Abstract
Seven Sm proteins, E, F, G, D1, D2, D3 and B/B′, assemble in a stepwise manner onto the single-stranded Sm site element (PuAU4–6GPu) of the U1, U2, U4 and U5 spliceosomal snRNAs, resulting in a doughnut-shaped core RNP structure. Here we show by UV cross-linking experiments using an Sm site RNA oligonucleotide (AAUUUUUGA) that several Sm proteins contact the Sm site RNA, with the most efficient cross-links observed for the G and B/B′ proteins. Site-specific photo-cross-linking revealed that the G and B/B′ proteins contact distinct uridines (in the first and third positions, respectively) in a highly position-specific manner. Amino acids involved in contacting the RNA are located at equivalent regions in both proteins, namely in loop L3 of the Sm1 motif, which has been predicted to jut into the hole of the Sm ring. Our results thus provide the first evidence that, within the core snRNP, multiple Sm protein–Sm site RNA contacts occur on the inner surface of the heptameric Sm protein ring.
Keywords: pre-mRNA splicing/RNA–protein interactions/site-specific UV cross-linking/Sm proteins/snRNPs
Introduction
The small nuclear ribonucleoproteins (snRNPs) U1, U2, U4/U6 and U5 are essential components of the spliceosome, which mediates catalysis of pre-mRNA splicing (for recent reviews see Will and Lührmann, 1997; Burge et al., 1999; Yu et al., 1999). The spliceosomal snRNPs consist of snRNA (U1, U2, U4/U6 and U5) as well as proteins that can be classified into two groups: the specific proteins that associate only with a certain snRNP particle, and the seven Sm proteins (B/B′, D1, D2, D3, E, F and G) that are common to each particle. Assembly of the Sm proteins onto snRNA is a critical step of the biogenesis of the snRNP particles. Following export from the nucleus to the cytoplasm, each of the U1, U2, U4 and U5 snRNAs associates with one set of the Sm proteins to generate the snRNP core particle (henceforth termed Sm core RNP). Formation of the Sm core RNP is essential for the metabolic stability of the snRNP, as well as for the subsequent hypermethylation of the 5′ cap from m7G to 2,2,7-trimethylguanosine, import of the particle into the nucleus and the recruitment of at least some particle-specific proteins (Plessel et al., 1994 and references therein).
In the absence of snRNA, the spliceosomal Sm proteins form three stable, RNA-free heteromeric complexes of E–F–G, D1–D2 and B/B′–D3 in vitro (Hermann et al., 1995; Raker et al., 1996). These Sm protein complexes associate with snRNA in at least two distinct steps. The D1–D2 and E–F–G complexes associate initially with the snRNA, creating the snRNP subcore particle. The subcore particle then provides a unique binding substrate for the B/B′–D3 complex, and the association of this completes Sm core assembly. While Sm core particle assembly in vitro is efficient in the absence of significant amounts of any additional, non-snRNP factors (Raker et al., 1999), the in vivo assembly appears to involve the non-snRNP factors of survival of motor neurons (SMN) and the SMN-interaction proteins (SIP) (Fischer et al., 1997; Liu et al., 1997).
The spliceosomal Sm proteins belong to a larger family of Sm and Sm-like (LSm) proteins, defined by the presence of a conserved motif, the Sm motif. The Sm motif is composed of two conserved regions, termed Sm1 and Sm2, which are separated by a non-conserved linker region (Hermann et al., 1995; Séraphin, 1995). The Sm motif encodes for a common folding domain (Sm domain) responsible for mediating the interactions among the Sm proteins. This was demonstrated biochemically by deletion mutagenesis of the B and D3 proteins (Hermann et al., 1995) and, conclusively, by the crystal structures of the B–D3 and D1–D2 protein complexes (Kambach et al., 1999).
The RNA determinants important for Sm core assembly appear to be highly complex. One prerequisite for Sm protein binding is the presence of the so-called Sm site element, a short, single-stranded region, which has the consensus sequence of PuAU4–6GPu and is usually flanked by stem–loop structures (Branlant et al., 1982; Liautard et al., 1982). The RNA binding specificity is provided directly by the single-stranded Sm site region, as demonstrated by the finding that an Sm site RNA oligonucleotide of AAUUUUUGA sufficed for the sequence-specific formation of an Sm core RNP particle that displayed biochemical and structural characteristics of native HeLa snRNP particles (Raker et al., 1999). In vivo studies in yeast and Xenopus oocytes revealed that the presence of the uridine stretch of the Sm site is essential for Sm protein binding, but these studies did not determine any base specificity for Sm protein recognition (Jones and Guthrie, 1990; Jarmolowski and Mattaj, 1993). In vitro analysis with a minimal Sm site RNA oligonucleotide revealed that the 5′ adenosine of the Sm site plays a critical role in Sm protein association, while the uridine bases and the 2′ hydroxyl groups collectively provide a binding determinant (Raker et al., 1999).
Very little is known about the manner in which the Sm proteins recognize and interact with the Sm site RNA element. A direct contact between the Sm G protein and the 5′ part of the Sm site element within HeLa U1 snRNP particles was demonstrated by cross-linking approaches (Heinrichs et al., 1992). Notably, neither the G protein nor any of the other Sm proteins contain established RNA binding motifs (Hermann et al., 1995), and no single Sm protein or heteromeric complex can directly interact with the U snRNA in a stable manner in vitro (Raker et al., 1996). This suggests that the RNA interaction surface and binding specificity of the Sm proteins are determined by interactions among the Sm protein complexes. A model based on biochemical studies and the crystallization of the D1–D2 and B–D3 Sm protein complexes proposed that, within the Sm core RNP, the Sm proteins form a ring-shaped structure (Kambach et al., 1999). The dimensions of the ring structure correlate well with the morphology of Sm proteins bound to snRNA as visualized by electron microscopy of negatively stained Sm core RNP particles, as well as by electron cryomicroscopy of the native U1 snRNP (Kastner et al., 1992; Stark et al., 2000). An intriguing possibility is that the Sm site RNA–Sm protein contacts occur on the inner surface of the proposed Sm ring, as suggested by the accumulation of the positively charged amino acid residues at the interior of the Sm ring (Kambach et al., 1999). However, as yet no data have been presented to support this idea.
We have now analysed the molecular basis of the RNA–protein interactions within spliceosomal Sm core RNP particles using UV cross-linking approaches combined with analysis of the cross-linking sites. We show that several Sm proteins contact the Sm site RNA, with the most efficient cross-links observed for the G and B/B′ proteins. Site-specific photo-cross-linking revealed that distinct uridine bases of the Sm site are contacted by the B/B′ and G proteins, whereby the first uridine was cross-linked to G and the third uridine to B/B′. Furthermore, using purified native snRNP particles for cross-linking, we determined that the cross-linked amino acids within the B/B′ and G proteins are in equivalent regions of the conserved Sm1 motif, namely, in the L3 loop, which has been predicted to jut into the interior of the proposed Sm protein ring structure. These results thus provide the first evidence that multiple Sm protein–Sm site RNA contacts occur on the inner surface of the Sm ring.
