Abstract
Arabidopsis COP1 is a photomorphogenesis repressor capable of directly interacting with the photomorphogenesis-promoting factor HY5. This interaction between HY5 and COP1 results in targeted deg radation of HY5 by the 26S proteasome. Here we characterized the WD40 repeat domain-mediated interactions of COP1 with HY5 and two new proteins. Mutational analysis of those interactive partners revealed a conserved motif responsible for the interaction with the WD40 domain. This novel motif, with the core sequence V-P-E/D-φ-G (φ = hydrophobic residue) in conjunction with an upstream stretch of 4–5 negatively charged residues, interacts with a defined surface area of the β-propeller assembly of the COP1 WD40 repeat domain through both hydrophobic and ionic interactions. Several residues in the COP1 WD40 domain that are critical for the interaction with this motif have been revealed. The fact that point mutations either in the COP1 WD40 domain or in the HY5 motif that abolish the interaction between COP1 and HY5 in yeast result in a dramatic reduction of HY5 degradation in transgenic plants validates the biological significance of this defined interaction.
Keywords: Arabidopsis/COP1/HY5/WD40 domain
Introduction
Arabidopsis seedlings follow two dramatically different developmental patterns depending on the presence or absence of light (Kendrick and Kronenberg, 1994). Light-grown seedlings undergo photomorphogenesis and have short hypocotyls with photosynthetically active and highly differentiated cotyledons. In contrast, seedlings grown in darkness are etiolated and develop elongated hypocotyls with unexpanded cotyledons. Genetic screens have identified both positive and negative regulators mediating this light control of seedling development in Arabidopsis (Deng and Quail, 1999; Nagy and Schafer, 2000; Neff et al., 2000).
Two of these regulators, HY5 and COP1, act as critical components of this developmental switch (Osterlund et al., 1999). The bZIP transcription factor HY5 promotes photomorphogenesis in the light, whereas COP1 represses photomorphogenic development in darkness. The COP1 protein contains three distinct domains, a Zn2+-binding RING finger domain, a coiled-coil domain and seven WD40 repeats in the C-terminal half of the protein (Deng et al., 1992; McNellis et al., 1994a). All characterized lethal cop1 alleles contain mutations within the WD40 domain, whereas the weak alleles either lack the WD40 domain or have mutations N-terminal to the domain (McNellis et al., 1994a). COP1 is nuclear localized in the dark, whereas its nuclear abundance decreases upon exposure to light (von Arnim and Deng, 1994, 1996). COP1 interacts directly with HY5, both genetically and physically, through its WD40 domain, negatively regulating HY5 activity (Ang and Deng, 1994; Ang et al., 1998; Torii et al., 1998). Recent evidence suggests that the interaction results in a dark-dependent degradation of HY5 (Osterlund et al., 2000). HY5 accumulation is very low in dark-grown seedlings and is ∼20-fold higher in the light. The level of HY5 protein correlates directly with photomorphogenic development and inversely with the nuclear abundance of COP1. The regulated HY5 degradation is likely to be mediated by the 26S proteasome and depends on its interaction with COP1 (Osterlund et al., 2000). Taken together, the results suggest that HY5 interacts with the WD40 domain of COP1 and that the interaction negatively regulates the abundance of HY5 in the dark.
Many regulatory proteins have WD40 domains that mediate protein–protein interactions. WD40 domains usually contain 4–8 WD40 repeats and were originally identified in the β-subunit of heterotrimeric G-proteins (Gβ) (Fong et al., 1986). The repeats have since been found in a large number of proteins, including transcriptional repressors, such as TUP1 and Groucho (Paroush et al., 1994; Komachi and Johnson, 1997), as well as several E3 ubiquitin ligases (Tyers and Jorgensen, 2000). Crystal structures of representative WD40 domains alone and in complex with interacting proteins have been determined (Wall et al., 1995; Gaudet et al., 1996; Lambright et al., 1996; Sondek et al., 1996; Sprague et al., 2000). The WD40 repeats form four antiparallel β-strands producing β-sheets, which in turn fold into a propeller-like structure, denoted the β-propeller (Sondek et al., 1996). The β-propeller gains rigidity from hydrophobic interactions between the β-sheets, and solvent-exposed residues in the loops between the β-sheets are free to interact with other proteins. Residues in the loops of Gβ interact with Gα and phosducin, as well as with several other effectors (Gaudet et al., 1996; Lambright et al., 1996; Ford et al., 1998). Furthermore, loop residues in TUP1 are important for the interaction between TUP1 and the DNA-binding protein α2 (Komachi and Johnson, 1997).
In this study, we characterize the interaction between the WD40 domain of COP1 and HY5 as well as with two new interaction partners, STO and STH. We define a novel WD40 domain-interacting motif conserved in all three COP1-interacting proteins. Furthermore, we identify residues important for the interaction both in the motif and in the WD40 domain of COP1, and reveal key features of the underlying interaction.
Results
Mutations in the COP1 WD40 domain abolish COP1–HY5 interaction in yeast
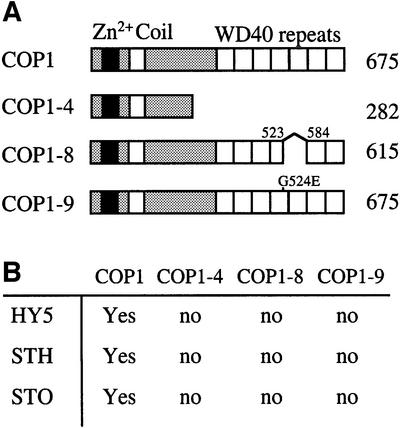
Deletion analyses have shown that the WD40 domain of COP1 is required for its interaction with the bZIP protein HY5 (Ang et al., 1998; Torii et al., 1998). In Arabidopsis, truncated COP1 proteins lacking the WD40 domain (cop1-4), containing a small deletion in the WD40 domain (cop1-8) or possessing a single amino acid substitution (cop1-9) in the WD40 domain cause a loss-of-function phenotypic defect (Figure 1A; McNellis et al., 1994a). Thus, we initially examined the ability of HY5 to interact with the COP1 proteins found in the three cop1 alleles above, using a yeast two-hybrid assay system. To this end, we fused amino acids 25–60 of HY5, sufficient for COP1 interaction in yeast (Hardtke et al., 2000), to the LexA DNA-binding domain (LexA-BD) and assayed the interaction with activation domain-fused wild-type COP1, COP1-4, COP1-8 and COP1-9 proteins. All these COP1 proteins were expressed to similar levels in yeast (data not shown). As shown in Figure 1, the COP1-interacting domain of HY5 interacts well with wild-type COP1 but is unable to interact with the COP1-4, COP1-8 and COP1-9 proteins, suggesting that these mutations in the WD40 domain interfere with the ability of COP1 to interact with HY5.
