Abstract
The hyperthermophilic archaeon Methanococcus jannaschii encodes two putative transcription regulators, Ptr1 and Ptr2, related to the bacterial Lrp/AsnC family of transcriptional regulators. We show that these two small helix–turn–helix proteins are specific DNA-binding proteins recognizing sites in their respective promoter regions. In vitro selection at high temperature has been used to isolate sets of high- affinity DNA sites that define a palindromic consensus binding sequence for each protein. Ptr1 and Ptr2 bind these cognate sites from one side of the DNA helix, as dimers, with each protein monomer making base- specific contacts in the major groove. As the first archaeal DNA-binding proteins with clearly defined specificities, Ptr1 and Ptr2 provide a thermostable DNA-binding platform for analysis of effector interactions with the core archaeal transcription apparatus; a platform allowing manipulation of promoter structure and examination of mechanisms of action at heterologous promoters.
Keywords: archaea/DNA-binding proteins/Lrp homologs/SELEX/transcriptional regulators
Introduction
Purification and characterization of RNA polymerases, the development of cell-free transcription systems and systematic genome sequencing have revealed that the basal components of the archaeal transcriptional apparatus resemble those of eukaryotic RNA polymerase II (for reviews see Langer et al., 1995; Thomm, 1996; Soppa, 1999). Archaeal promoters consist of an A+T-rich TATA-like segment referred to as the box A/TATA element. Like eukaryotic TATA box-binding protein (TBP), archaeal TBP (aTBP) recognizes the box A/TATA element, and the TFIIB-related transcription factor B (aTFB) binds to the aTBP–DNA complex, thereby directing the RNA polymerase to initiate transcription specifically at an initiator sequence located some 25 bp downstream of the TATA element. In addition to the initiator sequence and the TATA box, efficient pre-initiation complex assembly, and therefore promoter strength, depends on the adjacent purine-rich BRE element, which mediates sequence-specific DNA interactions by aTFB upstream of box A (Qureshi and Jackson, 1998), and ensures the unidirectionality of transcription complex assembly and initiation (Bell et al., 1999b).
Despite this emerging understanding of the basal archaeal transcription machinery, little is known about its regulatory mechanisms. The identification, from the growing body of archaeal genome sequences, of a large number of open reading frames (ORFs) encoding potential transcription regulators of bacterial type, including many helix–turn–helix (HTH) DNA-binding proteins, suggests a chimeric nature for the archaeal transcription apparatus (Bult et al., 1996; Klenk et al., 1997; Smith et al., 1997; Aravind and Koonin, 1999; Kyrpides and Ouzounis, 1999). For example, ORFs coding for putative homologs of Lrp, the leucine-responsive regulatory protein of Escherichia coli, have been identified in several archaea, including Pyrococcus species (Kyrpides and Ouzounis, 1999; Brinkman et al., 2000), Methanococcus jannaschii (Bult et al., 1996), Methanobacterium thermoautotrophicum (Smith et al., 1997), Archaeoglobus fulgidus (Klenk et al., 1997) and Sulfolobus species (Charlier et al., 1997; Napoli et al., 1999; Bell and Jackson, 2000; Enoru-Eta et al., 2000).
Lrp is the prototype, and by far the most studied member, of the Lrp/AsnC family of transcription regulators in Gram-positive and Gram-negative bacteria (for reviews see Calvo and Matthews, 1994; Newman and Lin, 1995; Newman et al., 1996). It is a global transcriptional regulator governing the expression of >75 genes in E.coli, with regulatory effects (positive or negative) that can require leucine, be abolished by leucine or be independent of it, leading to a total of six different modes of regulation. The detailed molecular mechanisms of regulation by E.coli Lrp are still largely unknown. Escherichia coli Lrp is a small, basic, abundant double-stranded DNA-binding protein composed of two identical 19 kDa subunits. Mutational analysis indicates that the N-terminal portion of Lrp, which contains a HTH motif, is responsible for DNA binding, whereas the middle and C-terminal parts are proposed to be involved in transcriptional activation and in leucine binding (Platko and Calvo, 1993).