Results
RNA–protein contacts within a minimal Smcore RNP
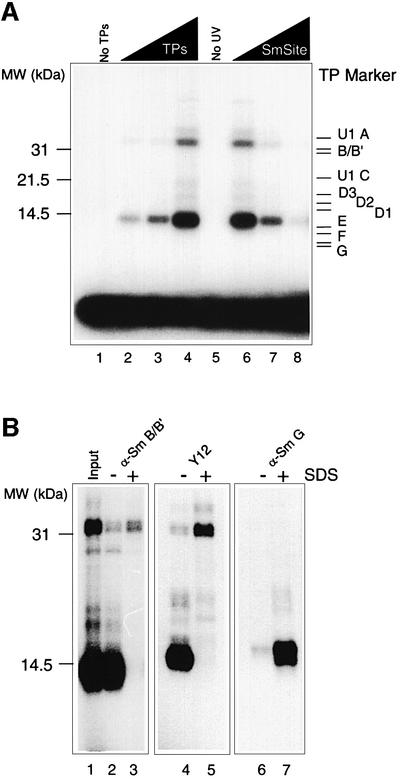
We demonstrated previously that all Sm proteins could be assembled onto a nonameric Sm site RNA oligonucleotide to form a stable minimal Sm core RNP particle that displayed biochemical and morphological characteristics of native HeLa snRNPs (Raker et al., 1999). We now addressed the question of which of the Sm proteins directly contact the Sm site RNA within the Sm core RNP, initially using this minimal system. For this, we UV irradiated minimal core RNP particles that had been reconstituted in vitro with a U4 RNA Sm site oligonucleotide (AAUUUUUGA) and Sm proteins (henceforth termed TPs, total snRNP proteins), isolated from HeLa snRNP particles. At the highest concentration of TPs, the Sm site oligonucleotide was observed in two major cross-linked products of a broad 14 kDa band and a doublet at 32 and 33 kDa (Figure 1A, lane 4). The 14 kDa band was the most prominent of these, containing ∼3% of the input Sm site RNA, and was likewise evident at the lower TP concentrations (Figure 1A, lanes 2 and 3). Several controls were performed to verify the authenticity of the cross-linking reaction. No cross-linked products were observed in the absence of either TPs (Figure 1A, lane 1) or UV irradiation (Figure 1A, lane 5). Furthermore, the cross-linked products could be efficiently competed out by the addition of a 100-fold molar excess of non-labelled Sm site oligonucleotide (Figure 1A, lanes 6–8), but not by a 1000-fold molar excess of a related RNA oligonucleotide containing cytidines rather than uridines (AACCCCCGA) (data not shown; see also Raker et al., 1999).
Fig. 1. UV cross-linking of Sm proteins. (A) UV cross-linking of multiple Sm proteins to the Sm site oligonucleotide. Reconstitution assays of TPs and radiolabelled Sm oligonucleotide (see Materials and methods) were subjected to UV irradiation at 254 nm for 10 min. Samples were separated on a high-TEMED, 12.5% SDS–polyacrylamide gel. Lane 1, no TPs; lanes 2–4, 50, 100 or 200 nM TPs, respectively; lane 5, no UV irradiation; lanes 6–8, 200 nM TPs and a 50-, 100- or 500-fold molar excess of unlabelled Sm oligonucleotide over the labelled Sm site oligonucleotide. (B) Immunoprecipitation of polypeptides cross-linked to the Sm site oligonucleotide. Reconstitution was performed with 5 nM radiolabelled Sm site oligonucleotide and 200 nM TPs. Following UV irradiation, half of the reconstitution sample was denatured by incubating for 2 min at 80°C in the presence of 1% SDS, and then diluted with PBS until an SDS concentration of 0.025% was reached (lanes 3, 5 and 7). The non-denatured half was diluted to an equal volume, and SDS was added to a final concentration of 0.025% SDS (lanes 2, 4 and 6). Samples were immunoprecipitated with mAb anti-B/B′ KSm3 (lanes 2 and 3), mAb Y12 (lanes 4 and 5) and polyclonal antibody against Sm G (lanes 6 and 7). The input UV irradiated sample is shown in lane 1. Samples were analysed by SDS–PAGE and autoradiography. The molecular weight markers (MW), indicated on the left, were visualized by Coomassie Blue staining. Note that gels with lanes 1–3 and 4–7 have been run for different times.
To identify the cross-linked proteins, a UV irradiated reconstitution mixture was split and tested in immunoprecipitation assays either following denaturation by SDS and heat, or under native conditions (Figure 1B). As estimated by their apparent molecular weights, the major cross-linked bands most likely contained the B/B′ proteins (for the 32/33 kDa product) and one of the small Sm proteins E, F or G (for the 14 kDa product). Indeed, under denaturing conditions, the monoclonal antibodies (mAbs) Y12 and KSm3 precipitated predominantly the cross-linked protein of apparent molecular weight 32/33 kDa (Figure 1B, lanes 3 and 5). While Y12 efficiently recognizes the B/B′ but also D1 and D3 Sm proteins, KSm3 exclusively recognizes B/B′ (Lerner et al., 1981; Williams et al., 1986). From these results we conclude that the 32/33 kDa band corresponds to a cross-link of B/B′ to the Sm site oligonucleotide. Importantly, under denaturing conditions, the major cross-linked protein (14 kDa band) was exclusively precipitated by an Sm G-specific rabbit antiserum (Figure 1B, lane 7), clearly demonstrating that the 14 kDa cross-link contained the G protein.
In the second half of the immunoprecipitation experiments, the irradiated minimal Sm core RNP particles were tested for immunoprecipitation under native conditions, i.e. without prior SDS treatment of the RNP particles. Significantly, the B/B′-specific KSm3 and Y12 antibodies efficiently co-precipitated the 14 kDa cross-linked protein (Figure 1B, lanes 2 and 4), demonstrating that cross-linking of the G protein had occurred within a fully assembled Sm core particle (see also below). The G-specific antiserum failed to precipitate either of the cross-linked products under non-denaturing conditions (Figure 1B, lane 6). This antiserum does not recognize the G protein in native snRNPs (Heinrichs et al., 1992), indicating that the cross-linked G protein is within Sm core particles. Note that both the B/B′ and G cross-links were co-precipitated by an anti-D2 antibody (data not shown), verifying that cross-linking of B/B′ had occurred within the context of the Sm core particle (see also Figure 3).

Fig. 3. UV cross-linking of various Sm protein subsets with the Sm site oligonucleotide. Radiolabelled U4-Sm site oligonucleotide (AAUUUUUGA) (∼5 nM) was incubated with the Sm protein subsets (∼200 nM) obtained by anti-B/B′ immunoaffinity chromatography (see Material and methods). Lane 1, the 500 mM KCl fraction (D1–D2); lane 2, eluate (B/B′–D3); lane 3, the flowthrough fraction (E–F–G); lane 4, the 150 mM KCl fraction (D1–D2 and E–F–G); lane 5, the 150 mM KCl eluate fractions (D1–D2, E–F–G and B/B′–D3). The position of the molecular weight markers on the left was determined by Coomassie Blue staining of the lane. Samples were analysed by SDS–PAGE and autoradiography.