Fig. 1. The WD40 domain of COP1 is required for interaction with STO, STH and HY5. (A) Diagrams of wild-type COP1 and three mutated versions. The total number of amino acids in each protein are indicated on the right. (B) Summary of the detected interactions between the four versions of COP1 and the three interactive partners. The STO and STH experiments were performed with Gal4-BD-fused COP1 proteins and the HY5 experiments were performed with AD-fused COP1 proteins. ‘Yes’ represents >50-fold activation over background for HY5 and 5- to 15-fold activation over background for STO and STH; ‘no’ represents background activation. See text for additional detail.
Identification of two new proteins interacting with the WD40 domain of COP1
To identify novel COP1-interacting proteins, we used full-length COP1 protein fused to the Gal4 DNA-binding domain (Gal4-BD) to screen a library made from 3-day-old dark-grown Arabidopsis seedlings (Kim et al., 1997). Fourteen of ∼170 000 independent transformants allowed auxotrophic growth and showed β-galactosidase activity in plate assays. The plasmids recovered from 10 of the transformants allowed COP1-dependent auxotrophic growth and β-galactosidase activity when re-introduced in yeast. Sequencing and restriction mapping revealed that the 10 plasmids were encoding five different cDNAs. One of the plasmids contained a truncated salt tolerance (STO) cDNA (Lippuner et al., 1996), whereas two independent plasmids contained a cDNA encoding a homolog of STO (STH) (Figure 2). The STO homolog has been sequenced by the Arabidopsis genome initiative and is located on chromosome II. The predicted protein (DDBJ/EMBL/GenBank accession No. AAD26481) is one amino acid shorter than the protein encoded by the STH cDNA due to an error in exon–intron boundary prediction. To confirm the interactions, we fused the coding regions of STO and STH to the Gal4 activation domain (Gal4-AD) in pGAD (a different two-hybrid vector, see Materials and methods). The STO and STH proteins expressed from the pGAD vector again interact well with wild-type COP1. However, like HY5, STO and STH were unable to interact with the COP1-4, COP1-8 and COP1-9 proteins (Figure 1), suggesting that the WD40 domain of COP1 is also required for interaction with the STO and STH proteins.
Fig. 2. Alignment of the STO and STH proteins. Identical amino acids are boxed and the tandem repeated cysteine-rich motifs are underlined. Arrows indicate the first amino acid of the protein encoded by pGAD-STO and the truncated STH cDNA in pACT-STH2, respectively. The respective DDBJ/EMBL/GenBank accession Nos are AF323666 (STH) and X95572 (STO).
The STO and STH proteins share 70% overall amino acid identity and contain two tandem cysteine-rich motifs in their N-termini (Figure 2). The cysteine motifs, predicted to form Zn2+ fingers, are homologous to motifs found in CONSTANS (CO) and resemble the DNA-binding Zn2+ finger domain in the GATA1 transcription factor (Putterill et al., 1995). The cysteine motifs are directly adjacent in CO but are spaced by nine amino acids in the STO and STH proteins. However, the STO protein and the truncated STH protein recovered in the screen lacked the cysteine motifs (Figure 2), indicating that this domain is dispensable for the interaction with COP1. Taken together, HY5, STO and STH interact with the WD40 domain of COP1 in yeast. The N-terminal half of HY5 and the C-terminal half of STO and STH are sufficient to mediate this interaction.
The mutations in strong and lethal cop1 alleles are likely to distort the structure of the WD40 domain
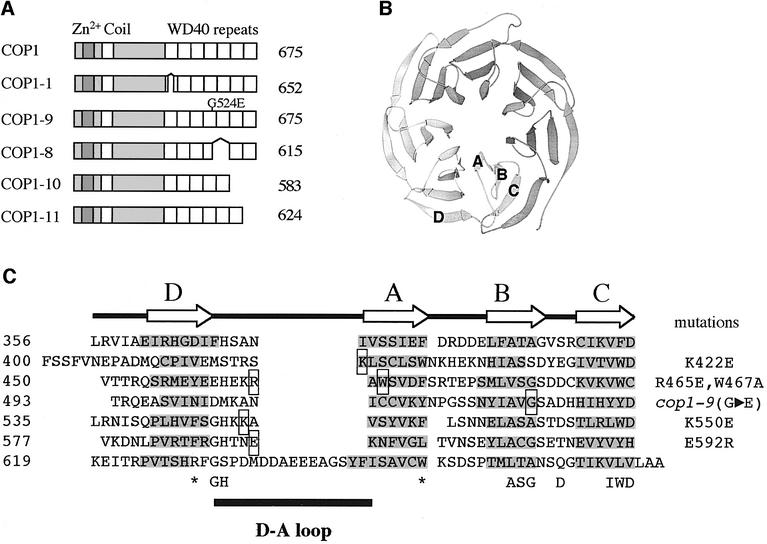
The mutations in the WD40 domain of COP1-8 and COP1-9 abolish interaction with the HY5, STO and STH proteins. So far, all characterized strong and lethal cop1 alleles that still accumulate COP1 protein contain mutations within the WD40 domain (Figure 3A; McNellis et al., 1994a). To gain insight into the molecular nature of the WD40 domain mutations in COP1, we modeled the COP1 WD40 repeats after the β-propeller of Gβ transducin (Sondek et al., 1996; Figure 3B). Secondary structure analysis of COP1 predicts 28 β-strands between amino acids 361 and 673 of COP1 (Figure 3C), suggesting that COP1, like Gβ, has seven β-sheets. The deletions in cop1-1, cop1-8, cop1-10 and cop1-11 remove multiple β-strands, whereas the substitution in cop1-9 (Figure 3A and C) replaces Gly524 with a glutamic acid. In Gβ, this conserved glycine is imbedded in the hydrophobic interface between two β-sheets. A large, charged residue in this position would interfere with the hydrophobic interactions and could potentially disrupt the β-propeller. Thus, it is likely that all mutations found in the strong and lethal cop1 alleles interfere with the formation of a β-propeller.
Fig. 3. Alignment of the WD40 repeats in COP1. (A) Schematic representation of cop1 alleles with WD40 domain mutations adapted from McNellis et al. (1994a). (B) Schematic representation of the β-propeller in Gβ adapted from Sondek et al. (1996). The positions of β-strands A–D are indicated. (C) Alignment of the WD40 repeats in COP1. The β-strands predicted by jpred (Cuff, 1998) are shaded and substituted amino acids are boxed. The coordinates of the amino acid positions are indicated on the left.
