Abstract
The evolution of reading frame maintenance must have been an early event, and presumably preceded the emergence of the three domains Archaea, Bacteria and Eukarya. Features evolved early in reading frame maintenance may still exist in present-day organisms. We show that one such feature may be the modified nucleoside 1-methylguanosine (m1G37), which prevents frameshifting and is present adjacent to and 3′ of the anticodon (position 37) in the same subset of tRNAs from all organisms, including that with the smallest sequenced genome (Mycoplasma genitalium), and organelles. We have identified the genes encoding the enzyme tRNA(m1G37)methyltransferase from all three domains. We also show that they are orthologues, and suggest that they originated from a primordial gene. Lack of m1G37 severely impairs the growth of a bacterium and a eukaryote to a similar degree. Yeast tRNA(m1G37)methyltransferase also synthesizes 1-methylinosine and participates in the formation of the Y-base (yW). Our results suggest that m1G37 existed in tRNA before the divergence of the three domains, and that a tRNA(m1G37)methyltrans ferase is part of the minimal set of gene products required for life.
Keywords: evolution/frameshift/1-methylguanosine/progenote/tRNA
Introduction
Following the chemical evolution of life, a condensation period may have occurred resulting in a population of primitive cells (progenotes) with a rudimentary translation apparatus (Woese, 1998). This translation apparatus was simple and presumably not able to maintain the reading frame for a long distance. Whereas most missense errors are not fatal for the activity and stability of a protein, frameshift errors are detrimental for the synthesis of proteins, which are required for a more evolved and efficient organism. Thus, an early feature to evolve would have been structures that were important for maintenance of the reading frame. Such structures may have been a prerequisite for the emergence of the three domains Archaea, Bacteria and Eukarya. Since tRNA is the primary decoding molecule in all organisms, some tRNA features, e.g. the presence of some early evolved modified nucleosides, may still play a pivotal role in reading frame maintenance and thus be found in tRNA of present-day organisms from all three domains.
Transfer RNA from all organisms contains modified nucleosides, which are derivatives of the four canonical nucleosides: adenosine (A), guanosine (G), uridine (U) and cytidine (C). At present, 79 different modified nucleosides have been characterized in tRNAs from various organisms (Rozenski et al., 1999). Not only are two positions in the tRNA, i.e. the wobble position (position 34) and the position adjacent to and 3′ of the anticodon (position 37), frequently modified, but a plethora of various modified nucleosides are also found in these two positions (Auffinger and Westhof, 1998; Björk, 1998). Of all the modified nucleosides present in tRNA, only eight are present in the same position and in the same subpopulation of tRNA in organisms from all domains (Björk, 1986), suggesting that they may have been present in the tRNA of the primitive organisms existing before the emergence of the three domains Archaea, Bacteria and Eukarya (Björk, 1986; Cermakian and Cedergren, 1998). If a convergent evolution has not occurred, derivatives of the ancestor genes, whose products catalyse the synthesis of these conserved modified nucleosides, will be present in the organisms of today and these modified nucleosides may still have the same function in all organisms. One of these eight conserved modified nucleosides is 1-methylguanosine (m1G37), which is present in position 37 in tRNAs specific for leucine (CUN codons, N being any of the four major nucleosides), proline (CCN) and one of the arginine tRNAs (CGG) from all three domains (Björk, 1998). In fact, among the >500 sequenced tRNAs, only three tRNA species [tRNAHisGUG from HeLa cells, tRNALeuIAG from Caenorhabditis elegans and tRNAArgPCU (P being pseudouridine Ψ) from Ascaris suum] have an unmodified G in position 37 (Sprinzl et al., 1999). Not only is m1G37 present in all organisms, including the organism with the smallest sequenced genome (Mycoplasma genitalium; Fraser et al., 1995), but it is also present in mitochondria and chloroplasts. Although the ‘universal’ code is used in most of the present-day organisms, the codes in Mycoplasma spp and in organelles are somewhat different (Watanabe and Osawa, 1995). tRNAs in organelles and in Mycoplasma spp are much less modified than cytosolic tRNAs (Sprinzl et al., 1999) and they often have a structure that is distinct from those of cytosolic tRNAs (Dirheimer et al., 1995). Still, the presence of m1G37 prevails in tRNA, even in Mycoplasma and in organelles (Björk, 1998). Interestingly, the presence of this conserved modified nucleoside has been shown to prevent frameshifting (Björk et al., 1989; Hagervall et al., 1993). Therefore, the conservation of m1G37 may fulfil the above suggestion that a structure important for the early evolving capacity to maintain the reading frame may still exist in present-day organisms. If indeed a gene whose product was responsible for the formation of m1G37 in tRNA was present in the population of primitive organisms existing before the three domains emerged, sequence similarities of the orthologues may still exist. Here this question is addressed by characterizing the orthologues that catalyse the synthesis of m1G in position 37 of the tRNA from the three domains.
The analysis of the DNA sequence of the smallest genome so far for an autonomously replicating organism (Fraser et al., 1995) initiated a discussion of what is the minimal set of genes required for cellular life. Theoretical considerations suggested that out of the 468 predicted protein-coding genes in M.genitalium, 256 were part of the minimal gene set (Mushegian and Koonin, 1996), and this suggestion was supported recently by transposon mutagenesis (Hutchison et al., 1999). In Salmonella typhimurium and Escherichia coli, the gene trmD, which encodes the enzyme catalysing the formation of m1G37 in tRNA, is the third gene in a four-cistron operon transcribed in the following order: rpsP (encoding ribosomal protein S16), rimM (RimM), trmD [tRNA(m1G37) methyltransferase; the TrmD protein] and rplS (ribosomal protein L19) (Byström et al., 1983). In M.genitalium, the operon organization is conserved except that the rimM gene is not present. Since the first and last genes of the M.genitalium operon encode proteins that are essential in E.coli (encoding S16 and L19, respectively; Persson et al., 1995), no transposon would be expected in the trmD gene even if it is not essential, since such an insertion would have a polarity effect on the expression of the downstream gene encoding the essential ribosomal protein L19. If the presence of m1G37 in tRNA is essential for growth, the trmD gene would be part of the minimal gene set. Therefore, it would contribute to our understanding of what is required as a minimal genome for free-living organisms if we know the influence that m1G37 in tRNA has on the growth capacity of organisms.
Here we characterize the trmD orthologues from organisms belonging to each of the three domains Archaea, Bacteria and Eucarya. We show that sequence similarities still exist between these orthologues, consistent with the suggestion that an ancestor gene encoding an enzyme with similar catalytic activity may have been present before the divergence occurred. We further show that in organisms belonging to either Bacteria or Eucarya, lack of m1G37 in tRNA severely impairs growth to a similar degree. Thus, this conserved modified nucleoside plays a pivotal role in the survival of the organisms in a competitive environment, and consequently belongs to the minimal set of genes required for cellular life. Since the presence of the conserved m1G37 prevents frameshifting (Björk et al., 1989; Hagervall et al., 1993), our results are also consistent with the suggestion that one of the early improvements to the translational apparatus was the ability to maintain the reading frame, thus allowing the synthesis of long peptides necessary for the evolution of present-day organisms.