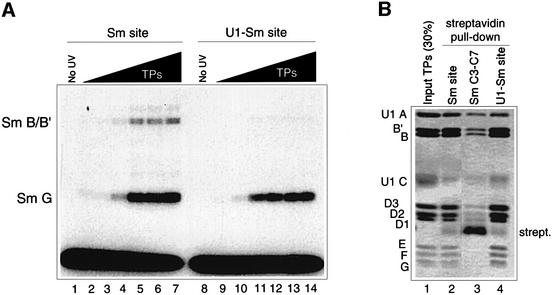
While previous cross-linking experiments revealed a direct cross-link between the G protein and the Sm site element within U1 snRNP particles, no significant B/B′ cross-link was identified at that time (Heinrichs et al., 1992). Notably, the human U1-Sm site differs from the U4-Sm site, used for the experiments in Figure 1, in that its uridine tract is interrupted by a guanosine (AAUUUGUGG). Thus, we analysed in vitro reconstituted minimal Sm core RNP particles that contained a U1-Sm site RNA oligonucleotide (AAUUUGUGG) by UV cross-linking. In contrast to the minimal core particles containing the U4-Sm site (Figure 2A, lanes 2–7), irradiation of the U1 minimal Sm core RNP particles resulted in an almost exclusive G protein cross-link product of 14 kDa and little or no B/B′ products at 32/33 kDa (Figure 2A, lanes 8–14, compared with lanes 1–7). Streptavidin pulldown experiments revealed that a biotinylated U1-Sm site oligonucleotide incubated with TPs was able to co-precipitate the full set of Sm proteins, including the B/B′ proteins, at a level comparable to that observed with the U4-Sm site oligonucleotide (Figure 2B, lanes 2 and 4, see also lane 1 for input). A biotinylated RNA oligonucleotide (SmC3-C7) in which the uridines were replaced with cytidines (AACCCCCGA), and which does not form stable Sm core RNP particles (Raker et al., 1999), did not co-precipitate Sm proteins (Figure 2B, lane 3). In summary, these data indicate that the nature of the Sm protein–Sm site RNA interactions in the minimal Sm core RNP particles closely resembles the situation observed in the purified native snRNP particles.
Fig. 2. Analysis of Sm protein binding to the U1-Sm site oligonucleotide. (A) Cross-linking of Sm protein to the Sm site and the U1-Sm site oligonucleotides. Radiolabelled U4-Sm site oligonucleotide (AAUUUUUGA) (lanes 1–7) or U1-Sm site oligonucleotide (AAUUUGUGG, lanes 8–14) was incubated with TPs (0, 25, 50, 100, 200, 400 or 600 nM), irradiated with UV light, and analysed by SDS–PAGE and autoradiography. Cross-linked Sm proteins are listed on the left. (B) Streptavidin pulldown assays. Assays contained TPs and either biotinylated U4-Sm site oligonucleotide (Sm site; lane 2), biotinylated SmC3-C7 oligonucleotide (Sm C3-C7; lane 3), or biotinylated U1-Sm site oligonucleotide (U1-Sm site; lane 4). Proteins that co-precipitated with the biotinylated RNA were analysed by SDS–PAGE and stained with Coomassie Blue (see also Raker et al., 1999).
Sm site RNA binding ability of the Sm proteins is dependent on their protein–protein contacts
Investigations of the core assembly pathway in vitro revealed that a stable subcore RNP complex is formed in the presence of the D1–D2 and E–F–G complexes but not with either the E–F–G, D1–D2 or B/B′–D3 complexes alone (Raker et al., 1996). We now analysed at which stage during Sm core RNP assembly less stable, initial RNA contacts might be, using the UV cross-linking method. Approximately equimolar concentrations of isolated Sm protein complexes (see Materials and methods) were incubated with the U4-Sm site oligonucleotide, and the reconstitution mixtures were subjected to UV irradiation. No specific cross-linking was observed after incubating D1–D2 or B/B′–D3 with the Sm site oligonucleotide (Figure 3, lanes 1 and 2, respectively). In contrast, UV irradiation of the mixture containing the E–F–G complex with the Sm site oligonucleotide led to multiple cross-linked products, the most prominent of which migrated at 14, 18 and 22 kDa (Figure 3, lane 3). Interestingly, formation of a subcore RNP, containing the E–F–G and D1–D2 complexes, dramatically changed the cross-linking pattern, now consisting mainly of the G cross-link at 14 kDa (Figure 3, lane 4). Finally, addition of the B/B′–D3 complex to the subcore RNP led to the B/B′ cross-link products at 32/33 kDa (Figure 3, lane 5). Thus, the E–F–G complex appears to have the capacity to form initial interactions with the Sm site RNA, which, however, require subsequent stabilization by the D1–D2 proteins. Formation of the subcore commits the particle to the core assembly pathway (Raker et al., 1996) and places the adenosine 5′ to the uridine tract into a chemical environment that resembles that of the mature core particle (Hartmuth et al., 1999). The lack of direct cross-linking of either the B/B′–D3 or the D1–D2 complex to the Sm site oligonucleotide reveals that these require the context of a core or subcore domain to establish the observed protein–RNA interactions.
Site-specific cross-linking reveals RNA position-specific contacts with individual Sm proteins
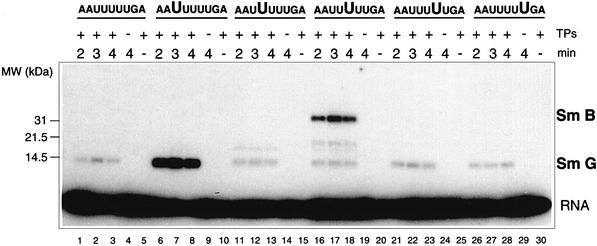
To determine whether the uridines within the Sm site were directly involved in contacting the Sm proteins, site-specific photo-cross-linking was performed using chemically synthesized Sm site oligonucleotides with single 5-bromouridine (br5U) substitutions at each position within the U tract of the U4 RNA Sm site oligonucleotide (AAUUUUUGA). Excitation of br5U at wavelengths of 308–312 nm results in specific photo-cross-linking of the C5 position of the uridine base to an amino acid residue when these are in close proximity to each other (Meisenheimer et al., 2000). Owing to the background emission of lower UV wavelengths, a 14 kDa G protein cross-link containing 1.5% of the input RNA was observed with the unmodified Sm site oligonucleotide (Figure 4, lanes 1–3). Strikingly, however, a large amount of the br5U1 oligonucleotide AAbr5UUUUUGA (23% of the input oligo) was cross-linked to the G protein, following a 3 min irradiation (Figure 4, lane 7). No br5U1 oligonucleotide cross-link was observed in the absence of either TPs (Figure 4, lane 9) or irradiation (Figure 4, lane 10). None of the remaining four br5U oligonucleotides were cross-linked to the G protein to a significant degree above the background levels observed with the non-br5U-containing Sm site oligonucleotide (Figure 4, lanes 11–30, as compared with lanes 1–5). Additionally, a significant amount (5% of the input) of the br5U3 RNA (AAUUbr5UUUGA) was incorporated into a 32/33 kDa B/B′ doublet. This cross-linking was specific for a br5U at the third position of the U tract, since none of the remaining br5U-containing oligonucleotides were incorporated into the 32/33 kDa cross-link in amounts significantly higher than that observed for the unmodified Sm site RNA (0.1%). A weaker cross-link to this oligonucleotide at 20 kDa was also observed (Figure 4, lanes 16–18). The identity of the protein involved could not be resolved by immunoprecipitation (it did not contain B/B′, D2, F or G) (data not shown). Finally, a weak cross-link product was observed for the br5U2 oligonucleotide (AAUbr5UUUUGA) (Figure 4, lanes 11–13), which was determined by immunoprecipitation to contain the F protein (data not shown). In summary, these results demonstrate that the major cross-linking occurs between G and B/B′ and the uridines at positions 1 and 3, respectively, of the U4 RNA Sm site U tract.
