Introduction
‘Black cat, white cat: whatever catches the mouse is a good cat’ (Chinese proverb).
Throughout the 3000 million years that bacteria have been on planet Earth, they have evolved amazing mechanisms for rapid adaptation to every imaginable type of environmental change. To give one extreme example, as little as 50 years after the introduction of the first antibiotics, very few of these drugs remain effective today at combating microbial infections (Davies, 1997). When bacteria are faced with antimicrobials, the only challenge they must overcome is to defeat the toxic effect of the compound through modification, cleavage or pumping the drug out of the cells; these processes generally involve one or just a few proteins. A far more complex environmental threat is that posed by organic chemicals, which have been discharged into many ecosystems through industrial and urban activities. Many of these chemicals are xenobiotic compounds (literally, alien to life), which include types of chemical ligatures (typically covalent C–Cl bonds) that have never been present in significant amounts in the biosphere and against which the housekeeping metabolic pathways of most microbes are generally useless (van der Meer et al., 1992). Unlike antibiotics, the challenge in this case is the construction of entire biodegradation pathways that endow bacteria with the ability to grow on these otherwise unpalatable chemicals. Such an outcome involves not just one protein, but sometimes dozens, which must adapt to entirely new substrates and intermediates. However, difficult as this might be, the successful assembly of a degradation pathway does not guarantee per se the survival of a particular strain. Bacteria that colonize polluted sites are subject to extremely tough competition from other microbial residents of the same niche. Transcriptional regulation of biodegradative genes and operons thus becomes a critical asset for the success of a newly assembled pathway to ensure its expression at only the right moment with a minimal waste of energy (de Lorenzo and Pérez-Martín, 1996). But how do bacteria learn to respond optimally to novel environmental signals and substrates?
Most of the known functional characteristics of prokaryotic promoters come from studies that use Escherichia coli as the test organism. Although far more complex than Buchnera (one of the simplest bacteria known so far; Shigenobu et al., 2000), the life cycle and the natural niches of E.coli can be relatively simple compared with those of not-so-distant relatives, such as pseudomonads, which thrive in soils polluted with toxic chemicals. Such rapidly adapting bacteria have become the experimental systems of choice in understanding how genes and pathways end up with regulated expression. In fact, since bacteria have been exposed to some such pollutants for only a few decades, it is possible, as discussed below, to find intermediate steps in the evolution process. In this review we summarize the features of the regulation of catabolic pathways for recalcitrant aromatic compounds that can help us to understand such a process. The conclusion is what we refer to as the ‘black cat/white cat principle’, which states that any regulatory mechanism is equally efficient provided that it ensures both a responsiveness to the new substrate and a suitable connection with the physiological state of the bacteria. A number of well studied cases to substantiate this notion are discussed below.
Transcriptional noise: promoters learning to respond to novel chemicals
The regulation of pathways for biodegradation of recalcitrant compounds by Gram-negative soil bacteria (mostly Pseudomonas, Alcaligenes, Bulkholderia and Acinetobacter) reveal some interesting mechanistic features by which operons acquire conditional promoters (Díaz and Prieto, 2000). The functioning of a new route depends on two major requirements that bacteria must attain to utilize the evolutionary advantage offered by the presence of fresh chemical species as potential carbon sources. One is the complement of genes encoding the whole suite of enzymes that build a pathway of reactions leading to metabolism of the compound to cardon dioxide and water. Operons destined for work in polluted sites need, in addition, an efficient transcriptional control system (de Lorenzo and Pérez-Martín, 1996). Regulated promoters are the key element that permit catabolic operons to be transcribed only when required and at levels adequate to guarantee a satisfactory metabolic return from the substrate. These two steps (assembly of a catabolic operon and acquisition of a substrate-responsive promoter) seem to be independent events, governed by different rules that operate at various times.
In general, isoenzymes that catalyse similar steps within a pathway tend to be alike at the sequence (DNA, protein) level also, even in cases where the initial substrates of the pathway are very different. This allows us to trace the origin of novel pathways to the patchwork assembly of pre-existing DNA segments bearing gene variants active on the novel substrates (van der Meer et al., 1992). Excellent examples of this include the adaptation of the entire set of catabolic genes of Pseudomonas putida for degradation of benzoate (involving the ben and cat genes) [Cl-benzoates (Parsek et al., 1994) or methyl-benzoates (van der Meer et al., 1992; Ramos et al., 1997)] or the recruitment of at least three catabolic segments for degradation of the very recalcitrant compound pentachlorophenol (Copley, 2000). There are no rules, however, to anticipate the type of regulator that may appear to respond to a novel chemical structure. New pathways generally start with low-level constitutive expression, on top of which increasingly specific promoters might develop. In contrast to the assembly of the enzymatic pathways, there is no specific requirement for a given type of regulator; on the contrary, it is common to find nearly identical catabolic operons preceeded by entirely different regulatory devices and proteins (de Lorenzo and Pérez-Martín, 1996). The bottom line is that a novel specificity may evolve through the leakiness of an earlier transcriptional control scheme (i.e. transcriptional noise). Any new control system should therefore start by recruiting the residual responsiveness of an already existing promoter/regulator to another signal, and then evolve by selecting changes such that residual responsiveness becomes predominant. As a consequence, the process from constitutive expression to response to a single inducer involves intermediate steps with various degrees of specificity, which can be found as remnants of the process in many catabolic pathways. Table I shows a number of cases that have been examined in detail. They include gratuitous induction (responsiveness to non-metabolizable compounds), cross-talk between structurally, but not functionally, related regulators, full replacement of one factor by another, or promiscuous activation among regulators of the same family. Pathways found in bacteria able to degrade substrates only very recently found in the environment (e.g. pentachlorophenol, polychlorobiphenyls, hexachlorocyclohexane, nitroaromatics, etc.) are often poorly regulated (Mouz et al., 1999; Copley, 2000; Watanabe et al., 2000), probably reflecting only an early step in the optimization of the corresponding metabolic route.
Table I. Levels of regulatory noise in catabolic promoters.
| Phenomenon | Examples | Reference |
|---|---|---|
| Minor/major gratuitous induction | Induction of the upper TOL pathway by o-xylene | Abril et al. (1989) |
| Induction of the lower NAH pathway by anthranylic acid | Cebolla et al. (1997) | |
| Induction of the alk pathway by DCPK | van Beilen et al. (1994) | |
| Induction of the lower TOL pathway by 2Br-benzoate | Ramos et al. (1986) | |
| Residual induction of σ54-dependent promoters by large collections of non-substrates | Abril et al. (1989); Garmendia and de Lorenzo (2000); Kahng et al. (2000); Jaspers et al. (2000) | |
| Cross-talk between akin regulators | Benzoate-responsive activation of the Pm promoter of thelower TOL pathway in the absence of XylS | Kessler et al. (1994) |
| Activation of the tfd gene cluster of Ralstonia eutropha JMP134 by TfdR in the absence of TfdT | Leveau and van der Meer (1996) | |
| Activation of clc genes by CatR in the absence of cognateregulator ClcR in P.putida AC27 | Parsek et al. (1994) | |
| Cross-regulation of toluene monooxygenases by TbmR andTbuT in Ralstonia pickettii PKO1 | Leahy et al. (1997) | |
| Regulatory takeover between related regulators | Phenol-dependent activation of upper TOL operon by DmpRand toluene-dependent activation of the dmp operon by XylR | Fernández et al. (1994) |
| Activation of the pheBA promoter of P.putida PaW85 by CatR | Parsek et al. (1995) | |
| Promiscuous activation | Activation from solution of σ54-dependent promoters by various regulators of the NtrC family | Pérez-Martín and de Lorenzo (1995) |
Physiological control of transcription: the phenomenon
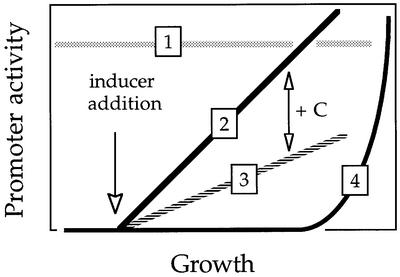
Bacterial regulation of catabolic pathways in the environment implies not just the ability to respond to a substrate, but also whether or not expression of the whole complement of enzymes is beneficial or detrimental to ecological performance. Bacteria thriving in a polluted niche receive a range of physical and chemical signals, other than just the presence of a substrate, which need to be processed to achieve a positive or negative outcome for each specific promoter (Cases and de Lorenzo, 1998). Such signals include nutrient availability, but also osmolarity, temperature, chaotropic agents, contact with surfaces, and interactions with other microorganisms. Under such tough conditions, it is of essence that expression of biodegradative operons becomes tightly coupled to the physiological and metabolic state of the cells. Figure 1 shows the various types of response found in promoters that drive expression of catabolic promoters. We would predict that biodegradative operons evolve from constitutive expression to substrate-responsive and metabolically controlled transcription. In this respect, pathways found in bacteria able to degrade man-made xenobiotic compounds, either totally or partially, frequently display a range of suboptimal, non-regulated expression profiles. In contrast, biodegradation of compounds that, despite being recalcitrant, have been available to bacteria for a long time is controlled through promoters endowed with sophisticated facets to ensure the processing of substrate-specific and general physiological signals. Typical promoters of this type are those that drive biodegradation of BTEX components of petroleum (benzene, toluene, ethylbenzene, xylenes), styrene, n-alkanes, or side-products of wood decay such as phenylacetate, phenols and benzoates (Díaz and Prieto, 2000). Some chloro-aromatic compounds (Cl-benzoates, Cl-catechols) may also be of natural origin, thus the cognate catabolic pathways frequently show responsiveness to both the substrate and the metabolic state.
