Abstract
Molecular motors move unidirectionally along polymer tracks, producing movement and force in an ATP-dependent fashion. They achieve this by amplifying small conformational changes in the nucleotide-binding region into force-generating movements of larger protein domains. We present the 2.8 Å resolution crystal structure of an artificial actin-based motor. By combining the catalytic domain of myosin II with a 130 Å conformational amplifier consisting of repeats 1 and 2 of α-actinin, we demonstrate that it is possible to genetically engineer single-polypeptide molecular motors with precisely defined lever arm lengths and specific motile properties. Furthermore, our structure shows the consequences of mutating a conserved salt bridge in the nucleotide-binding region. Disruption of this salt bridge, which is known to severely inhibit ATP hydrolysis activity, appears to interfere with formation of myosin’s catalytically active ‘closed’ conformation. Finally, we describe the structure of α-actinin repeats 1 and 2 as being composed of two rigid, triple-helical bundles linked by an uninterrupted α-helix. This fold is very similar to the previously described structures of α-actinin repeats 2 and 3, and α-spectrin repeats 16 and 17.
Keywords: α-actinin/Dictyostelium discoideum/lever arm/myosin/protein engineering
Introduction
As more high-resolution protein structures become available, possibilities for protein engineering are becoming increasingly feasible. Perhaps the most common example of such directed manipulation of protein structure and function is the mutation of amino acid side chains in order to achieve a particular effect; for example, more efficient enzymatic catalysis, increased protein stability or altered substrate specificity. Other examples of protein engineering include the addition of protein tags for purposes of protein purification or in vivo localization. The size and nature of these tags can range from the small poly-histidine tag used in Ni2+-affinity purification, to entire proteins, such as the use of green fluorescent protein to visualize the location of proteins inside living cells. More complicated examples of protein engineering are less common. One of the more interesting examples used an engineered complex between the F1-ATPase and an actin filament in order to visualize the rotary motion of the F1-ATPase (Noji et al., 1997). In another study, spectroscopic labels were bound to myosin light chains in a specific orientation in order to measure precise movements of the myosin lever arm during force production (Corrie et al., 1999). Yet another approach to protein engineering is the combination of functional and/or structural domains from different proteins in order to achieve new, altered or enhanced activities. In this study, we report the structure of an active molecular motor created by fusing the motor domain of myosin to an artificial lever arm composed of α-actinin. This engineered motor, while having similar kinetic activities to wild-type myosin, has specifically altered velocity and step size (Anson et al., 1996).
The myosin II lever arm consists of an ∼75 Å-long α-helix in complex with two calmodulin-like light chains (Rayment et al., 1993). It has been shown that myosin’s light chain binding domain can be replaced with one, two or three spectrin-like repeats (Manstein and Hunt, 1995). As each spectrin-like repeat is composed of a triple-helical bundle that forms an elongated, structurally rigid domain (Djinovic-Carugo et al., 1999), it is an ideal ‘building block’ to use when designing engineered proteins. In this study, we have determined the structure of the catalytic domain of Dictyostelium myosin II fused to 240 amino acids (266–505) of Dictyostelium α-actinin (Anson et al., 1996). In muscle and non-muscle cells, α-actinin is involved in crosslinking actin filaments to various cytoskeletal structural elements (Blanchard et al., 1989; Parry et al., 1992; Gilmore et al., 1994; Djinovic-Carugo et al., 1999). Functioning in vivo as a homodimer, α-actinin contains an N-terminal actin-binding domain, a central rod domain consisting of four 120 residue spectrin-like repeats, and a C-terminal Ca2+-binding domain. Due to the stability of the α-actinin domain, the artificial lever used in this study should form an elongated, rigid structure, similar in character to the natural myosin lever arm. Previous studies have shown this engineered motor to have normal kinetics and motile activities (Kurzawa et al., 1997); however, determination of its high-resolution structure was necessary in order to unambiguously determine the structure and conformation of the artificial lever.