Results
Lack of m1G in tRNA from S.typhimurium severely impairs growth
The trmD gene, which encodes tRNA(m1G37)methyl transferase (the TrmD peptide), is part of a tetra-cistronic operon transcribed in the following gene order: rpsP(S16)–rimM–trmD–rplS (L19) (Byström et al., 1983). Since deletions or nonsense codons in the trmD gene may also influence the expression of the downstream essential gene rplS (Persson et al., 1995), we isolated missense mutations that abolished the activity of the tRNA(m1G37)methyltransferase and did not have a polarity effect on the expression of the downstream rplS gene. To obtain such non-polar mutations in the trmD gene, we devised a selection procedure for strains deficient in m1G37, as described in Materials and methods. All mutations were shown to be in the trmD gene by complementation analysis using various plasmids (see Materials and methods). The various mutations were scattered all over the trmD gene (Table I). Note that no mutation was obtained that altered a sense codon to a nonsense codon, although the specificity of the mutagen used allows such mutations to occur. This fact suggests that such mutations may be lethal due to a polarity effect on the expression of rplS in addition to the phenotype mediated by the m1G37 deficiency.
Table I. m1G37 levels in tRNA and growth deficiency of S.typhimurium trmD mutants.
| trmD alleles | trmD+ | trmD23 | trmD27/31 | trmD30 | trmD28 | trmD25 | trmD26 | trmD3/24 | trmD33 | trmD29 | trmD32 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid substitution | P58L/L94F | S88L | G117S | G117N | G117Q | S165L | P184L | G199R | G214D | W217D | |
| m1G/Ψa | 0.12 | 0.006 | <0.005 | 0.005 | <0.005 | 0.014 | 0.005 | 0.047 | 0.006 | <0.005 | 0.040b |
| Relative colony size atc | |||||||||||
| 22°C | 1 (1.4 mm)d | No col.e | 0.3 | 0.6 | No col.e | 1.1 | –e | 1.1 | <0.1 | 1.1 | –e |
| 30°C | 1 (3 mm)d | 0.4 | 0.7 | 0.7 | 0.2 | 0.8 | <0.1 | 0.8 | 0.5 | 0.9 | <0.1 |
| 37°C | 1 (1.8 mm)d | 0.4 | 0.7 | 0.6 | <0.1 | 0.7 | 0.1 | 0.9 | 0.4 | 0.1–0.7 | <0.1 |
| 42.5°C | 1 (2.2 mm)d | <0.1 | <0.1 | 0.3 | No col.e | 0.4 | 0.4 | 0.5 | 0.3 | <0.1 | 0.2–0.4 |
| 44°C | 1 (2.5 mm)d | No col.e | No col.e | 0.2 | No col.e | 0.3 | 0.3 | 0.3 | 0.1 | <0.1 | 0.3 |
aThe level of m1G was determined by HPLC in bulk tRNA prepared from cells grown in a rich medium (LB) at 37°C and is expressed as the absorbance of m1G relative to that of pseudouridine (Ψ) at 254 nm.
bThis level is overestimated, since Gm was trailing into the region where m1G migrated.
cThe growth of the various mutants at the indicated temperatures was determined by single-cell outstreak on TYS plates. The relative colony size was determined as the average size of ∼5 colonies in relation to the size of the wild type. Thus, <0.1 indicates that the growth of the mutant was <10% of that of the wild type.
dAt 22 and 30°C, the diameters of the colonies were scored after 48 h and the sizes of the wild-type colonies were 1.4 and 3 mm, respectively. At 37, 42.5 and 44°C, the colony sizes were scored after 24 h and the sizes of wild-type colonies were 1.8, 2.2 and 2.5 mm, respectively.
eNo col. denotes that only weak growth at the primary streak was observed and no individual colonies were visible. – denotes no growth. Bold denotes severe impairment of growth compared with the wild type.
Many of the mutants (trmD23, trmD27, trmD30, trmD28, trmD26, trmD33 and trmD29) had only 4% or less of the wild-type level of m1G in their tRNA (Table I). Upon shift of the mutant trmD27 to 42.5°C, even less m1G was observed (data not shown). However, we cannot exclude the possibility that there was a residual level of m1G, which may be distributed unequally among the various tRNA species that normally contain m1G37. We conclude that some of these trmD mutations reduced the amount of m1G in tRNA to 4% or less of the wild- type level at 37°C and to an even lower level at higher temperature. Concomitant with this reduction in the level of m1G at 37°C, there was a 50 to >90% reduction in growth at 37°C (Table I). However, the reduction in growth became much more severe at a higher temperature (trmD23, trmD27 and trmD29). Since some amino acid substitutions in the TrmD peptide influence the substrate specificity (Li and Björk, 1999), the various mutant forms of the TrmD peptide may have different substrate specificities, resulting in various levels of m1G37 in different tRNA species and, therefore, different phenotypes. The m1G37 deficiency may also induce a temperature sensitivity of tRNA(s), resulting in a more pronounced growth defect. At less optimal temperatures, the growth was much more impaired (Table I). We conclude that the absence of m1G37 severely impairs the growth of S.typhimurium.
Characterization of the trmD orthologue of Methanoccocus vannielii and Methanococcus jannaschii
Plasmids harbouring non-bacterial genes lacking bacterial promoters and translational signals may be expressed in bacteria, albeit at low levels. However, even a low expression of a tRNA-modifying enzyme may be enough to complement a phenotype induced by an undermodified tRNA. To obtain the trmD orthologue from an Archaeon, we introduced a plasmid bank containing chromosomal fragments from M.vannielii into the temperature-sensitive S.typhimurium strain GT5337 (trmD27, purF2085). A plasmid, pUMV4, containing a 1.3 kb M.vannielii chromosomal insert rendered the temperature-sensitive strain GT5337 able to grow at high temperature (Table II). The plasmids also increased the level of m1G in the tRNA. Thus, the 1.3 kb HindIII fragment contains the trmD orthologue of the Archaeon M.vannielii.