Fig. 1. Expression profiles and development of promoters responding to novel environmental signals (e.g. latest carbon sources). Expression of genes and gene clusters encoding new catabolic abilities towards an evolutionarily recent substrate may be simply achieved through constitutive expression (type 1). This may evolve further into inducible expression (type 2). The presence of other nutrients in the medium that are easier to metabolize can later influence such induction and cause a C-source inhibition (type 3). Finally, both the presence of other nutrients, growth rate, and other environmental and physiological signals can be integrated for downregulation of the promoter during rapid growth, causing the so-called exponential silencing (type 4).
Table II shows a number of examples where the presence of a physiological control of biodegradative pathways has been observed as something superimposed on the substrate-dependent expression of the catabolic genes. Some of these will be examined in detail below. One typical environmental factor is the presence in the same niche of alternative carbon sources, the preferential consumption of which must be decided. This may or may not be related to the overall growth rate and growth phase, a major origin of signals that can be exploited for adjustment of promoter output. The most frequent induction pattern of highly evolved catabolic promoters is that referred to as type 4 in Figure 1, which exhibits a phenomenon termed ‘post-exponential induction’ or ‘exponential silencing’. Regardless of the mechanisms involved (discussed below), such conduct consists of a lack of transcriptional activity while bacteria grow rapidly on a nutrient-rich media, irrespective of the presence of the effector. This is then followed by rapid induction of the promoter once the growth rate of the bacteria decreases, they enter stationary phase, or cease to grow altogether. Effector-triggered post-exponential induction requires both the presence of a given effector and that cells reach a specific physiological stage. Such a stage might be defined not only by the depletion of a limiting nutrient or growth factor, but also by a particular overall energy state. To detect such a metabolic and physiological condition, and to connect it to the transcription of specific catabolic promoters, bacteria have evolved a diverse array of molecular artifices.
Table II. Biodegradative pathways subject to physiological control in pseudomonads.
| Pathway/operon and phenotype |
Substrate |
Reference |
| Degradation of the hydrocarbon by P.putida CA3 is inhibited by glutamate and citrate, not by glucose. | styrene | O’Connor et al. (1995) |
| Degradation by P.fluorescens ST subjected to inhibition by glucose, acetate andglutamate, and down-regulated by succinate and lactate. | Santos et al. (2000) | |
| Metabolic integration of 3 operons for catabolism of the aromatic substrate in afunctional unit (catabolon) of P.putida U. | phenyl acetate | Olivera et al. (1998) |
| Succinate inhibits consuption of the aromatic hydrocarbon in P.putida ML2. | benzene | Mason (1994) |
| Benzoate inhibits catabolism of phenol and acetate by Ralstonia eutropha. Succinateimpairs benzoate consumption by blocking one key enzyme for its metabolism. | phenol, benzoate | Ampe and Lindley (1995); Ampe et al. (1997, 1998) |
| Alk pathway of the OCT plasmid of P.oleovorans is inhibited in rich media and rapidgrowth. Repressed by C-sources. | n-alkanes | Yuste et al. (1998); Staijen et al. (1999) |
| TOL pathway of plasmid pWW0 of P.putida mt2. Repressed by rich media, rapidgrowth and some carbohydrates (i.e. glucose). | toluene, m-xylene,p-xylene | Cases et al. (1996); Marques et al. (1994); Duetz et al. (1994, 1996, 1997); Du et al. (1996) |
| Activity of the PnahG promoter of P.fluorescens HK44 is down-regulated by richmedia, glucose and toluene. | naphthalene, salicylate | Heitzer et al. (1994) |
| Catabolism of the aromatic substrate by P.fluorescens CA4 is inhibited by glutamate, but not by glucose or citrate. | ethyl benzene | Corkery et al. (1994) |
| clc pathway of P.putida AC27 down regulated by fumarate through inhibition of theregulator ClcR. | 3Cl-benzoate | McFall et al. (1997, 1998) |
| Downregulation of the metabolism of the xenobiotic compound by glutamate, glucoseand cellobiose in a Flavobacterium strain. | pentachlorophenol | Topp et al. (1988); Topp and Hanson (1990) |
| DMP pathway of plasmid pVI150 of Pseudomonas sp. CF600. Inhibited by rich mediaand C substrates allowing fast growth. | phenol, methyl-phenols | Sze et al. (1996) |
Assets for physiological control of catabolic operons
Unlike their eukaryotic counterparts, which seem to have a large number of transcription factors and controllable steps available to them, prokaryotic promoters have a very limited number of potential targets to integrate transcriptional co-regulation elements, namely specific regulators, sigma factors and promoter DNA. Such a paucity of molecular instruments has, however, been maximally combined and exploited with remarkable success. Of the many examples known (Table II) in which biodegradative pathways are subject to physiological regulation, the mechanisms involved have been studied to a significant degree in only a few instances. The diversity of regulatory strategies resulting in the same eventual phenotype is demonstrated in the cases that follow.
Metabolites that inhibit regulators
A remarkable example of how the transcriptional regulators of catabolic promoters can be subdued to the overall carbon metabolic and energetic status of cells is provided by the control of 3Cl-benzoate degradation in some strains of P.putida. The ortho-cleavage pathways of catechol and 3Cl-catechol are central catabolic pathways of P.putida that convert aromatic and chloroaromatic compounds (such as benzoate and 3Cl-benzoate) to tricarboxylic acid (TCA) cycle intermediates (McFall et al., 1997, 1998). They are encoded by the evolutionarily related catBCA and clcABD operons, respectively. Expression of the cat and clc operons requires the LysR-type transcriptional activators CatR and ClcR, and the inducer molecules cis,cis-muconate and 2Cl-cis,cis-muconate. Although the core transcriptional activation mechanisms of CatR and ClcR have been conserved in response to the presence of inducer, nature has provided some flexibility to respond to physiological signals. Transcriptional fusion studies demonstrated that the expression from the clc promoter is repressed when the cells are grown on succinate, citrate or fumarate, and that this repression is ClcR-dependent and occurs at the transcriptional level. The presence of these organic acids did not affect the expression from the cat promoter. In vitro transcription assays demonstrate that the TCA cycle intermediate, fumarate, directly and specifically inhibits the formation of the clcA transcript. No such inhibition was observed when CatR was used as an activator on either the cat or the clc template. Since both the catechol and the Cl-catechol pathways feed into the TCA cycle, but only the Cl-catechol pathway is inhibited by fumarate, there is a subtle difference in the regulation of these two pathways, where intracellular sensing of a TCA cycle intermediate leads to a reduction of chloroaromatic degradation. Titration studies of fumarate and 2-chloromuconate in vitro transcription assays show that the fumarate effect is concentration-dependent and reversible, indicating that fumarate and 2-chloromuconate most probably compete for the same binding site on ClcR (McFall et al., 1997, 1998). This is an interesting example of the transcriptional regulation of a biodegradative pathway through the sensing of the levels of one key metabolite of the TCA cycle. Unsophisticated as it may appear, this type of metabolic downregulation of a xenobiotic-degrading pathway (clc) caused by a side-metabolite from a substrate that is easier to consume is probably very frequent, since just a few mutations in the targeted protein makes it amenable to a degree of physiological control.