Myosin motors, the related kinesin motors and the G-protein family of molecular switches contain three conserved structural and sequential motifs in the region surrounding the nucleotide-binding site (Kull et al., 1998; Vale and Milligan, 2000). These motifs include the P-loop (conserved sequence GxxxxGKS/T), which binds to the α- and β-phosphates of the nucleotide, and switch I (NxxSSR) and switch II (DxxGxE), which sense the presence or absence of γ-phosphate. In myosin crystal structures, the relative positions of switch I and switch II change, switching between open and closed conformations (Fisher et al., 1995; Smith and Rayment, 1995; Rayment, 1996; Dominguez et al., 1998; Houdusse et al., 1999). This conformational change is linked to movement of the lever arm. When the switch region is closed, the lever arm is up, in the pre-power-stroke state. When the switch region is open, however, the lever arm and adjacent converter domain undergo a rigid-body 70° rotation to the down, post-power-stroke conformation (Geeves and Holmes, 1999). One feature of the closed conformation is the formation of a conserved salt bridge between an arginine residue in switch I and a glutamic acid residue in switch II. The formation of this salt bridge is essential for nucleotide hydrolysis and normal motility in both myosin and kinesin motors (Onishi et al., 1998; Furch et al., 1999; Rice et al., 1999; Endow, 2000). Mutation of Arg238 in myosin II to a glutamic acid residue reduces the basal ATPase rate 200-fold, and eliminates both actin activation of the ATPase and in vitro motility (Furch et al., 1999). Restoration of the salt bridge, by complementing the R238E mutant with Glu459 mutated to arginine, returns the basal ATPase, actin-stimulated ATPase, and in vitro motility to near wild-type levels (Furch et al., 1999). In order to demonstrate that elimination of the salt bridge prevents formation of the closed structure, we crystallized the R238E mutant of M761-2R in the presence of Mg-ADP-VO4.
The molecular motor presented in this paper represents a successful attempt at large-scale protein engineering. By attaching an artificial lever to the core region of a myosin, an active motor protein is produced, which has similar kinetic properties to wild-type myosin. However, as the length of the force-generating lever arm can be accurately changed, artificial motors can be produced with new motile properties that are optimized for specific in vitro or in vivo tasks.
Results and discussion
Overall structure
The fusion protein was expressed in a Dictyostelium expression system and purified following the normal procedure for Dictyostelium myosin head fragments (see Materials and methods for details). Briefly, cells were lysed and the ATP-depleted pellet of fusion protein and cytoskeletal elements was washed and resuspended. The addition of ATP then released active protein, which was subsequently purified by Ni2+-affinity chromatography. Once pure, the protein crystallized under conditions similar to that of the independent myosin motor domain. The structure was determined by molecular replacement methods, using Dictyostelium myosin residues 2–759 as a search model. The initial electron density map clearly showed the first α-actinin repeat. Following several rounds of crystallographic refinement, the second repeat was visible. The crystallographic data are presented in Table I.
Table I. Crystallographic data statistics.
| Data collection and phase determination by molecular replacement method | |
| Crystal space group | P21212 |
| Unit cell parameters | a = 135.42 Å |
| b = 155.42 Å | |
| c = 143.19 Å | |
| |
α = β = γ = 90° |
| Parameter |
Native data |
| Resolution (Å)a | 10.0–2.8 (2.9–2.8) |
| Wavelength (Å) | 0.784 |
| Completeness (%) | 97.7 (98.0) |
| Unique reflections | 73355 |
| Redundancy | 3.8 (3.9) |
| Rsym (%)b | 11.0 (32.4) |
| I/σ | 8.88 (3.09) |
| Refinement statistics | |
| resolution(Å) | 10.0–2.8 |
| reflections (work set/test set) | 67294/5989 |
| protein atoms | 16214 |
| ligand atoms | 56 |
| water molecules | 14 |
| average B factor (Å2) | 52.7 |
| Rwork (%)c | 23.2 |
| Rfree (%)d | 29.0 |
aValues in parentheses correspond to the highest resolution shell.
bRsym = ΣhΣi|I(h) – Ii(h)|/ΣhΣiIi|(h), where Ii(h) and I(h) are the ith and mean measurements of the intensity of reflection h.
cRwork = Σh|Fo – Fc<_UNICODE:9474>|/ΣhFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h.
dRfree is the same as Rwork, but calculated on the ∼9% of the data excluded from refinement.