COP1 proteins with substitutions of putative protein-interacting residues in the WD40 domain are partially functional in plants
A change in the overall structure of the WD40 domain, as predicted for the strong and lethal cop1 alleles, is likely to interfere with the ability of the domain to interact with multiple interacting proteins. To examine the interaction between the COP1 WD40 domain and the HY5, STO and STH proteins, we generated point mutations in the WD40 domain that could interfere specifically with the interactions of the domain without affecting its β-propeller structure. To this end, we generated five independent amino acid-substituted COP1 proteins (Figure 3C). The substitutions are located in four different D–A loops, thus potentially involved in protein–protein interactions and unlikely to interfere with the folding of the domain. One of the substitutions, W467A, replaces a large hydrophobic side chain with a single methyl group, whereas the others, K422E, R465E, K550E and E592R, are charge reversals.
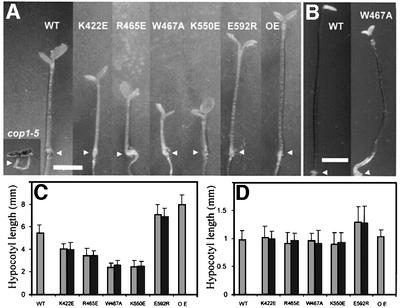
To examine the effect of the WD40 domain substitutions in vivo, we transformed constructs overexpressing each of the substituted COP1 proteins into heterozygous cop1-5 plants. Transgenes segregating as a single locus were selected and selfed to generate lines homozygous for both cop1-5 and the transgenes. At least three independent transgenic lines for each construct were examined, and western blot analysis indicated that the levels of mutated COP1 proteins were similar in all the lines and to that of the wild-type COP1 overexpressor control. All five amino acid-substituted COP1 proteins were able to rescue the lethal phenotype of the cop1-5 null mutation when overexpressed in plants (Figure 4). However, when the seedlings were grown in constant white light, the hypocotyl length differed between the lines. At higher light intensity, 200 µmol/m2/s, the E592R substitution results in slightly longer hypocotyls than wild type, whereas no significant difference was observed between wild-type seedlings and the seedlings overexpressing either COP1 proteins with the K422E, R465E, W467A and K550E substitutions or wild-type COP1, respectively (Figure 4D). At lower intensity light, 40 µmol/m2/s, the E592R substitution still results in a longer hypocotyl than wild type, comparable to seedlings overexpressing wild-type COP1, whereas the K422E, R465E, W467A and K550E substitutions display a significantly shorter hypocotyl than wild-type seedlings (Figure 4A and C). Dark-grown seedlings expressing the substituted COP1 proteins all showed elongated hypocotyls (Figure 4B and data not shown), but ∼70% of the seedlings expressing the W467A-substituted COP1 protein had open cotyledons (Figure 4B). The ability of the substituted COP1 proteins to partially rescue the cop1-5 null mutation indicates that the overexpressed COP1 proteins are functional in plants and that the WD40 domains are properly folded. Furthermore, the phenotypes observed for the substituted COP1 proteins are much weaker as compared with those of the genetically identified mutations, which are likely to affect the overall structure of the WD40 domain. This suggests that the substitutions might affect specific protein–protein interaction surfaces on the COP1 WD40 domain; thus, each substitution potentially may interfere with the interaction of only a subset of COP1 target proteins.
Fig. 4. The amino acid-substituted COP1 proteins can largely rescue the cop1-5 null mutation. (A) Six-day-old cop1-5 null mutant, wild-type (WT), cop1-5 seedlings homozygous for transgenes expressing COP1 proteins with the indicated substitution and seedlings overexpressing wild-type COP1 in a wild-type background (OE) grown in 40 µmol/m2/s constant white light. The scale bar represents 2 mm. Western analysis indicated that all COP1-overexpressing lines accumulated similar levels of COP1 protein. (B) A 7-day-olddark-grown wild-type (WT) seedling and a cop1-5 mutant with 35S::COP1W467A transgene. The scale bar represents 2 mm. (C) Graphical representation of the hypocotyl length of seedlings grown in 40 µmol/m2/s and (D) 200 µmol/m2/s constant white light. Error bars represent standard deviations, n = 30. Data from two representative lines from each construct are presented in (C) and (D). Similar results were observed in all three lines examined for each construct.
Substitutions of loop residues affect the interactions between COP1 and the three interaction partners
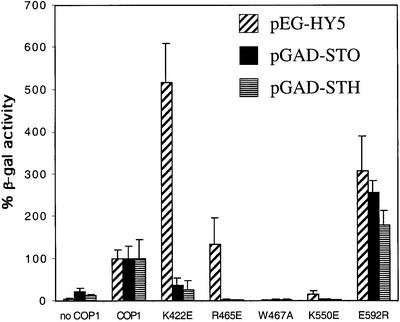
We examined the effect of the COP1 substitutions on the interactions with HY5 and the STO/STH proteins in yeast two-hybrid assays. Due to technical reasons (see Materials and methods), we assayed the COP1–HY5 interactions in the LexA system and the COP1–STO/STH interactions in the Gal4 system. The amino acid-substituted COP1 proteins expressed as well as wild-type COP1 in both two-hybrid systems (data not shown). The use of different two-hybrid systems prevents a direct comparison between the HY5 and the STO/STH proteins; however, the relative effect that the WD40 domain substitutions have on the interaction with each partner can still be compared (Figure 5). Two of the substituted COP1 proteins, K422E and R465E, interact differently with HY5 and the STO/STH proteins. The K422E substitution results in a >5-fold higher interaction with HY5 compared with wild-type COP1, but allows only a weak interaction with the STO and STH proteins. The R465E-substituted protein interacts at wild-type level with HY5 but fails to interact with the STO/STH proteins. The finding that the K422E substitution enhances the interaction with HY5 and decreases the interaction with the STO/STH proteins resembles results seen with Gβ. A study by Ford et al. (1998) has shown that the substitution of loop residues in Gβ could increase, decrease or abolish interactions with different effectors, indicating that a change in the WD40 interaction surface can have different effects on different target proteins.
Fig. 5. Substitutions of loop residues in COP1 affect its interaction with HY5 and STO/STH proteins. The HY5 experiments were performed with AD-fused COP1 proteins in the LexA system, and the STO and STH experiments were performed with Gal4-BD-fused COP1 proteins in the Gal4 system. To allow comparison between the different two-hybrid systems, we set the relative β-galactosidase activities between the wild-type COP1 and its partners to 100%. Error bars represent standard deviations, n = 6.
Interestingly, three of the substitutions have similar effects on the interactions with HY5 and the STO/STH proteins. The COP1E592R protein interacts slightly better than wild-type COP1 with HY5, STH and STO, whereas COP1W467A and COP1K550E are unable to interact with either HY5 protein or STO/STH. The inability of W467A and K550E to interact with HY5 or STO/STH suggests that these residues may be critical for the interaction of COP1 with all three proteins. The identical pattern of interactions between STO and STH with the substituted COP1 proteins suggests that these two proteins interact with a common surface on COP1. Furthermore, the finding that the interaction pattern of the STO and STH proteins overlaps with that of the HY5 protein suggests that STO/STH and HY5 interact with partially overlapping surfaces on the COP1 WD40 domain.