Parasitizing sigma factors
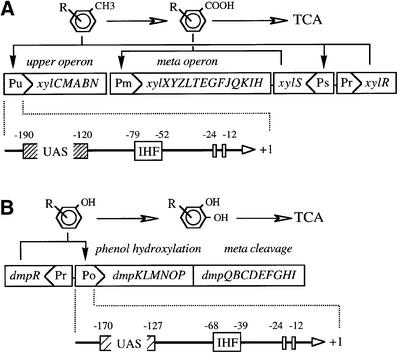
The general transcription machinery can also be used by degradation pathways to couple expression of biodegradative operons with different physiological signals. One remarkable example is the TOL plasmid of P.putida mt-2 for degradation of toluene, in which the interplay of two promoters, two regulators and four sigma factors provide a very efficient control mechanism. Pseudomonas putida cells harbouring the TOL plasmid pWW0 are able to grow on toluene and m-/p-xylene as the only carbon source, owing to expression of a two-step pathway for the complete mineralization of these hydrocarbons (Ramos et al., 1997). The first step (Figure 2A) involves the biotransformation of toluene/xylenes to their corresponding carboxylic acids through oxidation of one methyl group of the aromatic substrate. The second stage channels the benzoate (or toluate) into the Krebs’ cycle. This follows a complex pathway summarized in Figure 2A. The biochemical steps are reflected in two separate transcriptional units, the so-called upper operon (encoding enzymes for oxidation of the methyl group of toluene) and the lower (or meta) operon (responsible for the aromatic ring fission leading to pyruvate and acetaldehyde). Expression of the xyl genes is tightly regulated through a complex cascade of transcriptional controls (Ramos et al., 1997) that involve two regulators, the XylR and XylS proteins. These are responsible for the activation of the upper and meta operons, respectively, thus ensuring optimal expression of the degradative activities only in the presence of pathway substrates (Figure 2A).
Fig. 2. Organization of the TOL and dmp biodegradation pathways and their cognate Pu and Po promoters. (A) The regulatory cascade of the xyl genes in the TOL plasmid pWW0 of P.putida mt-2. In the presence of upper pathway substrates like m-xylene, the upper-operon promoter Pu and the xylS promoter Ps are activated by XylR in combination with the σ54-containing RNA polymerase (σ54-RNAP). Subsequently, an excess of XylS product or XylS bound to its effectors (i.e. substrates of the meta pathway) activate Pm. There is no physical continuity between the upper and the meta operons. Below the scheme of the pathway, the Pu promoter region is expanded, showing the boundaries of relevant DNA sequences: upstream binding sites (UAS) for XylR, the –12/–24 sequences recognized by σ54-RNAP, and a single IHF binding site located within the intervening region. (B) Regulation of the pVI150-encoded dmp-operon of Pseudomonas sp. CF600. The dmpR gene product that is responsive to phenol and cresols activates transcription of the divergently transcribed dmp-operon from the Po promoter. A subset of the dmp genes are involved in phenol hydroxylation, while the rest encode enzymatic activities of the meta-cleavage pathway for dissimilation of the catechol intermediate. The Po promoter region is expanded below the scheme of the dmp pathway. Relevant portions of the sequence are pinpointed.
The Pm promoter (also called OP2), which drives expression of the lower operon for metabolism of benzoate and toluates all the way to the TCA cycle intermediates, is expressed at a high level throughout the growth curve (Marqués et al., 1995). This transcription is dependent on the positive activator XylS (of the AraC family of prokaryotic regulators) activated by 3-methyl benzoate. Although from just inspecting the DNA sequence this promoter would be predicted to be dependent on the housekeeping sigma factor σ70, recent observations (Marqués et al., 1995, 1999; Miura et al., 1998) have revealed a most intricate sigma succession mechanism that ensures continuous Pm activity throughout all stages of growth. First, it appears that thermosensitive rpoD mutants that transiently lack σ70 are still able to support Pm activity at the non-permissive temperature, thereby suggesting that other sigmas may actually drive promoter activity. In fact, it could be shown that it is the heat shock factor σH, rather than σ70, that is required for Pm output following induction with 3-methyl benzoate. The surprising finding is that σH levels are generally very low unless a signal triggering the heat shock response occurs. When cells are challenged with the aromatic effector, however, it does trigger such a response, probably due to its effects on membrane properties. This is true mostly for cells that are exposed to 3-methyl benzoate (Marqués et al., 1999). When cells enter stationary phase, the starvation sigma σS seems to take over and replace σH as the factor that directs Pm activity. It thus seems that activation of Pm transcription is achieved through a switch between two stress-responsive factors: σH in exponential phase and σS in stationary phase. In both cases, Pm is dependent on the same activator, XylS, and starts transcription in the same point. The Pm/XylS system reveals a strategy of coupling transcription of a specific promoter to the cell physiology by ‘choosing’ general stress signals mediated by sigma factors. Since the whole TOL system is plasmid encoded, it is remarkable how expression of the biodegradation functions is the result of an interplay, if not a parasitism, of general host factors with system-specific, plasmid-encoded regulators.
The σ54 promoter Pu of the TOL plasmid
In 1986, a now classic paper (Dixon, 1986) reported that the Pu promoter of upper operon of the same TOL plasmid pWW0 of P.putida mt-2 mentioned above (Figure 2A) had features that made its expression dependent on the ntrA gene. This gene was later identified as the determinant of the sigma factor for nitrogen metabolism and thus called σN (or, more frequently, σ54; Merrick, 1993). Since then, the σ54-dependent promoter Pu has become a landmark for studies on the regulation of biodegradative pathways. Such extensive work has yielded detailed knowledge on both specific effector-mediated regulation and the devices that couple its performance in vivo to cell physiology. Pu is regulated by the XylR protein, which belongs to the family of prokaryotic enhancer-binding activators that act in concert with σ54 (Morett and Segovia, 1993; Shingler, 1996). In the presence of toluene, xylenes and other structural analogues, the XylR protein activates the Pu promoter of the upper TOL operon (Figure 2A), using a mechanism that is generally shared by other activators of the family. This involves the binding of the regulator to upstream activating sequences (UAS) and the looping-out of the complex into close proximity with the σ54-containing form or RNA polymerase bound to the –12/–24 region of the promoter (Morett and Segovia, 1993; North et al., 1993). This event is assisted by the presence of an integration host factor (IHF)-binding site at the intervening region between the UAS and the σ54-RNAP attachment site (Pérez-Martín and de Lorenzo, 1996a,b). Such an elaborate promoter architecture (Figure 2A) seems to be particularly well suited to integrating a repertoire of environmental signals.
