Abstract
Plant homeodomain (PHD) domains are found in >400 eukaryotic proteins, many of which are transcriptional regulators. Naturally occurring point mutations or deletions of this domain contribute to a variety of human diseases, including ATRX syndrome, myeloid leukemias and autoimmune dysfunction. Here we report the first structural characterization of a PHD domain. Our studies reveal that the PHD domain from KAP-1 corepressor binds zinc in a cross-brace topology between anti-parallel β-strands reminiscent of RING (really interesting new gene) domains. Using a mutational analysis, we define the structural features required for transcriptional repression by KAP-1 and explain naturally occurring, disease-causing mutations in PHD domains of other proteins. From a comparison of this PHD structure with previously reported RING and LIM (Lin11/Isl-1/Mec-3) structures, we infer sequence determinants that allow discrimination among PHD, RING and LIM motifs.
Keywords: KAP-1/LIM/PHD/RING/zinc finger
Introduction
Biological processes depend on the spatial and temporal targeting of specific macromolecular interactions. The presence of highly conserved amino acid sequence motifs and their structural arrangement within a novel protein can provide the first clue to the protein’s function. Modular motifs within multi-domain proteins often specify interacting partners and the identification of these interactions has been beneficial in defining biochemical functions of particular proteins and their cellular pathways. Furthermore, these modular domains are often targets of mutations that disrupt the cellular pathways in which they function. Identification of modular domain type through sequence homology studies alone can lead to ambiguous domain-type assignment, making prediction of function a difficult enterprise. Thus, biophysical characterization and determination of the three-dimensional (3D) structure are essential for understanding cellular and biological function.
The plant homeodomain (PHD) or leukemia associated protein (LAP) domain is a relatively small motif of ∼60 amino acids that is found in >400 eukaryotic proteins, many of which are believed to be involved in the regulation of gene expression, including the KAP-1/TIF1β, WCRF/WSTF, Mi-2 and CBP/p300 families (Aasland et al., 1995). It has been suggested that this domain is involved in protein–protein interactions related to a possible role in chromatin-mediated regulation of gene expression (Jacobson and Pillus, 1999). Although no direct role in transcriptional processes has been reported for this domain, recent reports indicate that the extended PHD domain of AF10 is necessary for homo-oligomerization, an event required for the ability of the AT hook motif in AF10 to bind DNA (Linder et al., 2000).
The biological importance of the PHD domain is underscored by its involvement in the pathogenesis of several human disorders. Clinically relevant missense mutations in the PHD domain of the ATRX protein result in α-thalassemia and mental retardation (Gibbons et al., 1997; Rinderle et al., 1999). Germline nonsense mutations in the AIRE gene result in truncated proteins where one or both of the PHD domains are deleted in patients with autoimmune polyglandular syndrome type 1 (APECED) (The Finnish–German APECED Consortium, 1997). Somatically acquired point mutations in the PHD domain of ING1 have been identified in head and neck squamous cell carcinomas (Gunduz et al., 2000). Chromosomal rearrangements that delete this motif in proteins such as MLL, CBP, MOZ and AF10 result in myeloid leukemias (Jacobson and Pillus, 1999). Furthermore, genes encoding PHD domain proteins have been identified in the critical deletion regions of several contiguous gene deletion syndromes such as Williams syndrome (WSTF) and the immunodeficiency syndrome ICF (DMNT3B) (Lu et al., 1998; Aapola et al., 2000). The prevalence of disease-causing mutations in PHD domains suggests that they play a basic and essential role in the normal function of human cells.
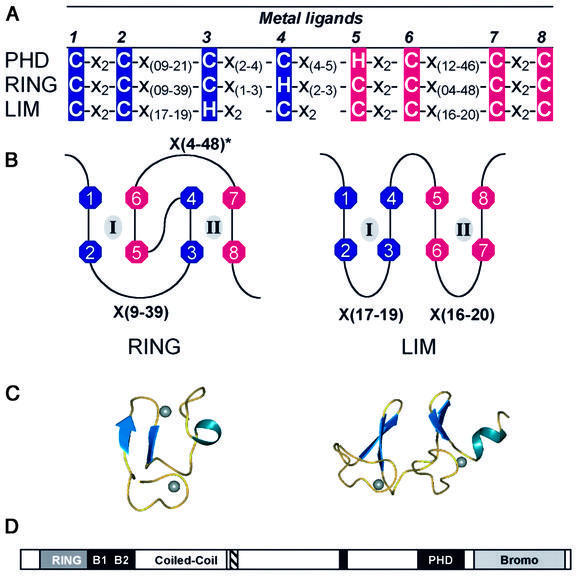
The PHD domain is a motif characteristically defined by seven cysteines and a histidine that are spatially arranged in a C4HC3 consensus with intervening sequences of varying length and composition (Aasland et al., 1995). This particular arrangement of amino acids is highly homologous to the RING (really interesting new gene) and LIM (Lin11/Isl-1/Mec-3) domains (Figure 1A) (Borden and Freemont, 1996; Dawid et al., 1998). In both the RING and the LIM domain, these conserved cysteine and histidine residues are utilized to bind two zinc atoms, a process cooperatively coupled to the folding of the domains. The LIM uses a sequential zinc ligation scheme, where the first and second pair of metal ligands create the first zinc-binding site, while the third and fourth pair form the second site (Figure 1B). In contrast, the RING domain utilizes a unique cross-brace ligation topology, where the first and third pair of metal ligands form the first zinc-binding site (site I), while the second and fourth pair form the second site (site II) (Figure 1B). The different arrangements of metal ligands result in dramatically different 3D structures (Figure 1C). The choice of ligation topology is undoubtedly critical for proper folding and domain functionality. Given the sequence similarity amongst PHD, RING and LIM domains, one is unable to predict the ligation scheme and fold by sequence analysis alone.
Fig. 1. (A) The consensus sequences that define the PHD, RING and LIM domains. C, cysteine; H, histidine; X, any residue. Above the consensus sequence the number of each metal ligand, ml, is given. The first and second pairs of sequential metal ligands are in dark blue, while the third and fourth pairs are in magenta. (B) Demonstration of zinc ligation patterns found in RING and LIM domains. The RING uses a cross-brace ligation scheme while the LIM uses a sequential ligation scheme. Numbers correlate to the number of the metal ligand as defined in (A). Zinc atoms are represented by gray ovals and the zinc-binding sites are denoted by roman numerals. (C) Ribbon diagrams of the three-dimensional structures. Zinc atoms are represented by spheres. Structures are from the RING of PML (1BOR) and for LIM (1A7I). (D) A schematic representation of the domains in the KAP-1 protein. RING, the RING domain; B1 and B2, two adjacent B-boxes; Coiled-Coil, a leucine coiled-coil domain; hatched box, the TIFF signature sequence; black box, the HP1-binding site; PHD, the PHD domain; Bromo, the bromodomain.
Here, we report the first solution structure of the PHD domain from the KAP-1 corepressor (also known as TIF1β or KRIP1), a universal corepressor for the KRAB-zinc finger superfamily of transcriptional repressors (Friedman et al., 1996). An N-terminal RBCC domain (RING domain, B-boxes and coiled-coil region) is both necessary and sufficient for interaction of KAP-1 with the KRAB repression module (Peng et al., 2000a,b). Amino acid sequences C-terminal to the RBCC domain represent at least two independent repression domains. A direct interaction between KAP-1 and the HP1 family of proteins suggests that KRAB-zinc finger proteins repress transcription through heterochromatin-mediated gene silencing (Ryan et al., 1999; Lechner et al., 2000). The PHD domain is found on the C-terminus of KAP-1, adjacent to a bromodomain defining an independent repressional unit (Figure 1D). These two domains are found together in ∼30 PHD-containing proteins, suggesting that they may functionally interact (Le Douarin et al., 1995, 1996; Friedman et al., 1996; Venturini et al., 1999; Bochar et al., 2000; Jones et al., 2000). We demonstrate that the PHD forms an autonomously folding domain that binds two zinc atoms in a cross-brace RING-like arrangement. Not only are the zinc ligation schemes identical, but also the tertiary topologies of the PHD and RING domains are remarkably similar. From the comparison of PHD, RING and LIM domains, it is possible to derive unique structural determinants. Structure-based site-directed mutational analysis reveals structural features of the PHD domain that are critical for the ability of KAP-1 to repress transcription. Furthermore, we are able to rationalize the structural consequences of mutations in the PHD domains of other proteins such as ATRX and ING1, where these mutations correlate with human disease.
