Abstract
Human CD81, a known receptor for hepatitis C virus envelope E2 glycoprotein, is a transmembrane protein belonging to the tetraspanin family. The crystal structure of human CD81 large extracellular domain is reported here at 1.6 Å resolution. Each subunit within the homodimeric protein displays a mushroom-like structure, composed of five α-helices arranged in ‘stalk’ and ‘head’ subdomains. Residues known to be involved in virus binding can be mapped onto the head subdomain, providing a basis for the design of antiviral drugs and vaccines. Sequence analysis of 160 tetraspanins indicates that key structural features and the new protein fold observed in the CD81 large extracellular domain are conserved within the family. On these bases, it is proposed that tetraspanins may assemble at the cell surface into homo- and/or hetero-dimers through a conserved hydrophobic interface located in the stalk subdomain, while interacting with other liganding proteins, including hepatitis C virus E2, through the head subdomain. The topology of such interactions provides a rationale for the assembly of the so-called tetraspan-web.
Keywords: CD81 extracellular domain structure/HCV receptor/human CD81/tetraspanins/tetraspan-web
Introduction
Tetraspanins form a widely distributed protein superfamily (Maecker et al., 1997; Levy et al., 1998) structurally characterized by four hydrophobic transmembrane regions (TM1–4) and two extracellular domains, known as large and small extracellular loops (LEL and SEL, respectively). The LEL domain, topologically located between TM3 and 4, contains four invariant Cys residues, two of which define a conserved Cys-Cys-Gly motif. In humans, tetraspanins are localized in different organs and tissues. CD9, CD63, CD81 and CD82 are nearly ubiquitous, whereas other tetraspanins seem to be restricted to specific tissues, such as the lymphoid cells (CD53) or mature B cells (CD37). Remarkably, many tetraspanins are being discovered as tumor-associated antigens related to tumor growth and proliferation. Such an observation is in line with an emerging body of evidence indicating that tetraspanins are key players in the regulation of cell adhesion, proliferation, activation and migration (Levy et al., 1998). Tetraspanins are known as molecular facilitators, associating in large cell-surface signaling complexes (referred to as the tetraspan-web), which include integrins (Hemler, 1998), co-receptors (CD4, CD8 and CD2) (Imai and Yoshie, 1993; Hammond et al., 1998) and other tetraspanins (Berditchevski et al., 1996; Rubinstein et al., 1996). Moreover, two members of the tetraspanin family serve as ligands for two important human pathogens: hepatitis C virus (HCV) (Pileri et al., 1998) and diphtheria toxin (Cha et al., 2000).
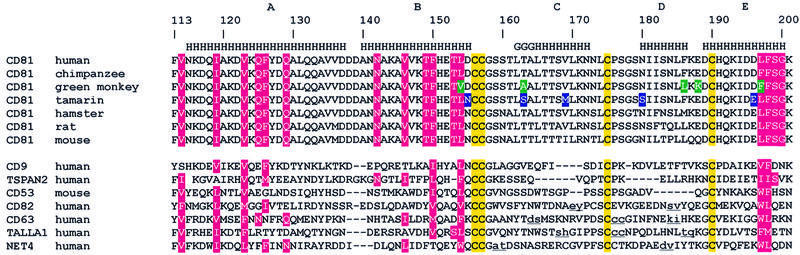
Sequence database surveys allow recognition that the tetraspanin homology superfamily is composed of several subfamilies [CD9, CD37, CD63, CD53, CD82, CD151 and others under characterization (Maecker et al., 1997)], giving a total of ∼160 family members presently known [see Figure 1 for a multi-sequence alignment of human CD81 large extracellular loop (hCD81-LEL) versus selected tetraspanins]. Among these, CD81 and CD9 families display close sequence (and therefore structural) homology (in man, 23% residue identities in their respective LEL domains), whereas hCD82-LEL is the most distantly related to hCD81-LEL (9% identical residues). Very little is known about structure–function relationships within the tetraspanin family. Recently, it has been shown that very high-affinity binding of hCD81 to the E2 envelope glycoprotein of HCV is strictly dependent on the integrity of the two disulfide bridges formed upon oxidation of the four conserved cysteines within the LEL domain (Petracca et al., 2000). Moreover, in studying E2–hCD81 interaction, Phe186 has been identified as a crucial CD81 residue mediating HCV binding, while only few additional residues, all belonging to the LEL domain, have been shown to participate in viral recognition (Higginbottom et al., 2000; Meola et al., 2000). On the other hand, the C-terminus proximal region of the CD151-LEL domain has been implicated in the interaction with the α3 subunit of α3β1 integrin (Yauch et al., 2000). Considering that anti-hCD81 conformational antibodies bind both the membrane-associated hCD81 and the purified soluble hCD81-LEL with high affinity, whereas they do not bind the reduced or denatured proteins (i.e. suggesting correct folding; Higginbottom et al., 2000), the recombinant hCD81 soluble form has been adopted for functional characterization studies (Higginbottom et al., 2000; Meola et al., 2000; Petracca et al., 2000).
