (Fe-S) clusters involves several pathways and in prokaryotes the mobilization of the sulfur (SUF) system is paramount for Fe-S biogenesis and repair during oxidative stress. The prokaryotic SUF system consists of six proteins: SufC is an ABC/ATPase that forms a complex with SufB and SufD, SufA acts as a scaffold protein, and SufE and SufS are involved in sulfur mobilization from cysteine. Despite the importance of Fe-S proteins in higher plant plastids, little is known regarding plastidic Fe-S cluster assembly. We have recently shown that Arabidopsis harbors an evolutionary conserved plastidic SufC protein (AtNAP7) capable of hydrolyzing ATP and interacting with the SufD homolog AtNAP6. Based on this and the prokaryotic SUF system we speculated that a SufB-like protein may exist in plastids. Here we demonstrate that the Arabidopsis plastid-localized SufB homolog AtNAP1 can complement SufB deficiency in Escherichia coli during oxidative stress. Furthermore, we demonstrate that AtNAP1 can interact with AtNAP7 inside living chloroplasts suggesting the presence of a plastidic AtNAP1·AtNAP6·AtNAP7 complex and remarkable evolutionary conservation of the SUF system. However, in contrast to prokaryotic SufB proteins with no associated ATPase activity we show that AtNAP1 is an iron-stimulated ATPase and that AtNAP1 is capable of forming homodimers. Our results suggest that AtNAP1 represents an atypical plastidic SufB-like protein important for Fe-S cluster assembly and for regulating iron homeostasis in Arabidopsis.
Iron-sulfur (Fe-S)1 clusters are important cofactors of Fe-S proteins involved in numerous vital biological processes in all organisms studied (1, 2). Although Fe-S clusters are derived from two of the most abundant elements on earth they are not formed spontaneously but arise by controlled biosynthesis requiring an intricate interplay of numerous proteins (3, 4). Most research on Fe-S cluster assembly has come from studies on bacteria, and it is clear that three bacterial systems exist termed NIF (nitrogen fixation), ISC (iron-sulfur cluster), and SUF (mobilization of sulfur) (5, 6). The NIF system is specifically involved in the assembly and maturation of Fe-S clusters in nitrogenase proteins found in nitrogen-fixing bacteria and ɛ-proteobacteria (7, 8), whereas the ISC system is more generally involved in the biosynthesis of numerous Fe-S proteins in both bacteria and higher eukaryotes (9–11).
The suf operon (sufABCDSE) represents the third Fe-S system and the relationship between the different Suf proteins during Fe-S cluster formation has recently been reported. SufB, SufC, and SufD seem to be evolutionary conserved and in bacteria SufC, which is an ATP binding cassette (ABC)/ATPase, forms a complex with SufB and SufD (6, 12, 13). Although it is thought that this SufC·SufB·SufD complex acts as an ATP-driven energizer during Fe-S cluster assembly the role of SufB and SufD remains unknown. SufE interacts with SufS involved in cysteine desulfuration to mobilize sulfur and although SufE is capable of forming homodimers, the binding of SufE to SufS stimulates the cysteine desulfurase activity of SufS ~50-fold (6). SufA acts as a scaffold protein involved in the assembly of Fe-S clusters and subsequent Fe-S cluster transfer to target apoproteins (14). It has been suggested that SufC is probably the most essential Suf protein in that SufC deficiency in E. coli results in a number of phenotypes related to oxidative stress and iron homeostasis, similar to mutants lacking the entire suf operon (13, 15).
Plant mitochondria harbor a Fe-S cluster biogenesis system involving the ABC protein Sta1 (16). Moreover, the recent observation that Arabidopsis harbors chloroplast-localized Nif proteins AtCpNIFS/AtNFS2, AtCnfU-V, and AtCnfU-IVb (17–19) and HCF101 (20) demonstrates that plant Fe-S cluster biogenesis occurs in both mitochondria and plastids. In chloroplasts Fe-S clusters are required for cytochrome b6/f complex, ferredoxin and photosystem I ensuring electron flow in the thylakoids (21, 22); however, to date Fe-S cluster biogenesis in plastid is poorly understood. As a step toward understanding SUF-mediated Fe-S cluster assembly in plastids we have recently provided evidence that Arabidopsis plastids most probably contain a complete SUF system (23). We have shown that the non-intrinsic ABC protein AtNAP7 is a functional ATPase and that it is a plastidic SufC-like protein (23). As in bacteria, AtNAP7 appears to be involved in the maintenance and repair of oxidatively damaged Fe-S clusters in that AtNAP7 can complement growth defects observed in SufC-deficient E. coli, because of damaged oxygen-labile Fe-S clusters, caused by oxidative stress. AtNAP7 clearly plays an essential role in plants as AtNAP7-deficiency in Arabidopsis leads to embryo lethality (23), suggesting that Fe-S proteins play vital roles during early stages of embryogenesis. Evidence for a SufC·SufD complex in plastids has come from protein interaction studies where we have shown that AtNAP7 can interact with the SufD homolog AtNAP6 (23). Because of the evolutionary conservation of the AtNAP7 mode of action and the fact that an AtNAP6·AtNAP7 complex exists we speculated that a SufB function may also be present in plastids.