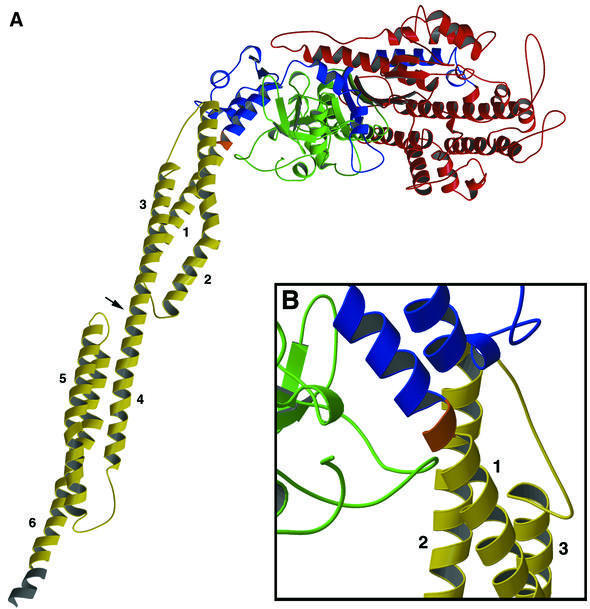
The structure of the M761-2R-R238E construct contains 1005 amino acid residues. The structure of the myosin motor domain is similar to those previously described in a number of different nucleotide conformations (Rayment et al., 1993; Fisher et al., 1995; Gulick et al., 1997), with the α-actinin lever arm leaving myosin’s converter domain in a manner identical to the wild-type lever arm (Figure 1A). The artificial lever arm in this study consists of repeats 1 and 2 of Dictyostelium α-actinin, each repeat being 65 Å long and 20 Å in diameter. A His8 purification tag extends the lever arm by 10 Å, resulting in a total length of 140 Å. Similar to the structure of the human α-actinin (Djinovic-Carugo et al., 1999), both repeats fold into a triple helix and the two repeats are connected by a continuous helical linker (Figure 1A). Remarkably, the engineered linker connecting myosin’s converter domain to α-actinin also forms an uninterrupted, structurally sound α-helix (Figure 1B). Although the crystallographic asymmetric unit contained two molecules, analysis of the various crystal contacts did not reveal any biologically significant dimerization region.
Fig. 1. Structure of M761-2R-R238E. Although two molecules are present in the crystallographic asymmetric unit, only one is shown here. The two molecules are essentially identical throughout the myosin motor domain (residues 2–761). However, upon leaving the converter domain, the lever arms assume slightly different orientations and deviate at the ends by 19.4 Å. (A) A complete molecule spanning amino acids 2–1010 is shown. No electron density was observed for five residues at the N-terminus, the loop region 205–208 and one residue at the C-terminus. The N-terminal domain (2–200) is shown in green; 50 kDa domain in red (201–613); C-terminal and converter domain in blue (614–761); linker region in orange (762–764); α-actinin lever arm in yellow (765–1003); and seven histidines from the His8 purification tag in gray (1004–1010). The linker region is composed of three residues (Leu-Gly-Arg) introduced during cloning. The observed lever arm is ∼140 Å long (measured from Cα of 761 to Cα of 1010). Each α-actinin repeat contributes ∼65 Å, and the histidine purification tag another 10 Å. Helices 1–3 make up the first α-actinin repeat, and 4–6 the second. The arrowhead indicates the α-helical region linking the two repeats. The disruptive kink in helix 2 is caused by the presence of two adjacent proline residues (see Figure 4A). (B) Detailed view of the linker region joining the myosin converter domain to helix 1 of α-actinin. The view is rotated 180° around a vertical axis from that in (A).
Salt bridge mutation
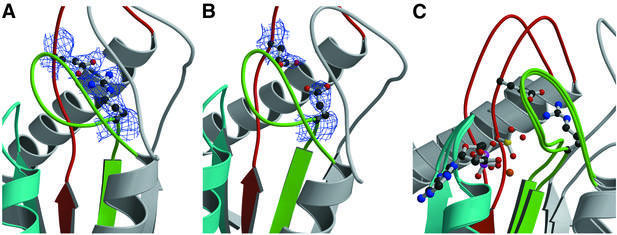
Existing crystal structures show that Mg-ADP-VO4 should mimic a transition state analog and produce a closed myosin conformation (Figure 2A) (Smith and Rayment, 1996). The crystal structure of the R238E mutant is clearly in the open form (Figure 2B). As previous kinetic studies indicate that the salt bridge is critical for the formation of the closed state, this mutant should never be able to form a stable, closed conformation. VO4 is not observed in the nucleotide-binding site. The absence of VO4 is likely to be a result of the inability of the open myosin conformation to bind the transition state complex. In the closed conformation, binding of VO4 or BeF3 is stabilized by a hydrogen bond to the amide nitrogen of Gly457 in the switch II loop. In the open structure, this loop moves away from the nucleotide by ∼5 Å, precluding formation of this hydrogen bond (Figure 2C) and perhaps destabilizing binding of VO4.