Table II. Complementation of the S.typhimurium trmD27 mutation by the trmD orthologues from M.vannieliia and S.cerevisiaea.
| S.typhimurium strains | Relative m1G levelb |
Growth ability atc |
|||
|---|---|---|---|---|---|
| m1G/Ψ | % of wt | 30°C | 37°C | 42.5°C | |
| GT3670 (trmD+) | 0.13 | 100 | 1.0 | 1.0 | 1.0 |
| GT5337 (trmD27) | <0.005 | <4 | 0.5 | 0.5 | <0.1 |
| GT5055 (trmD27/pUMV5a) | <0.005 | <4 | 0.2 | 0.2 | <0.1 |
| GT5054 (trmD27/pUMV4a) | 0.053 | 41 | 0.6 | 0.8 | 0.4 |
| GT5058 (trmD27/pUSC1a) | 0.072 | 60 | 0.7 | 0.8 | 0.4 |
aPlasmids pUMV4 and pUMV5 have a 1.3 and 1.5 kb chromosomal insert, respectively, from M.vannielii, and the pUSC1 plasmid has an insert from S.cerevisiae.
bThe level of m1G was determined as described in Materials and methods and is expressed as the ratio m1G/Ψ and as a percentage of the level in the wild-type strain. Bacteria were grown at 37°C.
cBacteria were grown at the indicated temperatures on TYS plates, or TYS plates containing carbenicillin for the plasmid-containing strains. The relative colony size was determined as described in Table I. The colony size of the wild-type strain GT3670 was 1.3, 2.2 and 2.2 mm at 30, 37 and 42.4°C, respectively.
The insert was partially sequenced and compared with the sequence of M.jannaschii. A 493 nucleotide sequence in the middle of the 1.3 kb insert was 68.8% similar to the MJ883 open reading frame (ORF) of M.jannaschii. We conclude that the MJ883 ORF of M.jannaschii is its trmD orthologue. The M.jannaschii TrmD orthologue showed 7% identity and 16% similarity to the E.coli TrmD protein.
Characterization of the yeast trmD orthologue
The ORF YHR070W encodes a protein of 499 amino acids with unknown function. This protein is 17% identical and 33% similar to the protein encoded by the MJ883 ORF of M.jannaschii. Thus, YHR070W is a potential yeast trmD orthologue. We cloned this gene into vector pYES2, which resulted in plasmid pUSC1. This plasmid complemented the temperature-sensitive growth of S.typhimurium strain GT5337 and raised the level of m1G in the tRNA to ∼40% of the wild-type level (Table II). The increased level of m1G was thus similar to that induced by the 1.3 kb HindIII fragment of M.vannielii. We conclude that YHR070W is the yeast trmD orthologue and we denote this gene TRM5 in accordance with the nomenclature for other yeast genes encoding tRNA methyltransferases.
Deletion of TRM5 in Saccharomyces cerevisiae severely impairs growth
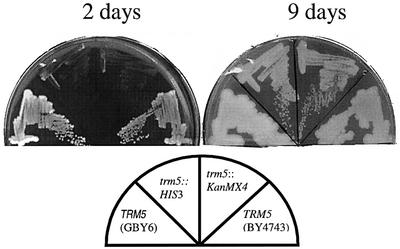
Having identified the TRM5 gene as the potential structural gene for the tRNA(m1G37)methyltransferase of S.cerevisiae, we deleted it by inserting the HIS3 gene between codons 19 and 463 of the 499 amino acid TRM5 gene in the diploid strain GBY1, generating strain GBY3 (TRM5/trm5::HIS3). Upon sporulation of the diploid strain GBY3, His+ and His– segregants were obtained at a ratio of 2:2. Moreover, extremely slow growing segregants co-segregated with the His+ phenotype, linking the slow-growing phenotype with the trm5::HIS3 allele. All slow-growing segregants from several tetrads analysed showed the same growth defect, ruling out that this residual growth was caused by an accumulation of some extragenic suppressors. Figure 1 shows that the TRM5 segregants (His–) formed large colonies after 2 days of incubation at 30°C, whereas the His+ segregants (trm5::HIS3) needed 9 days of incubation to grow to small colonies. Thus, a deletion of the TRM5 gene severely impaired growth.
Fig. 1. Growth of the S.cerevisiae congenic pairs TRM5 and trm5::HIS3, and TRM5 and trm5::kanMX4 (see key to strain at the bottom of the figure) on rich plates after 2 and 9 days, respectively, of incubation at 30°C. After 2 days of incubation, there were no visible colonies of the trm5 mutant.
A previously constructed mutant of the YHR070W ORF contained a substitution of a kanMX4 gene between the start and the stop codons. Analysis of this mutant showed that this gene was essential (Saccharomyces Genome Deletion Project); thus we found it necessary to clarify this discrepancy. The diploid strain BY4743 (TRM5/trm5::kanMX4; obtained from Research Genetics) was sporulated, and fast- and slow-growing segregants were obtained in a 2:2 ratio. All slow-growing segregants from several tetrads showed the same growth defect, suggesting, as above, that an extragenic suppressor did not cause this residual growth. The growth of the haploid deletion strain at 30°C was impaired to a degree similar to that of the deletion strain (trm5::HIS3) constructed by us (Figure 1). Thus, with both constructs, our analysis showed that the TRM5 gene is not essential, although a deletion of it severely impairs growth to a degree similar to that observed for the S.typhimurium trmD mutants.
The yeast tRNA(m1G37)methyltransferase, encoded by the TRM5 gene, catalyses the transfer of a methyl group if a G or an inosine is present in position 37
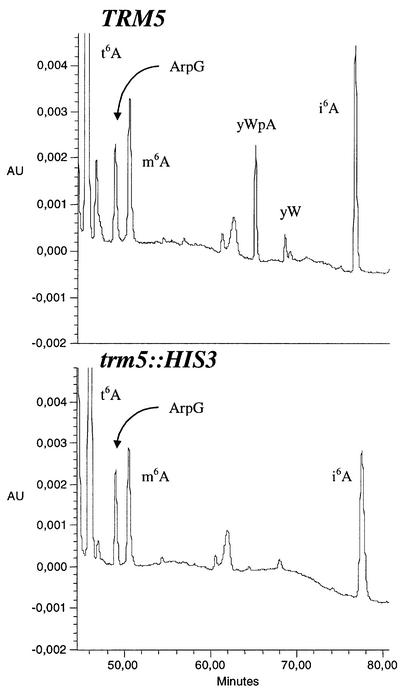
We next determined the m1G content in bulk tRNA from the haploid TRM5 and trm5::HIS3 strains. Table III shows that in the mutant strain the m1G level was reduced by only 21% and lacked yW (the nucleoside of the Y-base present in yeast tRNAPhe; Figure 2). It is known that eukaryotic tRNA species also have m1G in position 9 (Sprinzl et al., 1999), the formation of which is most probably catalysed by an enzyme distinct from tRNA(m1G37)methyltrans ferase. Unlike in E.coli, tRNAHisGUG, tRNAAspIGC and tRNALeuUAG from yeast also contain m1G37. Since the tRNA(m1G37) methyltransferase from E.coli methylates only tRNA specific for leucine (decoding CUN codons), proline (CCN) and arginine (CGG), the yeast Trm5p enzyme may also methylate the same subset of tRNAs exclusively. These considerations might explain the modest decrease of m1G in bulk tRNA of the mutant. To clarify the specificity of the Trm5p enzyme in yeast, we purified various tRNA species by hybridization to matrix-bound oligonucleotides complementary to the 3′ side of the tRNA from wild-type and mutant. The purified tRNA samples, which should only contain one tRNA species, were then analysed, and the modification pattern was determined by HPLC. The results are shown in Table IV.