We previously reported that the plastid localized LAF6 (AtABC1) protein showed good homology to annotated non-intrinsic ABC proteins from cyanobacteria and alga (24). As an extension of this analysis we show here that AtABC1 also exhibits high similarity to prokaryotic SufB proteins. Because of this and keeping in line with recent nomenclature of non-intrinsic ABC proteins (25) we have redesignated AtABC1 as AtNAP1. We demonstrate that AtNAP1 interacts with AtNAP7 inside chloroplasts, suggesting the presence of an AtNAP1·AtNAP7·AtNAP6 (SufB·SufC·SufD) complex in Arabidopsis plastids. We further show that AtNAP1 is able to complement the susceptibility of an E. coli SufB mutant to oxidative stress, suggesting that AtNAP1 is indeed an evolutionary conserved plastidic SufB protein. Despite this remarkable functional conservation, detailed biochemical characterization showed that in contrast to prokaryotic SufB proteins (12) AtNAP1 is an iron-stimulated ATPase. Furthermore, AtNAP1 is capable of forming homodimers, again in contrast to its bacterial counterparts. We suggest that depending on the interaction state of AtNAP1, diverse biological processes can be affected explaining the varied phenotypes observed in mutants deficient for either AtNAP1 (23) or AtNAP7 (24). From our findings we propose that AtNAP1 represents an atypical plastidic SufB protein in Arabidopsis involved in Fe-S cluster assembly and regulation of iron homeostasis.
EXPERIMENTAL PROCEDURES
Plasmid Construction for Protein Expression
All oligonucleotide primers used in this study are listed in Table I. A 1674-bp full-length AtNAP1 (At4g04770) cDNA was amplified by PCR using Pwo DNA polymerase (Roche Applied Science) and primers NAP1/1 and NAP1/2 and ligated into pRSETA (Invitrogen) to generate pRSETA-AtNAP1. To generate a non-functional AtNAP1 protein pRSETA-AtNAP1 was digested with EcoRV removing a 393-bp DNA fragment (nucleotide position 297 to 689 of AtNAP1 removing amino acids 99–229) resulting in pRSETA-AtNAP1Trun (Truncated AtNAP1). All constructs were verified by DNA sequencing.
Table I.
Sequence of oligonucleotides used for strains and plasmid constructions
| Oligonucleotide | 5′–3′ Sequence | |
|---|---|---|
| NAP1/1 | AAGGATCCATGGCGTCT CTTCTCGCAAA | BamHI underlined |
| NAP1/2 | AACTCGAGTTAACCCACTGATCCTTCAAGC | XhoI underlined |
| NAP1/4 | AAGAATTCATGGCGTCTCTTCTCGCAAA | EcoRI underlined |
| NAP1/5 | AAGGATCCTTAACCCACTGATCCTTCAAGC | BamHI underlined |
| 5NAP1GFP | TACTCGAGATGGCGTCTCTTCTCGCAAACGG | XhoI underlined |
| 3NAP1GFP | ATGGTACCACCCACTGATCCTTCAAGC | Kpn1 underlined |
| AtNAP1-F | GCGATAACTTGGAAGTACCC | |
| AtNAP1-R | GCTCTCTCGTGATCGATTCC | |
| NAP1-UC-L | ATTCTAGAGGCGTCTCTTCTCGCAAACG | XbaI underlined |
| NAP1-UC-R | ATGGTACCTTAACCCACTGATCCTTCAAGC | KpnI underlined |
| AtNAP7/1 | AAGAATTCATGGCCGGCGTTAACCTAC | EcoRI underlined |
| AtNAP7/2 | CTAACCGGATATCGCTTTGT | |
| AtSufC-L | AATCTCGAGATGGCCGGCGTTAACCTAC | XhoI underlined |
| AtSufC-R | AAGGTACCACCGGATATCGCTTTGAGCC | KpnI underlined |
| YnhEL | TCTCGTAATACTGAAGCAACTGACGATGTCAAAACCTGGAGTGTAGGCTGGAGCTGCTTC | |
| YnhER | CGCTGTGTTCAAGACTGATGGCGAGGAGTTTTTGTGCTTCCATATGAATATCCTCCTTAG |
Protein Expression and Purification
pRSETA-AtNAP1 and pRSETA-were transformed into E. coli strain BL21(DE3) and AtNAP1Trun 100-ml cultures were grown at 37 °C to a density of A600 = 0.6. Protein expression was induced using 1.5 mm isopropyl β-d-thiogalactoside for 3 h at 37 °C. To assess protein solubility 5 ml of each culture was harvested, resuspended in sodium phosphate buffer (50 mm sodium phosphate, 300 mm NaCl, pH 7.0) followed by sonication. The pellet and supernatant were separated by centrifugation, denatured in SDS loading buffer by heating to 95 °C, and used for SDS-PAGE analysis. Both AtNAP1 and AtNAP1Trun were insoluble and therefore dissolved with 1% Sarkosyl in sodium phosphate buffer and purified using Talon metal affinity resin (BD Biosciences) under denaturing conditions following the user manual for Talon metal affinity resins. Proteins were eluted by sodium phosphate buffer (45 mm sodium phosphate, 270 mm NaCl, and pH 7.0) containing 150 mm imidazole and the purity was verified by SDS-PAGE. Purified proteins were refolded by extended dialysis in dialysis buffer (50 mm sodium phosphate, 50 mm NaCl, 0.1 mm EDTA, 1.5 mm dithiothreitol, 10% glycerol, pH 7.2).
ATPase Assays
Each reaction mixture (40 μl) contained 50 mm Tris-Cl (pH 7.4), 50 mm NaCl, 0.1 mm EDTA, 1.5 mm dithiothreitol, 10 mm KCl, and 10–80 μm [γ-32P]ATP (specific activity 10 Ci/mmol) unless otherwise stated. 0.4 μm Protein was used in each reaction and all reactions were terminated using 1 μl of 1 m formic acid. For the time course experiments, 40-μl reactions were incubated at 37 °C for the specified time, aliquots were removed and spotted onto PEI-cellulose (POLYGRAM CEL 300 PEI, MACHEREY-NAGEL) TLC plates. Air-dried plates were developed using 0.5 m lithium chloride and 1 m formic acid. Radioactive nucleotides were visualized by autoradiography using x-ray film and for quantification purposes plates were scanned using a phosphorimager.