Results and discussion
The PHD domain requires zinc for folding
The conservation of eight potential metal ligands in the PHD family suggested that it is a zinc-binding protein. In order to analyze the chemical and physical characteristics of the PHD domain, we expressed the KAP-1 PHD (amino acids 618–679) in Escherichia coli and purified the protein to homogeneity using affinity and size exclusion chromatographies. Analytical ultracentrifugation experiments suggest that >90% of the PHD protein in solution is a monomer (data not shown). To determine the identity and stoichiometry of metal bound by the PHD domain, we used inductively coupled plasma (ICP) spectrometry. These measurements indicate that a 3.0 mM solution of KAP-1 PHD protein contains 5.2 mM (± 3.3%) zinc. This ratio of one protein molecule to 1.7 zinc atoms indicates that the PHD binds two zinc atoms per protein molecule, potentially utilizing its eight conserved cysteine and histidine residues for metal ligation.
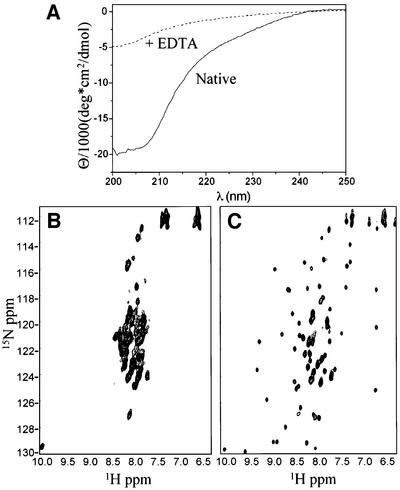
Based on this result, we investigated the role of zinc in structure formation of the PHD domain using circular dichroism (CD) and nuclear magnetic resonance (NMR) spectroscopies. The CD spectrum of KAP-1 PHD denotes a folded protein with substantial β-sheet and no significant helical content (Figure 2A). Introduction of the metal chelating agent EDTA results in complete loss of secondary structure. Addition of ZnCl2, in excess of EDTA, results in re-folding of the domain (data not shown). Similarly, 1H/15N-HSQC (heteronuclear single quantum correlation) NMR experiments indicate that the KAP-1 PHD domain requires zinc for proper folding (Figure 2B and C). In these experiments, the PHD domain was expressed in minimal media in the presence or absence of zinc and purified identically. The spectrum of the sample expressed in the absence of zinc shows poor dispersion of amide proton chemical shifts, indicative of an unfolded protein (Figure 2B). In contrast, the sample expressed in the presence of zinc produces a spectrum revealing a substantial increase in chemical shift dispersion, a decrease in line widths, and the appearance of virtually all expected backbone amide resonances; all hallmarks of a well-folded protein (Figure 2C). Thus, like RING and LIM domains, the binding of two zinc atoms is necessary for folding of the PHD domain.
Fig. 2. (A) CD spectrum of untagged KAP-1 PHD prior to addition of EDTA (solid line) and after addition (dashed line).(B and C) 1H/15N-HSQC of KAP-1 PHD produced in minimal media not supplemented (B) or supplemented with zinc (C).
The structure of the KAP-1 PHD domain reveals a cross-brace zinc ligation scheme
We determined the solution structure of the KAP-1 PHD domain using standard homonuclear and heteronuclear NMR techniques. Initial NMR studies were conducted on an N-terminal His-tagged construct of KAP-1 (amino acids 618–679) containing the entire PHD domain. A comparison of 2D 1H/15N HSQC and NOESY (nuclear Overhauser effect spectroscopy) spectra of tagged and untagged proteins revealed that the two spectra were identical apart from resonances corresponding to the tag (data not shown). Thus, the presence of this N-terminal His tag had no influence on the structure of the PHD domain.
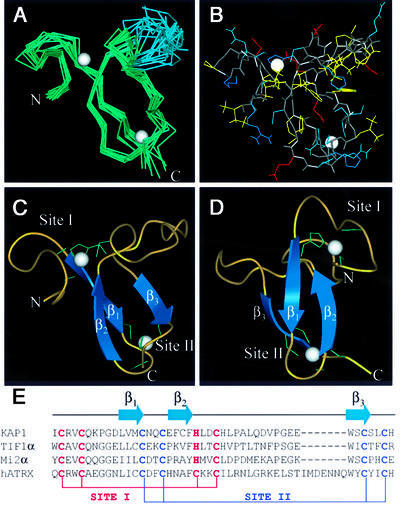
An initial set of structures was generated without reference to metal ligation in order to determine the residues involved in zinc binding. These structures indicated a cross-brace ligation scheme with residues C628, C631, H648 and C651 forming one zinc-binding site (site I) and C640, C643, C666 and C669 forming the other (site II). These sites were partially defined by the individual NOEs observed between residues implicated in zinc ligation. For instance, long-range NOEs were observed between C666 and C640, C631 and C651, and C640 and C669; in addition, several NOEs between residues adjacent to the zinc ligands were observed. Subsequent structure calculations included Zn atoms and additional constraints defining tetrahedral coordination. To ensure that a cross-brace was the correct zinc ligation scheme, alternative schemes were calculated but none satisfied the experimental constraints. The final ensemble is displayed in Figure 3A and all atoms are depicted in Figure 3B. Statistics of the ensemble are given in Table I.
Fig. 3. The KAP-1 PHD structure. (A) α-carbon overlay of 10 KAP-1 PHD domain structures. The residues N-terminal to I627 and C-terminal to H670 are disordered and not shown. The average position of the two zinc atoms is represented by white spheres. The blue portion of the ensemble represents the flexible hinge region described in the text. Structural statistics are given in Table I. (B) All-atom view of KAP-1 PHD in the same orientation as in (A). The main chain is colored in gray. Side chains are colored as follows: hydrophobic in yellow, polar (non-charged) in cyan, polar (acidic) in red, and polar (basic) in blue. (C and D) Ribbon diagram of the KAP-1 PHD domain. The β-strands are shown as arrows and zinc ligands in green. (E) Sequence alignment of PHD domains and a schematic depicting zinc ligation.
Table I. Structural statistics.
| Restraints for structure calculation (residues 627–670) | |
| total restraints | 385 |
| total NOE restraints | 317 |
| intra-residue | 78 |
| sequential | 118 |
| medium range (2 < |i–j| < 4) | 26 |
| long-range (|i–j| ≥ 5) | 95 |
| dihedral angle (φ) | 20 |
| H-bond restraints | 12 |
| zinc distance restraints | 20 |
| zinc angle restraints | 16 |
| Final energies (kcal/mol) | |
| Etotal | 37.1 ± 1.1 |
| ENOE | 0.30 ± 0.21 |
| Ecdih | 0.05 ± 0.04 |
| Coordinate precision (627–653, 664–670) | |
| r.m.s.d. of backbone atoms | 0.73 ± 0.16 Å |
| r.m.s.d. of all heavy atoms | 1.48 ± 0.18 Å |
| Procheck analysis (627–670) | |
| most favored regions | 36.0% |
| additionally allowed | 45.9% |
| generously allowed | 14.9% |
| disallowed region | 2.4% |
The structure of the KAP-1 PHD domain reveals a globular domain binding two Zn atoms in a distinguishing cross-brace fashion (Figure 3). In this arrangement, residues C628, C631, H648 and C651 form a single zinc-binding site (site I), while C640, C643, C666 and C669 form the second site (site II) (refer to Figure 3E). The formation of site I begins with an eight residue loop (residues 627–634), which continues into an extended region (residues 635–636) followed by the first β-strand (β1; residues 637–640). Site II is then created by a turn (residues 641–644) that ends in the second β-strand (β2; residues 645–647) running anti-parallel to β1. The first zinc-binding site is completed by another loop (residues 648–652). This portion is followed by an extended flexible region (residues 653–663; shown in blue, Figure 3A) demarcated by two proline residues (P654 and P660), which appear to act as a hinge relative to the rest of the domain (Figure 3A). This flexible region is followed by a third β-strand (β3; residues 664–666), leading to the completion of the second zinc-binding site (residues 666–670). Numerous hydrophobic interactions appear to stabilize this structure. F647 is at the center of the hydrophobic core packing against W664, V638, L656 and H652. Interestingly, the proton resonances for the aromatic ring of F647 are not degenerate, indicating that the phenyl group is not free to rotate in the core, suggesting that the core is tightly packed. Analysis of the electrostatic surface potential reveals two areas of discrete charge (Nicholls et al., 1991). Most notably, the flexible hinge (residues 653–663) is lined with several Glu and Asp residues, contributing to a negatively charged surface, and the presence of Arg629 and Lys633 results in a positively charged cluster around site I. Ongoing mutational analyses will be utilized to determine the importance of these features to KAP-1 PHD function. In terms of charge and topology, no structural features typical of nucleic acid binding proteins are observed, such as those present in the Kruppel-like or GATA-like zinc fingers (Klevit, 1991; Omichinski et al., 1993). This suggests that the PHD is not a nucleic acid binding domain.