Fig. 1. Amino acid sequence alignment of tetraspanin LEL domains from mammalian sources. Letters indicating the secondary structure [as obtained from PROCHECK (Laskowski et al., 1993): H, α-helix; G, 310 helix] are shown on the top line. On the left, protein names and sources are indicated. The Entrez accession Nos for the sequences used in the alignment are: human CD81, NP_004347; tamarin (Saguinus oedipus), CAB89875; rat, NP_037219; mouse, P35762; human CD9, NP_001760; human TSPAN2, NP_005716; mouse CD53, NP_031677; human CD82, NP_002222; human CD63, NP_001771; human TALLA1, AAF4412; human NET4, AAC17120. Amino acid sequences of CD81 from chimpanzee, green monkey and hamster have been obtained from Pileri et al. (1998). The last four sequences contain insertions, which have not been included in the alignment to avoid the introduction of long gaps, located between pairs of lower case underlined residues. In detail, the insertion stretches are: in CD82, eLMNRPEVTy and sLSVRKGFCEAPGNRTQSGNHPEDWPv; in CD63, dWEKIPs, cINVTVGc and kAi; in TALLA1, sPYFLEh, cMNETDc and tVAATKVNq; and in NET4, aFGADDWNLNIYFNCt and dVINTQCGYDARQKPEVDQQIv. The conserved Cys residues are enclosed in yellow boxes. Amino acids involved in the association interface are shown in pink boxes. The residues that differ between hCD81-LEL and agmCD81-LEL or tamCD81-LEL are boxed in green and blue, respectively. Note that residue 163 is mutated in both agmCD81-LEL and tamCD81-LEL.
With the aim of shedding light on the so far undiscovered structural bases of tetraspanins and of the hCD81–HCV interaction, we have undertaken crystallographic analysis of the hCD81 LEL domain, whose three-dimensional (3D) structure is reported here at 1.6 Å resolution.
Results and discussion
Structural bases of the CD81-LEL fold
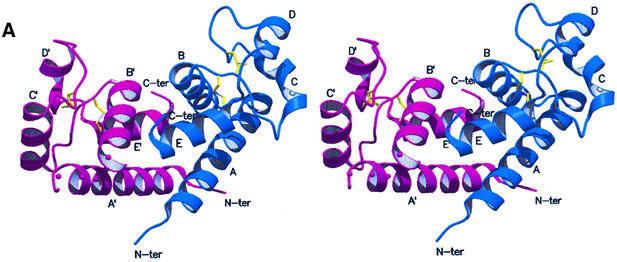
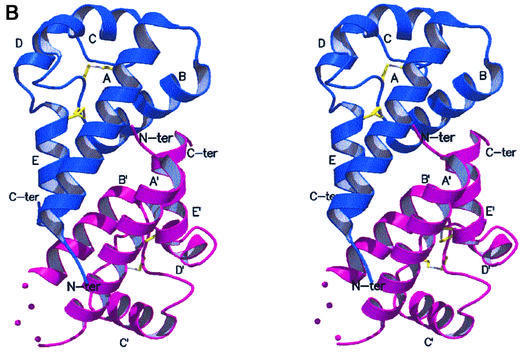
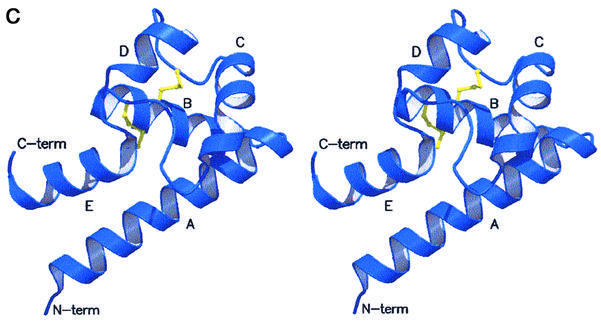
The crystal structure of hCD81-LEL was solved by means of multiple isomorphous replacement techniques, and subsequently refined at 1.6 Å resolution (general R-factor = 0.187, Rfree = 0.238, for the data in the 20.0–1.6 Å resolution range; see Materials and methods; Table I). As shown in Figure 2A and B, hCD81-LEL is organized in a non-crystallographic homodimeric structure, hosting 176 residues and 194 solvent molecules per asymmetric unit. Each subunit in the dimer is composed of five α-helices (A–E) (Figure 2C), spanning residues Asn115–Asp137, Ala140–Asp155, Leu165–Asn172, Asn180–Phe186 and Asp189–Gly200, respectively [amino acids are numbered according to the full hCD81 molecule sequence (Maecker et al., 1997)]. A short 310 helical segment covers residues Leu162–Ala164. The antiparallel A- and E-helices can be seen as the stalk of a mushroom-shaped molecule, whose head subdomain (∼60 residues) is essentially composed of the last two turns of the A-helix, the B-, C- and D-helices and their intervening segments, and the DE loop (loop regions are defined by the helices they connect, e.g. AB, DE). A DALI search through the database of protein 3D structures (Holm and Sander, 1993) does not highlight significant structural homology of the hCD81-LEL fold to any known protein tertiary structure.