Sulfolobus solfataricus Lrs14 is a homodimeric Lrp-like protein whose transcripts accumulate during late stages of growth (Napoli et al., 1999; Bell and Jackson, 2000). In electrophoretic mobility shift assays (EMSAs) and DNase I footprinting analyses, Lrs14 produced in E.coli was shown to bind specifically to multiple sites in its own promoter region (Napoli et al., 1999), and to repress transcription of its gene in a reconstituted in vitro transcription system (Bell and Jackson, 2000). Similarly, Sa-Lrp, the Sulfolobus acidocaldarius Lrp homolog and a homotetrameric HTH protein, binds to multiple sites in its own promoter region (Enoru-Eta et al., 2000); and the Pyrococcus furiosus LrpA protein binds to its promoter (Brinkman et al., 2000).
In the experiments reported here, we have characterized two Lrp-like proteins from the hyperthermophilic archaeon M.jannaschii. We show that these HTH proteins bind to sites within their respective promoter regions. Using a variation of the SELEX strategy (systematic evolution of ligands by exponential enrichment; Tuerk and Gold, 1990), we have isolated a set of high-affinity DNA sites that define a consensus binding sequence for each protein. We specify the mode of DNA binding by these two proteins on the basis of DNase I, hydroxyl radical and methylation interference footprinting, as well as site-directed mutagenesis and deletion analyses. We also present preliminary results of a genome-wide search for putative binding sites for these proteins and discuss their potential involvement in gene-specific and global regulation of transcription.
Results
Ptr1 and Ptr2: two M.jannaschii Lrp homologs
In the M.jannaschii genome (Bult et al., 1996), ORFs MJ0151 and MJ0723 encode two Lrp homologs: putative transcription regulators Ptr1 and Ptr2, respectively. Ptr1 (148 amino acids) and Ptr2 (140 amino acids) are small, basic proteins (molecular mass 16.6 and 15.9 kDa, respectively) exhibiting an N-terminal HTH motif that is characteristic of the bacterial Lrp/AsnC family of transcriptional regulators. A database search of both finished and unfinished microbial genome sequences shows that the predicted Ptr1 protein shares significant sequence similarity with Pyrococcus furiosus LrpA (43% identity), and with other archaeal hypothetical proteins from Pyrococcus abyssi [PAB0392 (41% identity), PAB1938 (37% identity), PAB0322 and PAB2257 (34% identity)], Pyrococcus horikoshii [PH1592 (40% identity), PH1519 (37% identity), PH0140 (34% identity) and PH1692 (30% identity)], M.thermoautotrophicum [MTH1193 (36% identity)], A.fulgidus [AF1148 (31% identity)] and S.solfataricus [SS-C01007 (35% identity)]. In pairwise alignments with members of the bacterial Lrp/AsnC family of transcription regulators, Ptr1 shares the most sequence similarity with E.coli AsnC (34% identity). Similarly, Ptr2 shows the most extensive sequence similarity to the P.furiosus LrpA (55% identity), and to four other archaeal hypothetical proteins: two from P.horikoshii [PH1592 (54% identity) and PH0140 (42% identity)], one from P.abyssi [PAB0392 (54% identity)] and one from A.fulgidus [AF1723 (47% identity)]. Ptr1 and Ptr2 are also significantly similar (33% identity). Figure 1 shows a sequence alignment of Ptr1 and Ptr2 with their above-mentioned homologs.
Fig. 1. Multiple sequence alignment of Ptr1, Ptr2 and their closest homologs from P.furiosus (PF-LrpA), P.abyssi (PAB0392, PAB1938, PAB0322 and PAB2257), P.horikoshii (PH1592, PH1519, PH0140 and PH1692), M.thermoautotrophicum (MTH1193), A.fulgidus (AF1148 and AF1723), S.solfataricus (SS-C01007) and E.coli (AsnC and Lrp). Secondary structure elements predicted by the program PHDsec (Rost and Sander, 1993) for the Ptr1 and Ptr2 wHTH DNA-binding domains (S1 and S2 representing the two β-strands that constitute the wing) are shown at the top. The conserved arginine in helix 3 (R38 in Ptr1 and R37 in Ptr2) is indicated by an asterisk.