From the early studies on the regulation of this promoter, it became evident that expression of the upper TOL operon was inhibited when cells grew exponentially in rich medium (Hugouvieux-Cotte-Pattat et al., 1990; de Lorenzo et al., 1993). This effect seemed not to require the activity of the whole complement of TOL genes, since it could be faithfully reproduced with only the regulatory elements that control transcriptional activity of Pu. Pseudomonas putida cells devoid of the TOL plasmid carrying a chromosomal insertion of the xylR gene and a Pu–lacZ fusion were unable to accumulate β-galactosidase when growing exponentially in Luria–Bertani (LB) medium, regardless of the presence or absence of m-xylene (de Lorenzo et al., 1993). However, as soon as the cells leave the exponential growth phase and enter the stationary phase, the same Pu–lacZ fusion becomes extremely responsive to the aromatic inducer (de Lorenzo et al., 1993; Cases et al., 1996). The data of the reporter fusion match faithfully the quantitative S1 protection assays with mRNA from induced cells, so the effect certainly occurs at the transcriptional level (Marqués et al., 1994). Since Pu is functional in vitro simply by mixing purified and pre-activated XylR with σ54-containing RNAP and IHF (Pérez-Martín and de Lorenzo, 1996b), it is clear that additional elements, when induced, adjust transcription to the physiological state that governs the cells. These initial observations triggered a large number of studies on the mechanisms involved in such a physiological inhibition of Pu activity.
Various reports from different perspectives have documented that Pu activity is downregulated in response to exponential growth in rich media, a phenomenon referred to as catabolite repression (Duetz et al., 1994, 1996, 1997; Holtel et al., 1994; Marqués et al., 1994), stationary-phase dependency (Hugouvieux-Cotte-Pattat et al., 1990) or, as we prefer to call it, exponential silencing (Cases et al., 1996) (Figure 1). At least in part, this effect can be traced to modulation of the activity of the sigma factor iself, because its overproduction shifts Pu derepression to an earlier growth stage (Cases et al., 1996). In addition, ftsH mutants of E.coli (encoding a membrane-bound protease involved in the turnover of σ32) fail to activate Pu (Carmona and de Lorenzo, 1999). Overproduction of σ54 defeats the inhibition caused by the loss of FtsH protein. Increased levels of FtsH not only restore promoter activity but also relieve the physiological inhibition of the promoter. The mechanism by which all this occurs has not yet been resolved, but may involve the action of an anti-sigma or a connection between the activity of σ54 and the turnover machinery of the heat shock factor σ32, which involves the DnaK chaperone and the FtsH protease (Muffler et al., 1997). The presence or absence of the stationary phase sigma, σS, does not appear to have a role in the growth dependency of Pu outcome. This is striking, since other metabolically regulated promoters (see below) benefit from this global starvation and growth phase signal for behaving in a fashion indistinguishable from that observed in Pu. Finally, it is remarkable that in spite of being strongly inhibited by casamino acids, the Pu promoter functions normally in a relA/spoT strain of E.coli that fails to accumulate the alarmone (p)ppGpp that is typical of the stringent response (Cashel et al., 1996; Carmona et al., 2000). Yet, overproduction of RelA (predicted to force an increase ppGpp levels) does increase Pu activity and causes a certain relief of its exponential silencing (Carmona et al., 2000).
Carbon sources versus metabolic state
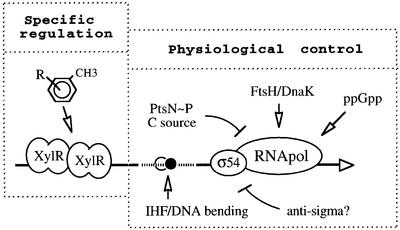
Besides the effect of growth rate and growth phase, the presence of certain carbon compounds such as glucose or gluconate also restrain Pu activity (Holtel et al., 1994; Cases et al., 1999). It should be noted that although a regulator analogue of the well characterized catabolite repression protein (CRP) of E.coli does exist in pseudomonads (named Vfr), it appears to play no role in catabolite repression, but instead is involved in quorum sensing control (Collier et al., 1996; Albus et al., 1997). Even transport of glucose (a nearly universal inducer of catabolic repression) is completely different in Pseudomonas (Collier et al., 1996). Catabolism of this sugar requires its predominant extracellular transformation into gluconic acid, followed by the induction of a specific energy-dependent gluconate uptake system. Once internalized, gluconate is phosphorylated and entered into the central metabolism as 6-phosphogluconate (Schleissner et al., 1997). These peculiarities in the carbon metabolism of P.putida make it unlikely that the inhibition of Pu activity by glucose and gluconate takes place through a typical cAMP/CRP-mediated mechanism of the sort so well known in E.coli (Kolb et al., 1993). Furthermore, the C-source dependent inhibition of Pu activity occurs within a different range than that caused by rapid growth. While exponential silencing in rich medium completely abolishes Pu activity, glucose does not inhibit promoter output by more than two-thirds of the maximal activity. The gene ptsN, encoding a new member of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) (Postma et al., 1993; Saier, 1996; Saier et al., 1996), has been related to this modulation, since its loss alleviates glucose inhibition of Pu in P.putida (Cases et al., 1999). Glucose assimilation is not affected in this mutant, suggesting that this gene participates in sensing rather than metabolizing the carbohydrate. Interestingly, ptsN mutants are still subject to exponential silencing. In the other direction, cells relieved of such silencing because of σ54 overproduction remain repressible by glucose (Cases and de Lorenzo, 2000). These observations indicate that the growth-rate inhibition of Pu and the downregulation by glucose can be separated genetically and, surely, are channeled into the promoter via different pathways (Figure 3). The mechanism by which PtsN exerts its inhibition on Pu in the presence of glucose is not yet known, but may involve other proteins of the PTS family such as PtsO (Powell et al., 1995; I.Cases and V.de Lorenzo, unpublished). The whole picture is by no means complete and will remain the subject of further investigation.
Fig. 3. Integration of specific and physiological signals on the outcome of the Pu promoter of the TOL plasmid pWW0. This archetypal promoter receives both inducer-specific and overall physiological inputs. The specific regulation that makes Pu respond to toluene and xylenes involves only the XylR regulator. On the contrary, metabolic inputs are channeled towards the transcription machinery through multiple molecular assets. These include the control of the activity or the turnover of the σ54 factor in vivo (perhaps through the FtsH/DnaK system, or an antisigma factor or both) (Cases et al., 1996; Carmona and de Lorenzo, 1999), the action of the phosphorylated form of PtsN (Cases et al., 1999) and also the influence of intrinsic or factor (IHF)-mediated DNA bending. The related Po promoter of catabolic plasmid pVI150 (see Figure 2B) seems to react positively to the intracellular levels of the alarmone ppGpp (Sze et al., 1999), whereas the response of Pu to the stringent response is far less pronounced (Carmona et al., 2000).
Variations on the σ54 promoter theme
Even promoters that are organized with the same control elements and that show similar physiological regulation patterns have evolved a different regulatory mechanism, as the Po/DmpR system ilustrates. Po, the phenol-responsive promoter that drives expression of the single dmp (dimethylphenol) operon of plasmid pVI150 harboured by Pseudomonas sp. CF600 (Powlowski and Shingler, 1994) (Figure 2B) is, as Pu, dependent on σ54 and is activated by a cognate protein named DmpR. Although responsive to phenol and methyl-phenols rather than xylenes, the sequence of this protein is largely similar to that of XylR (Shingler et al., 1993). In fact, it was soon proven that the major domains of the two proteins could be swapped without a loss of function and that either protein could activate the other cognate promoter and recognize their mutual UAS (Fernández et al., 1994; Shingler and Moore, 1994). This suggested that XylR and DmpR behaved more as variants of the same protein than as different proteins. When in vivo physiological studies of the Po promoter were carried out, it also became clear that Po was subjected to exponential silencing, i.e. no transcription could be detected while cells grew rapidly in rich LB medium (Sze et al., 1996). Although such behaviour would superficially be identical to that reported for Pu, further insights have revealed that the mechanisms involved differ significantly between the XylR/Pu and the DmpR/Po systems. For instance, Sze et al. (1996) reported that the exponential silencing observed in Po was, in general, inversely proportional to growth rate. Thriving in certain carbon sources thus inhibits promoter activity to the same extent that it increased growth rate. In a subsequent step, it was shown that cells lacking relA and spoT (Cashel et al., 1996; see above) failed to activate the σ54 promoter Po at any growth phase (Sze and Shingler, 1999). On the contrary, overexpression of relA (and the ensuing increase in ppGpp levels) entirely relieved exponential silencing. These observations suggested that Po activity was dependent on the intracellular accumulation of ppGpp related to growth rate, and provided a rationale to understand the coupling of this σ54 promoter to metabolic signals related to starvation. The notion has been strengthened by the observation that some RNAP variants, which bear mutations that mimic the response to ppGpp, overcome exponential silencing as well (Sze and Shingler, 1999). Such a subordination to ppGpp is in contrast to the lesser dependence of the XylR/Pu system to the same signal (Carmona et al., 2000; see above), and this difference is informative. In vitro transcription of the Po promoter with purified XylR was highly dependent on ppGpp, whereas Pu in the same experiment could be transcribed without any addition of the nucleotide (Carmona et al., 2000). On the basis of comparing the mechanisms that produce the physiological silencing of Pu/XylR with those of Po/DmpR, it appears that the class of metabolic signals that end up interweaving the control of such σ54 promoters is determined in part by their very DNA sequence.