The PHD domain structurally resembles a RING
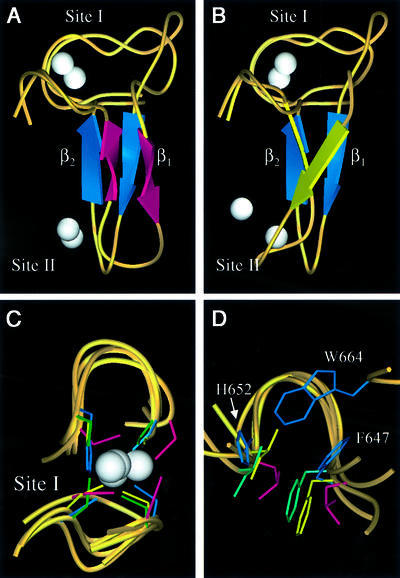
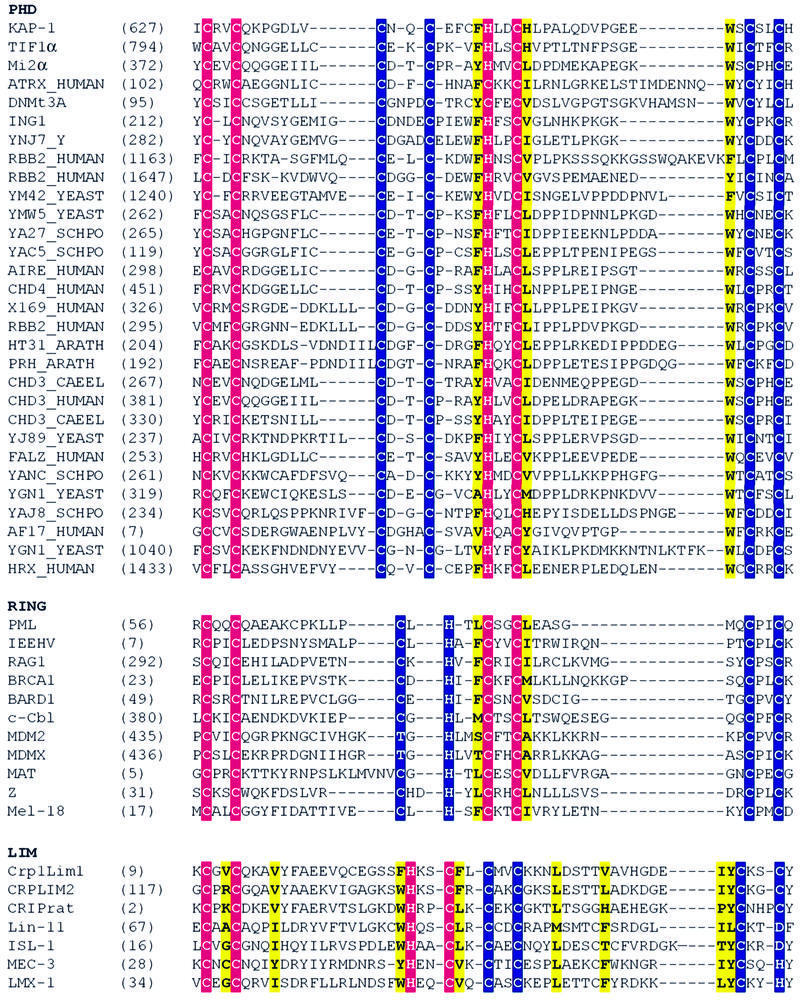
Unlike the sequential zinc ligation pattern used by the LIM domains, the KAP-1 PHD domain structure shows a zinc ligation pattern similar to RING domains. Therefore, we carried out a detailed comparison between the PHD domain and three previously reported RING domain structures, from the promyelocytic leukemia protein PML, the immediate early equine herpes virus protein IEEHV (also known as ICP0) and the recombination protein RAG1 (Protein Data Bank codes 1BOR, 1CHC and 1RMD, respectively; Barlow et al., 1994; Borden et al., 1995a; Bellon et al., 1997). These proteins have no sequence homology outside of conserved metal-binding residues. Strikingly, in all cases the inter-zinc distance is ∼14 Å, presumably because the central β-strand of these molecules is the same length (β2 in KAP-1 PHD) and all use the cross-brace ligation scheme. Because PHD and RING domains use different permutations of cysteines and histidines for metal ligation, for clarity we denote the ith conserved metal-ligating residue along the primary sequence by mli. Notably, a conserved hydrophobic core residue (F647) is located in β2, just N-terminal to ml5. Inspection of these structures indicates that the first zinc-binding site and anti-parallel β-strands (β1β2) of PHD overlay strikingly well with the three RING structures (Figure 4). An α-carbon backbone alignment of RING and KAP-1 PHD structures for site I overlay almost exactly, as do the zinc atoms. The root mean square difference (r.m.s.d.) of the α-carbon backbone for residues around site I is 1.4 Å when compared with PML, and 1.3 Å when compared with IEEHV or RAG1 (Figure 4C). PML, IEEHV and RAG1 RINGs have a large insertion along the β1 strand relative to KAP-1 PHD (Figure 5). Comparison of residues just N-terminal to ml1 through to just C-terminal to ml6, which include residues in the hydrophobic core, results in r.m.s.d. values between KAP-1 and PML of 2.0 Å, between KAP-1 and IEEHV of 2.1 Å, and between KAP-1 and RAG1 of 1.9 Å. The sequence identity in this region is 20% between PML and KAP-1, 22% between IEEHV and KAP-1, and 24% between RAG1 and KAP-1 (Figure 4A and B). Normally, r.m.s.d. values of ∼2 Å are expected for proteins with sequence identities >30% (Flores et al., 1993). Thus, these levels of similarity are striking considering there is virtually no sequence conservation between these domains outside of their conserved zinc ligands.
Fig. 4. Comparison of the KAP-1 PHD, PML RING, IEEHV RING and RAG1 RING. Superposition of KAP-1 PHD (with blue β-strands, aa 627–652) and PML RING (magenta β-strands, aa 56–81) (A) and RAG1 RING (yellow β-strands, aa 292–317) (B) from the first metal ligand to the sixth metal ligand. The white spheres represent zinc atoms with the upper zinc atom being site I. The orientation of KAP-1 PHD is the same in both panels. (C) Superposition of zinc-binding site I for KAP-1 (627–632, 647–652), PML (56–71, 76–81), IEEHV (7–12, 28–33) and RAG1 (292–297, 312–317). The metal ligands are colored according to protein: KAP-1 in blue, PML in magenta, IEEHV in green and RAG1 in yellow. (D) Superposition of the conserved hydrophobic core residues N-terminal to metal ligand 5 and C-terminal to metal ligand 6 (L76 and L81 in PML, F28 and I33 in IEEHV, and F312, I317, F647 and H652 in PHD). The side-chains are colored as in (C). The core residues from PHD are noted for clarity. The conserved tryptophan within the PHD family (W664 in KAP-1) is seen here inserting between the other core residues, repositioning the core.