Table III. Analysis of m1G content in S.cerevisiae TRM5 and the trm5::HIS3 mutant.
| Strains | Genotype | No. of analyses | m1G/Ψa | % of wt | yWb | i6Ab |
|---|---|---|---|---|---|---|
| GBY6,7 | TRM5 | 12 | 0.33 ± 0.04 | 100 | + | + |
| GBY8,9 | trm5::HIS3 | 6 | 0.265 ± 0.003 | 79 | – | + |
aThe level of m1G was determined as described in Materials and methods and is expressed as the m1G/Ψ ratio with the range in the determinations indicated.
bThe presence of yW and i6A was determined by HPLC analysis as shown in Figure 2. + indicates the presence and – the absence of the indicated modified nucleoside.
Fig. 2. Analysis of modified tRNA nucleosides from wild-type S.cerevisiae TRM5 (upper panel) and the trm5::HIS3 mutant (lower panel). Only the portion of the chromatogram between retention times 47 and 82 min is shown. Abbreviations: t6A, N6-threonylcarbamoyladenosine; Ar, 2′-O-ribosyladenosine; m6A, N6-methyladenosine; yW, wybutosine (nucleoside of the Y-base); i6A, N6-isopentenyladenosine.
Table IV. Content of modified nucleosides in various purified tRNA species in S.cerevisiae TRM5 and trm5::HIS3 strains.
| Modified nucleoside | Mutant/wild typea |
|||||||
|---|---|---|---|---|---|---|---|---|
| AlaIGC | AspGUC | LeuUAA | LeuUAG | HisGUG | ProNGG | ArgCCG | PheGmAA | |
| Ψ | 1.1 | 1.1 | 1.3 | 1.4 | 1.1 | 1.0 | 1.0 | 1.1 |
| ncm5U | –b | – | – | – | – | 0.91 | – | – |
| m1A | – | – | 3.7c | 0.97 | – | 1.0 | – | 1.0 |
| m5C | – | 1.0 | 1.1 | 0.83 | 1.3 | 1.0 | 1.4 | 1.1 |
| Cm | – | – | 1.0 | – | – | 1.0 | – | 0.96 |
| m7G | – | – | – | – | – | (incl. in I) | – | (incl. in I) |
| Id | 0.82 | 1.27 | 2.1 | 1.90 | 1.2 | 0.97 | 0.96 | 1.0 |
| m5U | 1.0 | 1.3 | 1.1 | 1.1 | 1.1 | 0.98 | 0.90 | 0.99 |
| Gm | – | – | 1.4 | 0.78 | 1.5 | – | – | 1.4 |
| m1I | <0.1 | – | – | – | – | – | – | – |
| m1G | 1.0 | <0.04 | <0.1 | <0.08 | <0.06 | 0.48 | <0.07 | – |
| ac4C | – | – | 1.3 | 1.1 | – | – | – | – |
| m2G | – | – | 1.0 | 1.1 | – | – | 1.6 | 1.0 |
| m22G | 1.0 | – | 1.0 | 1.0 | – | – | – | 1.0 |
| Am | – | – | – | – | 1.0 | – | – | – |
| yW | – | – | – | – | – | – | – | <0.1 |
aThe numbers given are the ratios of the various modified nucleosides in the indicated tRNA isoacceptor isolated from the trm5::HIS3 mutant and TRM5 wild type. The underlined modified nucleoside, which was used as internal standard, is, according to the sequence, present once in the tRNA species.
b– denotes that this modified nucleoside is, according to the sequence, not present in the wild-type tRNA species and, therefore, not expected to be present in the analysis of a particular tRNA. The absence of such a modified nucleoside demonstrates the purity of the tRNA analysed.
cThis high ratio depends on a low level of m1A in the wild-type tRNALeuUAA. The level of m1A in the trm5::HIS3 mutant was similar to that for all other tRNA species with m1A.
dInosine was found in all samples, regardless of whether the tRNA should contain I or not. This is most probably caused by chemical deamination of A to I in tRNA during the purification procedure.
The tRNALeuUAG and tRNAArgCCG were completely devoid of m1G37 in the trm5::HIS3 mutant. The level of m1G in tRNAProncm5UGG, which contains m1G37 and m1G9, was reduced to 50% of the wild-type level. These results show that the Trm5p enzyme methylates G37 in the same subset of tRNAs as the bacterial enzyme, but does not methylate G9. In addition, the yeast enzyme also methylates tRNAHisGUG, tRNAAspQUC and tRNALeuUAA (Table IV). Thus, the yeast Trm5p and the bacterial TrmD methylate the subset of tRNAs that have G37 and G36. In addition, the yeast enzyme also methylates tRNA species that have a G37, but also have a C36 (tRNAAspQUC) or an A36 (tRNALeuUAA).
The tRNAAlaIGC contains m1G9 and m1I37 (I, inosine) instead of m1G37. The synthesis of m1I37 occurs in two enzymatic steps: the formation of I37 by the Tad1p enzyme followed by a methylation step to form m1I37, catalysed by an unknown methyltransferase (Grosjean et al., 1996; Gerber et al., 1998). Table IV shows that tRNA from both the mutant and the wild type contains m1G, consistent with our suggestion that Trm5p does not mediate the formation of m1G9. However, the mutant lacks m1I37, showing that Trm5p catalyses the transfer of the methyl group to I37, thereby synthesizing m1I37.
The tricyclic nucleoside yW (Y-base nucleoside) is one of the most complex modified nucleosides characterized so far, and is found exclusively in tRNAPheGmAA of eukaryotic organisms, including yeast. Its presence in yeast tRNAPheGmAA, whose three-dimensional structure was the first to be established for a tRNA (Kim et al., 1974; Robertus et al., 1974), evoked much attention regarding not only its function but also its use as a natural fluorescent probe in molecular analysis of the mechanism of translation. The first step in the synthesis of this complex modified nucleoside in yeast is the formation of m1G37 (Droogmans and Grosjean, 1987). Table IV and Figure 2 show that tRNAPheGmAA from the trm5::HIS3 mutant was devoid of yW, suggesting that the tRNA(m1G37)methyl transferase encoded by the TRM5 gene mediates the first step in the synthesis of yW. This is the first gene to be identified whose product is involved in the synthesis of the Y-base.