A novel motif mediates interaction with the COP1 WD40 domain
If the HY5 and STO/STH proteins interact with partially overlapping surfaces on the COP1 WD40 domain, it is possible that they contain common sequence features able to mediate the interaction with COP1. Upon examining the amino acid sequence of the HY5 and STO/STH proteins, we found a short sequence within the COP1-interacting domain in HY5 that resembles the sequence in the very C-terminus of the STO and STH proteins (Figure 6A and B). The conserved sequence motif is a stretch of negative amino acids and a spacer of three amino acids followed by the motif V-P-E/D-Φ-G, where Φ designates a hydrophobic residue (Figure 6B). The motif is conserved in tomato HY5, STF proteins from soybean, in a bZIP protein from fava bean and in STO/STH homologs from rice (Figure 6A and B; Cheong et al., 1998; Song et al., 1998).
Fig. 6. HY5, STO and STH interact with COP1 through a novel motif. (A) Schematic representation of Arabidopsis HY5 and HY5 homologs from tomato (THY5; DDBJ/EMBL/GenBank accession No. AJ011914), soybean (STF1 and STF2; DDBJ/EMBL/GenBank accession Nos L28003 and L28004) and fava bean (Vf-bZIP; DDBJ/EMBL/GenBank accession No. X97904), and the Arabidopsis STO and STH proteins with their rice homologs (Os-zf; DDBJ/EMBL/GenBank accession Nos AB001883 and AB001885). The bar represents the position of the motif. Protein sizes (in amino acids) are indicated on the right. (B) Alignment of the motifs in the proteins in (A); the V-P-E/D-Φ-G motif and the cluster of negative amino acids are shaded. (C) Schematic representation of interactions between wild-type and amino acid-substituted COP1, STH and HY5 proteins. +++ represents >350-fold activation over the background level; ++ represents ∼70-fold activation for HY5 and 5- to 15-fold activation for STH over the background level; + represents 3- to 5-fold activation over background; and – represents background levels of activation. The STH experiments were performed with Gal4-BD-fused COP1 proteins and the HY5 experiments were performed with AD-fused COP1 proteins.
To examine whether this conserved motif is important for the interaction with COP1, we substituted the conserved valine–proline (VP) pair in the HY5 and STH protein with alanines. As shown in Figure 5C, alanine substitution of the VP pair completely abolishes the ability of both STH and HY5 to interact with wild type or any of the amino acid-substituted COP1 proteins, indicating that this motif is indeed important for COP1 interaction. The location of the motif, in the N-terminal half of HY5 and in the C-terminus of STO and STH, suggests that its function is position independent.
A salt bridge is involved in the interaction between COP1 and the conserved motif
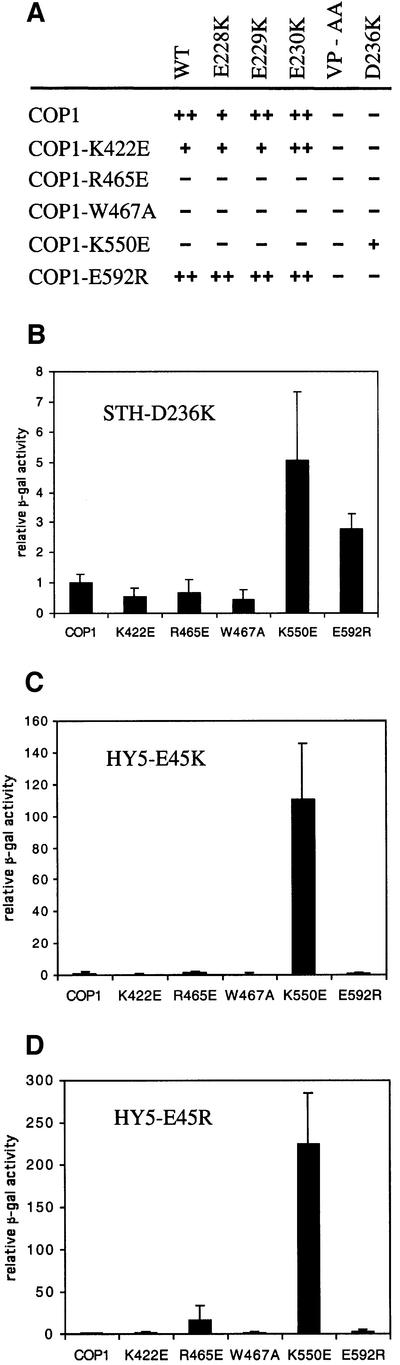
We have shown that the COP1R465E and COP1K550E proteins are unable to interact with the STO/STH proteins (Figure 5). Since both substitutions are positive to negative charge reversals, it is possible that an acidic amino acid in or close to the motif in the STO/STH proteins could form a salt bridge with either R465 or K550 in wild-type COP1. To test this possibility, we generated four STH proteins with single amino acid substitutions reversing the charge of amino acids 228, 229, 230 and 236. The substitutions are unlikely to be structurally disruptive since the motifs in both the STH and HY5 proteins are predicted to form a loop. Three of the amino acid-substituted STH proteins (E228K, E229K and E230K) were able to interact with the same set of COP1 proteins as wild-type STH (Figure 7A). Although the mutations have small quantitative effects on the interaction with wild-type COP1 and COP1K422E, no qualitative differences were observed (Figure 7A). This result suggested that each individual residue in this upstream negatively charged amino acid stretch is dispensable. However, as a whole, this stretch of negatively charged amino acids is essential for COP1–HY5 interaction (Ang et al., 1998). In contrast, the STH-D236K protein is unable to interact with wild-type COP1 but allows a weak interaction with COP1K550E (Figure 7A and B). Thus, the K550E-substituted COP1 protein is unable to interact with wild-type STH and the D236K-substituted STH protein is unable to interact with wild-type COP1. However, when the charge is reversed in both proteins, their interaction is restored, suggesting that amino acid K550 in COP1 interacts directly with amino acid D236 in STH. To test whether the motif in HY5 behaves similarly, we substituted the reciprocal amino acid (E45) with a lysine or an arginine. As shown in Figure 7C and D, the E45K and E45R substitutions in HY5 render the protein unable to interact with wild-type COP1 but allow a strong interaction with COP1K550E. Taken together, this new COP1-interacting motif found in the N-terminal half of HY5 and in the C-terminus of STO and STH requires the amino acids V-P-E/D-Φ-G. Furthermore, we identify direct amino acid contacts between K550 in COP1 and E45 in HY5 and D236 in STH.