To assess the effect of different cations on the AtNAP1 ATPase activity, 20-μl reactions were incubated at 37 °C for 30 min with 10 μm [γ-32P]ATP, and 10 mm KCl was replaced with either 10 mm CaCl2, MnSO4, MnCl2, MgCl2 or 100 μm FeSO4. A no enzyme control was included to assess the background. To test the pH dependence of the ATPase activity, 20-μl reactions were incubated at 37 °C for 40 min using 50 mm Tris buffer (pH 6.0–9.0), 50 mm NaCl, 0.1 mm EDTA, 1.5 mm dithiothreitol, 10 mm KCl, and 10 μm [γ-32P]ATP. For iron concentration experiments, 10 mm KCl was replaced by different concentrations of FeSO4, together with 10 μm [γ-32P]ATP in 20-μl reaction volumes. The reactions were incubated for 30 min at 37 °C.
Generation of an E. coli SufB Mutant and Complementation by AtNAP1
This method was adapted from Ref. 26. The SufB gene in E. coli strain MG1655 was replaced by the insertion of a chloramphenicol resistance cassette (1.0-kb fragment containing the chloramphenicol resistant gene was PCR amplified with primers ynhEL and ynhER using pKD3 as template), followed by resistance cassette removal using pCP20 to give rise to MG1665ΔsufB. A full-length AtNAP1 cDNA was PCR amplified with primers NAP1-UC-L and NAP1-UC-R, digested with XbaI and KpnI, and ligated into pUC19. The following construct was transformed into MG1665ΔsufB to generate strain MG1665ΔsufBAtNAP1. Complementation analysis in the presence of phenazine methosulfate (PMS) was conducted as described previously (23).
Yeast Two-hybrid Analysis
A full-length AtNAP1 cDNA was PCR amplified using Pwo DNA polymerase (Roche) and primers NAP1/4 and NAP1/5 and ligated into pGBKT7 (DNA binding domain; BD) and pGADT7 (DNA activation domain; AD) separately (Clontech, Matchmaker 3) to generate pGBK-AtNAP1 and pGAD-AtNAP1. A 1017-bp full-length AtNAP7 cDNA was PCR amplified as above using primers AtNAP7/1 and AtNAP7/2 and ligated into the EcoRI and SmaI of pGBKT7 and pGADT7 separately to generate pGBK-AtNAP7 and pGAD-AtNAP7. All constructs were verified by DNA sequencing.
The above four vectors including empty vector controls (pGBKT7 and pGADT7) were transformed into yeast strain HF7c. The different transformation combinations are shown in Fig. 4A. Single transformants were grown on minimal synthetic dropout media (SD medium) either lacking tryptophan (Trp) (pGBKT7 vectors) or lacking leucine (Leu) (pGADT7 vectors). Double transformants were selected for SD medium lacking both Trp and Leu. To test for protein-protein interactions, fresh colonies were streaked onto SD medium plates containing 10 mm 3-aminotrizole but lacking Trp, Leu, and His and allowed to grow for 4–6 days at 30 °C. Yeast growth was classified into 4 categories based on restoration of His auxotrophy from three independent experiments: +++, growth after 2 days; ++, growth after 3 days; +, growth after 5–6 days; −, normal background growth.
Fig. 4. Yeast two-hybrid and in planta analysis of protein-protein interactions.

A, HF7c yeast cells co-transformed with different vector combinations were plated on SD medium lacking Leu and Trp and positive interactions were scored on plates containing SD medium lacking Leu, Trp, and His. All plates were grown for 5 days at 30 °C. Controls and classification of yeast growth are described under “Experimental Procedures.” B, bimolecular fluorescence complementation in living chloroplasts showing that AtNAP1 not only forms homodimers but also interacts with AtNAP7.
In Planta Interaction Studies/Bimolecular Fluorescence Complementation
A full-length AtNAP1 cDNA was PCR amplified using Pwo polymerase (Roche) with primers 5NAP1GFP and 3NAP1GFP and inserted into pWEN-C-YFP (containing amino acids 155–238 of YFP) and pWEN-N-YFP (containing amino acids 1–154 of YFP). A full-length AtNAP7 cDNA was PCR amplified with primers AtSufC-L and AtSufC-R and inserted in pWEN-C-YFP. Two combinations of these plasmids, pWEN-AtNAP1-N-YFP/pWEN-AtNAP1-C-YFP or pWEN-AtNAP1-N-YFP/pWEN-AtNAP7-C-YFP, were co-bombarded and transiently expressed in tobacco leaves (27). The leaves were placed in darkness for 2 days followed by YFP fluorescence visualization using a Nikon TE-2000U inverted microscope (Nikon, Japan) and image analysis was performed using Openlab software (Improvision).
Expression Analysis of AtNAP1 in Iron-starved Seedlings
Wild-type (Ler) Arabidopsis seedlings were grown on Murashige and Skoog plates for 3 weeks followed by transfer to either fresh Murashige and Skoog plates or to Murashige and Skoog plates lacking FeSO4/EDTA but containing 50 μm ferrozine. Seedlings were harvested 3 days after transfer and total RNA was extracted using the Sigma RNA extraction kit (Sigma). The RNA was DNase I (Sigma) treated and 2 μg of RNA was reverse transcribed (Stratagene) according to the manufacturer’s instructions. 1 μl of first-strand cDNA was used for PCR amplification (25 cycles) with primers AtNAP1-F and AtNAP1-R and products analyzed on a 2% agarose gel.