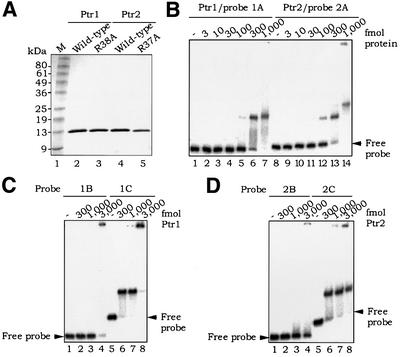
We have overproduced Ptr1 and Ptr2 in E.coli as fusions with an N-terminal histidine tag, and purified them by metal affinity chromatography (see Materials and methods). Figure 2A shows an SDS–PAGE analysis of the purified recombinant proteins. As judged by size exclusion chromatography (results not shown), recombinant Ptr1 is a homodimer in solution, as are E.coli Lrp (Willins et al., 1991) and S.solfataricus Lrs14 (Bell and Jackson, 2000). Ptr2, on the other hand, like Sa-Lrp from S.acidocaldarius (Enoru-Eta et al., 2000), is homotetrameric under similar conditions.
Fig. 2. Specific binding by Ptr1 and Ptr2 to their promoter regions. (A) SDS–PAGE analysis of recombinant wild-type and alanine substitution mutant Ptr1 and Ptr2 proteins (∼2 µg each), as indicated above each lane. (B) EMSA analysis of Ptr1 and Ptr2 binding to the promoter probes 1A (–125 to +118, relative to the putative Ptr1 promoter start site of transcription) (lanes 1–7) and 2A (Ptr2 promoter region –201 to +53) (lanes 8–14), respectively. Approximately 10 fmol of end-labeled probe were incubated in the absence (lanes 1 and 8) or presence of increasing concentrations of either protein (amounts indicated above each lane) for 20 min at 65°C, and subjected to an EMSA (as described in Materials and methods). The unbound DNA is indicated by an arrow. Protein quantities in this panel and throughout are stated in terms of the monomer. (C) EMSA analysis of Ptr1 binding to promoter probes 1B (–125 to –33) (lanes 1–4) and 1C (–34 to +118) (lanes 5–8). (D) Analysis of Ptr2 binding to promoter probes 2B (–201 to –104) (lanes 1–4) and 2C (–88 to +53) (lanes 5–8).
Ptr1 and Ptr2 are double-stranded DNA-binding proteins
Escherichia coli Lrp binds to specific sequences in the promoter regions of a variety of genes, including its own (Wang et al., 1994). Sulfolobus Lrp homologs Lrs14 and Sa-Lrp also bind to multiple sites in their respective promoter regions (Napoli et al., 1999; Bell and Jackson, 2000; Enoru-Eta et al., 2000), and it appeared likely that Ptr1 and Ptr2 would exhibit similar autoregulatory patterns. We therefore tested recombinant Ptr1 in an EMSA with the 243 bp DNA probe 1A, spanning bp –125 to +118 (relative to the putative Ptr1 start site of transcription as +1). As is evident in Figure 2B (lanes 1–7), Ptr1 binds to probe 1A. In order to narrow down the binding site for Ptr1, probe 1A was split at bp –33, yielding probes 1B (bp –125 to –33) and 1C (bp –34 to +118). Ptr1 bound to probe 1C (Figure 2C, lanes 5–8), but not to probe 1B (lanes 1–4), demonstrating a clear sequence specificity for Ptr1 binding in its promoter region. Similarly, when tested with the 254 bp probe 2A (encompassing bp –201 to +53 relative to the putative Ptr2 start site of transcription), recombinant Ptr2 yielded two distinct slowly migrating protein–DNA complexes (Figure 2B, lanes 8–14). Ptr2 was seen to bind to probe 2C (bp –88 to +53), forming a single protein–DNA complex (Figure 2D, lanes 5–8), but not to probe 2B (bp –201 to –104) (Figure 2D, lanes 1–4), demonstrating sequence specificity for Ptr2 binding (the presence of only one slowly migrating Ptr2–probe 2C DNA complex suggests that the intervening region between probes 2B and 2C contains, or overlaps with, a Ptr2-binding site). DNase I footprinting showed Ptr1 and Ptr2 protecting long stretches of their respective promoter regions against cleavage, with interspersed sites of increased sensitivity to DNase I (data not presented). Taken together, these results suggest multiple site occupancy patterns similar to those of E.coli Lrp (Wang et al., 1994), S.solfataricus Lrs14 (Napoli et al., 1999) and S.acidocaldarius Lrp (Enoru-Eta et al., 2000).