Fig. 5. Amino acid sequence alignment of the PHD, RING and LIM families. Site I metal ligands are colored in magenta and site II metal ligands in blue. Conserved hydrophobic core residues are colored in yellow. DDBJ/EMBL/GenBank accession Nos and PDB codes: KAP-1 (U78773), TIF1α (AAD17258), Mi2α (Q14839), ATRX (P46100), DNMt3A (BAA95556), ING1 (AAG02578), YNJ7_YEAST (P50947), RBB2_HUMAN (P29375), YM42_YEAST (Q03214), YMW5_YEAST (Q04779), YA27_SCHPO (Q09698), YAC5_SCHPO (Q09819), AIRE_HUMAN (O43918), CHD4_HUMAN (Q14839), X169_HUMAN (CAA89909), HT31_ARATH (Q04996), PRH_ARATH (P48785), CHD3_CAEEL (Q22516), CHD3_HUMAN (Q12873), YJ89_YEAST (P47156), FALZ_HUMAN (Q12830), YANC_SCHPO (Q10077), YGN1_YEAST (P53127), YAJ8_SCHPO (Q09908), AF17_HUMAN (Q09908), YGN1_YEAST (P53127), HRX_HUMAN (Q03164), PML (1bor), IEEHV (1chc), RAG1 (1RMD), BRCA1 (A58881), BARD1 (NP_00456), c-Cbl (P22681), MDM2 (CAA41684), MDMX (O15151), MAT (S60157), Z (P18541), Mel-18 (P35227), Crp1LIM1 and LIM2 (1B8T), CRIPrat (1IML), Lin-11 (CAA38240), ISL-1 (CAA3749), MEC-3 (S28390), LMX-1 (B46233).
Like the PHD family, the RING family shows substantial structural plasticity between ligands ml6 and ml7, where the chain completes site I and continues towards site II. In PHD domains, the spacing varies from 12 to 46 residues, and in RING domains, the spacing varies from 4 to 48 residues. The RINGs of IEEHV, RAG1 and BRCA1 have large inserts in this region, forming a helix. However, RINGs with shorter inserts, e.g. PML and BARD1, do not have a helix in this region (information for BRCA1 and BARD1; P.Brozvic and R.Klevit, personal communication; reviewed in Borden, 1998). In contrast, this region in the KAP-1 PHD contains the flexible hinge (residues 653–663). Although this insert is comparable in size to that of RINGs containing a helix, the propensity for this region to adopt a helical conformation is highly unlikely due to the presence of two proline residues (P654 and P660).
A major structural difference between PHD and RING domains is found in the hydrophobic core. Residues that form the hydrophobic core of the RING structures are N-terminal to ml5 and C-terminal to ml6 (L76 and L81 in PML, F28 and I33 in IEEHV, F312 and I317 in RAG1; see also Figures 4D and 5) (Barlow et al., 1994; Borden et al., 1995a; Borden, 1998). Conserved hydrophobic residues at the same positions within the amino acid sequence are also found in the PHD (F647, N-terminal to ml5; and H652, C-terminal to ml6). However, there exists an additional conserved hydrophobic residue in PHD: a tryptophan two residues N-terminal to ml7 (W664 in KAP-1; Figures 4D and 5). This tryptophan, which is highly conserved throughout the PHD family, inserts between the core residues N-terminal to ml5 and C-terminal to ml6 and thereby forms part of the hydrophobic core. In RINGs, no hydrophobic core residue exists at the position two residues before ml7 (Figures 4D and 5). Rather, in all RING structures reported, the amino acid at this position is actually solvent exposed (Borden, 1998). This additional core residue appears specific to the PHD family. The presence of W664 alters the arrangement of the core where H652, which is C-terminal to ml6, is not as central to the core as this residue is in the RING domain. Additionally, the trace of the main chain is virtually unaltered relative to the RING fold (Figure 4D). Ligands ml3 and ml4 in site II are positioned almost identically in both PHDs and RINGs. In summary, both RING and PHD domains have conserved hydrophobic residues that are in the same sequence position and are found in the core of the domains. However, PHD accommodates an additional core residue, W664.
Aside from differences in the hydrophobic core, there are significant differences in the main-chain trace between ml6 and ml7 in PHD compared with RING domains. At H652, just C-terminal to ml6, the main-chain trace deviates significantly from RING domains. This difference appears to result from the participation of W664 (two residues N-terminal to ml7) in the hydrophobic core. This alters the position of ml7 and ml8 relative to the RINGs. These differences suggest that the PHD domain, although structurally similar to the RING in part, could bind structurally distinct partners and display unique biological activities. We expect that insertion size and sequence variability between ml2 to ml3 and ml6 to ml7 result in substantial structural plasticity in PHD domains, imparting specificity onto a common structural scaffold stabilized by zinc.
Sequence determinants for RING, LIM and PHD domains
The structural similarity between PHD and RING domains, coupled to the dissimilarity with LIM domains, prompted us to examine whether there were any structure-based sequence determinants that would enable one to predict whether a given amino acid sequence is PHD-, RING- or LIM-like in structure. The major structural determinant appears to be based on the zinc ligation scheme utilized. Sequence alignments for selected members of the PHD, RING and LIM families are given in Figure 5. For RING and LIM domains, conserved hydrophobic core residues determined through inspection of the structures are shown. In these cases, residues that form the hydrophobic core are shown in yellow, site I in magenta and site II in blue. These alignments suggest that proteins with conserved hydrophobic residues at positions N-terminal to ml5 and C-terminal to ml6 utilize a cross-brace ligation scheme as in the PHD/RING structures, whereas proteins with hydrophobic residues N-terminal to ml3 and C-terminal to ml4 utilize a sequential LIM-like ligation scheme. Alignments indicate that in the PHD/RING cross-brace schemes, the presence of a tryptophan residue two residues N-terminal to ml7 may be diagnostic of a PHD fold. In contrast, LIMs have additional conserved hydrophobic core residues that are not found in PHD or RING (Figure 5). Furthermore, residue spacing in the LIM domain is much more highly conserved than in PHDs or RINGs (Figure 1A).
Analysis of this structure allows one to distinguish between conserved hydrophobic residues observed by sequence information and conserved hydrophobic residues that additionally form the core of the domain. Some hydrophobic residues conserved within the PHD family do not participate in formation of the hydrophobic core. Residues positioned one and two amino acids N-terminal to ml3, Leu 637 and Val 638, are conserved as hydrophobic residues in the family (Figure 5) but are not part of the core. Mutation of Leu637 to Ala does not disrupt the transcriptional repression activity of KAP-1 to the same extent as mutation of conserved hydrophobic core residues, which results in almost complete loss of transcriptional activity and an unfolded protein (Figures 6 and 7; Table II).
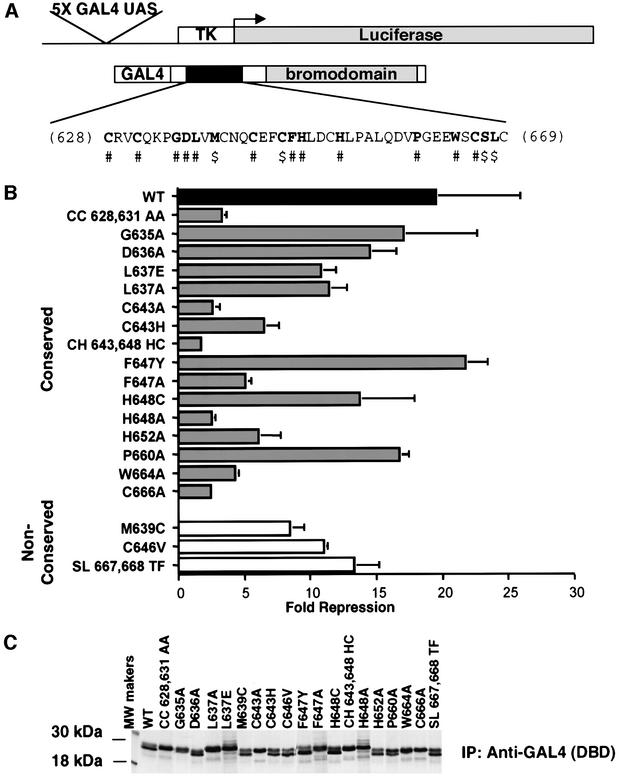
Fig. 6. Mutations in the PHD domain of KAP-1 significantly impair the intrinsic repression activity of KAP-1. (A) Schematic diagram illustrating the reporter plasmid (5×-GAL4-UAS-TK-luciferase) and effector plasmid (GAL4-KAP-1619–835). The primary amino acid sequence of the minimal KAP-1 PHD domain is illustrated. #, conserved amino acids that were mutated. $, non-conserved amino acids in KAP-1 that were mutated to match the corresponding amino acids in TIF1α/TIF1γ. Each mutation was made in the context of a GAL4-KAP-1 (619–835) expression construct. (B) Mutations in the KAP-1 PHD domain disrupt its role in transcriptional repression. All experiments were performed in NIH 3T3 cells with 5 µgof the indicated GAL4 fusion protein and 1 µg of a 5×-GAL4(UAS)-TK-luciferase reporter. A black bar represents wild type, gray bars represent mutations of conserved amino acid residues, and white bars represent substitutions at non-conserved amino acids. (C) Stable expression of each protein was determined via transfection into COS-1 cells followed by immunoprecipitation of [35S]methionine-labeled whole cell extracts with anti-GAL4 (DBD) antiserum (1 µg).