RESULTS
Sequence Analysis of AtNAP1
We previously reported the cloning and initial characterization of AtABC1, a plastid-localized 557-amino acid protein showing high similarity to annotated ABC-like proteins from cyanobacteria and alga (24). In agreement with our initial classification, subsequent in silico studies by Rea and colleagues (25) suggested that AtABC1 represents a soluble non-intrinsic ABC protein (NAP) belonging to a heterogeneous group of 15 single nucleotide binding fold proteins. In accordance with this new classification we now refer to AtABC1 as AtNAP1. Interestingly, more recent reports have suggested that AtNAP1 may represent a SufB homolog possibly involved in Fe-S cluster formation and/or repair (12, 28). In agreement with this we find that AtNAP1 shows ~60% similarity to SufB from both E. coli and Erwinia chrysanthemi (Fig. 1) indicating that AtNAP1 may indeed be a plastid-localized SufB protein. In bacteria, SufB physically interacts with SufC, an ABC/ATPase, and together with SufD this protein complex acts as an ATP-driven energizer for Fe-S cluster biogenesis and repair (9, 10). Although no ATPase activity has to date been demonstrated for prokaryotic SufB proteins, AtNAP1 contains degenerate Walker A and Walker B motifs (Fig. 1). Interestingly, both putative Walker domains are found in regions of limited homology between AtNAP1 and prokaryotic SufB proteins (Fig. 1) possibly indicating some functional divergence. Although AtNAP1 has been classified as a non-intrinsic ABC protein (25, 29) the absence of the conserved ABC signature motif suggests that AtNAP1 does not represent a classical ABC/ATPase but rather an ATPase with similarity to NAPs.
Fig. 1. Amino acid sequence alignment between AtNAP1 and SufB proteins from E. chrysanthemi (SufB Erw. CAC17125) and E. coli (SufB E. coli P77522).
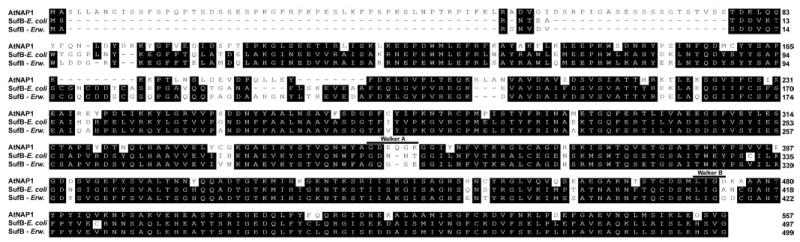
The degenerate Walker A and Walker B motifs are indicated.
Through extended BLAST searches of the Arabidopsis genome we discovered a second possible SufB gene (At5g44316) with high similarity to AtNAP1 (data not shown). Although the predicted amino acid sequence of At5g44316 shows 85% similarity to AtNAP1 we were unable to identify an At5g44316 transcript from extensive RT-PCR experiments and no EST has to date been deposited in any data base. This suggests that Arabidopsis contains only one SufB protein.
AtNAP1 Is an Fe2+-stimulated ATPase
Although AtNAP1 has been classified as a NAP with presumed ATPase activity we wanted to examine whether AtNAP1 could indeed bind and hydrolyze ATP. To this end we expressed a hexahistidine-tagged full-length AtNAP1 (WT) fusion protein in E. coli. Fractionation analysis showed that AtNAP1 was insoluble and the protein was therefore purified under denaturing conditions using Ni2+ affinity chromatography followed by refolding by dialysis. The purity of the refolded protein was analyzed by SDS-PAGE revealing a single protein band of the expected size (61 kDa) with no detectable contamination (Fig. 2A). Purified AtNAP1 protein (0.4 μm) was then incubated with radiolabeled [γ-32P]ATP at pH 7.4 and analyzed by thin layer chromatography for the release of radiolabeled inorganic phosphate (Pi). Analysis using two different protein batches showed clear AtNAP1-mediated ATP hydrolysis (Fig. 2B).
Fig. 2. Purified AtNAP1 and ATPase assays.
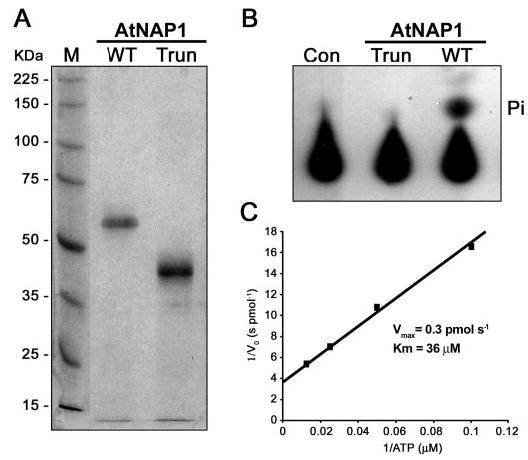
A, SDS-PAGE of purified full-length wild-type AtNAP1 (WT) and truncated AtNAP1 (Trun) stained with Coomassie Blue. B, autoradiography of ATP hydrolysis by purified wild-type AtNAP1 protein. Released radioactively labeled phosphate (Pi) is indicated. No Pi release is observed when using the purified truncated version (Trun) of AtNAP1. A no enzyme reaction is included as a control (Con). C, kinetic parameters of ATP hydrolysis by AtNAP1. A double-reciprocal plot of the rate of Pi formation (1/initial rate (Vo) s pmol−1) versus substrate concentration (1/[ATP] μm) is shown.
The AtNAP1 polypeptide was not present in the imidazole eluate from E. coli cells lacking the AtNAP1 expression plasmid when chromatographed in parallel with cells expressing At-NAP1 (data not shown). To ensure that the ATPase activity was attributable to AtNAP1 and not because of a contaminating E. coli protein, we expressed a truncated hexahistidine-tagged AtNAP1 fusion protein (AtNAP1Trun) lacking 131 amino acids (see “Experimental Procedures”) predicted to be nonfunctional in terms of ATPase activity. Similar to the full-length wild-type AtNAP1, the purified and refolded Atnap1trun protein (Trun) was analyzed by SDS-PAGE revealing a single protein band of the predicted size (47 kDa) with no detectable contamination (Fig. 2A). As expected no ATPase activity was detected when 0.4 μm Atnap1trun (Trun) was incubated with radiolabeled [γ-32P]ATP (Fig. 2B) demonstrating that AtNAP1Trun is non-functional in terms of ATPase activity but more importantly that the measured ATPase activity is attributable to AtNAP1.