Fig. 4. Site-specific cross-linking of Sm proteins to chemically synthesized Sm site oligonucleotides containing a br5U at a single position within the uridine tract (AAUUUUUGA), as indicated above each lane by large letters. Samples were irradiated at 312 nm for 2, 3 or 4 min, as indicated above. Control assays were performed either in the absence of TPs (lanes 4, 9, 14, 19, 24 and 29) or without irradiation (lanes 5, 10, 15, 20, 25 and 30). Samples were analysed by SDS–PAGE and autoradiography. Positions of the molecular weight marker (MW) as determined by Coomassie Blue staining are shown on the left. The identity of each of the cross-linked Sm protein that could be resolved by immunoprecipitation (data not shown) is indicated on the right.
Cross-linking at equivalent sites within the Sm1 motif of the G and B/B′ Sm proteins
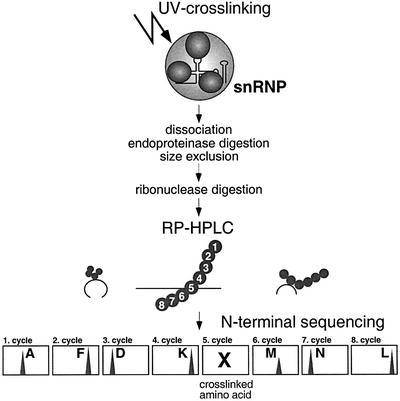
The relatively high yield of cross-linking of the U4 RNA Sm site oligonucleotide to the G and B/B′ proteins encouraged us to identify the amino acids involved in these interactions by protein biochemistry methods. Since these analytical investigations required a high amount of starting material, we used native U1 and [U4/U6⋅U5] tri-snRNP particles, purified from HeLa cells. Identification of the peptide regions within individual Sm proteins directly contacting the RNA was carried out biochemically based on a method that has been successfully used to identify peptide stretches involved in protein–RNA interactions within the ribosome (Urlaub et al., 1995, 1997). Briefly, UV irradiated snRNP particles were dissociated by SDS and heat, and the protein moieties were digested with endoproteinases (see Figure 5 for the scheme). Cross-linked peptide–snRNA complexes were enriched by size exclusion (SE) chromatography, which separates snRNA, both free and cross-linked to peptides, from the excess of non-cross-linked peptides. Subsequently, snRNA was digested by RNase A and/or T1. Cross-linked peptide– oligonucleotides were then isolated by C18 reversed phase (RP) high performance liquid chromatography (HPLC). In RP-HPLC, the RNA oligonucleotides are present in the flowthrough, whereas peptides and peptide– oligonucleotide cross-links are retained and separated during elution. Peptide fractions that exhibited a strong absorbance at both 220 and 260 nm were considered to contain a cross-linked peptide–oligonucleotide moiety and were subjected to automated N-terminal Edman sequencing. The cross-linked amino acid within the peptide cannot be identified by analysis of the Edman products due to its covalent modification, and is therefore identified by a gap in the sequence analysis (Urlaub et al., 1995).
Fig. 5. Purification strategy of cross-linked peptide–oligonucleotide complexes of UV irradiated snRNP particles (see text for details).
RP-HPLC separation of peptide–oligonucleotide heteromers derived from cross-linked tri-snRNP particles, digested with endoproteinase Lys-C and RNases A and T1, revealed a single, sharp peak that showed an absorbance at both 220 and 260 nm (Figure 6A, marked by an arrow). N-terminal sequencing of the peak fraction identified a peptide with the sequence AFDKXMNLILCDCDEFRK. The amino acid could be clearly identified in each cycle except the fifth, indicating that this contained the cross-linked amino acid (Figure 6B, denoted by an X). This sequence could be unambiguously assigned to the Lys-C fragment derived from the Sm B/B′ proteins of 33AFDKHMNLILCDCDEFRK50, which encompasses amino acid positions 33–50. Within the B/B′ peptide, His37 is in the position of the non-detectable amino acid in the fifth cycle of the Edman degradation. Since both non-modified and modified histidines are readily detected during analysis of the Edman degradation products (Grant and Crankshaw, 1997), this result demonstrates that His37 of the B/B′ protein is directly cross-linked to snRNA. Intriguingly, this cross-linked His37 is located within the evolutionarily conserved Sm1 motif.
Fig. 6. Isolation and identification of cross-linked Sm B/B′ and Sm G peptides derived from UV irradiated snRNP particles. (A) RP-HPLC of peptide–oligonucleotides from UV irradiated tri-snRNP particles. Irradiated tri-snRNP particles (800 µg) were treated with endoproteinase Lys-C and RNases A and T1, and injected onto a C18RP-HPLC column. The gradient applied is indicated within the chromatogram. The fraction that showed an absorbance at 220 (A220) and 260 nm (A260) (indicated by an arrow) was subjected to N-terminal sequencing [see (B), below]. The sequence of the Sm B/B′ Lys-C fragment within this fraction is outlined in the chromatogram. The highlighted amino acid (His37) indicates the cross-linked aminoacid that was identified by Edman degradation [see (B), below]. (B) N-terminal sequence analysis of the isolated Sm B/B′ Lys-C fragment cross-linked to snRNA. The analysis and identification of the Edman degradation products of the first seven residues of the Sm B/B′ peptide are shown. The first panel shows the elution profile of the PTH-amino acid standards (each 10 pmol). Diphenylthiourea (dptu) is an Edman degradation by-product. Amino acids corresponding to the first seven residues of the peptide are shown in bold in the subsequent panels. The PTH-amino acid derivative of residue 5 (His37) is missing (designated X), demonstrating that the amino acid in this position is cross-linked to the snRNA. The complete peptide sequence is listed in (A). (C) RP-HPLC of peptide–oligonucleotides derived from UV-irradiated U1 snRNP particles. MonoQ purified U1 snRNPs (500 µg) were UV irradiated, treated with trypsin and RNase T1, and directly injected onto a C18 RP-HPLC column. The gradient applied is indicated within the chromatogram. Fractions exhibiting an absorbance at 220 (A220) and 260 nm (A260) were N-terminal sequenced. The fraction marked by an arrow contained a tryptic Sm G fragment that could be sequenced from position 33 to 42 (33-GFDFMNLVI-42). The residues Phe37, Met38 and Asn39 could not by identified during analysis of the Edman degradation products (bold letters), indicating that this peptide region is cross-linked to the U1 snRNA (see text for details).