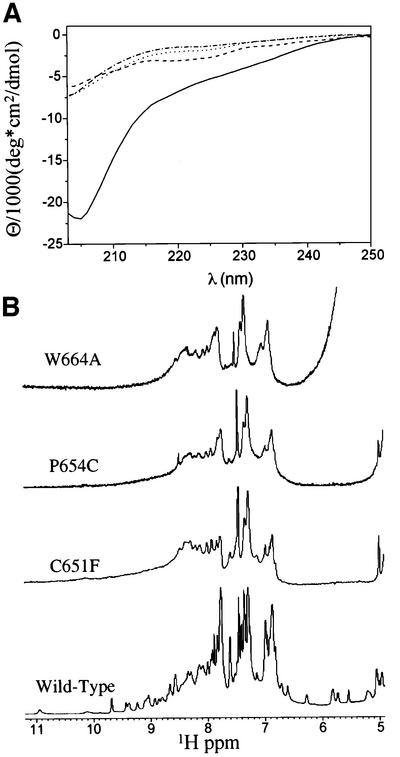
Fig. 7. Some mutations in the PHD domain of KAP-1 disrupt proper folding. (A) Comparison of CD spectra of wild-type KAP-1 PHD (solid line), W664A mutant (dashed/dotted line), P664C mutant (dotted line) and C651F mutant (dashed line) taken at pH 7.5, 25°C. (B) 1D 1H NMR spectra of wild-type and mutant KAP-1 PHD taken at pH 7.5, 30°C. The spectral regions containing the amide and aromatic proton resonances are shown.
Table II. PHD mutations and structural consequences.
| Mutations | Location in KAP-1 PHD structure | Transcriptionalrepressiona (%) | Structural consequences |
|---|---|---|---|
| CC628,631AA | Zn2+ ligands of site I | 17 | loss of metal binding, incorrect folding |
| G635A | surface, extended region before β1 | 87 | no obvious structural effect |
| D636A | surface, extended region before β1 | 74 | change in surface charge |
| L637A | surface, conserved hydrophobic | 58 | no obvious structural effect |
| L637E | surface, conserved hydrophobic | 55 | no obvious structural effect |
| M639C | β1, adjacent to metal ligand of site II (C640) | 43 | extra zinc ligand, may disrupt site II formation |
| C643A | Zn2+ ligands of site II | 13 | loss of metal binding, incorrect folding |
| C646V | β2, surface exposed | 56 | not a zinc ligand, no obvious structural effect |
| F647Y | core residue, β2 | 100 | conservative substitution |
| F647A | core residue, β2 | 26 | disrupts hydrophobic core |
| H648C | Zn2+ ligand of site I | 70 | conservative substitution |
| H648A | Zn2+ ligand of site I | 13 | loss of metal binding, incorrect folding |
| H652A | core residue | 31 | disrupts hydrophobic core |
| P660A | flexible hinge | 86 | no obvious structural effect |
| W664A | core residue, β3 | 22 | disrupts hydrophobic core |
| C666A | Zn2+ ligand of site II | 12 | loss of metal binding, incorrect folding |
| SL 667,668 TF | surface, site II | 68 | no obvious structural effect |
| ATRXb,c | |||
| C628R | Zn2+ ligand of site I | 15 | loss of metal binding, incorrect folding |
| V630S | surface, site I | 85 | may be important for protein–protein interactions |
| C651F | Zn2+ ligand of site I | 12 | loss of metal binding, incorrect folding |
| P654C | flexible hinge | 42 | extra zinc ligand causing incorrect folding |
| Q657E | surface, flexible hinge | 63 | change in surface charge |
| ING1b,d | |||
| C631S | Zn2+ ligand of site I | – | loss of metal binding, incorrect folding |
| N632S | surface, adjacent to metal ligand | – | may disrupt site I formation |
aPercentage compared with transcriptional repression by wild-type KAP-1 PHD.
bNumbers refer to the residue numbering for KAP-1.
cResults for ATRX mutations appear elsewhere (D.C.Schultz, J.R.Friedman and F.J.Rauscher,III, submitted).
dTranscriptional assays were not conducted for ING1 mutations.
For RING and LIM domains, flexibility in ligating residue type has been observed, e.g. His can replace Cys and even Asp can be used as a ligand. We examined the importance of specific types of zinc ligands to the fold of the PHD domain by examining the ability to repress transcription. Initially, we swapped the type of zinc ligand, e.g. His for Cys, at ligand position ml5 (648), and determined the ability of KAP-1 to act as a transcriptional repressor (Figure 6B; Table II). This mutation has little effect, indicating that the type of ligating residue, at least at this position, is not crucial. However, substitution of Ala for His at ml5 completely destroys the transcriptional repression properties of the KAP-1 PHD domain (Figure 6B; Table II). Intriguingly, in studies where KAP-1 was modified (Cys643His648 to His643Cys648) so that it used a RING-like zinc ligation scheme (Figure 1), there was a substantial loss of transcriptional activity relative to wild-type protein (Figure 6). Furthermore, mutation of Cys643 to His results in reduction in this activity but not to the same extent as the double mutation. Thus, ligand type may be critical at some positions within the sequence, whereas the choice of ligand type appears more flexible at other positions.
Structural integrity of the PHD domain is essential for KAP-1 to repress transcription
In order to define structural features of the PHD domain important for the transcriptional repression by KAP-1, mutational analysis was carried out on a GAL4-KAP-1 (619–835) expression construct (Figure 6A). This construct encodes both the PHD domain and the bromodomain of KAP-1, which together comprise an independent repression domain where both domains are required for this biological activity (Figure 6B and C; Table II; D.C.Schultz, J.R.Friedman and F.J.Rauscher,III, submitted). The positioning of the PHD domain adjacent to a bromodomain has been observed in several proteins; however, most PHD-containing proteins do not contain a bromodomain, suggesting that the PHD is an independent structural unit. Yet, in KAP-1, it is possible that the bromodomain could alter the fold of the PHD domain and thereby modulate its biological activity. In order to address whether the bromodomain could alter the structure of the PHD, we carried out additional NMR studies on a KAP-1 PHD–bromodomain construct. In these experiments, the chemical shifts of the resonances corresponding to the PHD domain appear largely identical to resonances in constructs containing only the PHD domain, indicating that the bromodomain does not substantially alter the fold of the PHD domain (our unpublished observations). Although the bromodomain does not appear to alter the fold of the PHD domain significantly in KAP-1, it may make crucial contacts that modulate its function or cause small-scale structural rearrangements, which may be critical for function.