Fig. 7. Amino acid K550 in COP1 may form a salt bridge withthe amino acids D236 in STH and E45 in HY5. (A) Schematic representation of interactions between wild-type and amino acid-substituted COP1 and STH. ++ represents 5- to 15-fold activation for STH; + indicates 3- to 5-fold activation over background; and – indicates background levels of activation. (B) Interactions between wild-type and amino acid-substituted COP1 proteins and the STH-D236K, (C) HY5-E45K and (D) HY5-E45R proteins, respectively. The STH experiments were performed with Gal4-BD-fused COP1 proteins and the HY5 experiments were performed with AD-fused COP1 proteins. Error bars represent standard deviation, n = 6.
Mutations affecting the COP1 and HY5 interaction also interfere with HY5 accumulation
A recent study showed that the HY5 protein is degraded in the absence of light, suggesting that the COP1–HY5 interaction mediates a COP1-dependent, negative regulation of the abundance of HY5 protein in the dark (Osterlund et al., 2000). If the residues W467 and K550 in the COP1 WD40 domain are important for the interaction between COP1 and the novel motif in vivo, it is expected that substitutions of W467, K550 or in the motif would interfere with the COP1-mediated degradation of HY5 in Arabidopsis. To test this assumption, we used two independent approaches. First, we assayed the stability of endogenous HY5 protein in the lines expressing the substituted COP1 proteins. Secondly, we examined the ability of endogenous COP1 protein to affect the stability of overexpressed HY5 protein with or without substitutions in the defined motif.
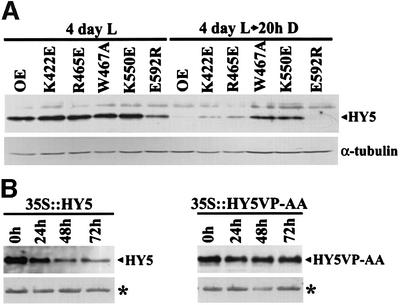
As shown in Figure 8A, HY5 is readily detectable in 4-day-old light-grown seedlings overexpressing wild-type COP1 or either of the five amino acid-substituted COP1 proteins in the cop1-5 null mutant background. Transfer of the seedlings to darkness for 20 h results in a reduction in the amount of endogenous HY5 protein as previously observed (Osterlund et al., 2000). Interestingly, the W467A and K550E substitutions result in a delayed dark-dependent HY5 reduction, suggesting that the COP1 proteins with these two substitutions are less efficient in targeting HY5 for degradation. The observed reduced degradation of HY5 in seedlings expressing the W467A- and K550E-substituted COP1 proteins corresponds well with the loss of interaction in yeast two-hybrid assays (Figure 5) and with the short hypocotyl seen when these seedlings were grown in low-intensity white light (Figure 4A and D). On the other hand, the E592R mutation resulted in a reduced HY5 level in the light, also consistent with its elongated hypocotyl under continuous white light (Figure 4A, C and D) and with the increased interaction in two-hybrid assays (Figure 5). Unexpectedly, while K422E has higher affinity for HY5 in yeast, the seedlings expressing K422E have slightly reduced HY5 degradation in darkness and shorter hypocotyls than seedlings overexpressing wild-type COP1 in low-intensity light. Although we do not know the cause of this result, it is plausible that this mutation has drastic effects on other unknown targets, whose effect on the hypocotyl is strong and dominant over that of HY5.
Fig. 8. Mutations in the COP1 WD40 domain or in the interacting motif of HY5 interfere with dark-dependent degradation of HY5 in vivo. (A) Seedlings homozygous for both cop1-5 and transgenes expressing the indicated substituted COP1 protein were grown in continuous white light (200 µmol/m2/s) for 4 days and then transferred to darkness for 20 h. OE represents 35S-driven overexpression of wild-type COP1 in wild-type Columbia background. Protein extracts were prepared at day 4 (4 Day L) and after 20 h in darkness (20 h D), and the protein concentration was normalized. The amounts of HY5 and α-tubulin proteins were assayed by immunoblotting. (B) The 35S::HY5 and 35S::HY5VP-AA seedlings were grown in white light for 3 days and then transferred to darkness. Protein extracts were prepared at day 3 and after 24, 48 and 72 h in darkness, and the protein concentration was normalized. The amount of HY5 was assayed by immunoblotting, and the asterisk marks a cross-reacting band (Osterlund et al., 2000) that serves as internal loading control. Two independent lines of each transgene were examined and similar results were observed.
We also generated homozygous transgenic lines overexpressing either wild-type HY5 (35S::HY5) or a HY5 protein with the critical VP pair substituted by alanines (35S::HY5VP-AA) in hy5-ks50, a null allele of hy5 (Oyama et al., 1997). Overexpression of both wild-type HY5 and HY5VP-AA complement the hypocotyl phenotype in hy5-ks50, indicating that both proteins are functional in plants (data not shown). The stability of the HY5 proteins was assayed by growing the 35S::HY5 and 35S::HY5VP-AA seedlings in white light for 3 days and subsequent transfer to the dark. Protein extracts were prepared at day 3 and after 24, 48 and 72 h in darkness. As shown in Figure 8B, 35S::HY5 displays clearly reduced HY5 protein levels after 1 day in the dark. In contrast, the HY5 protein levels remain unaffected during the course of the experiment in 35S::HY5VP-AA seedlings. Thus, substitution of the critical VP pair in the motif abolishes COP1 interaction in yeast and results in a dramatic reduction of dark-dependent HY5 degradation in plants. Taken together, the results support the finding that this motif is important for COP1–HY5 interaction and COP1-dependent degradation of HY5 in the dark.
Discussion
This study defines a novel motif found in HY5 and two new COP1-interacting proteins, STO and a STO homolog (STH), which are essential for their interaction with the WD40 domain of COP1. We identify several key residues important for the interaction both in the COP1 WD40 domain and in the motif. Finally, substitutions in the COP1 WD40 domain or in the motif that abolish interaction between HY5 and COP1 in yeast also dramatically reduce the dark-dependent degradation of HY5 protein in plants.