Consensus sequences for Ptr1 and Ptr2 binding to DNA
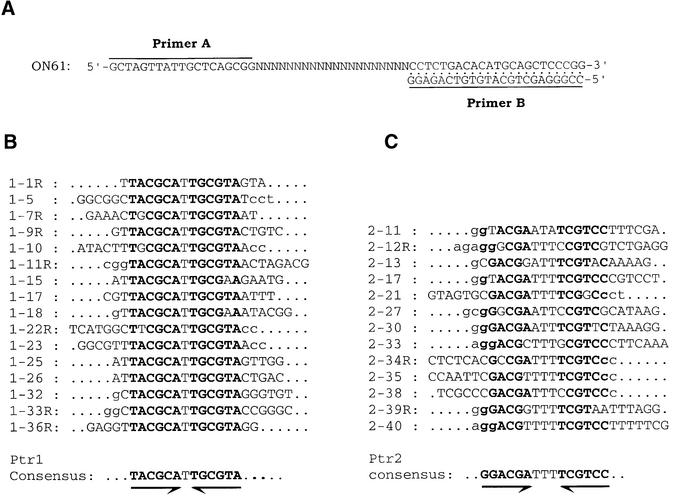
While the preceding analysis demonstrates a clear sequence specificity for DNA binding by Ptr1 and Ptr2, a close examination of DNA sequences in their promoter regions does not allow target sequences to be discerned with any degree of certainty. To that end, we have used a variation of the SELEX procedure (Tuerk and Gold, 1990) to select, from a large pool of double-stranded molecules initially randomized over 20 bp (Figure 3A), sets of DNA sites with high affinity for Ptr1 or Ptr2. Briefly, the randomized DNA was incubated (at 65°C for 15–20 min) with either Ptr1 or Ptr2 (Materials and methods) and separated by EMSA. DNA was eluted and amplified from gel slices corresponding to the position where a Ptr1– or Ptr2–DNA complex should migrate. This pattern of selection–amplification was repeated until the selection had enriched the population to a suitable percentage of high-affinity binders (five rounds for Ptr1, and four rounds for Ptr2), at which point a library of these operators was generated by standard cloning techniques, and individual clones were isolated and sequenced. Their analysis showed that, although each clone is unique, certain motifs appear multiple times in the population. Among 16 DNA sites selected for Ptr1 binding, 11 contain two inverted, perfect 6 bp repeats of TACGCA separated by one T–A base pair (Figure 3B). The remaining operators contain slightly imperfect variations of this motif. When aligned, the 13 ligands selected for Ptr2 binding exhibit more sequence variation at certain positions, although a consensus sequence is readily inferred, consisting of two inverted, perfect 6 bp repeats of GGACGA separated by three T–A base pairs (Figure 3C). The finding that specific motifs are present in multiple clones supports the conclusion that we have selected bona fide Ptr1- and Ptr2-binding sites. Furthermore, the palindromy within the deduced consensus sequences suggests symmetrical DNA binding by protein dimers.
Fig. 3. In vitro selection of Ptr1- and Ptr2-binding sites. (A) The single-stranded SELEX template (ON61) was annealed to 32P-labeled primer B and converted into duplex form. The sequences of primers A and B are shown. (B) Sequence alignment of 16 SELEX-derived Ptr1-binding sites. (C) Sequence alignment of 13 SELEX-derived Ptr2-binding sites. The consensus sequence is shown below each alignment. The DNA strand shown for each site was chosen arbitrarily for the presence of T at the consensus dyad axis of symmetry. For sites identified with an ‘R’ (shown on the left), the represented DNA strand corresponds to PCR primer A. Lower case letters identify non-randomized sequences derived from primers A and B.