We monitored the effects of mutations in the PHD domain on transcriptional activity (Figure 6; Table II). Mutations of zinc ligands in either site I (CC628, 631AA or H648A) or site II (C643A and C666A) abolished transcriptional repression, while mutation of the non-zinc-ligating cysteine (C646V) shows no effect. Furthermore, mutations that disrupt the hydrophobic core of the molecule are as debilitating as mutations that disrupt zinc ligation. Consistently, conservative non-disruptive mutations, such as F647Y, in the core do not alter transcriptional repression by KAP-1, whereas F647A abolishes this activity. Mutation of a conserved hydrophobic non-core residue, L637A, does not abolish transcriptional activity of the domain. A mutation that potentially introduces an extra zinc ligand (M639C or P654C) results in a lowered activity, presumably because this ‘new’ ligand may disrupt the formation of the cross-brace by competition for zinc during the folding process. In the wild-type structure, the M639 side chain is solvent exposed and P654 is part of the flexible hinge region. Reduction in transcriptional repression activity in the case of P654C or M639C could be due either to misfolding or to a disruption of PHD–bromodomain intermolecular interactions.
To determine the effects that some of these mutations impose on the structural properties of the domain, we expressed the PHD protein containing mutations of three different types. Based on the structure, W664A is expected to disrupt the hydrophobic core, C651F is expected to disrupt metal binding and P654C is expected to cause misfolding by disrupting metal binding through addition of an extra zinc ligand. Each protein was expressed and purified as previously described. As an initial evaluation of the biophysical consequences of these mutations on the structure of the PHD domain, we estimated the hydrodynamic size of each protein by gel filtration. Greater than 90% of the C651F mutant protein eluted from the gel filtration column immediately following the void volume, suggesting that the protein is likely to exist as a soluble aggregate (data not shown), consistent with being largely unfolded. A protein containing the P654C mutation eluted from the column in two distinct peaks. Approximately 50% of the protein eluted from the column just after the void volume, as observed for the C651F mutant protein. The remainder of the protein eluted with a Stokes radius consistent with the monomeric size observed with the wild-type protein (data not shown). The W664A mutant protein eluted from the column as a distribution of species, with a small but significant fraction of the protein eluting at a volume that overlaps with the monomeric size observed with the wild-type protein (data not shown). Fractions corresponding to the monomer for each mutant were collected and further analyzed to determine whether the proteins were folded. NMR and CD studies on the W664A, C651F and P654C proteins indicated that, as expected, disruption of the hydrophobic core or of zinc binding resulted in unfolded proteins (Figure 7). Analysis of CD spectra of each mutant indicates a substantial loss in molar ellipticity relative to wild type, indicating that the mutants are unfolded (Figure 7A). 1H NMR studies indicate that there is poor dispersion of the resonances in the amide and aromatic region of the spectra of all three mutants, indicative of an unfolded protein (Figure 7B). Furthermore, there are few NOEs observed in 2D 1H NOESY experiments, again demonstrating that these proteins are largely unstructured (data not shown). Thus, the ability of the KAP-1 PHD domain to bind zinc and maintain the hydrophobic core is required for its proper folding and ability to repress transcription.
The potential structural consequences of PHD domain mutations in other proteins
Naturally occurring mutations and deletions in the PHD domains of several proteins exemplify the biological importance of this domain. Germline missense mutations in the PHD domain of the ATRX protein predispose individuals to α-thalassemia and mental retardation (Gibbons et al., 1997). Somatic missense mutations found in the PHD domain of ING1 are detected in several types of carcinoma (Gunduz et al., 2000). Analysis of these mutations in light of the KAP-1 PHD structure reveals the basis for their devastating nature (Table II). In the ING1 PHD domain, two mutations of site I are found in squamous cell carcinomas (Gunduz et al., 2000). C631S (where these amino acids are numbered in the context of the KAP-1 PHD), like previous zinc ligand mutations, should destroy zinc binding and structure formation. N632S, adjacent to ml2, may indirectly interfere with zinc binding and, therefore, formation of site I. Two mutations in ATRX (C628R and C651F) disrupt the ability of this domain to ligate zinc. These mutations are expected to result in an unfolded protein, and equivalent mutations in the KAP-1 PHD domain result in a loss of repression (Table II) and unfolding (Figure 7). Another mutation in ATRX introduces an additional potential metal ligand (P654C), which disrupts biological activity through misfolding (Figure 7). Some surface mutations (V630S and Q657E) have no effect on transcriptional activity in these assays; however, these mutations are found in ATRX patients. Thus, these residues may be required by the ATRX protein for mediating specific protein–protein interactions. An interesting feature of the wild-type ATRX PHD domain is that it is a PHD domain with eight cysteines (His at ml5 is replaced by Cys). Mutation of this ligand in KAP-1 (H648C) resulted in only a slight loss of transcriptional activity (Figure 6B), suggesting that either a histidine or a cysteine can be tolerated as a metal ligand at this position. It is interesting to note that ATRX and ING1 proteins do not contain bromodomains, indicating that their disease potential does not arise because of disruption of a PHD–bromodomain interaction. Overall, the functional significance of any of these mutations will depend on context-specific biochemical functions of the given domain. None the less, this panel of mutations will serve as a useful tool in future studies in defining new macromolecular interactions and biological functions for the PHD domain.
The PHD domain is likely to be a protein–protein interaction domain
KAP-1 PHD participates in protein–protein interactions with Mi-2α (D.C.Schultz, J.R.Friedman and F.J.Rauscher, III, submitted), a component of the NuRD histone deacetylase complex (Wade et al., 1998; Brehm et al., 1999). Therefore, KAP-1 PHD may be important for proper spatial and temporal scaffolding of the repressor complex. This association is likely to be critical for transcriptional repression by KAP-1. Consistent with our results, recent work indicates that the AIRE PHD domain is required for formation of nuclear protein complexes (Rinderle et al., 1999). Furthermore, the AF10 PHD domain is reported to be a homo-oligomerization module (Linder et al., 2000). Like RINGs, the PHD domain appears to function as a protein–protein interaction motif. Presumably, disruption of the structural integrity of the PHD domain by mutation of critical residues fundamentally disrupts this action. Recent studies show that some RING domains act as E3 ubiquitin ligases through interactions with the ubiquitin machinery (reviewed in Borden, 2000). Because of the similarity to the RING, we tested KAP-1 in this system. Neither the PHD alone nor the PHD–bromodomain together has detectable E3 ligase activity (data not shown). It will be necessary to test several other PHD domains before this activity can be ruled out for the family as a whole.
Conclusions
We present the first structure of a PHD domain and show that it is an autonomously structured domain that requires zinc for folding. From sequence analysis alone it is impossible to determine how the eight conserved metal ligands are utilized to bind zinc. This uncertainty is evident from a recently modeled structure of the DNMT3L PHD domain where the ligation scheme was assumed to be LIM-like (Aapola et al., 2000). The actual ligation of zinc by the PHD domain is accomplished by a cross-brace motif like that used by the RING family. Not only are the ligation schemes similar between the two domains, but also the structures are topologically identical from ml1 to ml6, which includes the first zinc-binding site and the anti-parallel β-strands. We identify structural determinants that distinguish ligation schemes of RING, LIM and PHD domains. In the structure of the KAP-1 PHD domain, there exists a flexible hinge region between ml6 and ml7 characterized by a discrete negatively charged surface. Amongst the RING domains, this same region shows structural variability, forming an extended strand or a helix. The plasticity of this region may underlie the biological specificity of these rather similar domains. The PHD domain is required for transcriptional repression by KAP-1 and we present structural determinants required for this activity, which elucidate the structural basis of disease-causing missense mutations in ATRX and ING1.
Materials and methods
Preparation of the PHD domain
A DNA fragment encoding the PHD domain of human KAP-1 (amino acids 618–679) was subcloned into the pQE30 expression vector (Qiagen) and expressed in E.coli BL21(DE3) cells (Novagen). The His-tagged protein contained an additional 17 N-terminal amino acids, MRGSHHHHHHGSDIIDE, amino acids 618–679 of KAP-1, and nine C-terminal amino acids, VDLQACKLN. For the untagged KAP-1 PHD domain protein, a DNA fragment encoding amino acids 618–679 was subcloned into the expression vector pQE50 (Qiagen) and expressed in E.coli SG13009 cells (Qiagen). The expressed protein contained four N-terminal amino acids, MRGS, followed by amino acids 618–679 of KAP-1. His-tagged or untagged protein was prepared from logarithmically growing bacteria cultured in either 2YT media or in minimal media, containing 15NH4Cl as the sole nitrogen source and supplemented with 100 µM zinc acetate (for uniformly labeled 15N protein). In both types of medium, protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C. Induced bacterial pellets were lysed in 20 mM NaH2PO4 pH 7.5, 500 mM NaCl, 5 mM dithiothreitol (DTT). The His-tagged PHD domain was purified by affinity chromatography on a Ni2+-NTA–agarose column (Qiagen), followed by fractionation through a Superdex 75 size exclusion column (Pharmacia). The untagged PHD domain was purified sequentially by ion exchange chromatography (DEAE–cellulose), hydrophobic interaction chromatography (butyl–Sepharose) and size exclusion chromatography (Superdex 75). NMR experiments utilized both constructs, while metal binding analysis (i.e. ICP and CD) used only the untagged PHD construct. W664A, P654C and C651F mutant proteins were produced as His-tagged proteins and purified as described above.