Optimizing DNA sequence as a regulatory framework
A final facet of the physiological control of some σ54 systems is the role of the very promoter DNA sequence as a possible port of entry of additional signals from cell metabolism. Since the early observation that Pu bore a functional binding site for IHF (Figure 2A) that is essential to maintain a suitable promoter architecture, DNA bending was considered a potential regulatory obstacle for the system (Pérez-Martín and de Lorenzo, 1995, 1997a). It is known that IHF accumulation in E.coli is growth-phase-dependent (Aviv et al., 1994), so the bend produced by the factor could contribute to the enhanced activity of Pu in stationary phase. However, this notion was discarded because Pu promoter variants in which the IHF binding site had been replaced by a statically bent DNA sequence, and was thus IHF independent, were still subject to exponential silencing (Cases et al., 1996). In addition, it appeared that in E.coli, the nucleoid-associated protein HU can replace the DNA-bending function of IHF (Pérez-Martín and de Lorenzo, 1997b), so that the effect of the loss is more difficult to monitor in vivo. But following these observations in E.coli, it was noticed that a P.putida IHF knockout mutant failed entirely to support any Pu activity (Calb et al., 1996). This was surprising, since there is a functional analogue of HU in P.putida (F.Bartels and V.de Lorenzo, unpublished), and thus some promoter function should be expected. A clue to understanding this phenomenon was the realization that IHF was necessary to recruit the σ54-RNAP holoenzyme to the promoter DNA through a mechanism that involved the interactions of the C-terminal domain of the α subunit of the RNAP (αCTD) with an upstream DNA, UP-like sequence (Bertoni et al., 1998). Under limiting concentrations of σ54-RNAP, such a recruitment becomes a kinetic barrier to promoter activity, more limiting than XylR activation itself (Carmona et al., 1999). On the basis of these observations, the role of IHF in physiological control will have to be re-assessed. Whether such a possible regulatory device is also relevant in other σ54-dependent promoters that contain IHF sites (Dworkin et al., 1998; Wassem et al., 2000) remains unknown.
All the data collected so far on the physiological control of σ54 promoters suggests that they are particularly well suited to group a whole range of metabolic symptoms into one or more steps of the transcription initiation process. With only two constant players (the substrate-responsive UAS-binding regulator and σ54), these promoters afford a stunning amalgam of the effector-specific transcription with the cell physiology. The specific choice of physiological signals (coupling to the stringent response, to the turnover of other sigma factors, to the PTS system of C-sensing, to the physiological levels of IHF, to growth phase-dependent DNA bending, etc.) seems to be mostly determined by the diversity of DNA sequences at the promoter. Not unlike a music score instructing the entrance of specific instruments to play a melody, in the promoters discussed above, it becomes manifest that distinct instruments can end up performing the same theme.
Promoters responding to gasoline?
While the Pu/XylR and Po/DmpR systems have developed virtually the same behaviour by working upon variations of the σ54 theme, a sophisticated degree of physiological control can be generated as well with quite different molecular assets. The OCT (octane degradation) plasmid borne by Pseudomonas oleovorans Gpo1 allows this microorganism to grow on medium chain-length n-alkanes (van Beilen et al., 1994). This is due to a plasmid-borne metabolic pathway, whose induction is regulated by the AlkS protein (of the LuxR/MalT family of transcriptional regulators) and the expression of which is fully σ54 independent. In the presence of alkanes or the gratuitous inducer dicyclopropyl ketone (DCPK; see Table I), AlkS activates the AlkB promoter, which drives expression of most genes of the pathway. The study of the behaviour of chromosomally inserted PalkB–lacZ fusions and S1 nuclease protection assays revealed that the promoter remains silent when cells grow exponentially in rich LB medium, even if cultures are exposed to the inducer (Yuste et al., 1998). A milder downregulation occurred also when cells were grown in various carbon sources such as lactate or succinate (Yuste et al., 1998; Staijen et al., 1999). Phenomenologically, these inhibitory effects linked to exponential growth and carbon sources were not unlike those seen in the Pu and Po promoters above. However, given the very different promoter structures, it could be anticipated that the mechanisms involved ought to be different. An intriguing similarity with DmpR and PhlR, though (see above), is that overproduction of AlkS seemed to overcome the silencing imposed by rapid growth in rich medium (Müller et al., 1996; Yuste et al., 1998).
An interesting edge to the control of PalkB expression came through the realization that alkS transcription was itself regulated by growth phase. This is because the PalkS promoter seems to have a preference for the RNAP holoenzyme bearing the starvation sigma σS (Canosa et al., 1999). Although this could be related to the physiological control of alk genes, it might deal instead with a separate aspect of the system. More recent studies by Rojo’s group have shown that the PalkS promoter is more complex (Canosa et al., 2000). The promoter region includes PalkS1, which is fully σS-dependent and thus silent during exponential growth and active in the stationary phase. In the absence of alkanes, the AlkS protein represses PalkS1 moderately, thus achieving a reasonable level of expression of the regulator. But when cells face the inducer, AlkS both completely represses PalkS1 and activates a second, dowstream promoter (PalkS2). In this manner, activation of PalkS2 allowed a very efficient transcription of alkS when alkanes are in the medium. On this basis, it is plausible that AlkS levels are controlled by a positive feedback mechanism, which leads to a rapid increase in alkS transcription when alkanes are present and causes a fast off-switch when alkanes are depleted. Yet, it happens that PalkS2 itself is inhibited by exponential growth in rich medium and is downregulated by some carbon sources such as lactate, a phenomenon that still deserves further investigation (Canosa et al., 2000).
Black cat/while cat: evolutionary convergence of physiological regulation
In spite of the very different regulatory schemes present in the alk system of the OCT plasmid as compared with the σ54-dependent routes of pWW0 and pVI150, it is amazing how both have evolved via entirely disparate mechanisms to achieve the same result: (i) responding to specific effectors; (ii) reacting to the presence of other carbon sources in the medium; and (iii) weaving promoter activity to the growth phase. The first is achieved through different regulators (AlkS and XylR/DmpR) belonging to completely unrelated families of proteins, the second through a connection of an undisclosed catabolite repression mechanism to proteins of the PTS system, and the third by recruiting stationary phase or stress sigmas, by coupling polymerase activity to ppGpp or by checking the activity of the sigmas by additional growth-phase-dependent factors. But these considerations, as discussed above, could also be extended to each of the other catabolic promoters where the issue has been studied and that involve other strategies for coupling promoter activity to the cell physiology.