To determine the peptide region within the G protein that is cross-linked to an snRNA Sm site, we switched to native U1 snRNPs as the starting material, mainly since these were more abundant at preparative levels relative to the tri-snRNP particle. To favour the identification of cross-linked G peptide–oligonucleotides, the UV irradiated U1 snRNPs were digested with trypsin, which was expected to yield G peptides of lengths more suitable for isolation by RP-HPLC than those yielded by digestion with Lys-C. As shown in Figure 6C, the RP-HPLC elution profile contained several peaks with a strong absorbance at both 220 and 260 nm. N-terminal sequencing revealed that all of these peaks contained peptides derived from the U1 70K protein cross-linked to U1 snRNA (data not shown; Urlaub et al., 2000). After subtracting the co-eluting U1 70K peptide sequence, we were able to identify an additional sequence from one peak (Figure 6C, indicated by an arrow) with the sequence GFDPXXXLVI (sequencing data not shown). This sequence could be clearly assigned to a tryptic fragment from the G protein encompassing positions 33–42 (33GFDPFMNLVI42). Co-migration of a cross-linked Sm G peptide–oligonucleotide with a U1 70K peptide–oligonucleotide is consistent with the broadness of the eluting peak at 220 and 260 nm, as seen in Figure 6C. Three amino acids within the Sm G peptide, namely Phe37, Met38 and Asn39, could not be identified during analysis of the Edman degradation products, suggesting that this peptide region is cross-linked to U1 snRNA. In the absence of cross-linking, Phe37 was an easily identifiable residue, whereas Met38 and Asn39 were more difficult to identify due to technical reasons. That is, co-migration of a sequencing by-product obscured Asn39 during the analysis of the Edman degradation products, and methionine residues in general are weak PTH-amino acids that give low signals that are difficult to detect over background. We thus conclude that the G protein is cross-linked to the RNA via the peptide region 37FMN39, with the most likely cross-linking site at Phe37. However, we cannot rigorously exclude that the peptide is cross-linked to the RNA via several adjacent amino acids. Significantly, this cross-linked tri-peptide is located within the same region of the Sm1 motif of the G protein as the cross-linking site within the B/B′ protein. Moreover, Phe37 in Sm G and His37 in B/B′ are located at equivalent amino acid positions. In summary, our data show that both the B/B′ and G proteins contact snRNA via equivalent peptide regions within loop L3 of the Sm1 motif of the Sm domain.
The G protein is cross-linked to the first uridine (U127) of the Sm site U tract within native U1 snRNPs
The combined results described so far strongly suggest that the loop L3 regions of the Sm G and B/B′ proteins are cross-linked to the first and third uridines of the Sm site U tract, respectively. In order to provide more direct evidence for this idea, we decided to determine the exact cross-linking site within the U1 snRNA Sm site for the G protein, using UV irradiated native HeLa U1 snRNPs and employing an immunoprecipitation/primer extension approach. This method has been described in detail elsewhere (Urlaub et al., 2000). Initially, RNA–protein cross-linking sites on U1 snRNA within U1 snRNP particles were analysed by primer extension analysis, using a primer complementary to positions 134–152 of U1 snRNA. In U1 snRNA derived from UV irradiated snRNP particles, a strong stop was observed at U128 in the Sm site and a weaker one at G106 (Figure 7A, lane 4). Since these stops are underrepresented or absent in the irradiated naked U1 snRNA (Figure 7A, lane 2), they are probably caused by protein-specific cross-linking to the corresponding nucleotides (U127 and U105, respectively).
Fig. 7. Identification of the Sm G cross-linking site on U1 snRNA within native U1 snRNPs. (A) Primer extension analysis of U1 snRNA derived from 5 µg of UV cross-linked U1 snRNPs (lane 4) compared with UV irradiated naked U1 snRNA (lane 2). Lanes 1 and 3, no UV irradiation (Ctr); lanes 2 and 4, 2 min UV irradiation at 254 nm (UV). C, U, A and G, dideoxy sequence markers. Reverse transcriptase stops at positions U128 and G106 in U1 snRNA derived from UV irradiated U1 snRNP particles are denoted on the right. Sm site nucleotides are listed on the left. Primer extension analysis was carried out as described by Hartmuth et al. (1999). The primer oligonucleotide was complementary to nucleotides 134–152 of U1 snRNA. (B) Primer extension analysis of cross-linked U1 snRNA after immunoprecipitation of cross-linked U1 snRNPs with antibodies specific for Sm G and Sm B/B′. Lanes 1 and 3, no UV irradiation (Ctr); lanes 2 and 4, 2 min UV irradiation at 254 nm (UV). C, U, A and G, dideoxy sequence markers. The nucleotide position of the reverse transcriptase stop at U128 in lane 2 can be deduced from the marker.
To determine which of the Sm proteins were responsible for the cross-links to these nucleotides, we immunoprecipitated the denatured, UV irradiated U1 snRNP particles with antibodies specific for the Sm G and B/B′ proteins and analysed co-precipitated U1 snRNA by primer extension. Importantly, a strong reverse transcriptase stop at U128 was observed only after immunoprecipitation with the anti-Sm G antibody (Figure 7B, lane 2), clearly demonstrating that the Sm G protein was cross-linked to the U127 in the Sm site (AAUUUGUGG; Figure 7B). It is interesting to note that, consistent with our U1 Sm site oligonucleotide cross-linking data (Figure 2), no significant cross-link with the B/B′ proteins was detected (Figure 7B, lane 4). The finding that the first uridine within both the minimal U4 RNP core particles and native U1 snRNP particles was cross-linked to the G protein suggests that the interaction of the G protein with the 5′-most uridine within the Sm site is a conserved contact within core domains.