To more fully characterize the catalytic activity of AtNAP1 we determined key kinetic parameters of AtNAP1-mediated ATP hydrolysis. Using input [γ-32P]ATP concentrations in the range of 10 to 80 μm We Quantified The Release Of Radiolabeled Pi as a function of time in separate time course experiments (Supplementary Materials Fig. 1). From the measured initial reaction rates we generated a double-reciprocal plot and calculated a Km of 36 μm ATP and a Vmax of 0.3 pmol s−1 (Fig. 2C Further kinetic analysis also showed that ATP hydrolysis was optimal at pH 7.5 (Fig. 3A) although hydrolysis under acidic conditions (pH 6–7) was significantly higher than in a more alkaline environment (pH 8–9).
Fig. 3. Kinetic and expression analysis of AtNAP1.

A, effect of pH on ATP hydrolysis by AtNAP1. Activity peaked at pH 7.5. B, the effect of cations on AtNAP1 ATPase activity. FeSO4 had a marked effect on AtNAP1-mediated ATP hydrolysis, whereas MnCl2, MnSO4, and KCl had a modest effect on activity. A no enzyme control (Con) is included. C, the effect of different FeSO4concentrations on AtNAP1 ATPase activity. The 0 FeSO4 reaction only contains 50 mM NaCl. D, semiquantitative RT-PCR analysis of AtNAP1 transcripts in seedlings grown in the presence (+Fe) and absence (−Fe) of iron. AtNAP1 was significantly down-regulated in seedlings subjected to iron starvation. Actin was used as a control.
As divalent cations have been shown to influence the ATPase activity of bacterial ABC/ATPases (30–32), we analyzed the effect of different cations on the ATP hydrolyzing ability of AtNAP1. Interestingly, addition of CaCl2 and MgCl2 had little effect on AtNAP1 activity, whereas MnCl2, MnSO4, and KCl lead to a moderate stimulation of activity (Fig. 3B). By contrast, the ATPase activity of AtNAP1 was dramatically stimulated by FeSO4 showing more than a 2-fold increase in ATPase activity compared with KCl and MgSO4 (Fig. 3B). More detailed analysis demonstrated further that FeSO4 supported AtNAP1 activity over a broad range of concentrations (5 3m to 5 mm) with an optimal FeSO4 concentration of 50 mM (Fig. 3C).
To gain further insight into the relationship between iron and the ATPase activity of AtNAP1 we tested whether the expression of AtNAP1 in Arabidopsis was influenced by the iron status. From our ATPase activity data (Fig. 3, B and C) we predicted that AtNAP1 may either be up-regulated in response to iron or down-regulated in response to iron starvation. Wild-type Arabidopsis seedlings were grown on Murashige and Skoog media for 3 weeks followed by transfer to fresh iron-containing Murashige and Skoog media or to media lacking iron. Semiquantitative RT-PCR analysis demonstrated that after 3 days of iron starvation the expression of AtNAP1 was significantly down-regulated (Fig. 3D). Interestingly, expression analysis of the Arabidopsis Suf homologs SufA, SufC, SufD, SufE, and SufS showed no substantial differences in expression upon iron starvation under our conditions (data not shown), highlighting the novel characteristics of AtNAP1. These combined results demonstrate that iron does not only stimulate AtNAP1 ATPase activity but also AtNAP1 expression at the transcriptional level.
AtNAP1 Can Form Homodimers and Interact with AtNAP7 in Yeast and in Planta
Most ABC proteins have been shown to act as dimers (33) and we therefore tested whether AtNAP1 could dimerize in the yeast two-hybrid system. We generated an AD-AtNAP1 construct and a BD-AtNAP1 construct and assayed for restoration of His auxotrophy in HF7c yeast cells co-transformed with both vectors. After a 2-day incubation period His auxotrophy was restored in cells containing both AD-AtNAP1 and BD-AtNAP1 demonstrating that AtNAP1 has the ability to dimerize (Fig. 4A). As a control for autoactivation and false His auxotrophy restoration we analyzed the growth of yeast cells containing either the empty BD or AD vectors in combination with AD-AtNAP1 and BD-AtNAP1, respectively. His auxotrophy was not restored using either of these vector combinations (Fig. 4A).
In prokaryotes it has been demonstrated that SufB interacts with SufC and that this protein complex is involved in Fe-S cluster formation and repair (12, 13). The finding that AtNAP1 shows high similarity to prokaryotic SufB proteins (Fig. 1) suggested that AtNAP1 may interact with the SufC homolog AtNAP7 in plastids. We have recently shown that the Arabidopsis ABC/ATPase AtNAP7 is a plastid-localized SufC protein essential for plant development (23) and we therefore assayed whether AtNAP1 could interact with AtNAP7 at the protein level. We generated both AD- and BD-AtNAP7 constructs and co-transformed HF7c yeast cells with these constructs together with either BD-AtNAP1 or AD-AtNAP1, respectively. After a 2-day incubation period His auxotrophy was restored in cells containing both constructs demonstrating that AtNAP1 can interact with AtNAP7 (Fig. 4A). As a control we analyzed the growth of yeast cells containing either the empty BD or AD vectors in combination with AD-AtNAP7 and BD-AtNAP7, respectively. His auxotrophy was not restored using either of these vector combinations (Fig. 4A). Interestingly, we found no evidence of His auxotrophy restoration for yeast cells expressing both AD-AtNAP7 and BD-AtNAP7 (Fig. 4A), suggesting that AtNAP7 is unable to form homodimers.