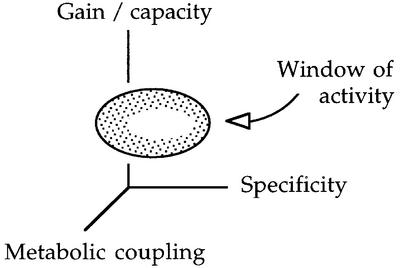
On the basis of the comparisons between the regulatory systems presented in this review, we entertain the notion that catabolic pathways for recalcitrant compounds (or, by extension, for any new compound) evolve in three major planes that define its window of activity in the enviroment (Figure 4). First, it is the assembly of the complement of catabolic enzymes that are required to obtain a suitable energy return out of the metabolism of a given chemical. Since the number of biochemical strategies to break bonds is limited, this step will generally follow quite strict rules imposed by the very chemistry of the biodegradation process. Secondly, newly developed metabolic operons mature from constitutive or semi-constitutive expression to conditional transcription responsive to the presence of the substrate in the medium (Figure 1). The unfolding of substrate specificity may start by enrolling the residual responsiveness of an already existing promoter–regulator pair, so the early control stages are necessarily relaxed. The choice of a given promoter–regulator pair may depend much more on the evolutionary story of the degradation operon(s) than on the specific properties of the regulatory proteins; it is indeed frequent to find very similar biodegradative operons controlled by very different promoter types (van der Meer et al., 1992). But thirdly, in the highly competitive environment that predominates in sites polluted with recalcitrant compounds, promoters must be able to process a range of simultaneous environmental signals so that expression of a degradation pathway for one compound present in the medium does not turn against the overall ecological performance of the host. To this end, it is of essence that the transcription machinery of specific promoters are able to sense the physiological state of the cells as a whole and react to it. That refining specific regulation and coupling it to cell physiology are frequently overlapping processes in time can be noted by comparing Tables I and II. Whether sensing of that kind occurs through responding to a metabolite of the TCA cycle, to a nutritional alarmone, to a stress signal or to proteins responding to extra carbon sources does not really seem to matter, provided that it links one thing to the other.
Fig. 4. Evolutionary optimization of promoter performance. The scheme illustrates the black cat/white cat principle discussed in the text. Promoters recruited to respond to novel enviromental signals, such as unfamiliar carbon sources, gravitate necessarily towards an optimal window of activity. Such a window is defined as the combination of: (i) the various working ranges of promoter gain/capacity (inducibility versus absolute promoter output); (ii) the specificity of the regulator/promoter pair for a given chemical; and (iii) the connection of promoter activity to the cell physiology. The drift towards such an optimal window is independent of the molecular mechanism to reach it. The presence of specific DNA/promoter/regulators combinations in front of given biodegradative operons thus reflect much more the evolutionary history of the system than a requirement for a distinct type of transcriptional factor.
This molecular black cat/white cat principle applied to biodegradative pathways has repercussions that extend beyond the mere basic understanding of bacterial promoters. It has major consequences in the way novel expression systems might be designed for biotechnological processes (Schroetckh et al., 1999; Farmer and Liao, 2000), particularly in those applications involving metabolic engineering projects of bacteria destined for the environment as bioremediation agents (de Lorenzo, 1994; Timmis et al., 1994; Matin et al., 1995; Timmis and Pieper, 1999).
Acknowledgments
Acknowledgements
The authors are indebted to K.N.Timmis, S.Marqués, J.L.Ramos, J.Pérez-Martín, F.Rojo and G.Bertoni for many inspirational discussions. V.Shingler and S.Busby are gratefully acknowledged also for their critical reading of the manuscript and many useful suggestions. The work of the authors’ laboratory is supported by contracts BIO4-CT97-2040 and QLRT-99-00041 of the EC, and by Grant BIO98-0808 of the Spanish Comisión Interministerial de Ciencia y Tecnología (CICYT).
REFERENCES
- Abril M.A., Michan,C., Timmis,K.N. and Ramos,J.L. (1989) Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol., 171, 6782–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albus A.M., Pesci,E.C., Runyen-Hanecky,L.J., West,S.E. and Iglewski,B.H. (1997) Vfr controls quorum sensing in Pseudomonas aeruginosa.J. Bacteriol., 179, 3928–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampe F. and Lindley,N.D. (1995) Acetate utilization is inhibited by benzoate in Alcaligenes eutrophus: evidence for transcriptional control of the expression of acoE coding for acetyl coenzyme A synthetase. J. Bacteriol., 177, 5826–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampe F., Uribelarrea,J.L., Aragao,G.M. and Lindley,N.D. (1997) Benzoate degradation via the ortho pathway in Alcaligenes eutrophus is perturbed by succinate. Appl. Environ. Microbiol., 63, 2765–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampe F., Leonard,D. and Lindley,N.D. (1998) Repression of phenol catabolism by organic acids in Ralstonia eutropha. Appl. Environ. Microbiol., 64, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv M., Giladi,H., Schreiber,G., Oppenheim,A.B. and Glaser,G. (1994) Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol. Microbiol., 14, 1021–1031. [DOI] [PubMed] [Google Scholar]
- Bertoni G., Fujita,N., Ishihama,A. and de Lorenzo,V. (1998) Active recruitment of σ54-RNA polymerase to the Pu promoter of Pseudomonas putida: role of IHF and αCTD. EMBO J., 17, 5120–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calb R. et al. (1996) Structure and function of the Pseudomonas putida integration host factor. J. Bacteriol., 178, 6319–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canosa I., Yuste,L. and Rojo,F. (1999) Role of the alternative sigma factor σS in expression of the AlkS regulator of the Pseudomonas oleovorans alkane degradation pathway. J. Bacteriol., 181, 1748–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canosa I., Sanchez-Romero,J.M., Yuste,L. and Rojo,F. (2000) A positive feedback mechanism controls expression of AlkS, the transcriptional regulator of the Pseudomonas oleovorans alkane degradation pathway. Mol. Microbiol., 35, 791–799. [DOI] [PubMed] [Google Scholar]
- Carmona M. and de Lorenzo,V. (1999) Involvement of the FtsH (HflB) protease in the activity of σ54-promoters. Mol. Microbiol., 31, 261–270. [DOI] [PubMed] [Google Scholar]
- Carmona M., de Lorenzo,V. and Bertoni,G. (1999) Recruitment of RNA polymerase is a rate-limiting step for the activation of the σ54 promoter Pu of Pseudomonas putida. J. Biol. Chem., 274, 33790–33794. [DOI] [PubMed] [Google Scholar]
- Carmona M., Rodríguez,M.J., Martínez-Costa,O. and de Lorenzo,V. (2000) In vivo and in vitro effects of (p)ppGpp on the σ54 promoter Pu of the TOL plasmid of Pseudomonas putida. J. Bacteriol., 182, 4711–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I. and de Lorenzo,V. (1998) Expression systems and physiological control of promoter activity in bacteria. Curr. Opin. Microbiol., 1, 303–310. [DOI] [PubMed] [Google Scholar]
- Cases I. and de Lorenzo,V. (2000) Genetic evidence of distinct physiological regulation mechanisms in the σ54Pu promoter of Pseudomonas putida. J. Bacteriol., 182, 956–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases I., de Lorenzo,V. and Pérez-Martín,J. (1996) Involvement of σ54 in exponential silencing of the Pseudomonas putida TOL plasmid Pu promoter. Mol. Microbiol., 19, 7–17. [DOI] [PubMed] [Google Scholar]
- Cases I., Pérez-Martín,J. and de Lorenzo,V. (1999) The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the σ54-dependent Pu promoter of the TOL plasmid. J. Biol. Chem., 274, 15562–15568. [DOI] [PubMed] [Google Scholar]
- Cashel M., Gentry,D.R., Hernández,V.J. and Vinella,D. (1996) The stringent response. In Neidhart,F.C. (ed.), E. coli and Salmonella, Cellular and Molecular Biology. Vol. 1. American Society for Microbiology, Washington, DC, pp. 1458–1496. [Google Scholar]