Analysis of individual Ptr1- and Ptr2-binding sites
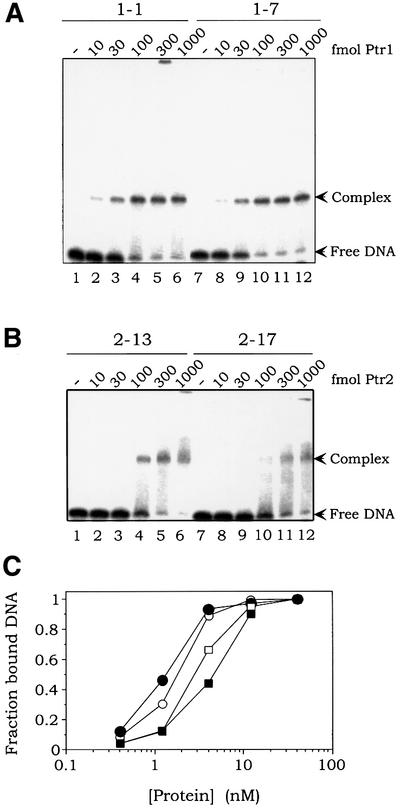
DNA binding and EMSAs were performed with selected sites in order to examine affinities for their target proteins. Figure 4A shows Ptr1 binding to sites 1-1 (lanes 1–6) and 1-7 (lanes 7–12). By measuring complex formation over a range of Ptr1 concentrations in excess over DNA, apparent equilibrium dissociation constants (in this case, the concentrations of Ptr1 that generate 50% complex formation) were found to be in the range 1–2 nM for sites 1-1 and 1-7 (Figure 4C). Figure 4B shows a similar analysis for binding of Ptr2 to sites 2-13 (lanes 1–6) and 2-17 (lanes 7–12), with apparent dissociation constants in the range 3–5 nM (Figure 4C). We conclude that the in vitro selection has led to the isolation of very high affinity sites for Ptr1 and Ptr2. Efficient binding of Ptr1 and Ptr2 to these SELEX-derived sites is maintained over a wide range of temperatures (from 25°C to the DNA probe melting point; results not shown).
Fig. 4. Quantitative analysis of Ptr1 and Ptr2 binding to SELEX-derived DNA sites. (A) DNA sites 1-1 (lanes 1–6) and 1-7 (lanes 7–12) were incubated in the absence (lanes 1 and 7) or presence of increasing amounts of Ptr1 (indicated above each lane) and subjected to EMSA. (B) DNA sites 2-13 (lanes 1–6) and 2-17 (lanes 7–12) were incubated in the absence (lanes 1 and 7) or presence of increasing amounts of Ptr2 and analyzed as described above. (C) The fraction of bound DNA in each lane of (A) and (B) (defined as the fraction of DNA migrating above the free DNA band and including the specific protein–DNA complex, taking into account dissociation of Ptr–DNA complexes during gel electrophoresis) is plotted as a function of protein concentration. Ptr1 binding to probes 1-1 and 1-7 is represented by open and closed circles, respectively. Ptr2 binding to probes 2-13 and 2-17 is represented by open and closed squares, respectively.
DNase I and hydroxyl radical footprinting of Ptr1– and Ptr2–DNA complexes
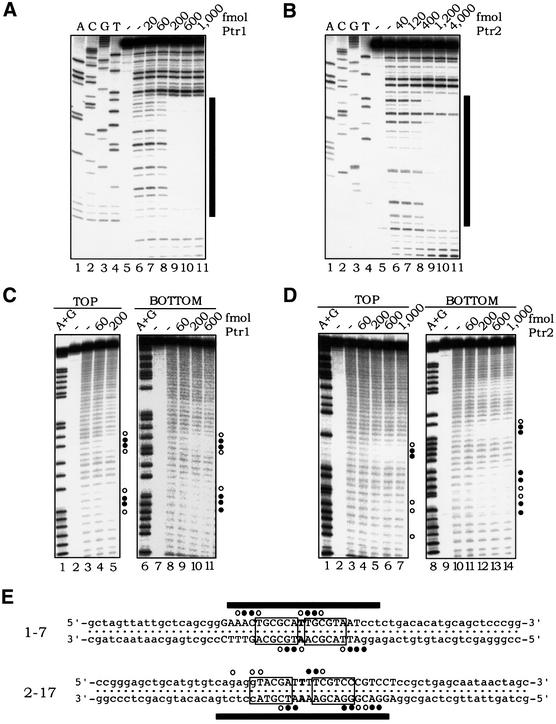
In order to characterize their mode of DNA binding further, Ptr1– and Ptr2–DNA complexes were formed at sites 1-7 and 2-17, respectively, at 65°C with increasing concentrations of either protein and then probed with DNase I. As shown in Figure 5A, Ptr1 binding to operator 1-7 generates almost complete protection against DNase I cleavage of an ∼22 nucleotide segment centered on the consensus DNA-binding site (Figure 5E). Similarly, Ptr2 protects an ∼25 nucleotide segment centered on the dyad axis of site 2-17 (Figure 5B and E). Also noteworthy are sites of increased accessibility to DNase I at the outer edges of the regions protected by Ptr1 and Ptr2 binding, suggesting protein-induced distortions of DNA.