To test whether AtNAP1 and AtNAP7 protein interactions also occur in planta we conducted bimolecular fluorescence complementation studies (34) in living plant cells. We made use of the fact that separate, non-fluorescent N-terminal and C-terminal YFP protein domains can associate to form a functional fluorescent bimolecular complex when brought into proximity by interacting fusion proteins. To verify AtNAP1 dimerization we generated two plasmid constructs containing AtNAP1 either fused to a non-fluorescent N-terminal YFP polypeptide (amino acids 1–154; AtNAP1-N-YFP) or a C-terminal YFP polypeptide (amino acids 155–238; AtNAP1-C-YFP). Transient expression of co-transformed AtNAP1-N-YFP and AtNAP1-C-YFP in wild-type tobacco cells showed clear fluorescence demonstrating that AtNAP1 is able to form homodimers in living chloroplasts (Fig. 4B). To verify the observed AtNAP1/AtNAP7 interaction we generated a plasmid construct containing AtNAP7 fused to the C-terminal YFP polypeptide (amino acids 155–238; AtNAP7-C-YFP) and co-expressed this construct with AtNAP1-N-YFP in tobacco cells. As for the AtNAP1/AtNAP1 interaction studies we observed clear fluorescence in living chloroplasts thus further confirming that AtNAP1 and AtNAP7 can interact in planta (Fig. 4B).
AtNAP1 Complements Oxidative Stress Susceptibility in a ΔsufB E. coli Mutant
In bacteria SufB and SufC are necessary for the protection of enzymes with oxygen-labile Fe-S clusters (13). As a consequence SufB-deficient E. coli cells are unable to grow on minimal media under oxidative stress conditions. To test whether AtNAP1 represents a true SufB homolog in Arabidopsis we examined whether AtNAP1 could complement the growth defect in a SufB-deficient E. coli strain during oxidative stress. We generated a sufB knockout strain (MG1655ΔsufB) in E. coli and tested its growth characteristics compared with wild-type (MG1665). Under normal growth conditions both strains showed identical growth characteristics (Fig. 5A), whereas in the presence of the oxidative agent PMS, MG1655ΔsufB, unlike the wild-type strain, exhibited only limited growth (Fig. 5, A and B). We then tested the growth of MG1655ΔsufB expressing AtNAP1 (MG1655ΔsufBAtNAP1) in the presence of PMS. Under these conditions MG1655ΔsufBAtNAP1 showed similar growth characteristics to the wild-type MG1655 strain (Fig. 5, A and B) demonstrating that AtNAP1 can complement SufB deficiency in E. coli. These results demonstrate that AtNAP1 represents an evolutionary conserved SufB-like protein and that AtNAP1 has retained its ability to repair of oxidatively damaged Fe-S clusters.
Fig. 5. AtNAP1 can complement SufB deficiency in E. coli under oxidative stress.
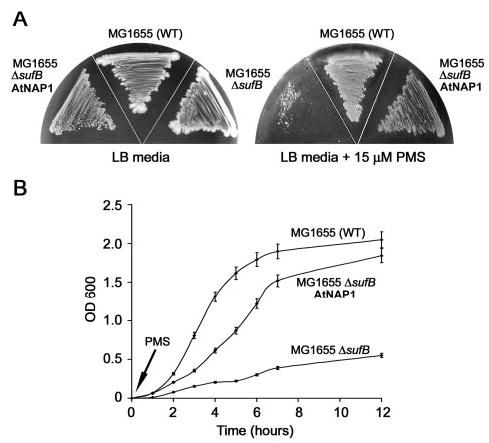
A, WT E. coli (MG1655), an E. coli SufB mutant (MG1655ΔsufB), and E. coli MG1655ΔsufB expressing AtNAP1 (MG1655ΔsufBAtNAP1) were grown on LB media and LB media supplemented with the oxidative agent PMS. All strains grew equally well on LB but in the presence of PMS strain MG1655ΔsufB exhibited limited growth. By contrast, MG1655ΔsufBAtNAP1 grew similar to WT. B, MG1655, MG1655ΔsufB, and MG1655ΔsufBAtNAP1 were grown in liquid culture. PMS was added and the growth of each strain measured (A600) over a 12-h time course. MG1655ΔsufB showed minimal growth after the addition of PMS. In contrast, after an initial lag period MG1655ΔsufBAtNAP1 showed similar growth characteristics to MG1655. Experiments were performed in triplicate and standard deviations are shown.
DISCUSSION
It is well established that the biosynthesis and repair of Fe-S clusters requires the coordinated interplay of numerous proteins (3, 4) and extensive studies in bacteria have revealed that Fe-S assembly involves three pathways termed ISC (iron-sulfur cluster), NIF (nitrogen fixation), and SUF (mobilization of sulfur) (5, 6). In plants, NIF (17–19), HCF101 (20), and NFU proteins (35) have been shown to be involved in Fe-S assembly and we recently suggested that Arabidopsis most probably also contains a complete plastid-localized SUF system (23). In support of this notion we have demonstrated that the essential ABC/ATPase AtNAP7 can restore growth defects in a SufC-deficient E. coli mutant demonstrating that Arabidopsis harbors an evolutionary conserved SufC protein (23). As SufC acts in concert with SufB in bacteria this further suggests the presence of a functional SufB protein in plants.
Here we show that the plastid-localized AtNAP1 protein in Arabidopsis has high similarity to prokaryotic SufB proteins (Fig. 1). In prokaryotes SufB interacts with SufC which in turn interacts with SufD (13) and it has been proposed that this SufB·SufC·SufD complex acts as an energizer, fueled by the ATPase activity of SufC, for iron acquisition, Fe-S cluster assembly, and/or transfer of Fe-S clusters to apoproteins (6). Interestingly, no ATPase activity has been associated with prokaryotic SufB proteins and their mode of action remains somewhat obscure (12). Although AtNAP1 can restore growth defects in SufB-deficient E. coli under oxidative stress conditions (Fig. 5), demonstrating clearly that AtNAP1 has retained its SufB function, we show here that AtNAP1, in contrast to its bacterial counterpart, has clear ATPase activity (Figs. 2 and 3). This suggests that although AtNAP1 represents an evolutionary conserved SufB protein involved in Fe-S cluster assembly, AtNAP1 has probably acquired additional roles during plant development.