Model for the interaction between the motif and the WD40 domain
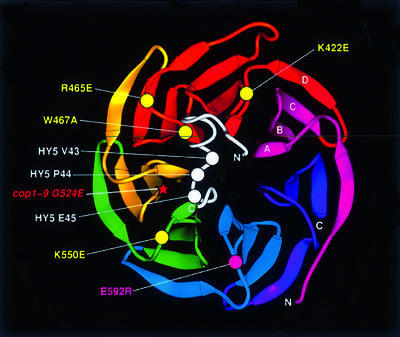
We substituted residues in the COP1 WD40 domain based on known effector-interacting residues in Gβ, assuming that the COP1 domain also forms a β-propeller. The differential interactions observed between the amino acid-substituted COP1 proteins and the HY5/STH proteins support this assumption and suggest that at least four of the five substituted residues, K422, R465, W467 and K550, are important for COP1 and target protein interactions. To understand further the interaction between the motif and COP1, we extended this modeling approach. Figure 9 shows a picture of the Gβ β-propeller where, based on the alignment of the COP1 WD40 repeats, we have indicated the relative position of the substituted COP1 residues. Although the predicted loop sizes in COP1 differ slightly from Gβ, the modeling provides a good estimate of the distances between, and the relative position of, the substituted residues. The residues important for motif interaction cluster on one side of the structure, whereas R592 on the other side had little or no effect. Importantly, W467 would be positioned so that it extends its large hydrophobic side chain towards the center of the β-propeller. This is interesting since, on the one hand, COP1W467A is unable to interact with HY5, STO and STH (Figure 5). On the other hand, a valine–proline pair in the motif of HY5 and STH is critical for COP1 interaction (Figure 6C). Together this could indicate a possible hydrophobic interaction between W467 in COP1 and the VP pair in the conserved motif. Secondary structure analyses suggest that the motif region in the HY5 and STO proteins forms a loop. Furthermore, the sequence immediately preceding the core motif in the HY5 protein is similar to the sequence 216-SDEEIRRDP, which forms a short helix, essentially a loop, on the surface of glutaconate coenzyme A-transferase (GCT, Protein Data Bank file 1POI; Jacob et al., 1997).
Fig. 9. A structural model for the interaction between the HY5 motif and the WD40 domain of COP1. The ribbons diagram (Carson, 1997) of the Gβ β-propeller was used to align the COP1 WD40 repeats. The carbon alpha backbone (Cα) positions of the substituted COP1 residues are marked by circles. Yellow circles indicate residues important for motif interaction whereas the magenta circle marks the mutation that had little or no effect. The star marks the position of the cop1-9 mutation. The COP1-interacting motif from HY5 (in white) is modeled based on the structure of homologous residues 213–239 in GCT (Jacob et al., 1997). The three key residues (V43, P44 and E45) are marked by white circles.
Based on these findings, we propose a model for the defined interaction (Figure 9). In the model, the interaction between the motif and COP1 is mediated in part by a hydrophobic interaction between W467 in COP1 and the VP pair in the motif, and in part by a salt bridge between K550 and the negatively charged residue following the proline in the motif. The distance between the VPE residues is shorter than the distance between W467 and K550, suggesting that the motif may span the hole of the β-propeller. This model could indicate a general mechanism for how the COP1 WD40 domain interacts with its partners. A core hydrophobic interaction complemented by formation of salt bridges with different residues in the loops of the COP1 WD40 domain would then provide multiple interacting surfaces on the domain. The spatial arrangement of the interaction motif on the β-propeller also suggests that even though the WD40 domain of COP1 could have multiple partners, each domain can only interact with one partner at a time.
Short motifs have previously been shown to be important for interactions with WD40 domains. The interaction between the Drosophila WD40 domain-containing transcriptional repressor Groucho and the transcription factor Hairy requires the sequences WRPW found in the C-terminus of Hairy (Paroush et al., 1994). This finding facilitated identification of the transcription factors Runt and Huckebein as targets of Groucho-mediated repression; Runt contains the sequence VWRPY in the C-terminus and Huckebein contains the sequence FRPW in the N-terminus (Aronson et al., 1997; Goldstein et al., 1999). The COP1-interacting motif is present in HY5 homologs in tomato and soybean and in STO homologs in rice, suggesting that the motif has a conserved function.
A possible modulating role of the sequence preceding the motif
Interestingly, the sequence ESDEE preceding the COP1 interaction motif in the HY5 and HY5-like proteins constitutes a consensus casein kinase II (CKII) phosphorylation site (Figure 6B; Oyama et al., 1997). Recent results suggest that this site can be phosphorylated by a CKII-like activity in vitro and that the site is phosphorylated in vivo (Hardtke et al., 2000). In vitro, phosphorylation of this residue reduced HY5’s ability to interact with COP1. Furthermore, when expressed from the native HY5 promoter, a HY5 protein with the phospho-acceptor serine substituted by an alanine, HY5S36A, not only rescues the phenotypic traits of the hy5 mutation, but also compensates them beyond wild-type levels (Hardtke et al., 2000). The mechanism for a phosphorylation-mediated modulation of a WD40 domain interaction was described recently for the Gβ–phosducin interaction. In this case, phosphorylation of Ser73 in phosducin was shown to disrupt the interaction interface between phosducin and Gβγ (Gaudet et al., 1999). If phosphorylation of the CKII site indeed modulates the interaction between HY5 and COP1 in vivo, it would provide a mechanism for fine-tuning of the COP1–HY5 interaction and thereby presumably the COP1-mediated degradation of HY5. The small quantitative differences seen with the 228, 229 and 230 substituted STH proteins (Figure 7A) support the notion of a modulating role for the cluster of negative residues in front of the motif.
Identification of two new COP1-interacting proteins
COP1 has previously been found to interact with CIP1, CIP7 and CIP8; however, none of these proteins interacts with the WD40 domain of COP1 (reviewed in Holm and Deng, 1999). It is interesting that STO and STH interact with the WD40 domain of COP1 since it suggests that STO and STH, like HY5, may be targets of COP1-mediated protein degradation. The STO protein was identified originally by its ability to rescue the salt intolerance phenotype in calcineurin (CN)-deficient yeast (Lippuner et al., 1996). CN is a Ca2+-dependent serine/threonine phosphatase, identified from a variety of eukaryotic organisms. In mammals, CN is an essential component of the Ca2+-dependent signaling pathway leading to T-cell activation (Rao et al., 1997). In plants, the role of CN is less defined; however, a calcineurin-like activity was found to play a role in regulating K+ channels in Vicia faba guard cells in plants (Luan et al., 1993). Expression of the Arabidopsis STO protein in CN-deficient yeast not only complemented the Na+ intolerance but could partially complement all tested CN phenotypes, suggesting that STO might modulate a CN-dependent pathway in yeast (Lippuner et al., 1996). Pharmacological results suggest that both G-protein-coupled and Ca2+-mediated signals are involved in light signaling in plants (Bowler et al., 1994). However, the connection between these results and the genetically identified proteins regulating light signaling remains obscure. It is interesting that COP1 can interact with STO, a putative transcription factor able to modulate Ca2+ signaling in yeast, since it suggests a direct link between a genetically identified repressor of light signals and a downstream target of Ca2+ signals. If the COP1–STO interaction results in STO degradation, it would provide a mechanism by which COP1 could modulate Ca2+-dependent transcription. Further experiments are needed to test this hypothesis.
WD40 domains are key structural signatures in a large number of cellular regulators involved in diverse processes such as signal transduction, transcriptional regulation and cell cycle control. The definition and characterization of a novel COP1-interacting motif could facilitate the identification of novel COP1-interacting proteins and aid in the characterization of WD40 domain-interacting proteins in general.