We conclude that the tRNA(m1G37)methyltransferase encoded by the TRM5 gene in S.cerevisiae catalyses the synthesis of m1G37 in the subset of tRNAs [Leu(CUN), Pro(CCN) and Arg(CGG)] for which m1G37 is conserved. This fact demonstrates that the yeast Trm5p enzyme has a similar and overlapping substrate specificity to that of the bacterial TrmD enzyme. In addition, it also mediates the formation of m1I37 in tRNAAspQUC and yW in tRNAPheGmAA. Unlike the bacterial TrmD enzyme, it is not dependent on the nature of the nucleoside 5′ of the target nucleoside (position 36 can be at least A, G and C). Since the Trm5p from yeast also methylates I37, it does not require an amino group in position 2, as found in G, of the target purine. However, an amino group at position 6, as in adenine, seems to inhibit Trm5p, since several tRNAs in yeast have an unmodified A in position 37. Our results establish furthermore that there is only one tRNA(m1G37) methyltransferase in yeast, since all of the tRNAs analysed that have G37 were shown to be the substrate of the Trm5p enzyme (Table IV).
Discussion
We have identified experimentally the orthologues encoding the tRNA(m1G37)methyltransferase from the three domains Archaea, Bacteria and Eukarya (Table II and Figure 3). The ubiquitous presence of m1G37 in the same subset of present-day tRNAs suggests that this modification may be pivotal for the survival and evolution of different organisms in a competitive environment. Results presented here support this view, since a lack of m1G37 severely impaired the growth of both a bacterium and a eukaryote (Table I and Figure 1). The growth defect in mutants lacking m1G37 in combination with the high conservation of this modified nucleoside suggest that a gene (trmD orthologue) responsible for the synthesis of m1G37 must be part of a minimal gene set. Sequence similarities between these orthologues suggest that organisms present before the three domains emerged possessed an ancestor gene responsible for the formation of m1G37 in their tRNA. Furthermore, our results suggest that the yeast tRNA(m1G37)methyltransferase also catalyses a step in the formation of the tricyclic modified nucleoside yW37 (nucleoside of the Y-base) and m1I37 (Figure 2 and Table IV). Unlike the bacterial enzyme, the yeast enzyme is not dependent on the nature of the nucleoside positioned 5′ and adjacent to the target nucleoside. In the latter case, the target nucleoside can be either G or I.
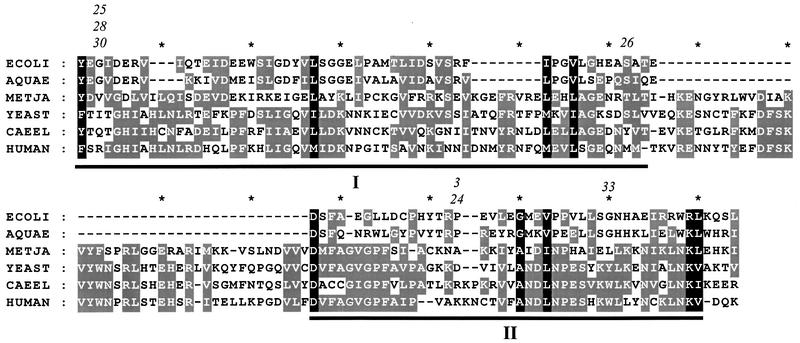
Fig. 3. Sequence of trmD orthologues from E.coli, Aquifex aeolicus, M.jannaschii, S.cerevisiae (yeast), C.elegans and human. The trmD orthologues of C.elegans, A.aeolicus and human were identified by sequence similarities. White letters on a black background indicate similar amino acids in all six species, and white letters on a grey background indicate similar amino acids in at least half of the species. A phylogenetic analysis based on several orthologues of each phylogenic domain revealed that the trmD orthologues fell into the respective domains in a manner typical of macromolecular synthesis of proteins. Numbers above the E.coli sequence are allele numbers for those seven trmD mutations that are located within blocks I or II.
In yeast, 10 tRNAs have introns and three of them (tRNAPheGmAA tRNALeuUAG and tRNALeuCAA) have m1G or yW in position 37 (Westaway and Abelson, 1995). Since these modified nucleosides are located next to the anticodon, they are synthesized after the splicing reaction. Indeed, the tRNA(m1G37)methyltransferase, which is encoded by the TRM5 gene (Table IV) and which participates in the synthesis of yW (Droogmans and Grosjean, 1987), is inhibited by the presence of an intron (Jiang et al., 1997). Thus, the tRNA(m1G37)methyltransferase is expected to be localized to the cytoplasm or on the outside of the nuclear membrane to which the spliced tRNA is transported. Accordingly, no typical nuclear localization signal (NLS)—either a lysine-rich stretch or a bipartite NLS—is present in yeast Trm5p. Several mitochondrial tRNAs also contain m1G37, which may be synthesized by the nuclear-encoded Trm5p provided it is transported to this organelle. The mitochondrial targeting signal (MTS) is usually located in the N-terminal end of the protein and contains basic and hydrophobic amino acids (Stanford et al., 2000). In the case of two other tRNA-modifying enzymes, the nuclear-encoded Trm1p and Mod5p enzymes, the MTS is located in the N-terminus of these proteins. Moreover, the genes for these proteins have multiple translation start sites, which generate different sized proteins transported to various compartments. The TRM5 gene also contains in its 5′ end multiple ATG codons, which may act as alternative translation starts and thus generate, similarly to Trm1p and Mod5p, different sized proteins that may be targeted to the mitochondria. The various N-termini contain basic and hydrophobic amino acids as required for an MTS, suggesting that some of these peptides may contain an MTS. Future experiments will reveal whether Trm5p is transported to the mitochondria
We have shown earlier that a 50% deficiency of m1G37 in S.typhimurium reduces the growth rate by 9–24% depending on the growth medium (Li and Björk, 1995). A 50% reduction of m1G37, as caused by the trmD3 mutation, induces frameshift errors (Björk et al., 1989; Hagervall et al., 1993). We know that deficiency of m1G37 in a frameshift suppressor tRNA increases its frameshifting activity, resulting in lethality (Li and Björk, 1999). Therefore, we suggest that the severe growth impairment in mutants lacking m1G37 is caused by extensive frameshifting by some, if not all, of the tRNAs that normally have m1G37. The yeast trm5 mutant lacked, besides m1G37 in the same subset of tRNAs as the bacterial mutant, m1G37 in tRNAAspQUC, m1I37 in tRNAAlaIGC and yW37 in tRNAPheGmAA. Therefore, these modification deficiencies may also contribute to the growth impairment of the yeast mutant in addition to the suggested frameshifting imposed by the lack of m1G37 in the same subset of tRNAs as in the bacterial mutant. The tad1 mutant, which is mutated in the A37 deaminase, completely lacks m1I37 in tRNAAlaIGC but shows no growth defect (Gerber et al., 1998), suggesting that lack of m1I37 in this tRNA does not contribute to the growth defect of the trm5 mutant. The presence of m1G37 in tRNAAspQUC prevents mischarging by arginine in vitro (Putz et al., 1994); however, since lack of m1G37 does not influence the charging of the cognate amino acid, it is questionable if arginine mischarging occurs under competitive in vivo conditions. The yW modification of yeast tRNAPheGmAA seems not to have any major influence on the aminoacylation reaction (Thiebe and Zachau, 1968), but yW influences –1 frameshifting at the human immunodeficiency virus (HIV) site (Carlson et al., 1999). Lack of the bulky hydrophobic hypermodified nucleoside, ms2io6A37, in S.typhimurium tRNAPheGmAA promotes +1 frameshifting (Qian, 1997; Schwartz and Curran, 1997). Since yW is also a bulky hydrophobic nucleoside, lack of it in the trm5 mutants may also cause frameshifting and thus contribute to the growth defect observed. However, the similarity of the growth defect between the bacterial and the yeast mutant suggests that the major cause of the growth defect is the same, and thus frameshifting may occur by the subset of tRNAs that in both organism contain m1G37, i.e. tRNA specific for leucine, proline and arginine.