Fig. 2. Detailed view of the conserved salt bridge linking switch I and switch II. The conserved nucleotide binding/sensing elements found in all myosins, kinesins and G-proteins are highlighted for the P-loop in blue, switch I in green and switch II in red. (A) The structure of Dictyostelium myosin II motor complexed with Mg-ADP-BeF3 (F.J.Kull and K.C.Holmes, unpublished results). As in Mg-ADP-VO4 (Smith and Rayment, 1996) and Mg-ADP-BeF3 (Dominguez et al., 1998) structures, switch I and switch II are closed. The conserved salt bridge between residues R238 and E459 is shown as a ball-and-stick model surrounded by 2.6 Å experimental 2Fo – Fc electron density (blue wire-frame), contoured at 1σ. As expected for a salt bridge, the electron density is continuous between the residues, which point toward each other. (B) The same region as observed in the crystal structure of M761-2R-R238E. The electron density was calculated from a model with alanines at positions 238 and 459 in order to eliminate model bias. Electron density for two glutamic acid residues is clearly visible, but the side chain of E238 now points away from E459 and the switch II loop has moved away from switch I. (C) The same region showing a superposition of the M761-2R-R238E structure with a structure of Dictyostelium myosin II motor complexed with Mg-ADP-VO4 (PDB code 1VOM) (Smith and Rayment, 1996). The nucleotide and R238–E459 salt bridge are shown as ball-and-stick models. Both the P-loop and switch I regions are in essentially identical conformations in both structures. However, the switch II region (red) shifts to the right, towards the nucleotide, by ∼5 Å in the Mg-ADP-VO4 structure, allowing the formation of the R238–E459 salt bridge.
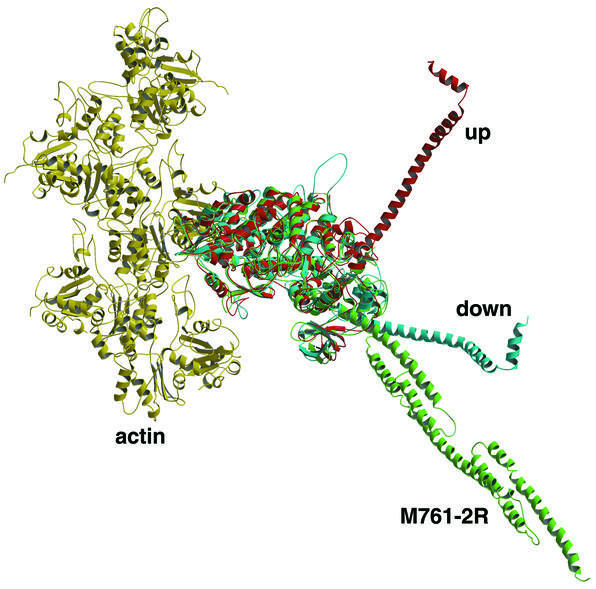
A further consequence of the disrupted salt bridge in the switch I/II region is seen in the position of the α-actinin lever arm. Comparison with lever-up and lever-down structures shows the artificial lever arm in M761-2R to be in the down position, as expected with an open switch region (Figure 3). Although the M761-2R lever arm helix emerges from the converter domain in a position and an orientation identical to that of wild-type myosin, the position of the far end of the lever differs slightly due to its α-helical bend. As the bending of helix 1 in the first α-actinin repeat dictates the position of the end of the lever arm, changing the helical register in the motor-lever linker region would allow accurate repositioning of the C-terminal end of the lever arm.
Fig. 3. Orientation of the myosin lever arm. Five molecules of actin making up part of a helical actin filament are shown in yellow. Modeled on to this are myosin in the ‘pre-power-stroke’ up/closed orientation in red, the ‘post-power-stroke’ down/open orientation in blue, and the M761-2R-R238E structure in green. The up, down and actomyosin complex structures were modeled as described previously (Geeves and Holmes, 1999). The M761-2R-R238E structure was then aligned to the core domain of the down/open structure via residues 160–200, which includes the highly conserved P-loop region. Note that in the M761-2R-R238E structure, the helix leaving the converter domain initially superposes with the down/open structure, but then deviates due to the different helical bend of the α-actinin.
Structure of α-actinin
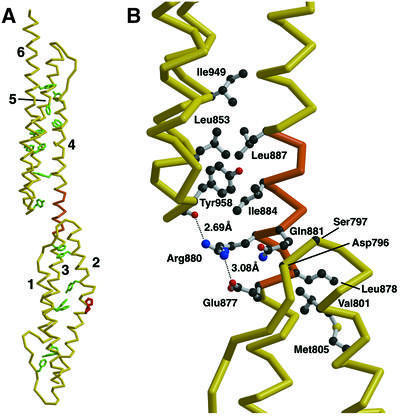
A high-resolution structure has recently been determined for repeats 2 and 3 of the rod domain of human skeletal muscle α-actinin (Djinovic-Carugo et al., 1999), and repeat 16 and 17 of human α-spectrin (Grum et al., 1999). Comparison with Dictyostelium α-actinin repeats 1 and 2 shows a similar fold consisting of two triple helical repeats connected by an α-helical linker. Interestingly, helix 2 in repeat 1 contains two adjacent proline residues, which produce a sharp kink in the helix without actually breaking it (Figure 4A). Within each repeat, the three α-helices are held together via hydrophobic contacts along their entire lengths. This hydrophobic core is remarkable as it is made up not only of smaller residues common in leucine zipper motifs (leucine, isoleucine and valine), but also of 17 large, aromatic side chains (phenylalanine, tyrosine and tryptophan) (Figure 4A). The huge surface area buried by these hydrophobic contacts contributes greatly to the structural rigidity of the α-actinin structure.