Materials and methods
Bacterial strains, yeast strains, plant material and plant transformation
The Escherichia coli strain BNN123 was used for library excision, DH5α for subcloning, and the Agrobacterium strain GV3101 pMP90 was used for plant transformation. The yeast strain Y190 (Kim et al., 1997) was used for the two-hybrid screen and for assays with the pAS and pGAD constructs. The yeast strain EGY48-0 (Ausubel et al., 1994) was used for the assays with the pEGHY5(25–60) and pJGCOP1 constructs. The transgenic line overexpressing wild-type COP1 has been described previously (McNellis et al., 1994b). The point-mutated 35S::COP1 constructs were transformed into cop1-5 (Deng et al., 1992) and the 35S::HY5 and 35S::HY5VP-AA constructs into hy5-ks50 (Oyama et al., 1997) plants by the floral dip method (Clough and Bent, 1998). Homozygous lines were then established from primary transformants. At least three independent lines with similar COP1 protein levels for each construct were selected and used for detailed analysis. For analysis of the HY5 protein in the overexpressors, two or more independent transgenic lines per construct were analyzed and the same result was confirmed.
Recombinant plasmids
Plasmid pJGCOP1, fusing the B42 activation domain to the N-terminus of COP1, has been described elsewhere (Ang et al., 1998). All other Gal4-BD and COP1-AD constructs were created by subcloning respective COP1 cDNA to the pAS1 (Kim et al., 1997) and pJG4-5 (Ausubel et al., 1994) vectors, respectively. The pACT-STO plasmid contains a truncated STO cDNA (DDBJ/EMBL/GenBank accession No. X95572) beginning at nucleotide 431. The pACT-STH1 and pACT-STH2 plasmids contain the cDNA for the predicted protein, DDBJ/EMBL/GenBank accession No. AAD26481; the sequence is identical except that the cDNA encodes an additional glutamic acid at position 160. The STH1 sequence begins 82 bp 5′ of the predicted start methionine, whereas the truncated STH2 cDNA encodes amino acids 95–238 of STH. The pACT-STO and full-length pACT-STH, but not the truncated pACT-STH2, contain frameshifts between the Gal4-AD and the cDNA, precluding the translation of AD-fused STO and STH proteins. The pGAD vectors, fusing the Gal4-AD to amino acid 1 of STH and to amino acid 95 of STO, were therefore generated and used in all β-galactosidase assays. The STH coding region, PCR amplified from pACT-STH1, and the STO cDNA from pACT- STO were inserted in pGAD424 (Roder et al., 1996). Plasmid pEGHY5(25–60) and the mutated derivatives fuse the LexA-BD to amino acids 25–60 of HY5. The plasmids were made by PCR amplifying the region of the HY5 cDNA with primers including an in-frame EcoRI restriction site; the fragments were subsequently inserted in the EcoRI and XhoI sites of pEG202. The COP1 cDNAs for transgenic overexpression were generated by site-directed mutagenesis and cloned into pPZP222 (Hajdukiewicz et al., 1994) for expression under control of the 35S cauliflower mosaic virus (CaMV) promoter and terminator. The HY5 cDNAs designed by site-directed mutagenesis were cloned into pBAR-A (DDBJ/EMBL/GenBank accession No. AJ251013) for expression under control of the full-length 35S CaMV promoter and the nopaline synthase terminator. All amino acid substitutions were made by PCR-based mutagenesis. The open reading frames were sequenced and it was confirmed that no mutations were introduced other than the desired ones.
Yeast two-hybrid experiments
The λACT cDNA expression library (Arabidopsis Biological Resource Center # CD4-22) was converted to a pACT library according to Durfee et al. (1993). The two-hybrid screen was performed essentially as described in Kim et al. (1997). The STO/STH–COP1 interactions were assayed with Gal4-BD-COP1 and Gal4-AD-STO and -STH in the Y190 strain, and the HY5–COP1 interactions with LexA-HY5 and AD-COP1 in the EGY48 strain. Different two-hybrid systems were used due to the behavior of COP1 and HY5 proteins in yeast. In the LexA system, AD-fused COP1 and LexA-BD-fused HY5 were readily detectable in immunoblots but we were unable to detect LexA-BD-fused full-length COP1. In the Gal4 system, Gal4-BD-fused full-length COP1 expresses well, but introduction of Gal4-AD-fused HY5 caused a severe reduction in yeast growth. Transformation of Y190 with COP1 bait and STO or STH prey constructs and of EGY48-0 with HY5(25–60) bait and COP1 prey constructs was performed as previously described (Ang et al., 1998). The β-galactosidase assays were performed according to McNellis et al. (1996), with the exception that the galactose induction was omitted in the Y190 assays since the expression from pAS and pGAD vectors is driven by constitutive promoters. Expression of Gal4-BD-COP1 and AD-COP1 fusion proteins was examined by immunoblotting using anti-Gal4-BD and anti-HA antibodies, respectively.
Acknowledgments
Acknowledgements
We thank Drs Mark Osterlund, James Deane and Matthew Terry for reading and commenting on this manuscript, and Matthew Terry for helpful discussions. This work was supported by an NIH grant (GM47850) and in part by a grant from the Human Frontier Science Program Organization (to X.-W.D.). M.H. was supported in part by a postdoctoral fellowship from the Wenner-Gren foundation and the Swedish Medical Research Council, C.S.H. was a Human Frontier Science Program Organization postdoctoral fellow, and X.-W.D. is an NSF Presidential Faculty Fellow.
REFERENCES
- Ang L.-H. and Deng,X.-W. (1994) Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell, 6, 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L.-H., Chattopadhyay,S., Wei,N., Oyama,T., Okada,K., Batschauer,A. and Deng,X.-W. (1998) Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell, 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Fisher,A.L., Blechman,K., Caudy,M. and Gergen,J.P. (1997) Groucho-dependent and -independent repression activities of Runt domain proteins. Mol. Cell. Biol., 17, 5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Saccharomyces cerevisiae. In Current Protocols in Molecular Biology (Suppl.). John Wiley and Sons, New York, NY, pp. 13.6.2–13.6.4.