Site-directed mutagenesis
Site-directed point mutations in the KAP-1 PHD domain were engineered by standard overlap extension PCR-mediated mutagenesis procedures. The mutagenic primers for the described mutations (Figure 6) contained the following codons: CC628,631AA, TGC to GC; C628R, TGC to CGC; V630S, GTC to AGC; G635A, GGC to GCC; D636A, GAT to GCT; M639C, ATG to TGC; C643A, TGT to GCT; C646V, TGT to GTT; F647Y, TTC to TAC; H648C, CAC to TGC; C651F, TGT to TTT; P654C, CCG to TGC; Q657E, CAG to GAG; P660A, CCA to GCA; W664A, TGG to GCG; C666A, TGC to GCC; SL667,668TF, TCA and CTC to ACA and TTC. Appropriate reading frame fusions and integrity of flanking sequences for all constructs created by PCR were confirmed by DNA sequence analysis of both strands.
Transient transfections
Protein expression from all plasmids was confirmed by transient transfection of COS-1 cells followed by immunoprecipitation of [35S]methionine-labeled cell extracts. All transcription assay transfections were carried out as previously described (Ryan et al., 1999).
ICP spectrometry
ICP spectrometry was conducted using the untagged PHD sample at Galbraith Laboratories, Inc. (Knoxville, TN). Twenty-five microliters of a 3.0 mM PHD sample (20 mM NaH2PO4, 500 mM NaCl, 5 mM DTT pH 7.5) were prepared for ICP analysis using a wet ash digestion procedure as previously described (Bock, 1979). Measurements were made on a Perkin-Elmer P2000 using a primary wavelength of 213.856 nm (Wallace, 1981).
CD spectroscopy
Spectra were collected on a Jasco 810 spectropolarimeter at 25°C using a 1 mm path-length cuvette. For the unfolding experiments (Figure 2), the concentration of the untagged PHD sample was 30 µM in 20 mM NaH2PO4, 50 mM NaCl pH 7.5. After an initial spectrum was collected, a 6-fold molar excess of EDTA was added and pH was adjusted to 7.5. The sample was then left at room temperature for 16 h to ensure it had reached equilibrium. Spectra were then recorded. For the comparison of KAP-1 PHD mutants (Figure 7), the concentration of PHD wild type and PHD mutants ranged from 15 to 30 µM in 20 mM NaH2PO4, 50 mM NaCl pH 7.5 at 25°C. The buffer contribution was subtracted and signals normalized to protein concentrations.
NMR spectroscopy
For NMR, KAP-1 PHD domain protein concentration ranged from 1.5 to 3.0 mM in 20 mM NaH2PO4, 500 mM NaCl, 5 mM DTT pH 7.5. No spectral changes were observed in this sample concentration range. For 1H NMR of KAP-1 PHD mutants (Figure 7), protein concentrations ranged from 1.1 to 2.5 mM in 20 mM NaH2PO4, 500 mM NaCl, 5 mM DTT pH 7.5. Spectra were recorded at 30°C on a Bruker DRX500 spectrometer. Sequential assignments were obtained using 3D 1H/15N NOESY-HSQC, 3D 1H/15N HMQC-NOESY-HSQC and 3D 1H/15N total correlated spectroscopy (TOCSY)-HSQC spectra. Side-chain resonances were assigned using 3D 1H/15N TOCSY-HSQC, 2D 1H-TOCSY and 2D 1H double quantum filter (DQF)-COSY spectra. Aromatic protons were assigned though 2D 1H-TOCSY, 2D 1H-NOESY and 2D 1H DQF-COSY spectra recorded in 2H2O. 2D data were collected and analyzed as described (Borden et al., 1995a,b). For side-chain and aromatic assignments, a construct with no His tag was used. 3D 1H/15N NOESY-HSQC experiments (with mixing times, τm, of 75 and 150 ms) and 2D 1H-NOESY experiments (τm of 75 and 150 ms) were also used for obtaining interproton distance restraints. φ angle restraints were determined based on the 3JHN,Hα coupling constants measured in a 3D HNHA experiment. Slowly exchanging amide protons were identified from 2D 15N-HSQC spectra recorded 12 h after the 1H2O buffer was exchanged for 2H2O. Detailed descriptions of these experiments along with their original references have been reviewed elsewhere (Clore and Gronenborn, 1994; Cavanagh, 1996). All NMR data were processed using the NMRpipe software system (Delaglio et al., 1995) and analyzed with the program NMRview (Johnson et al., 1994).
Structure calculations
NOE-derived distance restraints were obtained from 3D 1H/15N NOESY-HSQC spectra and 2D 1H-NOESY spectra. The intensities of NOE cross-peaks assigned in the 2D 1H-NOESY and the 3D 1H/15N NOESY-HSQC spectra were classified as strong (1.8–3.0 Å), medium (1.8–4.0 Å) and weak (1.8–5.0 Å) using secondary structure elements for calibration. Pseudo-atom corrections were added to the upper distance limit when appropriate; for example, 2.4 and 1.0 Å were added to methyl and β methylene protons, respectively (Wüthrich, 1986). Dihedral φ angle restraints were estimated from 3JHN,Hα coupling constants obtained from a 3D HNHA spectrum. 3JHN,Hα <6 Hz were given φ angle restraints of –60 ± 40°, while 3JHN,Hα >8 Hz were given φ angle restraints of –120 ± 40°. Additionally, 3JHN,Hα coupling constants were obtained from highly digitized DQF-COSY spectrum in 1H2O. Values obtained for coupling constants from the two methods were in agreement. Structures were calculated with a distance geometry/simulated annealing protocol in X-PLOR v. 3.851 (Brunger, 1996). Initial structure calculations were performed without any assumptions about zinc coordination. Initial structures indicated a cross-brace zinc ligation scheme (see Results and discussion). Subsequent structure calculations included zinc atoms with additional distance and angle constraints to maintain the tetrahedral bonding geometry of the sites and appropriate bond lengths as previously described by Neuhaus et al. (1992). Other alternative ligation schemes were tested but were unable to satisfy the experimentally derived constraints. Structures from these calculations were used for an automated, iterative assignment of remaining NOEs using ARIA (Nilges, 1995; Nilges et al., 1997). In addition to NOE data, calculations used a total of six hydrogen bonds and 20 φ dihedral restraints. Quality of the final structures was assessed using PROCHECK-NMR (Laskowski et al., 1996). Figures were prepared using the program PREPI. The coordinates for the KAP-1 PHD domain have been deposited in the Protein Data Bank under accession code 1FP0. Chemical shift information is available upon request.
Sequence and structure comparisons
Structural comparisons and r.m.s.d. calculations between PHD, PML (1bor) and IEEHV (1chc) were performed using the least squares fitting algorithm LSQFIT. The graphical overlays were viewed using PREPI. PREPI and LSQFIT were kindly provided by S.Islam and M.Sternberg, Imperial Cancer Research Fund. Sequence alignments were carried out using Clustal W 1.7.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful for critical reading of the manuscript by Aneel Aggarwal, Graeme Carlile, Alex Kentsis, Pauline McIntosh and Larry Shapiro. We thank Lei Zeng for assistance in setting up NMR experiments. PREPI and LSQFIT programs were a kind gift of Suhail Islam and Mike Sternberg (Imperial Cancer Research Fund). A.D.C. is supported by an NCI predoctoral training grant. D.C.S. is supported in part by the Wistar Basic Cancer Research Training Grant CA 09171 and DAMD 17-98-1-8269 grant. F.J.R. is supported in part by National Institutes of Health grants CA 52009, Core grant CA 10815, DK 49210, GM 54220, DAMD 17-96-1-6141, ACS NP-954, the Irving A.Hansen Memorial Foundation, the Mary A.Rumsey Memorial Foundation, and the Pew Scholars Program in the Biomedical Sciences. K.L.B.B. is a scholar of the Leukemia and Lymphoma Society. Financial support for K.L.B.B. was provided by National Institutes of Health grant RO1 CA 80728-01.