Fig. 4. The structure of α-actinin repeats 1 and 2. (A) An α-carbon chain trace of the six helices making up repeats 1 (helices labeled 1–3) and 2 (helices labeled 4–6) is shown in yellow. The 17 hydrophobic aromatic amino acid residues stabilizing the triple-helical packing are shown in green (seven tyrosines, six phenylalanines and four tryptophans). Two adjacent proline residues are shown in red, which cause a kink but not a break in α-helix 2 of repeat 1. The uninterrupted α-helix linking repeats 1 and 2 is shown in orange. (B) Detailed view of the linker region, highlighting the stabilizing hydrophobic and hydrogen bonding interactions. Colors and orientation are identical to those in (A). Side chains are shown as ball-and-stick models, with the exception of Asp796 and Ser797, in which only the α-carbon atoms involved in hydrophobic contacts are shown for clarity. The salt bridge between Arg880 and Glu877, and the hydrogen bond between Arg880 and the carbonyl oxygen of Leu956 (also shown as a ball-and-stick model), are shown as dashed lines.
The linker connecting repeats 1 and 2 of the Dictyostelium α-actinin is also similar to the linkers in human α-actinin and human α-spectrin. The region maintains an uninterrupted α-helix, stabilized by a number of interactions between the linker region and adjacent loop regions. Although these interactions are predominantly hydrophobic, Arg880 is involved in two stabilizing hydrogen bonds, which contribute to the helical stability of this region (Figure 4B). One of these interactions is an intra-helical salt bridge between Arg880 and Glu877. As these two residues are one helical turn apart from each other, in order for the salt bridge to form, the protein chain must adopt an α-helical conformation. Additionally, Arg880 forms a hydrogen bond with the carbonyl oxygen of Leu956, a residue located in the loop region connecting helices 5 and 6 in repeat 2. Although a bond of this type is less strong than the salt bridge, it nevertheless contributes to the overall stability and fold of the linker region.
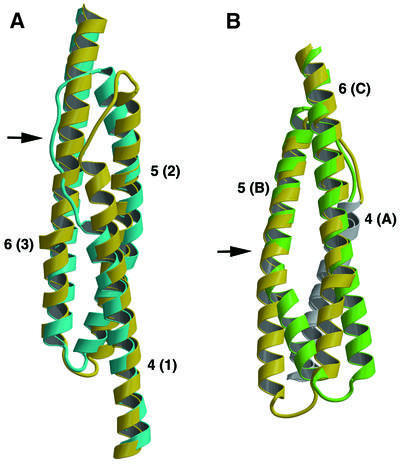
The overall fold of Dictyostelium α-actinin is similar to that of human α-actinin. However, a detailed comparison of the overlapping repeat 2 regions shows significant differences. Although most of helix 4 and all of helix 5 overlap closely with the human α-actinin structure, the position of the connecting loop differs, resulting in an overlap of 14 amino acid residues from the human α-actinin loop 4–5 with helix 6 from Dictyostelium (Figure 5A). Furthermore, beyond the 10 amino acid residues at the beginning of helix 6, the positions of these helices are substantially different. Comparison of our structure with repeat 16 of α-spectrin (Grum et al., 1999) shows a much higher degree of structural similarity, with all three helices, as well as the loop 4–5 region, superposing. The major difference between these two structures is in the loop region connecting helices 5 and 6, where the α-spectrin structure, due to a proline-induced kink in helix 5, is shifted in respect to the Dictyostelium structure (Figure 5B).