Table I. Data collection, structure solution and refinement statistics for hCD81-LEL.
| Native1 | LuCl3 | Hg(Ac)2 | K2PtCl6 | Native2 | |
|---|---|---|---|---|---|
| Diffraction data | |||||
| wavelength (Å) | 1.54 | 1.54 | 1.54 | 1.54 | 0.93 |
| resolution (Å) | 2.7 | 2.7 | 3.0 | 3.3 | 1.6 |
| unique reflections | 5136 | 4979 | 3619 | 2521 | 21557 |
| Rmerge (%)a | 5.8 | 6.0 | 8.4 | 4.4 | 3.8 |
| completeness (%)b | 99.7 (99.4) | 98.0 (93.4) | 98.1 (97.1) | 91.8 (87.5) | 98.0 (93.1) |
| redundancy | 3.4 | 2.0 | 3.2 | 8.2 | 6.9 |
| MIR phasing | |||||
| phasing powerc/Rcullisd | |||||
| acentric | 1.49/0.78 | 1.25/0.84 | 1.30/0.87 | ||
| centric | 1.16/0.82 | 0.98/0.85 | 0.90/0.89 | ||
| anomalous | 0.95/0.97 | 0.66/0.99 | 0.65/0.99 | ||
| overall FOM (acentric/centric) | 0.62/0.58 | ||||
| Refinement | |||||
| resolution range (Å) | 20.0–1.6 | ||||
| Rfactore/Rfreef (%) | 18.7/23.8 | ||||
| no. of protein atoms | 1345 | ||||
| no. of solvent atoms | 194 | ||||
| Ramachandran distribution (% core, allowed, generous, disallowed) | 92.4, 7.6, 0, 0 | ||||
| r.m.s. bonds (Å), angles (°) | 0.006, 1.2 | ||||
| average B value (Å2) | 35.2 |
aRmerge = Σ|Ii – <Ii>|/Σ<Ii>, where Ii is the observed intensity and <Ii> is the average intensity over symmetry equivalent measurements.
bCompleteness for all reflections and for the highest resolution shell in parentheses.
cPhasing power = Σ|FH|/Σ||FPH(obs)| – |FPH(calc)||, where FPH and FH are the derivative and calculated heavy-atom structure factors, respectively.
dRcullis = Σ│||FPH ± FP| – FH(calc)|/Σ|FPH – FP|, where FPH, FP and FH are the derivative, native and calculated heavy-atom structure factors, respectively.
eRfactor = Σ||Fo| – |Fc||/Σ|Fo|
fRfree is calculated as Rfactor, but on 5% of all reflections that were never used in crystallographic refinement.
Fig. 2. Stereoview of the dimeric hCD81-LEL. The view, generated with MOLSCRIPT and RASTER3D (Kraulis, 1991; Merritt and Murphy, 1994), shows the two subunits in blue and purple. The helix labels are distinguished in the two protomers by a prime. The molecular 2-fold axis is close to vertical (A) and located between the N- and C-termini of the two chains. Solid dots (in the purple subunit) trace an approximate path for residues 238–241 not observed in the electron density maps. In (B), the 2-fold axis is orthogonal to the drawing plane. (C) Stereoview of the hCD81-LEL isolated protomer. The view highlights the head subdomain localization relative to the N- and C-terminal helices (α-helices A and E, respectively), and the labeling of secondary structure elements. The two disulfide bridges are shown in yellow.