Discussion
Multiple Sm proteins contact the Sm site RNA element within Sm core RNP particles in a highly site-specific manner
Here we have investigated at the molecular level the Sm protein–Sm site RNA interactions within spliceosomal snRNP core particles using UV cross-linking approaches. Within the context of a minimal Sm core RNP, two major cross-links were observed between the U4 RNA Sm site oligonucleotide and the G as well as the B/B′ protein (Figure 1). The site of interaction for each protein with the Sm site RNA was unique, since cross-linking with br5U-substituted oligonucleotides demonstrated that the G protein was covalently linked to the first uridine of the Sm site U tract (i.e. AAUUUUUGA), and the B/B′ protein to the third uridine (AAUUUUUGA). Furthermore, a weak cross-link between the second uridine and the F protein was observed (data not shown). Cross-linking of the G protein to the first uridine (AAUUUUUGA) (Figure 4) within a minimal Sm core RNP correlated nicely with the results from UV cross-linking and primer extension/reverse transcriptase analysis of native HeLa U1 snRNP particles, which mapped the site of G protein interaction to the equivalent uridine position of the U1 RNA Sm site element (AAUUUGUGG) (Figure 7). We conclude that the Sm site RNA element is specifically positioned with respect to the Sm proteins, and that this positioning is a conserved feature of Sm core RNP particles.
It is important to note that although we did not identify Sm proteins efficiently cross-linked to uridine residues 4 and 5 of the U4-Sm site, this does not necessarily imply that these nucleotides do not interact with Sm proteins in the Sm core. Rather, these results may indicate that the nature of the protein–RNA interactions at these sites are not favourable for photo-cross-linking. Indeed, intimate contacts of the entire Sm site RNA with Sm proteins is indicated by our previous finding that all uridines within the Sm site element of U2 RNA were found to be protected from chemical modification in native U2 snRNP particles (Hartmuth et al., 1999). Similar considerations apply to our finding that, within Sm core RNP particles, the B/B′ protein could not be cross-linked to the U1-Sm site, although it was cross-linked efficiently in the context of a U4-Sm site sequence. Thus, the guanosine at position 4 of the U1-Sm site may influence the protein contact at the neighbouring uridine at position 3, such that the B/B′ protein in the U1 Sm core RNP no longer contacts the third uridine in a manner favourable for UV cross-linking.
Sm site RNA is contacted by equivalent regions within the Sm1 motif of the B/B′ and G proteins
Mapping of the UV cross-linked amino acids within native HeLa snRNP particles revealed that equivalent regions of the G and B/B′ proteins were involved in interactions with the Sm site RNA. Specifically, the His37 residue of B/B′ and the peptide region of Phe37–Asn39 of G (with Phe37 as the most likely cross-linked amino acid) were shown to directly contact the snRNA within [U4/U6⋅U5] tri-snRNP and U1 snRNP particles, respectively. In both proteins, the cross-linked amino acids lie within loop L3 of the Sm1 region of the conserved Sm folding domain (Figure 8A and B). Our further finding that the G protein was cross-linked to the first uridine of the Sm site in both minimal core RNPs and native U1 snRNPs (see above) strongly argues that the purified cross-linked oligopeptide products contain peptides cross-linked to the Sm site RNA.
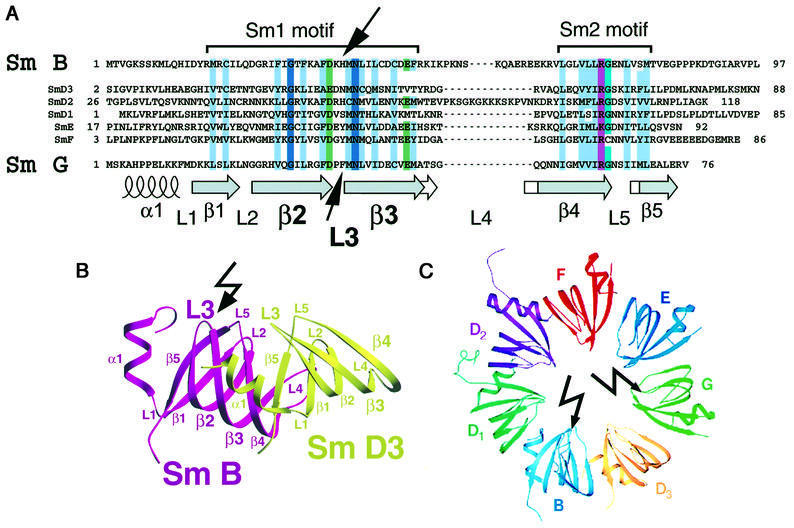
Fig. 8. Cross-linking sites within the Sm1 motif of the Sm B/B′ and G proteins. (A) Amino acid sequence alignment of the Sm1 and Sm2 motifs of the human Sm B/B′, D2, D1, D3, E, F and G proteins according to Hermann et al. (1995). The topology of the secondary structure was adapted from the crystal structure of the Sm B–D3 and Sm D1–D2 protein complexes (Kambach et al., 1999). Conserved amino acids are highlighted as follows: uncharged, hydrophobic residues (L, I, V, A, F, W, Y, C, M) are in light blue; acidic amino acids (D, E) in green; basic amino acids (R, K) in purple; the 80% conserved glycine in turquoise; and 100% conserved amino acids in dark blue. Arrows mark the cross-linked amino acids in the protein sequences, i.e. His37 in the Sm B/B′ protein and Phe37, Met38 or Asn39 in the Sm G protein, as identified by N-terminal sequencing (see Figure 6 and text for details). Note that the cross-linking sites are located in both proteins within loop L3 of the Sm1 motif. (B) Location of the Sm B cross-linking site within the 3D structure of the RNA-free Sm B–D3 complex (Kambach et al., 1999). The cross-linking site in loop L3 within the Sm1 motif of the Sm B protein is indicated by an arrow. The figure was produced with SETOR (Evans, 1993) according to Kambach et al. (1999) by using the coordinates of the complex available in the PDB database. (C) Location of the Sm G and Sm B cross-linking sites in the proposed higher order structure of the assembled Sm proteins. The model was adapted from Kambach et al. (1999). Arrows mark the cross-linking sites located in loop L3 of both proteins.
The conserved Sm domain is known to be responsible for the protein–protein interactions among the Sm proteins (Hermann et al., 1995; Kambach et al., 1999). Our finding that the loop L3 region of both the B/B′ and G proteins is additionally involved in RNA–protein interactions suggests that the Sm domain is also directly responsible for mediating RNA interactions. Crystal structures of the D1–D2 and B–D3 Sm protein complexes revealed that the β-strands from the Sm2 region were directly involved in the protein–protein contacts, while the β-strands from the Sm1 region formed a hydrophobic core in each protein (Figure 8; Kambach et al., 1999). Intriguingly, in the model of the heptameric Sm protein ring, loops L2, L3, L4 and L5 of the Sm domain jut into the central hole of the ring (Figure 8C), and amino acid residues near loops L3 and L5 were proposed to be involved in RNA binding (Kambach et al., 1999). Our cross-linking results provide the first direct evidence in support of such a model of Sm protein–RNA interactions, in which the Sm site RNA element interacts with multiple Sm proteins along the surface of the inner ring (Figure 8C).