Fig. 5. Comparison of Dictyostelium α-actinin with human α-actinin and human α-spectrin. (A) The overlapping repeat 2 region of Dictyostelium (yellow) and human (blue) α-actinin are shown as ribbon diagrams. Helices are numbered as described above for Dictyostelium α-actinin and, in parentheses, as described previously for human α-actinin (Djinovic-Carugo et al., 1999). The largest differences occur in the loop region connecting helices 4 and 5, indicated by an arrow, where the human α-actinin structure would seriously overlap with Dictyostelium helix 6. (B) The alignment of Dictyostelium repeat 2 (yellow) with repeat 16 human α-spectrin (green) is shown as ribbon diagrams. Helices are numbered as described above for the Dictyostelium protein and, in parentheses, as described previously for the human protein (Grum et al., 1999). Dictyostelium helix 4 and α-spectrin helix A, which are in the background, are colored white for clarity. In general, the two structures align more closely than the human/Dictyostelium alignment described above. The largest difference occurs in the loop region connecting helices 5 and 6, indicated by an arrow, where the human α-spectrin structure is moved in respect to the Dictyostelium α-actinin structure as a result of a proline-induced kink in helix B.
Structural and functional characterization
The structure of the artificial lever determined in this paper correlates very well with the known motile properties of these motor constructs. Previous functional studies have shown a linear relationship between lever arm length and in vitro sliding velocity in artificial motors with levers composed of one or two α-actinin repeats (Anson et al., 1996). As ATP turnover remained essentially constant in these constructs, the increased velocity was attributed to a longer lever arm moving at a constant cycle rate. Recently, single molecule measurements have been used to measure directly the step size of myosin constructs with a variety of natural and engineered lever arm lengths (Ruff et al., 2001). Again, a linear relationship was observed between step size and lever arm length for both wild-type and engineered lever arms. Models based on these experiments predict the total length of the lever arms to be 8 or 14 nm for motors containing either one or two α-actinin repeats, respectively. This length corresponds to ∼2 nm of the lever contained within the myosin plus one or two 6 nm α-actinin repeats. The actual length of the artificial lever visualized in this study verifies these experimental predictions, with the two α-actinin repeats making up 13 nm of a 14 nm long lever. It is clear that removing single α-actinin repeat would shorten the lever arm by 6.5 nm. If this shorter lever were coupled to an identical rotational displacement, the predicted step size would be ∼55% of that of the dual repeat construct. This is in close agreement with the measured step size ratio of ∼53% (Ruff et al., 2001).
A system for rapid structure determination
With the rapidly accelerating number of sequenced genomes, there is a growing backlog of high-resolution crystal structures that need to be determined (Burley et al., 1999; Service, 2000). The myosin fusion system described here is likely to be useful in this endeavor. As myosin II is produced in large quantities in wild-type Dictyostelium, overexpression of recombinant myosin is extremely high, resulting in amounts of 10–50 mg of protein per liter of expressing cells. As the myosin catalytic domain retains wild-type activity when expressed in this manner, fusion proteins can be purified by binding to the endogenous actin found in Dictyostelium. Correctly folded protein can then be released from the actin via the addition of ATP, myosin’s natural substrate. Although purity of the protein is quite high following the release step, subsequent Ni2+-affinity purification via an engineered poly-histidine tag produces protein of purity suitable for crystallization.
The conditions for crystallizing the catalytic domain of Dictyostelium myosin have been known for some time (Fisher et al., 1995; Smith and Rayment, 1995; Rayment, 1996; Dominguez et al., 1998; Houdusse et al., 1999). Remarkably, we found that the α-actinin fusion protein crystallized under conditions very near to those of the catalytic domain alone. As the fusions in this system are connected to myosin via a flexible converter domain, the resulting constructs have higher conformational variability than a fusion between two rigid protein domains, leading to more efficient crystallization. Following collection of a complete native X-ray data set, the structure of the unknown portion was determined using molecular replacement techniques. As the known region of the structure is large (765 amino acids), the resulting phase information was sufficient to solve the unmodeled region of structure. Although the α-actinin rod solved in this study was only 26 kDa, theoretically it is possible to solve the structure of proteins of equal size (or even slightly larger) to the myosin domain.
Materials and methods
Expression, purification and crystallization
Replacement of the 2 kb SalI–BstXI fragment of pDH12-2R with the corresponding fragment from pDH20 (R238E) was used to generate the expression vector for the production of M761-2R R238E. The protein was overproduced in Dictyostelium AX3-ORF+ cells and purified by Ni2+-chelate affinity chromatography as described previously (Manstein and Hunt, 1995; Manstein et al., 1995), followed by anion-exchange chromatography (Super Q, Supelco). The protein solution was concentrated to 5 mg/ml and dialyzed against 30 mM HEPES–NaOH pH 7.3, 0.5 mM EDTA, 2 mM dithiothreitol (DTT), 1 mM benzamidine, 1 mM MgCl2 and 3% sucrose. For crystallization, the protein solution was used immediately or stored at –70°C. Crystals were grown by the hanging drop method at 7°C. The drops contained equal volumes (2.2 µl) of the protein solution and the mother liquor. The mother liquor contained 12% PEGM 5K, 170 mM NaCl, 50 mM HEPES–NaOH pH 7.2, 5 mM MgCl2, 5 mM DTT, 0.5 mM EGTA and 2% 2-methyl-1,3-propanediol. The protein solution (5 mg/ml) contained additionally 200 µM ADP and 200 µM vanadate, and was incubated on ice for 1 h before setting up the drops. Crystals normally appeared after 7–8 days and reached maximum dimensions of 0.1 × 0.3 × 0.9 mm. Crystals were transferred to a solution of mother liquor plus 30% glycerol and frozen in liquid nitrogen for storage and data collection. To date, diffraction-quality crystals of the unmutated, artificial lever arm construct, M761-2R, have not been obtained.