The subunit fold is stabilized by a number of specific interactions. The stalk region is held together by hydrophobic contacts along the A- and E-helices (residues Ile119, Ala120, Val123, Ile194, Leu197, Phe198) and by two salt bridges (Lys124–Asp195 and Asp128–His191). The antiparallel pairing of the N- and C-terminal α-helices allows the two termini of the expressed hCD81-LEL to fall spatially close to each other, in keeping with their topological location between TM3 and -4 segments within the intact tetraspanin (Levy et al., 1998).
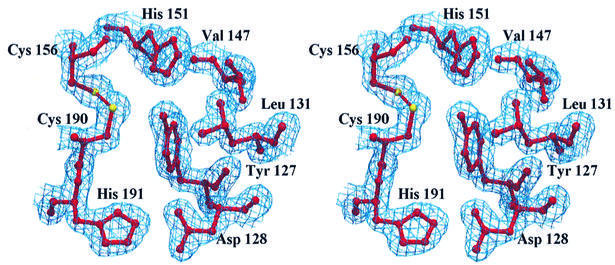
The head subdomain is stabilized by three major structural motifs. First, by the two tetraspanin-invariant disulfide bridges, which originate from two adjacent (156 and 157) Cys residues and stretch approximately in opposite directions within the domain (Figure 2C), connecting the C-terminal region of the B-helix (Cys156) to the first turn of the E-helix (Cys190), and the BC segment (Cys157) to the CD loop (Cys175). Secondly, by the Gly158 and Pro176 residues, which display unique backbone conformations (as indicated by their Ramachandran φ,ψ pairs: 87°,173° and –47°,145°, respectively) and are strategically located to accommodate the structural constraints imposed by the covalent Cys157–Cys175 disulfide connection on the CD loop. Thirdly, by Tyr127, fully buried and surrounded by Leu131, Val147, Phe150, His151 and by the Asp128–His191 hydrogen-bonded salt bridge (Figure 3). The Tyr127 hydroxyl group is hydrogen bonded to the His151 NE2 atom (2.71 Å) and falls 3.61 Å from the Cys190 Sγ atom. The imidazole ring of His151 is nestled between the two disulfide bridges, and is at hydrogen bonding distance from Cys190 Sγ (3.56 Å). Such a hydrogen-bonding network involving Tyr127, His151 and Cys190 may play a role in regulating the redox properties of one or both disulfide bridges, a key property in a cell-surface-exposed domain. Tyr127 is strictly conserved in CD81 and in all the CD9 sequences known to date; residue 151 is either His or Gln in the amino acid sequences representing the different tetraspanin families in Figure 1.
Fig. 3. Stereoview of part of the residues packing against the Tyr127 side chain at the core of the head subdomain, together with the Cys156–Cys190 disulfide. The Cys157–Cys175 disulfide has not been drawn for clarity. The electron density of the refined model (2Fo – Fc map) is represented at the 1σ level.
Inspection of the electrostatic potential at the molecular surface reveals two extended low-polarity regions. A first large hydrophobic cluster has an area of 986 Å2 and mediates the association of the dimeric structure shown in Figure 2A and B. The two hCD81-LEL chains assemble around a local 2-fold axis, displaying intermolecular contacts mainly at the A:A′ helix interface (the prime identifies the partner subunit) and along the B-helix, which is in contact with the C-terminal region of the facing protomer. The A:A′ interface segment involves primarily residues Val114, Ile119, Val123, Gln125, Phe126 and Gln129 in both antiparallel helices. The B-helix interface includes residues Asn142, Val146, Thr149, Phe150, Thr153 and Leu154, which contact Leu197′, Phe198′, Ser199′ and Gly200′. No water molecules are buried at the association interface. Owing to the dimer internal symmetry, the N- and C-termini of both protomers fall in a restricted, central region, but on opposite faces of the assembled dimer (Figure 2). Such a quaternary structure is compatible with the formation of similar dimeric assemblies also by the intact CD81 at the cell surface when transmembrane segments of the two tetraspanin partners are present.