The TrmD enzyme from the bacterium E.coli (and most likely S.typhimurium) is specific for position 37, presumably because of the structure of the anticodon loop (Redlak et al., 1997; Qian et al., 1998). Therefore, the lack of m1G37 in this subset of tRNAs causes the impaired growth of such mutants (Table I). Introduction of a plasmid harbouring an M.vannielii ORF complemented the growth phenotype of such an S.typhimurium trmD mutant, showing that the enzyme produced from the archaeal ORF most probably methylates G in position 37 of at least the tRNA(s) that was growth limiting. Since an archaeal gene complemented the growth phenotype and concomitantly increased the level of m1G, it is unlikely that the bacterial TrmD peptide has a second function mediating the growth phenotype. The yeast Trm5p is specific for position 37 (Table IV) and thus has a similar specificity to that of the bacterial enzyme. Thus, sequence similarities and experimentally verified enzymatic specificity demonstrate that the ORFs identified in M.vannielii and in yeast are the S.typhimurium tRNA(m1G37)methyltransferase orthologues. The m1G in position 37 is conserved and present in tRNA from all three domains, Archaea, Bacteria and Eucarya, and also in tRNAs of organelles. Figure 3 shows the regions of TrmD peptides from all three domains that are most similar. Two stretches are particularly similar, which we denote blocks I and II. These conserved sequences are also important for an active enzyme, as evidenced by the fact that seven out of the 12 mutations characterized in the trmD orthologue of S.typhimurium were located in either block I or II (Table I). It has been found that many in vitro induced mutations in block II result in catalytically inactive proteins (M.W.Holmes, personal communication). Although a convergent evolution cannot be ruled out, the sequence similarities that are still present in these proteins suggest that these genes have evolved from a common ancestor. During evolution, there has been a considerable divergence, especially between the bacterial enzyme compared with the Archaea and Eucarya enzymes. Since many components of the translation apparatus are more similar between Archaea and Eucarya (Bult et al., 1996), the observed similarity between the trmD orthologues from Archaea and Eucarya is expected, as the tRNA-modifying enzyme may be considered to be part of the translation apparatus.
The improvement in reading frame maintenance probably occurred already at the progenote stage and before the divergence into the three domains (see Introduction). The presence of m1G37 is important for reading frame maintenance (Björk et al., 1989; Hagervall et al., 1993), and the present work further demonstrates that lack of m1G37 severely impaired the growth of a bacterium and a eukaryote to a similar degree. We suggest that m1G37 represents an example of a feature evolved to improve maintenance of the reading frame before the divergence into the three domains occurred.
Materials and methods
Strains
Bacterial strains used were derivatives of S.typhimurium LT2. Strain GT3670 (purF2085) was obtained from D.Downs (University of Wisconsin, Madison, WI). Strains GT3534 (zff-2551::Tn10dCm) and GT5337 (trmD27, purF2085, zff-2551::Tn10dCm) were constructed in our laboratory. The various strains used in Table I contain, besides the various trmD alleles, the zff-2521::Tn10dCm insertion.
The S.cerevisiae strain GBY1 is a diploid strain (MATα/ MATa his3Δ200/his3Δ200 trp1Δ63/trp1Δ63 uraΔ0/ura3Δ0 met15 Δ0/MET15 ade2Δ::hisG/ADE2) used to construct the strain GBY3, which has the same genetic lesions as GBY1 and, in addition, trm5::HIS3/TRM5. Sporulation of the latter strain resulted in the haploid strains GBY6–9, of which GBY6 and 7 are TRM5, and GBY7 and 8 are trm5::HIS3. Strain BY4743 (MATa/MATα, TRM5/trm5::kanMX4, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, ura3Δ0/ura3Δ0, MET15/met15Δ0, LYS2/lys2Δ0) was obtained form Research Genetics.
Isolation of trmD mutants of S.typhimurium
Thiamine and purine pathways share a common intermediate, 4-aminoimidazolribotide (AIR) (Newell and Tucker, 1967). This intermediate is downstream from the action of the purF gene product and therefore a purF mutant is unable to synthesize thiamine. We have recently discovered that the trmD3 mutation, which results in a deficiency of m1G37, can induce thiamine-independent growth in a purF deletion background (G.R.Björk and K.Nilsson, unpublished). We have utilized this fact and characterized several mutations in the trmD gene that induced a thiamine-independent growth phenotype in a purF deletion strain at 37°C. Phage P22 was grown on strain GT3534, which contains the transposon zff-2521::Tn10dCm, which is closely linked to the trmD gene. This phage stock was treated with hydroxylamine (Hong and Ames, 1971) and used to transduce strain GT3670 (purF2085) selecting for chloramphenicol-resistant (CmR) transductants. Among the CmR transductants, Thi+ clones were selected. All these CmR-Thi+ transductants were mutated in the trmD gene as shown by complementation analysis using plasmids described earlier (Björk et al., 1989). The amino acid alterations caused by the various mutations in the trmD gene are shown in Table II.
Identification of the M.vannielii and S.cerevisiae trmD orthologues
An M.vannielii chromosomal bank in vector pBR322 (kindly provided by Professor August Böck, Munich) was introduced into strain GT5337 (trmD27, purF2085), which grows poorly at 42.5°C on rich medium. Carbenicillin-resistant clones were selected at 30°C and replicated to rich agar plates, which were incubated at 42.5°C. Even at 30°C, large and small colonies were obtained and the large colonies grew well at 42.5°C. By transferring the plasmids back to the parent strain GT5337, it was established that the temperature-resistant growth was associated with the plasmids. Three such clones were saved and one, which contained plasmid pUMV1, was analysed further. Digestion with HindIII resulted in four DNA fragments: a fragment containing the vector and, in addition, 3, 1.5 and 1.3 kb fragments. The 1.5 and 1.3 kb fragments were subcloned into plasmid pUC19, and the resulting plasmids pUMV4 and pUMV5, respectively, were transferred to strain GT5337 (purF2085, trmD27) for complementation tests. It was shown that only plasmid pUMV4 with the 1.3 kb insert, and not plasmid pUMV5 with the 1.5 kb insert, complemented the Ts phenotype conferred by the trmD27 mutation.