Crystallography and structure refinement
Diffraction data were collected at ESRF beamline ID-13 on a MarCCD detector and integrated and scaled using the program XDS (Kabsch, 1993), producing a data set 97.7% complete to 2.8 Å, with 4-fold redundancy and an Rsym of 11.0%. The M761-2R-R238E crystals belonged to spacegroup P21212 with two molecules in the asymmetric unit. Molecular replacement was performed with the program AMoRe (Navaza, 1994) using the crystal structure of Dictyostelium myosin residues 2–759 complexed with Mg-ADP-BeFx (PDB code 1mmd) (Fisher et al., 1995) as a starting model (the nucleotide and the side chains beyond Cβ of residues 238 and 459 were excluded). Initial maps showed clear helical density for the first repeat of the α-actinin lever arm, which was built as a poly-alanine model using the program O (v7.0 for WindowsNT). Following several rounds of simulated annealing refinement using torsional dynamics and a maximum likelihood target with the program CNS v0.9a (Brunger et al., 1998), the second α-actinin repeat was visible and built. Subsequent rounds of model building and refinement (including bulk solvent correction) produced the final structure of two M761-2R-R238E molecules containing 1005 residues each, two molecules of Mg-ADP and 14 water molecules (R-factor, 24.1%; Rfree, 29.9%). Ramachandran analysis shows all non-glycine residues to be in allowed regions. Figures were made using the programs Bobscript (Esnouf, 1997) and Raster3D (Merritt and Bacon, 1997). Atomic coordinates have been deposited in the Protein Data Bank under accession code 1G8X.
Acknowledgments
Acknowledgements
We would like to thank A.Perrakis and C.Riekel at ESRF beamline ID-13 for their help with data collection; W.Kabsch and D.Madden for crystallographic advice; A.Scherer and S.Zimmermann for expert technical assistance; J.Wray for scientific insight; and K.C.Holmes for discussions and continuous support. This work was supported by Deutsche Forschung Gemeinschaft grants KU1288/2-1 (F.J.K.) and MA1081/5-1 (D.J.M.), and the Max-Planck Society.
References
- Anson M., Geeves,M.A., Kurzawa,S.E. and Manstein,D.J. (1996) Myosin motors with artificial lever arms. EMBO J., 15, 6069–6074. [PMC free article] [PubMed] [Google Scholar]
- Blanchard A., Ohanian,V. and Critchley,D. (1989) The structure and function of α-actinin. J. Muscle Res. Cell Motil., 10, 280–289. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burley S.K. et al (1999) Structural genomics: beyond the human genome project. Nature Genet., 23, 151–157. [DOI] [PubMed] [Google Scholar]
- Corrie J.E. et al. (1999) Dynamic measurement of myosin light-chain-domain tilt and twist in muscle contraction. Nature, 400, 425–430. [DOI] [PubMed] [Google Scholar]
- Djinovic-Carugo K., Young,P., Gautel,M. and Saraste,M. (1999) Structure of the α-actinin rod: molecular basis for cross-linking of actin filaments. Cell, 98, 537–546. [DOI] [PubMed] [Google Scholar]
- Dominguez R., Freyzon,Y., Trybus,K.M. and Cohen,C. (1998) Crystal structure of a vertebrate smooth muscle motor domain and its complex with the essential light chain: visualisation of the pre-power stroke state. Cell, 94, 559–571. [DOI] [PubMed] [Google Scholar]
- Endow S.A. (2000) Molecular motors—a paradigm for mutant analysis. J. Cell Sci., 113, 1311–1318. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model., 15, 132–134, 112–113. [DOI] [PubMed] [Google Scholar]
- Fisher A.J., Smith,C.A., Thoden,J.B., Smith,R., Sutoh,K., Holden,H.M. and Rayment,I. (1995) X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry, 34, 8960–8972. [DOI] [PubMed] [Google Scholar]
- Furch M., Fujita-Becker,S., Geeves,M.A., Holmes,K.C. and Manstein,D.J. (1999) Role of the salt-bridge between switch-1 and switch-2 of Dictyostelium myosin. J. Mol. Biol., 290, 797–809. [DOI] [PubMed] [Google Scholar]