- Bowler C., Neuhaus,G., Yamagata,H. and Chua,N.H. (1994) Cyclic GMP and calcium mediate phytochrome phototransduction. Cell, 77, 73–81. [DOI] [PubMed] [Google Scholar]
- Carson M. (1997) Ribbons. Methods Enzymol., 277, 493–505. [PubMed] [Google Scholar]
- Cheong Y.H., Yoo,C.M., Park,J.M., Ryu,G.R., Goekjian,V.H., Nagao,R.T., Key,J.L., Cho,M.J. and Hong,J.C. (1998) STF1 is a novel TGACG-binding factor with a zinc-finger motif and a bZIP domain which heterodimerizes with GBF proteins. Plant J., 15, 199–209. [DOI] [PubMed] [Google Scholar]
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cuff J.A., Clamp,M.E., Siddiqui,A.S., Finlay,M. and Barton,G.J. (1998) Jpred: a consensus secondary structure prediction server. Bioinformatics, 14, 892–893. [DOI] [PubMed] [Google Scholar]
- Deng X.-W. and Quail,P. (1999) Signalling in light-controlled development. Semin. Cell Dev. Biol., 10, 121–129. [DOI] [PubMed] [Google Scholar]
- Deng X.-W., Matsui,M., Wei,N., Wagner,D., Chu,A.M., Feldmann,K.A. and Quail,B.H. (1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell, 71, 791–801. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer,K., Chen,P.L., Yeh,S.H., Yang,Y., Kilburn,A.E., Lee,W.H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Fong H.K., Hurley,J.B., Hopkins,R.S., Miake-Lye,R., Johnson,M.S., Doolittle,R.F. and Simon,M.I. (1986) Repetitive segmental structure of the transducin β subunit: homology with the CDC4 gene and identification of related mRNAs. Proc. Natl Acad. Sci. USA, 83, 2162–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford C.E. et al. (1998) Molecular basis for interactions of G protein βγ subunits with effectors. Science, 280, 1271–1274. [DOI] [PubMed] [Google Scholar]
- Gaudet R., Bohm,A. and Sigler,P.B. (1996) Crystal structure at 2.4 Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell, 87, 577–588. [DOI] [PubMed] [Google Scholar]
- Gaudet R., Savage,J.R., McLaughlin,J.N., Williardson,B.M. and Sigler,P.B. (1999) A molecular mechanism for the phosphorylation-dependent regulation of heterotrimeric G proteins by phosducin. Mol. Cell, 3, 649–660. [DOI] [PubMed] [Google Scholar]
- Goldstein R.E., Jimenez,G., Cook,O., Gur,D. and Paroush,Z. (1999) Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development, 126, 3747–3755. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab,Z. and Maliga,P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol., 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda,K., Osterlund,M.T., Oyama,T., Okada,K. and Deng,X.W. (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J., 19, 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M. and Deng,X.W. (1999) Structural organization and interactions of COP1, a light-regulated developmental switch. Plant Mol. Biol., 41, 151–158. [DOI] [PubMed] [Google Scholar]
- Jacob U., Mack,M., Clausen,T., Huber,R., Buckel,W. and Messerschmidt,A. (1997) Glutaconate CoA-transferase from Acidaminococcus fermentans: the crystal structure reveals homology with other CoA-transferases. Structure, 5, 415–426. [DOI] [PubMed] [Google Scholar]
- Kendrick R.E. and Kronenberg,G.M.H. (1994) Photomorphogenesis in Plants. Kluwer, Dordrecht, The Netherlands.
- Kim J., Harter,K. and Theologis,A. (1997) Protein–protein interactions among the Aux/IAA proteins. Proc. Natl Acad. Sci. USA, 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komachi K. and Johnson,A.,D. (1997) Residues in the WD repeats of Tup1 required for interaction with α2. Mol. Cell. Biol., 17, 6023–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambright G.D., Sondek,J., Bohm,A., Skiba,N.P., Hamm,H.E. and Sigler,P.B. (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature, 379, 311–319. [DOI] [PubMed] [Google Scholar]
- Lippuner V., Cyert,M.S. and Gasser,C.S. (1996) Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem., 271, 12859–12866. [DOI] [PubMed] [Google Scholar]
- Luan S., Li,W., Rusnak,F., Assmann,S. and Schriber,S. (1993) Immunosuppressants implicate protein phosphatase regulation of K+ channels in guard cells. Proc. Natl Acad. Sci. USA, 90, 2202–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T., von Arnim,A.G., Araki,T., Komeda,Y., Misera,S. and Deng,X.-W. (1994a) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell, 6, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim,A.G. and Deng,X.-W. (1994b) Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell, 6, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., Torii,K.U. and Deng,X.-W. (1996) Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell, 8, 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F. and Schafer,E. (2000) Nuclear and cytosolic events of light-induced, phytochrome-regulated signaling in higher plants. EMBO J., 19, 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Frankhauser,C. and Chory,J. (2000) Light: an indicator of time and place. Genes Dev., 14, 257–271. [PubMed] [Google Scholar]
- Osterlund M.T., Ang,L.H. and Deng,X.W. (1999) The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol., 9, 113–118. [DOI] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke,C.S., Wei,N. and Deng,X.W. (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature, 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura,Y. and Okada,K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev., 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z., Finley,R.L.,Jr, Kidd,T., Wainwright,M.S., Ingham,P.W., Brent,R. and Ish-Horowicz,D. (1994) Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell, 79, 805–815. [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson,F., Lee,K., Simon,R. and Coupland,G. (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell, 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Rao A., Luo,C. and Hogan,P.G. (1997) Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol., 15, 707–747. [DOI] [PubMed] [Google Scholar]
- Roder K.H., Wolf,S.S. and Schweizer,M. (1996) Refinement of vectors for use in the yeast two-hybrid system. Anal. Biochem., 241, 260–262. [DOI] [PubMed] [Google Scholar]
- Sondek J., Bohm,A., Lambright,D.G, Hamm,H.E. and Sigler,P.B. (1996) Crystal structure of a G protein βγ dimer at 2.1 Å resolution. Nature, 379, 369–374. [DOI] [PubMed] [Google Scholar]
- Song J., Yamamoto,K., Shomura,A., Itadani,H., Zhong,H.S., Yano,M. and Sasaki,T. (1998) Isolation and mapping of a family of putative zinc-finger protein cDNAs from rice. DNA Res., 5, 95–101. [DOI] [PubMed] [Google Scholar]
- Sprague E.R., Redd,M.J., Johnson,A.D. and Wolberger,C. (2000) Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J., 19, 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U., McNellis,T.W. and Deng,X.-W. (1998) Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J., 17, 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M. and Jorgensen,P. (2000) Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev., 10, 54–64. [DOI] [PubMed] [Google Scholar]
- von Arnim A.G. and Deng,X.-W. (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell, 79, 1035–1045. [DOI] [PubMed] [Google Scholar]
- von Arnim A. and Deng,X.-W. (1996) A role for transcriptional repression during light control of plant development. BioEssays, 18, 905–910. [DOI] [PubMed] [Google Scholar]
- Wall M.A., Coleman,D.E., Lee,E., Iniguez-Lluhi,J.A., Posner,B.A., Gilman,A.G. and Sprang,S.R. (1995) The structure of the G protein heterotrimer Giα1β1γ2. Cell, 83, 1047–1058. [DOI] [PubMed] [Google Scholar]