The potential yeast trmD orthologue, which was identified by sequence similarities to the M.jannaschii sequence (see Results), was cloned after PCR amplification using one primer covering the start codon of the ORF YHR070W and with a BamHI restriction site at the 5′ end (primer J1: AA-GGATCC-ATGAAAATCGCACTGCCA; the BamHI site is italicized and the start codon of TRM5 underlined) and another primer (primer J2: AA-GAATTC-TGCAAGAGCAGATCGCT; the EcoRI site is italicized) having an EcoRI restriction site at the 5′ end and its 3′ end at the stop codon UGA. The resulting PCR fragment was cloned into plasmid pYES2, which is able to replicate in both S.typhimurium and yeast (Invitrogen, Groningen, The Netherlands). The resulting plasmid pUSC1 was introduced into strain GT5337 (purF2085, trmD27) and was found to complement the Ts phenotype of this strain and induce the synthesis of m1G in its tRNA. Thus, in accordance with other yeast genes coding for tRNA methyltransferases, we denote this gene TRM5.
Construction of an in-frame deletion in TRM5
Primers UptrmD and DowntrmD were constructed to have a 40 nucleotide TRM5 sequence at their 5′ ends and a 20 nucleotide sequence at their 3′ ends, being complementary to the pRS vectors (Brachmann et al., 1998). UptrmD starts at the fifth codon of the TRM5 gene and runs to the 19th codon at which the 20 nucleotide RS sequence starts. The RS sequence is inserted 5′ of the various yeast genes, of which we used the HIS3 gene. The DowntrmD starts at codon 477 and runs to codon 463 at which the RS sequence 3′ of the plasmid inserts starts. The HIS3 gene from plasmid pRS303 (HIS3) (Brachmann et al., 1998) was amplified by PCR and transformed into the diploid His– strain GBY1, generating His+ strain GBY3. Thus, the amplified DNA fragment obtained has a 40 nucleotide TRM5 sequence on each end and the HIS3 gene inserted between codons 19 and 463. Integration at the TRM5 locus was confirmed by PCR. Strain GBY3 was allowed to sporulate and, out of eight tetrads analysed, all were found to co-segregate the His+ phenotype and a very slow growth (Figure 2) in a 2:2 ratio. Thus, the slow growth was always linked to the His+ phenotype. Segregants from one such tetrad were denoted GBY6–9 (Table I).
Analysis of modified nucleosides in tRNA
Bacterial strains were grown in LB medium at 37°C to ∼4–6 × 108 cells/ml (100–150 Klett units). Cells were lysed and total RNA was prepared (Emilsson and Kurland, 1990). Total RNA was dissolved in buffer R200 (10 mM Tris–H3PO4 pH 6.3, 15% ethanol, 200 mM KCl) and applied to a Nucleobond column equilibrated with the same buffer. Transfer RNA was eluted with the same buffer except that the KCl concentration was raised to 600 mM. The tRNA was precipitated with 2.5 vols of cold ethanol, washed twice with 80% ethanol, and dried. The dried tRNA was dissolved in water and a portion of it was degraded to nucleosides as described below. The detection limit of m1G was set to 0.005 m1G/Ψ, since this amount was quantified faithfully. However, in some mutants, this level of m1G was reduced further at high temperature although it was not possible to quantify such a low level.
Yeast cells were grown in YEPD medium (Adams et al., 1997) at 30°C to ∼150 Klett units. Cells were washed and resuspended in 0.9% NaCl and 2 vols of phenol were added. The mixture was shaken for 30 min at room temperatures and then 0.1 vol. of chloroform was added and the mixture was shaken for an additional 15 min. The water and phenol phases were separated by centrifugation for 20 min at 10 000–15 000 r.p.m. in a Sorvall GS3 rotor. Adding 2.5 vols of ethanol and 0.1 vol. of 20% potassium acetate precipitated the tRNA in the water phase. tRNA was purified with 2 M LiCl extraction as described previously (Avital and Elson, 1969). The tRNA precipitate was washed twice with 80% ethanol to remove salt, and dissolved either in water or in 2.5 M tetraethylammonium chloride (TEACl; see below). In all cases, the cultures used for tRNA preparations were homogenous and no revertants or contaminants were observed, as judged from single-cell streak on agar plates at the time of harvest.
To analyse the modification pattern of the various tRNA samples, the tRNA was degraded to nucleosides by nuclease P1 followed by treatment with bacterial alkaline phosphatase (Gehrke et al., 1982). This hydrolysis procedure results in complete digestion to nucleosides except for the wybutosine (yW) for which two products are generated: yWpA and yW. The former product is caused by incomplete hydrolysis at the linkage between yW37 and A38 in tRNAPheGmAA (Gehrke and Kuo, 1990). The hydrolysate was analysed by HPLC (Gehrke and Kuo, 1990).
Purification of specific tRNA species for analysis of tRNA modification pattern
To purify individual tRNA species for analysis of their modification pattern, the desired tRNA species was hybridized to matrix-bound oligonucleotides. These were biotinylated and complementary to the tRNA to be purified, from position 72 and up to a position in the anticodon stem before position 37 at which m1G is present. Thus, these probes were 35–37 nucleotides long. They were attached to Dynabeads M-280, which contains streptavidin, according to the manufacturer’s recommendation (Dynal A.S., Oslo, Norway). The hybridization procedure used is described by Tsurui et al. (1994), except that we used Dynabeads M-280-attached oligonucleotides. The tRNA was detached from the beads by heating at 60°C for 3 min, and was concentrated and desalted by centrifugation using Centricon 10 columns (Amicon, MA). The tRNA was digested to nucleosides, and its modification pattern was analysed as described above. Only those modified nucleosides that should be present in the tRNA species were found, as judged by HPLC analysis (Table IV). This suggests that the tRNA samples were pure and contain no, or only a small amount of, contaminating tRNA species.
Acknowledgments
Acknowledgements
The critical reading of the manuscript by Drs Kerstin Stråby, Bernt Eric Uhlin and Mikael Wikström is gratefully acknowledged. This work was supported by grants from the Swedish Cancer Foundation (Project 680 to G.R.B. and project 3516 to A.S.B.), the Swedish Natural Science Research Council (Project B-BU 2930 to G.R.B., BU-04856 and FO 04856 to A.S.B., and B650–19981122/2000 to O.P.P.), the Swedish Foundation for Strategic Research (BU 04856 to A.S.B.) and Carl Trygger (to O.P.P.).
References
- Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Auffinger P. and Westhof,E. (1998) Location and distribution of modified nucleotides in tRNA. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 569–576.
- Avital S. and Elson,D. (1969) A convenient procedure for preparing transfer ribonucleic acid from Escherichia coli. Biochim. Biophys. Acta, 179, 297–307. [DOI] [PubMed] [Google Scholar]
- Björk G.R. (1986) Transfer RNA modification in different organisms. Chem. Scripta, 26B, 91–95. [Google Scholar]
- Björk G.R. (1998) Modified nucleosides in positions 34 and 37 of tRNAs and their predicted coding capacities. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 577–581.