The UV cross-linking results described here are also interesting for the following reason. Within the current heptameric Sm ring model, the protein subunits are arranged in the order G–E–F–D2–D1–B–D3 (Figure 8C), as supported by the combined results from previous Sm protein–protein interaction studies (Kambach et al., 1999 and references therein). It was proposed that the Sm site RNA binds in a 5′ to 3′ polarity along the inner surface of the Sm G–E–F–D2–D1 proteins, with each of the five uridines contacting a neighbouring protein in a sequential manner (Kambach et al., 1999). However, our finding that the first uridine contacts the G protein, and the third uridine the B/B′ proteins, indicates that an alternative, more irregular, conformation of the RNA with respect to the Sm proteins has to be considered.
Recently, the three-dimensional (3D) structure of the HeLa U1 snRNP particle has been obtained by electron cryomicroscopy at 10 Å resolution, which supports the proposed ring-like organization of the Sm proteins in the Sm core domain (Stark et al., 2000). Of interest is the location in the U1 snRNP structure of the 4-way junction and the 3′-terminal stem–loop of U1 RNA, which mark the 5′ and 3′ ends of the single-stranded Sm site RNA element. These are placed beneath and on top of the Sm protein ring domain, respectively. This finding, combined with the results reported here, indicates that the Sm site RNA traverses the interior of the Sm ring. Since the hole of the Sm ring is too narrow to accommodate a double-stranded stem–loop (Kambach et al., 1999), these results provide an explanation as to why the Sm proteins assemble directly and in a step-wise manner onto the Sm site RNA, rather than as a pre-formed ring (see Introduction).
The canonical spliceosomal Sm proteins belong to a large family of Sm and Sm-like (LSm) proteins, all of which contain an Sm motif (Hermann et al., 1995; Séraphin, 1995; He and Parker, 2000). These proteins are involved in various aspects of RNA biogenesis. For example, a subset of the LSm proteins (2–8) associates with the U6 snRNA in both yeast (Mayes et al., 1999; Salgado-Garrido et al., 1999) and human (Achsel et al., 1999). In the absence of RNA, the LSm2–8 proteins form a heptameric ring structure, which appears to bind as such to the uridine-rich, 3′ end of the U6 snRNA (Achsel et al., 1999). It is suggestive that interactions of LSm proteins with the uridines occur within the interior of the LSm ring. An interesting difference is that the LSm proteins undergo a one-step assembly onto the U6 snRNA, at least in vitro (Achsel et al., 1999). Since the Sm site appears to traverse the Sm ring, assembly must occur directly at the Sm site and in multiple steps (see above), whereas the single-stranded 3′-terminal U tract of the U6 snRNA can freely pass into the preformed LSm ring. Our results presented here on the RNA–protein interaction surface within spliceosomal Sm core RNP particles may thus also provide a basis for understanding the nature of the RNA binding of the spliceosomal LSm proteins.
Materials and methods
UV cross-linking of in vitro reconstituted minimal Sm core RNPs and isolated HeLa snRNPs
Native TPs and Sm protein subsets were prepared essentially as described previously (Raker et al., 1999). Minimal Sm core RNPs were reconstituted from chemically synthesized Sm site RNA oligonucleotides and TPs as previously described (Raker et al., 1999). Reconstitution samples contained ∼5 nM radiolabelled Sm site oligonucleotides and 200 nM TPs (unless stated otherwise) in a 10 µl volume. For UV cross-linking, samples were irradiated within microfuge tubes (at a distance of ∼2.5 cm) with UV light at 254 nm for 10 min at 4°C. Samples containing the br5U-substituted Sm site oligonucleotides were irradiated at 312 nm for 1.5 min (or the time indicated) at the same distance.
For UV cross-linking of native U1 snRNPs and [U4/U6⋅U5] tri-snRNPs isolated from HeLa cells, ∼0.8–1 mg of purified snRNP particles at a concentration of 0.1 mg/ml in buffer A [20 mM HEPES–KOH pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithioerythritol (DTE), 250 mM NaCl] were irradiated at 254 nm for 2 min on ice at a distance of 3–4 cm in glass dishes.
Minimal Sm core RNP immunoprecipitation
UV irradiated reconstitution assay mixtures were diluted to 100 µl and then incubated together with 10 µl of pre-washed protein A–Sepharose–antibody slurry, in 400 µl of phosphate-buffered saline (PBS) pH 8.0 containing 1% (w/v) RNase-free bovine serum albumin and 0.025% (w/v) SDS. The following antibodies and amounts were used for coupling with 10 µl of protein A–Sepharose: 5 µl of mAb Y12 (Lerner et al., 1981), 5 µl of KSm3 (Williams et al., 1986) or 20 µl of polyclonal anti-G or anti-F antibody (Hermann et al., 1995). Samples were incubated for 90 min at 4°C with gentle, end-over-end rotation. The beads were washed extensively with IP buffer [10 mM Tris–HCl pH 8.0, 0.1% (v/v) NP-40 and 150 mM KCl], vacuum dried and then boiled in 30 µl of SDS sample buffer. For denaturation of the cross-linked samples prior to immunoprecipitation, SDS was added to a final concentration of 1% (w/v) and heated for 2 min at 80°C, and 400 µl of PBS pH 8.0 were added to dilute the SDS concentration to 0.025% (w/v). The non-denatured samples were diluted to an equal volume, and SDS was added to a final concentration of 0.025%.
Isolation and identification of cross-linked peptide–oligonucleotide complexes
UV irradiated snRNPs were ethanol precipitated, dissolved in buffer (50 mM Tris–HCl pH 7.5, 5 mM DTE) containing 8 M urea (U1 snRNPs) or 1% SDS (w/v) (tri-snRNPs), heated for 10 min at 90°C and cooled down to room temperature. The U1 snRNP solution was diluted with 50 mM Tris–HCl pH 7.5, 1 mM CaCl2 until the urea concentration was below 1 M, and the tri-snRNP solution was diluted with 50 mM Tris–HCl pH 7.5 until the SDS concentration was 0.1%. Endoproteolytic cleavage of snRNP proteins was carried out with trypsin in the case of U1 snRNPs, or Lys-C in the case of tri-snRNP, with an enzyme:substrate ratio of 1:20 for 16 h at 37°C. After endoproteinase digestion, the samples were ethanol precipitated and dissolved in an appropriate volume of buffer B (20 mM Tris–HCl pH 7.8, 150 mM NaCl, 5 mM EDTA) containing either 8 M urea (U1 snRNP experiment) or 0.1% SDS (tri-snRNP experiment). The samples were injected onto a Superose 12 column (10/30; Amersham-Pharmacia-Biotech), with a flow rate of 400 µl/min for buffer B. The absorbance was monitored at 254 nm. SnRNA-containing fractions were pooled, ethanol precipitated and dissolved in buffer containing 25 mM Tris–HCl pH 7.8, 2.5 mM EDTA. SnRNAs were digested with 10 µg of RNase A and/or 10 µg of RNase T1 for 2 h at 50°C and then subjected to a second incubation with 1 µg of endoproteinase trypsin or Lys-C for 16 h at 37°C. Digestion was stopped by injection onto a RP-HPLC C18 column (Vydac). Solvent A was water, 0.1% trifluoroacetic acid (TFA), and solvent B was acetonitrile, 0.085% TFA. Cross-linked peptide–oligonucleotide heteromers were isolated at a flow rate of 100 µl/min with the following gradients: 30 min isocratic elution at 5% (U1 snRNP experiment) or 10% solvent B (tri-snRNP experiment), 5 or 10% solvent B to 45% solvent B in 120 min, and 45–90% solvent B in 20 min. An additional isocratic step was performed at 10% solvent B when the gradient was started at 5% solvent B. The absorbance was monitored at 220 and 260 nm. Fractions with an absorbance at both 220 and 260 nm were dried under vacuum and applied to automated N-terminal sequence analysis.