As observed in prokaryotes we have demonstrated that AtNAP1 interacts with the Arabidopsis SufC homolog AtNAP7 inside living chloroplasts (Fig. 4) and from previous studies we have shown that AtNAP7 is involved in the maintenance and repair of oxidatively damaged Fe-S clusters (23). In addition, we have demonstrated that AtNAP7 interacts at the protein level with the SufD homolog AtNAP6 in Arabidopsis. This, together with the fact that AtNAP1 can efficiently complement SufB deficiency in E. coli under oxidative stress conditions (Fig. 5), suggests that an AtNAP1·AtNAP7·AtNAP6 (SufB·SufC·SufD) complex exists in plastids and furthermore, that this complex has retained its ability to repair damaged oxygen-liable Fe-S clusters. A recent study proposed that the SUF system in E. coli is adapted to synthesize and maintain Fe-S clusters when iron or sulfur metabolism is disrupted, for example, by oxidative stress, whereas the ISC system is responsible for housekeeping Fe-S cluster assembly (36). The oxidative stress that occurs in chloroplasts because of the production of reactive oxygen species may therefore have provided a driving force for the retention of a specialized prokaryotic-like Fe-S assembly/repair system within the chloroplast. It is also of interest to note that iron is an important co-factor of superoxide dismutase, which clears reactive oxygen species during oxidative stress in chloroplast and it is possible that the AtNAP1·AtNAP7·AtNAP6 protein complex is involved in ensuring superoxide dismutase activation by fueling the acquisition of iron as hypothesized for the SufC·SufB·SufD complex in bacteria (6).
If AtNAP1, as part of an AtNAP1·AtNAP7·AtNAP6 protein complex in plastids, contributes to iron acquisition, Fe-S cluster assembly, or transfer of Fe-S clusters to apoproteins it would be reasonable to assume that AtNAP1 activity could be regulated by iron. We have demonstrated here that the ATPase activity of AtNAP1 is strongly induced by iron (Fig. 3B) and that the optimal iron concentration for maximum AtNAP1 ATPase activity (Fig. 3C) lies within the physiological range of in planta iron concentrations. It has been speculated that in bacteria SufB and SufC may form subunits of a iron-dependent ABC transporter protein (12). Although it is tempting to speculate that AtNAP1 and AtNAP7 may act as interacting ATPase subunits involved in iron transport into or within the chloroplast and subsequently to sites of Fe-S biosynthesis and repair we have no evidence to support this notion. Based on our data we favor the idea that AtNAP1, in conjunction with AtNAP7, acts as a iron-stimulated sensor fueling Fe-S assembly and repair in response to changes in iron levels in plastids. Consistent with this notion AtNAP1 transcription is down-regulated in response to iron starvation in Arabidopsis (Fig. 3D), suggesting that AtNAP1 activity in response to iron is regulated both at the transcriptional and post-translational level.
Because AtNAP1 expression and activity appears sensitive to changes in iron concentration, AtNAP1 could act as a regulator of iron levels within plastids. With respect to this abnormal iron, homeostasis is one feasible explanation for the photomorphogenic phenotype and reduced chlorophyll content of AtNAP1-deficient laf6 mutant seedlings (24) as suggested previously (12). In laf6 chloroplasts compromised Fe-S cluster formation will lead to increased iron levels that in turn may shift the equilibrium between the heme and chlorophyll pathways during tetrapyrrole biosynthesis. The phenotype of laf6 may also be because of tetrapyrrole biosynthetic enzymes requiring Fe-S clusters. Together with the fact that tetrapyrrole intermediates are important for the communication between chloroplasts and the nucleus in plants, disordered tetrapyrrole biosynthesis caused by abnormal iron homeostasis may explain the photomorphogenic phenotype of AtNAP1-deficient plants (24).
Both previous data and findings presented here suggest that through its mode of action AtNAP1 ultimately affects several processes underpinned by its involvement in Fe-S cluster assembly/repair. It is well documented that ABC/ATPases are promiscuous and that their mode of action often depends on their interacting protein partner (33). We have shown that AtNAP1 cannot only interact with AtNAP7 forming an energizer complex for Fe-S cluster assembly and repair but also that AtNAP1 can form homodimers (Fig. 4). In prokaryotes SufB does not dimerize as part of Fe-S cluster assembly (13) and this again underlines the different properties of AtNAP1 compared with its prokaryotic counterpart. Based on these data it is possible that AtNAP1 homodimers have a different mode of action compared with an AtNAP1·AtNAP7·AtNAP6 protein complex and this may explain the different phenotypic effects observed between plants deficient for AtNAP1 and AtNAP7. Further studies are needed to understand the exact assembly combinations of AtNAP1, AtNAP7, and AtNAP6 and how these different subcomplexes affect diverse biological processes in plants.
It is clear that the plastidic SUF system in plants has not only retained key prokaryotic functions but has acquired new properties. At present therefore we can only speculate on the exact mechanism by which the plastidic SUF system operates in Arabidopsis. However, our analysis of AtNAP1 has increased our understanding of the SUF system and laid a solid foundation for future studies.
Supplementary Material
Acknowledgments
We thank Clive R. Bagshaw for advice on enzyme kinetics. We also thank the E. coli Genetics Stock Center at Yale University (CGSC) for providing MG1655/pKD46, BW25141/pKD3, and BT340/pCP20.