- Geeves M.A. and Holmes,K.C. (1999) Structural mechanism of muscle contraction. Annu. Rev. Biochem., 68, 687–728. [DOI] [PubMed] [Google Scholar]
- Gilmore A.P., Parr,T., Patel,B., Gratzer,W.B. and Critchley,D.R. (1994) Analysis of the phasing of four spectrin-like repeats in α-actinin. Eur. J. Biochem., 225, 235–242. [DOI] [PubMed] [Google Scholar]
- Grum V.L., Li,D., MacDonald,R.I. and Mondragon,A. (1999) Structures of two repeats of spectrin suggest models of flexibility. Cell, 98, 523–535. [DOI] [PubMed] [Google Scholar]
- Gulick A.M., Bauer,C.B., Thoden,J.B. and Rayment,I. (1997) X-ray structures of the MgADP, MgATPγ-S, and Mg AMPPNP complexes of Dictyostelium discoideum myosin motor domain. Biochemistry, 36, 11619–11628. [DOI] [PubMed] [Google Scholar]
- Houdusse A., Kalabokis,V.N., Himmel,D., Szent-Gyorgyi,A.G. and Cohen,C. (1999) Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell, 97, 459–470. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Kull F.J., Vale,R.D. and Fletterick,R.J. (1998) The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J. Muscle Res. Cell Motil., 19, 877–886. [DOI] [PubMed] [Google Scholar]
- Kurzawa S.E., Manstein,D.J. and Geeves,M.A. (1997) Dictyostelium discoideum myosin II: characterization of functional myosin motor fragments. Biochemistry, 36, 317–323. [DOI] [PubMed] [Google Scholar]
- Manstein D.J. and Hunt,D.M. (1995) Overexpression of myosin motor domains in Dictyostelium: screening of transformants and purification of the affinity tagged protein. J. Muscle Res. Cell Motil., 16, 325–332. [DOI] [PubMed] [Google Scholar]
- Manstein D.J., Schuster,H.P., Morandini,P. and Hunt,D.M. (1995) Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene, 162, 129–134. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D Version 2: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Noji H., Yasuda,R., Yoshida,M. and Kinosita,K.,Jr (1997) Direct observation of the rotation of F1-ATPase. Nature, 386, 299–302. [DOI] [PubMed] [Google Scholar]
- Onishi H., Kojima,S., Katoh,K., Fujiwara,K., Martinez,H.M. and Morales,M.F. (1998) Functional transitions in myosin: formation of a critical salt-bridge and transmission of effect to the sensitive tryptophan. Proc. Natl Acad. Sci. USA, 95, 6653–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D.A., Dixon,T.W. and Cohen,C. (1992) Analysis of the three-α-helix motif in the spectrin superfamily of proteins. Biophys. J., 61, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I. (1996) The structural basis of the myosin ATPase activity. J. Biol. Chem., 271, 15850–15853. [DOI] [PubMed] [Google Scholar]
- Rayment I., Rypniewski,W.R., Schmidt-Base,K., Smith,R., Tomchick,D.R., Benning,M.M., Winkelmann,D.A., Wesenberg,G. and Holden,H.M. (1993) Three-dimensional structure of myosin subfragment-1: a molecular motor. Science, 261, 50–58. [DOI] [PubMed] [Google Scholar]
- Rice S. et al. (1999) A structural change in the kinesin motor protein that drives motility. Nature, 402, 778–784. [DOI] [PubMed] [Google Scholar]
- Ruff C., Furch,M., Brenner,B., Manstein,D.J. and Meyhöfer,E. (2001) Single-molecule tracking of myosins with genetically engineered amplifier domains. Nature Struct. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Service R.F. (2000) Structural genomics offers high-speed look at proteins. Science, 287, 1954–1956. [DOI] [PubMed] [Google Scholar]
- Smith C.A. and Rayment,I. (1995) X-ray structure of the magnesium(II)-pyrophosphate complex of the truncated head of Dictyostelium discoideum myosin to 2.7 Å resolution. Biochemistry, 34, 8973–8981. [DOI] [PubMed] [Google Scholar]
- Smith C.A. and Rayment,I. (1996) X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry, 35, 5404–5417. [DOI] [PubMed] [Google Scholar]
- Vale R.D. and Milligan,R.A. (2000) The way things move: looking under the hood of molecular motor proteins. Science, 288, 88–95. [DOI] [PubMed] [Google Scholar]