- Cebolla A., Sousa,C. and de Lorenzo,V. (1997) Effector-specificity mutants of the transcriptional activator NahR of naphthalene-degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J. Biol. Chem., 272, 3986–3992. [DOI] [PubMed] [Google Scholar]
- Collier D.N., Hager,P.W. and Phibbs,P.V. (1996) Catabolite repression control in the Pseudomonads. Res. Microbiol., 147, 551–561. [DOI] [PubMed] [Google Scholar]
- Copley S.D. (2000) Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem. Sci., 25, 261–265. [DOI] [PubMed] [Google Scholar]
- Corkery D.M., O’Connor,K.E., Buckley,C.M. and Dobson,A.D. (1994) Ethylbenzene degradation by Pseudomonas fluorescens strain CA4. FEMS Microbiol. Lett., 124, 23–27. [DOI] [PubMed] [Google Scholar]
- Davies J.E. (1997) Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found. Symp., 207, 15–27. [PubMed] [Google Scholar]
- de Lorenzo V. (1994) Designing microbial systems for gene expression in the field. Trends Biotechnol., 12, 365–371. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V. and Pérez-Martín,J. (1996) Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol. Microbiol., 19, 1177–1184. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Cases,I., Herrero,M. and Timmis,K.N. (1993) Early and late responses of TOL promoters to pathway inducers: identification of growth-phase dependent promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol., 175, 6902–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz E. and Prieto,M.A. (2000) Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol., 11, 467–465. [DOI] [PubMed] [Google Scholar]
- Dixon R. (1986) The xylABC promoter from the Pseudomonas putida TOL plasmid is activated by nitrogen regulatory genes in Escherichia coli.Mol. Gen. Genet., 203, 129–136. [DOI] [PubMed] [Google Scholar]
- Du Y., Holtel,A., Reizer,J. and Saier,M. (1996) Sigma 54-dependent transcription of the Pseudomonas putida xylS operon is influenced by the IIANtr protein of the phosphotransferase system in Escherichia coli.Res. Microbiol., 147, 129–132. [DOI] [PubMed] [Google Scholar]
- Duetz W.A., Marqués,S., de Jong,C., Ramos,J.L. and van Andel,J.G. (1994) Inducibility of the TOL catabolic pathway in Pseudomonas putida (pWW0) growing on succinate in continuous culture: evidence of carbon catabolite repression control. J. Bacteriol., 176, 2354–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duetz W.A., Marqués,S., Wind,B., Ramos,J.L. and van Andel,J.G. (1996) Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWW0 under various conditions of nutrient limitation in chemostat culture. Appl. Environ. Microbiol., 62, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duetz W.A., Wind,B., Kamp,M. and van Alden,J.G. (1997) Effect of growth rate, nutrient limitation and succinate on expression of TOL pathway enzymes in response to m-xylene in chemostat cultures of Pseudomonas putida (pWW0). Microbiology, 143, 2331–2338. [DOI] [PubMed] [Google Scholar]
- Dworkin J., Ninfa,A.J. and Model,P. (1998) A protein-induced DNA bend increases the specificity of a prokaryotic enhancer-binding protein. Genes Dev., 12, 894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer W.R. and Liao,J.C. (2000) Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol., 18, 533–537. [DOI] [PubMed] [Google Scholar]
- Fernández S., Shingler,V. and de Lorenzo,V. (1994) Cross-regulation by XylR and DmpR activator of Pseudomonas putida suggests that transcriptional control of biodegradative operons evolves independently of catabolic genes. J. Bacteriol., 176, 5052–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia J. and de Lorenzo,V. (2000) The role of the inter-domain B linker in the activation of the XylR protein of Pseudomonas putida. Mol. Microbiol., 38, 401–410. [DOI] [PubMed] [Google Scholar]
- Heitzer A., Malachowsky,K., Thonnard,J.E., Bienkowski,P.R., White,D.C. and Sayler,G.S. (1994) Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol., 60, 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtel A., Marqués,S., Möhler,I., Jakubzik,U. and Timmis,K.N. (1994) Carbon source-dependent inhibition of xyl operon expression of Pseudomonas putida TOL plasmid. J. Bacteriol., 176, 1773–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Köhler,T., Rekik,M. and Harayama,S. (1990) Growth-phase-dependent expression of the Pseudomonas putida TOL plasmid pWW0 catabolic genes. J. Bacteriol., 190, 6651–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers M.C., Suske,W.A., Schmid,A., Goslings,D.A., Kohler,H.P. and van der Meer,J.R. (2000) HbpR, a new member of the XylR/DmpR subclass within the NtrC family of bacterial transcriptional activators, regulates expression of 2-hydroxybiphenyl metabolism in Pseudomonas azelaica HBP1. J. Bacteriol., 182, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahng H.Y., Byrne,A.M., Olsen,R.H. and Kukor,J.J. (2000) Characterization and role of tbuX in utilization of toluene by Ralstonia pickettii PKO1. J. Bacteriol., 182, 1232–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler B., Marques,S., Kohler,T., Ramos,J.L., Timmis,K.N. and de Lorenzo,V. (1994) Cross talk between catabolic pathways in Pseudomonas putida: XylS-dependent and -independent activation of the TOL meta operon requires the same cis-acting sequences within the Pm promoter. J. Bacteriol., 176, 5578–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Busby,S., Buc,H., Garges,S. and Adhya,S. (1993) Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem., 62, 749–795. [DOI] [PubMed] [Google Scholar]
- Leahy J.G., Johnson,G.R. and Olsen,R.H. (1997) Cross-regulation of toluene monooxygenases by the transcriptional activators TbmR and TbuT. Appl. Environ. Microbiol., 63, 3736–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau J.H. and van der Meer,J.R. (1996) The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134. J. Bacteriol., 178, 6824–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqués S., Holtel,A., Timmis,K.N. and Ramos,J.L. (1994) Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J. Bacteriol., 176, 2517–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqués S., Gallegos,M.T. and Ramos,J.L. (1995) Role of σS in transcription from the positively controlled Pm promoter of the TOL plasmid of Pseudomonas putida.Mol. Microbiol., 18, 851–857. [DOI] [PubMed] [Google Scholar]
- Marqués S., Manzanera,M., González-Pérez,M.M., Gallegos,M.T. and Ramos,J.L. (1999) The XylS-dependent Pm promoter is transcribed in vivo by RNA polymerase with σ32 or σ38 depending on the growth phase. Mol. Microbiol., 31, 1105–1113. [DOI] [PubMed] [Google Scholar]
- Mason J.R. (1994) The induction and repression of benzene and catechol oxidizing capacity of Pseudomonas putida ML2 studied in perturbed chemostat culture. Arch. Microbiol., 162, 57–62. [DOI] [PubMed] [Google Scholar]
- Matin A., Little,C.D., Fraley,C.D. and Keyhan,M. (1995) Use of starvation promoters to limit growth and selectively enrich expression of tricholoethylene- and phenol-transforming activity in recombinant Escherichia coli.Appl. Environ. Microbiol., 61, 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall S.M., Abraham,B., Narsolis,C.G. and Chakrabarty,A.M. (1997) A tricarboxylic acid cycle intermediate regulating transcription of a chloroaromatic biodegradative pathway: fumarate-mediated repression of the clcABD operon. J. Bacteriol., 179, 6729–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall S.M., Chugani,S.A. and Chakrabarty,A.M. (1998) Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme. Gene, 223, 257–267. [DOI] [PubMed] [Google Scholar]
- Merrick M.J. (1993) In a class of it own: the RNA polymerase sigma factor σ54 (σN). Mol. Microbiol., 10, 903–909. [DOI] [PubMed] [Google Scholar]
- Miura K., Inouye,S. and Nakazawa,A. (1998) The rpoS gene regulates OP2, an operon for the lower pathway of xylene catabolism on the TOL plasmid and the stress response in Pseudomonas putida mt-2. Mol. Gen. Genet., 259, 72–78. [DOI] [PubMed] [Google Scholar]
- Morett E. and Segovia,L. (1993) The σ54-bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol., 175, 6067–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouz S., Merlin,C., Springael,D. and Toussaint,A. (1999) A GntR-like negative regulator of the biphenyl degradation genes of the transposon Tn4371. Mol. Gen. Genet., 262, 790–799. [DOI] [PubMed] [Google Scholar]