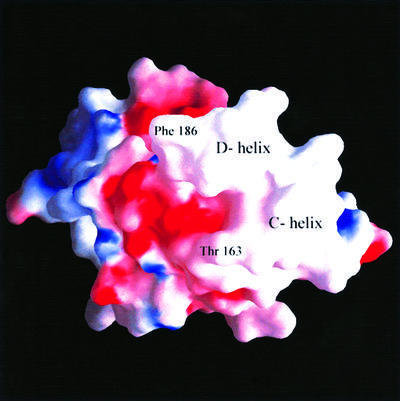
The second low-polarity patch (Figure 4) is located in a solvent-exposed region comprising the C- and D-helices. In the crystal, this region is virtually solvent inaccessible due to extended lattice-packing contacts. Such a hydrophobic region appears peculiar since its solvent-exposed location is energetically unfavorable, and therefore prone to mutations by natural selection. On the contrary, residues Ile181, Ile182, Leu185 and Phe186 are highly conserved throughout different CD81 amino acid sequences (Figure 1). These uncommon structural properties are indicative of a molecular region designed and conserved for the purpose of protein–protein recognition, in keeping with CD81’s propensity to interact with several membrane-associated proteins, as is the case for the highly stable interaction with the α4 subunit of α4β1 integrin (Serru et al., 1999). As an alternative hypothesis, not explored so far by any experimental approach, this might be a site of intramolecular contact with the CD81 short extracellular loop. Consideration of the tetraspanin topology, of the small size of the hCD81-SEL region (30 amino acids) and of its mostly polar nature does not support such a hypothesis. Indeed, as described below, this low-polarity patch is the hCD81-LEL region held to interact with the HCV-E2 envelope glycoprotein.
Fig. 4. View of the molecular surface of hCD81-LEL. The dimer (GRASP view; Nicholls et al., 1991) is in an orientation rotated by ∼90° around the vertical axis relative to that of Figure 2A. Such an orientation brings α-helices C and D into the foreground; they are labeled in the exposed low-polarity region (white) of the molecular surface. The two amino acid residues shown to play a role in hCD81-LEL–HCV-E2 protein interaction are also indicated.
Inspection of the crystal structure shows that in the dimeric hCD81-LEL the distance between the two low-polarity patches, each comprising C- and D-helices, is ∼30 Å, remarkably matching the distance between antigen-binding sites in an assembled antibody molecule. Higher affinity of bivalent proteins, such as specific antibodies, capable of recognizing hCD81, relative to isolated monomeric E2, has been reported (Higginbottom et al., 2000), stressing the occurrence of hCD81 as a homodimeric species at the cell surface. Recent theoretical modeling studies indicate that the HCV-E2 glycoprotein may indeed exist as a head-to-tail homodimer, with CD81-binding sites located at opposite ends of the assembled dimer (Yagnik et al., 2000).
CD81–HCV-E2 interaction
The proposed role of the head subdomain exposed low-polarity patch as the CD81 contact site to the viral glycoprotein is supported by analysis of the natural or engineered mutations affecting CD81-LEL–HCV-E2 interaction. In fact, two animal models from primates not infected by HCV are available (Houghton, 1996; Meola et al., 2000). However, their CD81-LEL domains display opposing trends for what concerns HCV–CD81 binding and infectivity. On one hand, according to predictions, African green monkey (Chlorocebus aethiops) CD81 (agmCD81) does not bind HCV-E2 (Pileri et al., 1998); on the other hand, tamarin (S.oedipus) CD81 (tamCD81) binds to E2 with very high affinity (Meola et al., 2000). Such apparently contrasting observations indicate that full virulence may be based on several cooperating factors, such as the availability of a high-affinity HCV/hepatocyte docking protein (active CD81), as well as of efficient coreceptor(s) for virus internalization (Petracca et al., 2000).
As shown in Figure 1, mutations occur at five residue sites in both agmCD81-LEL and tamCD81-LEL domains, relative to hCD81-LEL. Nevertheless, single-site mutagenesis experiments have so far been limited to only three sites, indicating, most prominently, that Phe186 is the critical residue affecting E2 binding. In fact: (i) the Phe186Ala mutation engineered in hCD81-LEL fully impairs binding to E2 and to specific antibodies; (ii) the Leu186Phe mutation engineered in agmCD81-LEL restores E2 binding; and (iii) tamCD81, which binds E2 with high affinity, naturally displays Phe186 (Pileri et al., 1998; Higginbottom et al., 2000; Meola et al., 2000). On the other hand, the Thr163Ala and Asp196Glu mutations affect hCD81 binding to E2 only marginally (Higginbottom et al., 2000).