Footnotes
This work was supported in part by the Biotechnology and Biological Sciences Research Council Grants 91/P16510, 91/C17189, and 91/REI18421 (to S. G. M.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The abbreviations used are: Fe-S, iron-sulfur; SUF, mobilization of sulfur; ABC, ATP-binding cassette; NIF, nitrogen fixation; ISC, iron-sulfur cluster; LAF6, long after far-red 6; NAP, non-intrinsic ABC protein; YFP, yellow fluorescence protein; PMS, phenazine methosulfate; BD, DNA binding domain; AD, activation domain; RT, reverse transcriptase.
References
- 1.Beinert H, Holm RH, Munck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Beinert H, Kiley PJ. Curr Opin Chem Biol. 1999;3:152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 3.Lill R, Kispal G. Trends Biochem Sci. 2000;25:352–356. doi: 10.1016/s0968-0004(00)01589-9. [DOI] [PubMed] [Google Scholar]
- 4.Frazzon J, Dean DR. Curr Opin Chem Biol. 2003;7:166–173. doi: 10.1016/s1367-5931(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi Y, Tokumoto U. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 6.Loiseau L, Ollagnier-de-Choudens S, Nachin L, Fontecave M, Barras F. J Biol Chem. 2003;278:38352–38359. doi: 10.1074/jbc.M305953200. [DOI] [PubMed] [Google Scholar]
- 7.Jacobsen MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Mol Gen Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 8.Olson JW, Agar JN, Johnson MK, Maier RJ. Biochemistry. 2000;39:16213–16219. doi: 10.1021/bi001744s. [DOI] [PubMed] [Google Scholar]
- 9.Zheng L, Cash VL, Flint DH, Dean DR. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi Y, Nakamura MJ. Biochem J (Tokyo) 1999;126:917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. Proc Natl Acad Sci U S A. 2000;97:9009–9014. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rangachari K, Davis CT, Eccleston JF, Hirst EM, Saldanha JW, Strath M, Wilson RJ. FEBS Lett. 2002;514:225–228. doi: 10.1016/s0014-5793(02)02369-4. [DOI] [PubMed] [Google Scholar]
- 13.Nachin L, Loiseau L, Expert D, Barras F. EMBO J. 2003;22:427–437. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ollagnier-de-Choudens S, Lascoux D, Loiseau L, Barras F, Forest E, Fontecave M. FEBS Lett. 2003;555:263–267. doi: 10.1016/s0014-5793(03)01244-4. [DOI] [PubMed] [Google Scholar]
- 15.Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F. Mol Microbiol. 2001;39:960–972. doi: 10.1046/j.1365-2958.2001.02288.x. [DOI] [PubMed] [Google Scholar]
- 16.Kushnir S, Babiychuk E, Storozhenko S, Davey MW, Papenbrock J, De Rycke R, Engler G, Stephan UW, Lange H, Kispal G, Lill R, Van Montagu M. Plant Cell. 2001;13:89–100. doi: 10.1105/tpc.13.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilon-Smits EA, Garifullina GF, Abdel-Ghany S, Kato S, Mihara H, Hale KL, Burkhead JL, Esaki N, Kurihara T, Pilon M. Plant Physiol. 2002;130:1309–1318. doi: 10.1104/pp.102.010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leon S, Touraine B, Briat JF, Lobreaux S. Biochem J. 2002;366:557–564. doi: 10.1042/BJ20020322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yabe T, Morimoto K, Kikuchi S, Noshio K, Terashima I, Nakai M. Plant Cell. 2004;16:993–1007. doi: 10.1105/tpc.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lezhneva L, Amann K, Meurer J. Plant J. 2004;37:174–185. doi: 10.1046/j.1365-313x.2003.01952.x. [DOI] [PubMed] [Google Scholar]
- 21.Raven JA, Evans MCW, Korb RE. Photosynth Res. 1999;60:111–150. [Google Scholar]
- 22.Kapazoglou A, Mould RM, Gray JC. Eur J Biochem. 2000;267:352–360. doi: 10.1046/j.1432-1327.2000.01006.x. [DOI] [PubMed] [Google Scholar]
- 23.Xu XM, Møller SG. Proc Natl Acad Sci U S A. 2004;101:9143–9148. doi: 10.1073/pnas.0400799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Møller SG, Kunkel T, Chua NH. Genes Dev. 2001;15:90–103. doi: 10.1101/gad.850101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Fernandez R, Davies TG, Coleman JO, Rea PA. J Biol Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kost B, Spielhofer P, Chua NH. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- 28.Ellis KE, Clough B, Saldanha JW, Wilson RJ. Mol Microbiol. 2001;41:973–981. doi: 10.1046/j.1365-2958.2001.02588.x. [DOI] [PubMed] [Google Scholar]
- 29.Garcia O, Bouige P, Forestier C, Dassa E. J Mol Biol. 2004;343:249–265. doi: 10.1016/j.jmb.2004.07.093. [DOI] [PubMed] [Google Scholar]
- 30.Morbach S, Tebbe S, Schneider E. J Biol Chem. 1993;268:18617–18621. [PubMed] [Google Scholar]
- 31.Nikaido K, Liu PQ, Ames GF. J Biol Chem. 1997;272:27745–27752. doi: 10.1074/jbc.272.44.27745. [DOI] [PubMed] [Google Scholar]
- 32.Sarin J, Aggarwal S, Chaba R, Varshney GC, Chakraborti PK. J Biol Chem. 2001;276:44590–44597. doi: 10.1074/jbc.M105401200. [DOI] [PubMed] [Google Scholar]
- 33.Holland IB, Blight MA. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- 34.Walter M, Chaban C, Schutze K, Batistic O, Weckerman K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 35.Leon S, Touraine B, Ribot C, Briat JF, Lobreaux S. Biochem J. 2003;371:823–830. doi: 10.1042/BJ20021946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Outten FW, Djaman O, Storz G. Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