- Muffler A., Barth,M., Marschall,C. and Hengge-Aronis,R. (1997) Heat shock regulation of sS turnover: a role DnaK and relationship between stress responses mediated by σS and σ32 in Escherichia coli.J. Bacteriol., 179, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C., Petruschka,L., Cuypers,H., Burchhardt,G. and Herrmann,H. (1996) Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J. Bacteriol., 178, 2030–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North A.K., Klose,K., Stedman,M. and Kustu,S. (1993) Prokaryotic enhancer-binding proteins reflect eukaryotic-like modularity: the puzzle of nitrogen regulatory protein C. J. Bacteriol., 175, 4267–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor K., Buckley,C.M., Hartmans,S. and Dobson,A.D. (1995) Possible regulatory role for nonaromatic carbon sources in styrene degradation by Pseudomonas putida CA3. Appl. Environ. Microbiol., 61, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera E.R., Minambres,B., García,B., Muniz,C., Moreno,M.A., Ferrández,A., Diaz,E., García,J.L. and Luengo,J.M. (1998) Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl Acad. Sci. USA, 95, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M.R., McFall,S., Shinabarger,D. and Chakrabarty,A.M. (1994) Interaction of two LysR-type regulatory proteins CatR and ClcR with heterologous promoters: Functional and evolutionary implications. Proc. Natl Acad. Sci. USA, 91, 12393–12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M.R., Kivisaar,M. and Chakrabarty,A.M. (1995) Differential DNA bending introduced by the Pseudomonas putida LysR-type regulator, CatR, at the plasmid-borne pheBA and chromosomal catBC promoters. Mol. Microbiol., 15, 819–828. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J. and de Lorenzo,V. (1995) Integration host factor (IHF) suppresses promiscuous activation of the σ54-dependent promoter Pu of Pseudomonas putida. Proc. Natl Acad. Sci. USA, 92, 7277–7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martín J. and de Lorenzo,V. (1996a) ATP binding to the σ54-dependent activator XylR triggers a protein multimerization cycle catalysed by UAS DNA. Cell, 86, 331–339. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J. and de Lorenzo,V. (1996b) In vitro activities of an N-terminal truncated form of XylR, a σ54-dependent promoter Pu of Pseudomonas putida. J. Mol. Biol., 258, 575–587. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín, J. and de Lorenzo,V. (1997a) Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol., 51, 593–628. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J. and de Lorenzo,V. (1997b) Co-activation in vitro of the σ54 dependent promoter Pu of the TOL plasmid of Pseudomonas putida by the prokaryotic histone HU and the mammalian HMG-1 protein. J. Bacteriol., 179, 2757–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P.W., Lengeler,J.N. and Jacobson,R. (1993) Phosphoenolpyruvate carbohydrate phosphotransferase system in bacteria. Microbiol. Rev., 57, 543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell B.S., Court,D.L., Inada,T., Nakamura,Y., Michotey,V., Cui,X., Reizer,A., Saier,M.H. and Reizer,J. (1995) Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli.J. Biol. Chem., 270, 4822–4839. [DOI] [PubMed] [Google Scholar]
- Powlowski J. and Shingler,V. (1994) Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation, 5, 219–236. [DOI] [PubMed] [Google Scholar]
- Ramos J.L., Stolz,A., Reineke,W. and Timmis,K.N. (1986) Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc. Natl Acad. Sci. USA, 83, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J.L., Marqués,S. and Timmis,K.N. (1997) Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors an plasmid-encoded regulators. Annu. Rev. Microbiol., 51, 341–373. [DOI] [PubMed] [Google Scholar]
- Saier M.H. (1996) Cyclic AMP-independent catabolite repression in bacteria. FEMS Microbiol. Lett., 138, 97–103. [DOI] [PubMed] [Google Scholar]
- Saier M.H., Ramseier,T.M. and Reizer,J. (1996) Regulation of carbon utilization. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, 2nd ed., pp. 1325–1343.
- Santos P.M., Blatny,J.M., Di Bartolo,I., Valla,S. and Zennaro,E. (2000) Physiological analysis of the expression of the styrene degradation gene cluster in Pseudomonas fluorescens ST. Appl. Environ. Microbiol., 66, 1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleissner C., Reglero,A. and Luengo,J.M. (1997) Catabolism of d-glucose by Pseudomonas putida U occurs via extracellular transformation in d-gluconic acid and induction of a specific gluconate transport system. Microbiology, 143, 1595–1603. [DOI] [PubMed] [Google Scholar]
- Schroetckh V., Wenderoth,R., Kujau,M., Knupfer,U., Riesenberg,D. (1999) The use of elements of the E. coli Ntr system for the design of an optimized recombinant expression system for high cell density cultivations. J. Biotechnol., 75, 241–250. [DOI] [PubMed] [Google Scholar]
- Shigenobu S., Watanabe,H., Hattori,M., Sakaki,Y. and Ishikawa,H. (2000). Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature, 407, 81–86. [DOI] [PubMed] [Google Scholar]
- Shingler V. (1996) Signal sensing by σ54-dependent regulators: derepression as a control mechanism. Mol. Microbiol., 19, 409–416. [DOI] [PubMed] [Google Scholar]
- Shingler V. and Moore,T. (1994) Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J. Bacteriol., 176, 1555–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingler V., Bartilson,M. and Moore,T. (1993) Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J. Bacteriol., 175, 1596–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staijen I.E., Marcionelli,R. and Witholt,B. (1999) The PalkBFGHJKL promoter is under carbon catabolite repression control in Pseudomonas oleovorans but not in Escherichia coli alk+ recombinants. J. Bacteriol., 181, 1610–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze C.C. and Shingler,V. (1999) The alarmone (p)ppGpp mediates physiological-responsive control at the σ54-dependent Po promoter. Mol. Microbiol., 31, 1217–1228. [DOI] [PubMed] [Google Scholar]
- Sze C.C, Moore,T. and Shingler,V. (1996) Growth-phase dependent transcription of the σ54-dependent Po promoter controlling the Pseudomonas-derived (methyl) phenol dmp operon of pVI150. J. Bacteriol., 178, 3727–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K.N. and Pieper,D.H. (1999) Bacteria designed for bioremediation. Trends Biotechnol., 17, 200–204. [DOI] [PubMed] [Google Scholar]
- Timmis K., Steffan,R. and Unterman,R. (1994) Designing microorganisms for the treatment of toxic wastes. Annu. Rev. Microbiol., 48, 525–557. [DOI] [PubMed] [Google Scholar]
- Topp E. and Hanson,R.S. (1990) Degradation of pentachlorophenol by a Flavobacterium species grown in continuous culture under various nutrient limitations. Appl. Environ. Microbiol., 56, 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp E., Crawford,R.L. and Hanson,R.S. (1988) Influence of readily metabolizable carbon on pentachlorophenol metabolism by a pentachlorophenol degrading Flavobacterium sp. Appl. Environ. Microbiol., 54, 2452–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beilen J.B., Wubbolts,M. and Witholt,B. (1994) Genetics of alkane oxidation by Pseudomonas oleovorans.Biodegradation, 5, 161–164. [DOI] [PubMed] [Google Scholar]
- van der Meer J.R., de Vos,W.H., Harayama,S. and Zehnder,A. (1992) Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev., 56, 677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassem R., De Souza,E.M., Yates,M.G., Pedrosa,F.D. and Buck,M. (2000) Two roles for integration host factor at an enhancer-dependent nifA promoter. Mol. Microbiol., 35, 756–764. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Inoue,R., Kimura,N. and Furukawa,K. (2000) Versatile transcription of biphenyl catabolic bph operon in Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem., 275, 31016–31023. [DOI] [PubMed] [Google Scholar]
- Yuste L., Canosa,I. and Rojo,F. (1998) Carbon-source dependent expression of the PalkB promoter from the Pseudomonas oleovorans alkane degradation pathway. J. Bacteriol., 180, 5218–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]