- Björk G.R., Wikström,P.M. and Byström,A.S. (1989) Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science, 244, 986–989. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies,A., Cost,G.J., Caputo,E., Li,J., Hieter,P. and Boeke,J.D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Bult C.J. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- Byström A.S., Hjalmarsson,K.J., Wikström,P.M. and Björk,G.R. (1983) The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J., 2, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson B.A., Kwon,S.Y., Chamorro,M., Oroszlan,S., Hatfield,D.L. and Lee,B.J. (1999) Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology, 255, 2–8. [DOI] [PubMed] [Google Scholar]
- Cermakian N. and Cedergren,R. (1998) Modified nucleosides always were: an evolutionary model. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 535–541.
- Dirheimer G., Keith,G., Dumas,P. and Westhof,E. (1995) Primary, secondary and tertiary structures of tRNAs. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC. pp. 93–126.
- Droogmans L. and Grosjean,H. (1987) Enzymatic conversion of guanosine 3′ adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: dependence on the anticodon sequence. EMBO J., 6, 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V. and Kurland,C.G. (1990) Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J., 9, 4359–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.M. et al. (1995) The minimal gene complement of Mycoplasma genitalium. Science, 270, 397–403. [DOI] [PubMed] [Google Scholar]
- Gehrke C.W. and Kuo,K.C. (1990) Ribonucleoside analysis by reversed-phase high performance liquid chromatography. In Gehrke,C.W. and Kuo,K.C.T. (eds), Chromatography and Modification of Nucleosides. Part A. Analytical Methods for Major and Modified Nucleosides. J. Chromatography Library. Elsevier, Amsterdam, Vol. 45A, pp. A3–A71.
- Gehrke C.W., Kuo,K.C., McCune,R.A., Gerhardt,K.O. and Agris,P.F. (1982) Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J. Chromatogr., 230, 297–308. [PubMed] [Google Scholar]
- Gerber A., Grosjean,H., Melcher,T. and Keller,W. (1998) Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J., 17, 4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. et al. (1996) Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie, 78, 488–501. [DOI] [PubMed] [Google Scholar]
- Hagervall T.G., Tuohy,T.M., Atkins,J.F. and Björk,G.R. (1993) Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol., 232, 756–765. [DOI] [PubMed] [Google Scholar]
- Hong J.S. and Ames,B.N. (1971) Localized mutagenesis of any specific small region of the bacterial chromosome. Proc. Natl Acad. Sci. USA, 68, 3158–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.A., Peterson,S.N., Gill,S.R., Cline,R.T., White,O., Fraser,C.M., Smith,H.O. and Venter,J.C. (1999) Global transposon mutagenesis and a minimal Mycoplasma genome. Science, 286, 2165–2169. [DOI] [PubMed] [Google Scholar]
- Jiang H.Q., Motorin,Y., Jin,Y.X. and Grosjean,H. (1997) Pleiotropic effects of intron removal on base modification pattern of yeast tRNA(Phe): an in vitro study. Nucleic Acids Res., 25, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Suddath,F.L., Quigley,G.J., McPherson,A., Sussman,J.L., Wang,A.H., Seeman,N.C. and Rich,A. (1974) Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science, 185, 435–440. [DOI] [PubMed] [Google Scholar]
- Li J.N. and Björk,G.R. (1995) 1-Methylguanosine deficiency of tRNA influences cognate codon interaction and metabolism in Salmonella typhimurium. J. Bacteriol., 177, 6593–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.N. and Björk,G.R. (1999) Structural alterations of the tRNA(m1G37)methyltransferase from Salmonella typhimurium affect tRNA substrate specificity. RNA, 5, 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian A.R. and Koonin,E.V. (1996) A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA, 93, 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell P.C. and Tucker,R.G. (1967) New pyrimidine pathway involved in the biosynthesis of the pyrimidine of thiamine. Nature, 215, 1384–1385. [DOI] [PubMed] [Google Scholar]
- Persson B.C., Bylund,G.O., Berg,D.E. and Wikström,P.M. (1995) Functional analysis of the ffh–trmD region of the Escherichia coli chromosome by using reverse genetics. J. Bacteriol., 177, 5554–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz J., Florentz,C., Benseler,F. and Giege,R. (1994) A single methyl group prevents the mischarging of a tRNA. Nature Struct. Biol., 1, 580–582. [DOI] [PubMed] [Google Scholar]
- Qian Q. (1997) Transfer RNA Modification and Translational Frameshifting. PhD thesis, Umeå University, Umeå, Sweden. Solfjädern Offset AB, Umeå.
- Qian Q., Li,J.N., Zhao,H., Hagervall,T.G., Farabaugh,P.J. and Björk,G.R. (1998) A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell, 1, 471–482. [DOI] [PubMed] [Google Scholar]
- Redlak M., Andraos-Selim,C., Giege,R., Florentz,C. and Holmes,W.M. (1997) Interaction of tRNA with tRNA (guanosine-1)methyltransferase: binding specificity determinants involve the dinucleotide G(36)pG(37) and tertiary structure. Biochemistry, 36, 8699–8709. [DOI] [PubMed] [Google Scholar]
- Robertus J.D., Ladner,J.E., Finch,J.R., Rhodes,D., Brown,R.S., Clark,B.F. and Klug,A. (1974) Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature, 250, 546–551. [DOI] [PubMed] [Google Scholar]
- Rozenski J., Crain,P.F. and McCloskey,J.A. (1999) The RNA modification database: 1999 update. Nucleic Acids Res., 27, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. and Curran,J.F. (1997) Analyses of frameshifting at UUU-pyrimidine sites. Nucleic Acids Res., 25, 2005–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Vassilenko,K.S., Emmerich,J. and Bauer,F. (1999) tRNA sequences and tRNA genes database. http://www.uni-bayreuth.de/departments/biochimie/trna/
- Stanford D.R., Martin,N.C. and Hopper,A.K. (2000) ADEPTs: information necessary for subcellular distribution of eukaryotic sorting isozymes resides in domains missing from eubacterial and archaeal counterparts. Nucleic Acids Res., 28, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebe R. and Zachau,H.G. (1968) A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur. J. Biochem., 5, 546–555. [DOI] [PubMed] [Google Scholar]
- Tsurui H., Kumazawa,Y., Sanokawa,R., Watanabe,Y., Kuroda,T., Wada,A., Watanabe,K. and Shirai,T. (1994) Batchwise purification of specific tRNAs by a solid-phase DNA probe. Anal. Biochem., 221, 166–172. [DOI] [PubMed] [Google Scholar]
- Watanabe K. and Osawa,S. (1995) tRNA sequences and variations in the genetic code. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 225–250.
- Westaway S.K. and Abelson J. (1995) Splicing of tRNA precursors. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 79–92.
- Woese C. (1998) The universal ancestor. Proc. Natl Acad. Sci. USA, 95, 6854–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]