From a structural viewpoint, the residues naturally mutated in agmCD81- and tamCD81-LELs map in the head subdomain, with only the exception of residues 196 and 197, which are conservatively substituted next to the C-terminus (Figure 1), Leu197 being a subunit interface residue in hCD81-LEL. In the absence of specific mutational data, it can be noted that residues at sites 155, 180 and 188 are solvent exposed, such that their conservative replacement should not alter the domain conformation, but can affect intermolecular recognition. The substitutions of residues 154 and 169, on the contrary, are subject to polarity and residue size restrictions, since they occur at buried locations next to both disulfide bridges. Finally, in hCD81-LEL, Thr163 and Phe186, shown to participate in E2 binding, are located in the short 310 segment preceding the C-helix and in the last turn of the D-helix, respectively, both residues being solvent exposed (Figure 4).
Therefore, two main aspects can be considered concerning hCD81–E2 recognition. On one hand, the agmCD81-LEL residue mutations focus the attention on the C-, D- and E-helices, and their intervening segments, i.e. on residues that are mostly comprised within the two disulfide bridges and include the 179–193 sequence stretch previously recognized as the minimal epitope for E2 binding (Higginbottom et al., 2000). On the other hand, given the small size of the head subdomain and the presence of intramolecular structural constraints, conformational integrity of the hCD81-LEL region proposed to bind E2 is strictly related to maintenance of both disulfide bridges in the oxidized state. Reduction of hCD81-LEL disulfides has indeed been shown to impair E2 binding and antibody recognition completely (Higginbottom et al., 2000; Petracca et al., 2000). Altogether, these data support a primary role for the head subdomain low-polarity patch in mediating the hCD81–E2 interaction and suggest that additional residues, such as Ile181, Ile182 and Leu185, solvent exposed and clustering around Phe186 in the D-helix, may contribute to the virus recognition surface.
The tetraspanin family: implications from the CD81-LEL structure
Inspection of the tetraspanin amino acid sequence alignments suggests that the hCD81-LEL overall structural organization and the hydrophobic dimerization interface are basic structural features that may be largely conserved within this protein homology family. This hypothesis is supported by the invariance in all tetraspanins of the 4-Cys motif, together with substantial conservation of head subdomain key residues such as Tyr127, His151, Gly158, Pro176 and Ile194, indicating the likely presence of an α-helical LEL head subdomain throughout the family. Moreover, the conservation of seven apolar residues located in the A-, B- and E-helices (Figure 1) is indicative of a preserved hydrophobic association interface, leading to a conserved homodimeric assembly within the homology superfamily; it may also hint at the possible assembly of heterodimeric tetraspanin species.
Inspection of the available sequence data also shows that the tetraspanin LEL domains display enhanced residue variability (occasionally with substantial insertions or deletions) in protein segment 158–189, comprising the low-polarity patch identified above in the head subdomain. The exposed structural location and hypervariability of this segment (through the tetraspanin subfamilies), as opposed to a generally conserved protein association interface and head subdomain core, may indicate that in different subfamilies this surface region is indeed involved in species- or tetraspanin-specific recognition processes.
In the last few years, a large body of evidence has accumulated on the role of tetraspanins in the lateral association of large cell-surface signaling complexes. However, very little is known about how the different proteins interact to build up the so-called tetraspan-web. The two main structural motifs revealed by our crystallographic investigation on hCD81-LEL (i.e. the dimer association interface and the head subdomain low-polarity cluster centered around the C- and D-helices) provide the rationale for a tetraspan-web model based on the association of cell-surface tetraspanin homo- (or hetero-) dimers with integrins and other membrane proteins. Through dimeric assembly, tetraspanins could provide the topological organization to guide clustering of the different tetraspanin-binding proteins on the cell surface, allowing the proper signaling of kinase-mediated phosphorylation to take place (Woods and Couchman, 2000). According to such an assembly model, the formation of an extended bi-dimensional tetraspan-web would occur either through the involvement of other facilitator molecules, such as syndecans known to link integrins together (Hammond et al., 1998), or via as yet undiscovered tetraspanin-liganding proteins displaying two binding sites.
The 3D structure of hCD81-LEL reported here sheds first light on the structural bases of the new tetraspanin LEL fold. Moreover, it provides evidence that an evolutionarily conserved association interface may support assembly of the cell-surface tetraspan-web through homo- (or possibly hetero-) dimeric tetraspanin association, leading to both intra- and inter-cellular signaling and adhesion events. In this framework, a smaller, solvent-exposed apolar cluster, comprising hCD81-LEL C- and D-helices, is identified as the CD81 binding site for the viral E2 glycoprotein, based on the present structural data and mutational experiments as well as antibody binding studies. This surface region is, therefore, the primary target for future detailed characterization of the molecular association processes upon which HCV binding to the hepatocyte is based.