Immunoprecipitation of U1 snRNP protein–RNA cross-links
Two 25 µl droplets each containing 2.5 µg of purified U1 snRNPs were UV cross-linked as above. The cross-linked sample was incubated with 2% (w/v) SDS for 10 min at 70°C. After cooling to room temperature, Triton X-100 was added to a final concentration of 5% (v/v), and immunoprecipitation of U1 snRNP protein–RNA using antibodies against Sm proteins (see above) was carried out as previously described (Urlaub et al., 2000).
Acknowledgments
Acknowledgements
We thank Volker Kruft, Ivan Vidovic, Nick Watkins and Cindy Will for helpful discussions. We are grateful to Axel Badouin for preparation of snRNPs and Gerlinde Grelle for peptide sequencing. This work was supported by the Gottfried Wilhelm Leibniz Program and grants from the Deutsche Forschungsgemeinschaft (SFB 397 and SFB 523) to R.L.
REFERENCES
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol,A., Ebel,J.P., Lazar,E., Haendler,B. and Jacob,M. (1982) U2 RNA shares a structural domain with U1, U4 and U5 RNAs. EMBO J., 1, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Evans S.V. (1993) SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Fischer U., Liu,Q. and Dreyfuss,G. (1997) The SMN–SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell, 90, 1023–1029. [DOI] [PubMed] [Google Scholar]
- Grant G.A. and Crankshaw,M.W. (1997) Identification of PTH-amino acids by high-permormance liquid chromatography. In Smith,J.B (ed.), Methods in Molecular Biology. Vol. 64. Humana Press, Totowa, NJ, pp. 197–213. [DOI] [PubMed]
- Hartmuth K., Raker,V.A., Huber,J., Branlant,C. and Lührmann,R. (1999) An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J. Mol. Biol., 285, 133–147. [DOI] [PubMed] [Google Scholar]
- He W. and Parker,R. (2000) Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol., 12, 346–350. [DOI] [PubMed] [Google Scholar]
- Heinrichs V., Hackl,W. and Lührmann,R. (1992) Direct binding of small nuclear ribonucleoprotein G to the Sm site of small nuclear RNA. Ultraviolet light cross-linking of protein G to the AAU stretch within the Sm site (AAUUUGUGG) of U1 small nuclear ribonucleoprotein reconstituted in vitro. J. Mol. Biol., 227, 15–28. [DOI] [PubMed] [Google Scholar]
- Hermann H., Fabrizio,P., Raker,V.A., Foulaki,K., Hornig,H., Brahms,H. and Lührmann,R. (1995) snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J., 14, 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A. and Mattaj,I.W. (1993) The determinants for Sm protein binding to Xenopus U1 and U5 snRNAs are complex and non-identical. EMBO J., 12, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.H. and Guthrie,C. (1990) Unexpected flexibility in an evolutionarily conserved protein–RNA interaction: genetic analysis of the Sm binding site. EMBO J., 9, 2555–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C., Walke,S., Young,R., Avis,J.M., de la Fortelle,E., Raker,V.A., Lührmann,R., Li,J. and Nagai,K. (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 96, 375–387. [DOI] [PubMed] [Google Scholar]
- Kastner B., Kornstadt,U., Bach,M. and Lührmann,R. (1992) Structure of the small nuclear RNP particle U1: identification of the two structural protuberances with RNP-antigens A and 70K. J. Cell Biol., 116, 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E.A., Lerner,M.R., Janeway,C.A.,Jr and Steitz,J.A. (1981) Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl Acad. Sci. USA, 78, 2737–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liautard J.P., Sri-Widada,J., Brunel,C. and Jeanteur,P. (1982) Structural organization of ribonucleoproteins containing small nuclear RNAs from HeLa cells. Proteins interact closely with a similar structural domain of U1, U2, U4 and U5 small nuclear RNAs. J. Mol. Biol., 162, 623–643. [DOI] [PubMed] [Google Scholar]
- Liu Q., Fischer,U., Wang,F. and Dreyfuss,G. (1997) The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell, 90, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Mayes A.E., Verdone,L., Legrain,P. and Beggs,J.D. (1999) Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J., 18, 4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenheimer K.M., Meisenheimer,P.L. and Koch,T.H. (2000) Nucleoprotein photo-cross-linking using halo-pyrimidine-substituted RNAs. Methods Enzymol., 318, 88–104. [DOI] [PubMed] [Google Scholar]
- Plessel G., Fischer,U. and Lührmann,R. (1994) m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol. Cell. Biol., 14, 4160–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Plessel,G. and Lührmann,R. (1996) The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J., 15, 2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Hartmuth,K., Kastner,B. and Lührmann,R. (1999) Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol., 19, 6554–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bradago-Nilsson,E., Kandels-Lewis,S. and Séraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B. (1995) Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J., 14, 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H., Dube,P., Lührmann,R. and Kastner,B. (2000) The 3-D arrangement of RNA and proteins in the spliceosomal U1 snRNP. Nature, in press. [DOI] [PubMed] [Google Scholar]
- Urlaub H., Kruft,V., Bischof,O., Müller,E.C. and Wittmann-Liebold,B. (1995) Protein–rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J., 14, 4578–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub H., Thiede,B., Müller,E.C., Brimacombe,R. and Wittmann-Liebold,B. (1997) Identification and sequence analysis of contact sites between ribosomal proteins and rRNA in Escherichia coli 30S subunits by a new approach using matrix-assisted laser desorption/ionization-mass spectrometry combined with N-terminal microsequencing. J. Biol. Chem., 272, 14547–14555. [DOI] [PubMed] [Google Scholar]
- Urlaub H., Hartmuth,K., Kostka,S., Grelle,G. and Lührmann,R. (2000) A general approach for identification of RNA–protein crosslinking sites within native human spliceosomal snRNPs: analysis of RNA–protein contacts in native U1 and [U4/U6⋅U5] snRNPs. J. Biol. Chem., 275, 41458–41468. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- Williams D.G., Stocks,M.R., Smith,P.R. and Maini,R.N. (1986) Murine lupus monoclonal antibodies define five epitopes on two different Sm polypeptides. Immunology, 58, 495–500. [PMC free article] [PubMed] [Google Scholar]
- Yu Y.-T., Scharl,E.C., Smith,C.M. and Steitz,J.A. (1999) The growing world of small nuclear ribonucleoproteins. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 487–524.