REFERENCES
- Aapola U. et al. (2000) Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics, 65, 293–298. [DOI] [PubMed] [Google Scholar]
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Barlow P.N., Luisi,B., Milner,A., Elliott,M. and Everett,R. (1994) Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol., 237, 201–211. [DOI] [PubMed] [Google Scholar]
- Bellon S.F., Rodgers,K.K., Schatz,D.G., Coleman,J.E. and Steitz,T.A. (1997) Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nature Struct. Biol., 4, 586–591. [DOI] [PubMed] [Google Scholar]
- Bochar D.A., Savard,J., Wang,W., Lafleur,D.W., Moore,P., Cote,J. and Shiekhattar,R. (2000) A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl Acad. Sci. USA, 97, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. (1979) Decomposition Methods in Analytical Chemistry. T & A Constable Ltd, Edinburgh, UK.
- Borden K.L.B. (1998) RING fingers and B-boxes: zinc-binding protein–protein interaction domains. Biochem. Cell Biol., 76, 351–358. [DOI] [PubMed] [Google Scholar]
- Borden K.L.B. (2000) RING domains: master builders of macromolecular scaffolds? J. Mol. Biol., 295, 1103–1112. [DOI] [PubMed] [Google Scholar]
- Borden K.L.B. and Freemont,P.S. (1996) The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol., 6, 395–401. [DOI] [PubMed] [Google Scholar]
- Borden K.L.B., Boddy,M.N., Lally,J., O’Reilly,N.J., Martin,S., Howe,K., Solomon,E. and Freemont,P.S. (1995a) The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J., 14, 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K.L.B., Lally,J.M., Martin,S.R., O’Reilly,N.J., Etkin,L.D. and Freemont,P.S. (1995b) Novel topology of a zinc-binding domain from a protein involved in regulating early Xenopus development. EMBO J., 14, 5947–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Nielsen,S.J., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1999) The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J., 18, 2449–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A. (1996) XPLOR (Version 3.843): A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Cavanagh J., Fairbrother,W.J., Palmer,A.G.I. and Skelton,N.J. (1996) Protein NMR Spectroscopy. Academic Press, San Diego, CA.
- Clore G.M. and Gronenborn,A.M. (1994) Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol., 239, 349–363. [DOI] [PubMed] [Google Scholar]
- Dawid I.B., Breen,J.J. and Toyama,R. (1998) LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet., 14, 156–162. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Flores T.P., Orengo,C.A., Moss,D.S. and Thornton,J.M. (1993) Comparison of conformational characteristics in structurally similar protein pairs. Protein Sci., 2, 1811–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Fredericks,W.J., Jensen,D.E., Speicher,D.W., Huang,X.P., Neilson,E.G. and Rauscher,F.J.,III (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev., 10, 2067–2078. [DOI] [PubMed] [Google Scholar]
- Gibbons R.J. et al. (1997) Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nature Genet., 17, 146–148. [DOI] [PubMed] [Google Scholar]
- Gunduz M., Ouchida,M., Fukushima,K., Hanafusa,H., Etani,T., Nishioka,S., Nishizaki,K. and Shimizu,K. (2000) Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res., 60, 3143–3146. [PubMed] [Google Scholar]
- Jacobson S. and Pillus,L. (1999) Modifying chromatin and concepts of cancer. Curr. Opin. Genet. Dev., 9, 175–184. [DOI] [PubMed] [Google Scholar]
- Johnson M., Correia,J.J., Yphantis,D.A. and Halvorson,H.R. (1994) NMR VIEW—a computer program for the visualization and analysis of NMR data. J. Biomol. NMR, 6, 603–614. [DOI] [PubMed] [Google Scholar]
- Jones M.H., Hamana,N., Nezu,J. and Shimane,M. (2000) A novel family of bromodomain genes. Genomics, 63, 40–45. [DOI] [PubMed] [Google Scholar]
- Klevit R.E. (1991) Recognition of DNA by Cys2, His2 zinc fingers. Science, 253, 1367–1393. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Lechner M.S., Begg,G.E., Speicher,D.W. and Rauscher,F.J.,III (2000) Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain–KAP-1 corepressor interaction is essential. Mol. Cell. Biol., 20, 6449–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B. et al. (1995) The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J., 14, 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B., Nielsen,A.L., Garnier,J.M., Ichinose,H., Jeanmougin,F., Losson,R. and Chambon,P. (1996) A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J., 15, 6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Linder B., Newman,R., Jones,L.K., Debernardi,S., Young,B.D., Freemont,P., Verrijzer,C.P. and Saha,V. (2000) Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J. Mol. Biol., 299, 369–378. [DOI] [PubMed] [Google Scholar]
- Lu X., Meng,X., Morris,C.A. and Keating,M.T. (1998) A novel human gene, WSTF, is deleted in Williams syndrome. Genomics, 54, 241–249. [DOI] [PubMed] [Google Scholar]
- Neuhaus D., Nakaseko,Y., Schwabe,J.W. and Klug,A. (1992) Solution structures of two zinc-finger domains from SWI5 obtained using two-dimensional 1H nuclear magnetic resonance spectroscopy. A zinc-finger structure with a third strand of β-sheet. J. Mol. Biol., 228, 637–651. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nilges M. (1995) Calculation of protein structures with ambiguous distance restraints. Automated assignment of ambiguous NOE crosspeaks and disulphide connectivities. J. Mol. Biol., 245, 645–660. [DOI] [PubMed] [Google Scholar]
- Nilges M., Macias,M.J., O’Donoghue,S.I. and Oschkinat,H. (1997) Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from β-spectrin. J. Mol. Biol., 269, 408–422. [DOI] [PubMed] [Google Scholar]
- Omichinski J.G., Clore,G.M., Schaad,O., Felsenfeld,G., Trainor,C., Appella,E., Stahl,S.J. and Gronenborn,A.M. (1993) NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science, 261, 438–446. [DOI] [PubMed] [Google Scholar]
- Peng H., Begg,G.E., Harper,S.L., Friedman,J.R., Speicher,D.W. and Rauscher,F.J.,III (2000a) Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J. Biol. Chem., 275, 18000–18010. [DOI] [PubMed] [Google Scholar]
- Peng H., Begg,G.E., Schultz,D.C., Friedman,J.R., Jensen,D.E., Speicher,D.W. and Rauscher,F.J.,III (2000b) Reconstitution of the KRAB–KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein–protein interactions. J. Mol. Biol., 295, 1139–1162. [DOI] [PubMed] [Google Scholar]
- Rinderle C., Christensen,H.M., Schweiger,S., Lehrach,H. and Yaspo,M.L. (1999) AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: altered sub-cellular distribution of mutants lacking the PHD zinc fingers. Hum. Mol. Genet., 8, 277–290. [DOI] [PubMed] [Google Scholar]
- Ryan R.F., Schultz,D.C., Ayyanathan,K., Singh,P.B., Friedman,J.R., Fredericks,W.J. and Rauscher,F.J.,III (1999) KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol., 19, 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Finnish–German Autoimmune Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy Consortium (1997) An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. The Finnish–German APECED Consortium. Autoimmune Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy. Nature Genet., 17, 399–403. [DOI] [PubMed] [Google Scholar]
- Venturini L. et al. (1999) TIF1γ, a novel member of the transcriptional intermediary factor 1 family. Oncogene, 18, 1209–1217. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Jones,P.L., Vermaak,D. and Wolffe,A.P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol., 8, 843–846. [DOI] [PubMed] [Google Scholar]
- Wallace G. (1981) Analytical Methods for Inductively Coupled Spectrometry. Perkin Elmer Corp., Norwalk, CT.
- Wüthrich K. (1986) NMR of Proteins and Nucleic Acids. Wiley, New York, NY.