Materials and methods
Expression, purification and crystallization
His-tagged hCD81-LEL was purified from a recombinant Escherichia coli strain as a fusion protein with the IgG binding domain of the Staphylococcus aureus protein A (Nilsson et al., 1987). Briefly, the fusion protein gene, inserted into pEZZ 18 plasmid (Pharmacia), was designed by assembling: (i) a synthetic DNA fragment coding for a tandem duplication of the protein A-Z domain (derived from the plasmid vector); (ii) a hinge region encoding the peptide Leu-Val-Pro-Arg-Gly-Ser carrying the thrombin cleavage site; (iii) the sequence encoding the LEL domain from amino acid 112 to 202 (CD81 sequence); (iv) a His6-tag coding sequence. After purification by IgG affinity chromatography (Pharmacia), the fusion protein was cleaved with thrombin (Pharmacia), and hCD81-LEL was further purified using metal ion affinity chromatography (Pharmacia). The purified protein was fully active as judged by the recombinant E2–CD81 inhibition of binding assay (Rosa et al., 1996). Crystals were grown by equilibrating a hCD81-LEL solution, at 10 mg/ml, against a reservoir containing 15% (w/v) PEG 4000, 0.1 M sodium chloride and 0.1 M MES buffer pH 6.0, at 293 K, in sitting-drop vapor-diffusion set-ups. The crystallization droplets consisted of 2 µl of protein and 2 µl of reservoir solutions, with 500 µl of reservoir solution; triangle-shaped tabular crystals appeared within a few days and grew to maximum dimensions of ∼0.3 × 0.3 × 0.1 mm3 (Kitadokoro et al., 2001).
Data collection, MIRAS phasing and refinement
Crystals were flash-cooled at 100 K using 20% glycerol as added cryoprotectant. Native and derivative diffraction data were collected in-house on a MAR 345 imaging plate. Additional native data (at 1.6 Å resolution, completeness 98.0%, Rmerge 3.8%) were collected on beamline ID14 (ESRF, Grenoble, France) on a MAR CCD detector. All data were processed using DENZO and SCALEPACK (Otwinoski and Minor, 1997) and merged using the CCP4 (1994) program suite (see Table I). The crystals belong to the monoclinic space group P21 (a = 31.5 Å, b = 77.2 Å, c = 38.5 Å, β = 107.4°), with two molecules per asymmetric unit (Vm = 2.16 Å3/Da).
More than 50 different heavy-atom compounds were tested and diffraction data collected. Suitable derivatives used were LuCl3 (soaking concentration 2.5 mM), HgAc2 (soaking concentration 10 mM) and K2PtCl6 (soaking concentration 5 mM), which yielded an overall figure of merit of 0.62 at 3.2 Å resolution (Table I). Crystallographic phases were calculated with CCP4 programs and refined using SHARP (de La Fortelle and Bricogne, 1997) and SOLOMON (Abrahams and Leslie, 1996). The resulting electron density maps allowed tracing of ∼80% of the two independent molecules. Model building and inspection were based on the O suite (Jones et al., 1991). The structure was refined using CNS (Brünger et al., 1998) and REFMAC (Murshudov et al., 1997); 5% of the unique reflections were used to monitor Rfree. The final values for general R-factor and Rfree are 18.7 and 23.8%, respectively (all reflections in the 20.0–1.6 Å resolution range). The refined model consists of 176 amino acids, with 194 water molecules; stereochemistry checks indicate that the refined model is in quite good agreement with expectations for models within this resolution range (Laskowski et al., 1993) (Table I). The two hCD81-LEL chains are numbered Phe113–His202 and Phe213–His302 (His202 and His302 are part of the C-terminal His-tag). Residues 238–241 are disordered in the A′B′ loop. Atomic coordinates for hCD81-LEL have been deposited with the Protein Data Bank, with accession code 1g89 (Berman et al., 2000).
Acknowledgments
Acknowledgements
We are grateful to ESRF (Grenoble) for granting access to ID14 beamline and to EMBL/DESY (Hamburg). K.K. is a member of the Sakabe TARA project of the University of Tsukuba. We are indebted to Dr Rino Rappuoli for discussion and support throughout the work and for critical reading of the manuscript. We also thank Dr Camillo Rosano for helpful assistance in different computational stages of this investigation and Dr Alessandro Muzzi for artwork. This work was partially funded by the Italian Ministry of University and Scientific and Technological Research (MURST), and ASI grant I/R/28/00.
REFERENCES
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Berditchevski F., Zutter,M.M. and Hemler,M.E. (1996) Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembrane domains (TM4 proteins). Mol. Biol. Cell, 7, 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system (CNS): a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cha J.H., Brooke,J.S., Ivey,K.N. and Eidels,L. (2000) Cell surface monkey CD9 antigen is a coreceptor that increases diphtheria toxin sensitivity and diphtheria toxin receptor affinity. J. Biol. Chem., 275, 6901–6907. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Hammond C., Denzin,L.K., Pan,M., Griffith,J.M., Geuze,H.J. and Cresswell,P. (1998) The tetraspan protein CD82 is a resident of MHC class II compartments where it associates with HLA-DR, -DM and -DO molecules. J. Immunol., 161, 3282–3291. [PubMed] [Google Scholar]
- Hemler M.E. (1998) Integrin associated proteins. Curr. Opin. Cell Biol., 10, 578–585. [DOI] [PubMed] [Google Scholar]
- Higginbottom A. et al. (2000) Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol., 74, 3642–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Houghton M. (1996) In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott-Raven, Philadelphia, PA, pp. 1035–1058.
- Imai T. and Yoshie,O. (1993) C33 antigen and M38 antigen recognized by monoclonal antibodies inhibitory to syncytium formation by human T cell leukemia virus type 1 are both members of the transmembrane 4 superfamily and associate with each other and with CD4 or CD8 in T cells. J. Immunol., 151, 6470–6481. [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kitadokoro K., Galli,G., Petracca,R., Falugi,F., Grandi,G. and Bolognesi,M. (2001) Crystallization and preliminary crystallographic studies on the large extracellular domain of human CD81, a tetraspanin receptor for hepatitis C virus. Acta Crystallogr. D, in press. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Levy S., Todd,S.C. and Maecker,H.T. (1998) CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol., 16, 89–109. [DOI] [PubMed] [Google Scholar]
- Maecker H.T., Todd,S.C. and Levy,S. (1997) The tetraspanin superfamily: molecular facilitators. FASEB J., 11, 428–442. [PubMed] [Google Scholar]
- Meola A. et al. (2000) Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol., 74, 5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nilsson B., Moks,T., Jansson,B., Abrahmsen,L., Elmblad,A., Holmgren,E., Henrichson,C., Jones,T.A. and Uhlen,M. (1987) A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng., 1, 107–113. [DOI] [PubMed] [Google Scholar]
- Otwinoski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Petracca R. et al. (2000) Structure–function analysis of hepatitis C virus envelope–CD81 binding. J. Virol., 74, 4824–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri P. et al. (1998) Binding of hepatitis C virus to CD81. Science, 282, 938–941. [DOI] [PubMed] [Google Scholar]
- Rosa D. et al. (1996) A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl Acad. Sci. USA, 93, 1759–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E., Le Naour,F., Lagaudriere-Gesbert,C., Billard,M., Conjeaud,H. and Boucheix,C. (1996) CD9, CD63, CD81 and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur. J. Immunol., 26, 2657–2665. [DOI] [PubMed] [Google Scholar]
- Serru V., Le Naour,F., Billard,M., Azorsa,D.O., Lanza,F., Boucheix,C. and Rubinstein,E. (1999) Selective tetraspan–integrin complexes (CD81/α4β1, CD151/α3β1, CD151/α6β1) under conditions disrupting tetraspan interactions. Biochem. J., 340, 103–111. [PMC free article] [PubMed] [Google Scholar]
- Woods A. and Couchman,J.R. (2000) Integrin modulation by lateral association. J. Biol. Chem., 275, 24233–24236. [DOI] [PubMed] [Google Scholar]
- Yagnik A.T., Lahm,A., Meola,A., Roccasecca,R.M., Ercole,B.B., Nicosia,A. and Tramontano,A. (2000) A model for the hepatitis C virus envelope glycoprotein E2. Proteins, 40, 355–366. [DOI] [PubMed] [Google Scholar]
- Yauch R.L., Kazarov,A.R., Desai,B., Lee,R.T. and Hemler,M.E. (2000) Direct extracellular contact between integrin α(3)β(1) and TM4SF protein CD151. J. Biol. Chem., 275, 9230–9238. [DOI] [PubMed] [Google Scholar]