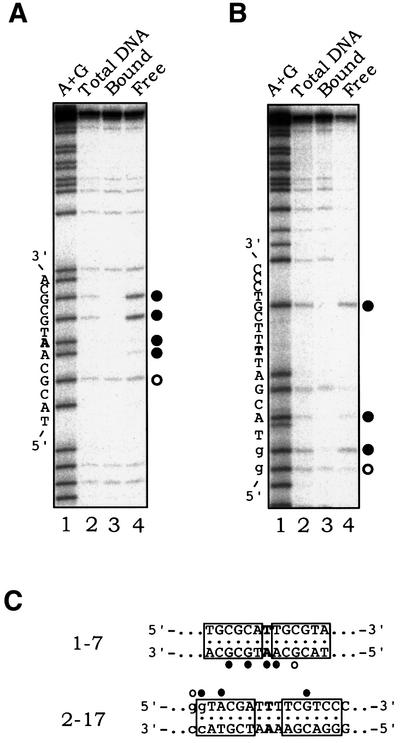
Fig. 5. Footprinting analyses of Ptr1– and Ptr2–DNA complexes. DNA sites 1-7 (A) and 2-17 (B) were incubated in the absence (lane 6, in each panel) or presence of increasing concentrations of Ptr1 and Ptr2 (lanes 7–11), respectively, and subjected to DNase I treatment. Lanes 1–4 of each panel show a dideoxynucleotide sequencing ladder of the analyzed DNA strand. The bars on the right mark the extent of the footprint, whose sequence is specified in (E). (C) Hydroxyl radical footprinting of Ptr1–DNA complexes. 1-7 DNA, 5′-end-labeled on either the top or bottom strand, was incubated in the absence (lanes 3 and 8) or presence of increasing amounts of Ptr1 (indicated above each of lanes 4–5 and 9–11) and subjected to hydroxyl radical cleavage for 30 s at 65°C. On the right of each panel, the positions of strong protection by Ptr1 are indicated by closed circles, and those exhibiting weaker protection are indicated by open circles. Lanes 1 and 6 show A+G chemical sequencing ladders. (D) Hydroxyl radical footprinting of Ptr2–DNA complexes. Ptr2 binding to 2-17 DNA probes, 5′-end-labeled on either the top or bottom strand, was analyzed as described above for Ptr1. (E) Summary of the DNase I and hydroxyl radical protection patterns. The complete nucleotide sequences of DNAs 1-7 and 2-17 are shown, with the consensus half-site hexamers boxed. On each probe, the region that is protected from DNase I cleavage upon binding to its cognate protein is indicated by black bars. The positions of protection against hydroxyl radical cleavage are indicated by circles, as above.
The mode of DNA binding and the proximity of operator-bound Ptr1 and Ptr2 to the sugar–phosphate DNA backbone were analyzed further by hydroxyl radical (⋅OH) footprinting. Hydroxyl radicals cleave DNA from the minor groove, virtually independently of sequence (Tullius et al., 1987). A bound protein creates a footprint of protection from ⋅OH cleavage wherever it comes into close contact with the DNA backbone or significantly diminishes minor groove width.
Operator 1-7 DNA was incubated at 65°C in the absence (Figure 5C, lanes 3 and 8) or presence of increasing amounts of Ptr1 (lanes 4–5 and 9–11), and was then subjected to ⋅OH cleavage (at 65°C) to introduce, on average, no more than a single gap per target DNA molecule. On each strand, the presence of Ptr1 produced two regions of protection, separated by ∼9 nucleotides (center-to-center), or approximately one turn of the helix (Figure 5E), indicating that Ptr1 lies on one face of the DNA double helix. The strongest protection by Ptr1 on each strand was seen near the dyad of the operator, offset from each other in the 3′ direction by 2–3 bp, suggesting equivalent protein interactions across the DNA minor groove (Dixon et al., 1991; Papavassiliou, 1995). Similarly, Ptr2 binding to operator 2-17 (Figure 5D) produced two regions of protection against hydroxyl radical cleavage on each strand, separated by ∼10 bp, suggesting that Ptr2 also binds to one side of the DNA. As in the case of Ptr1, the strongest protection on each strand was seen near the dyad of the operator sequence (Figure 5E), with a pattern suggesting protein contacts across the DNA minor groove.
Most of the protein–DNA backbone contacts on one strand could be related, by inversion around the dyad, to a corresponding contact on the opposite strand (Figure 5E). Thus, the symmetry in the Ptr1 and Ptr2 consensus operator sequences, derived from the SELEX experiment, clearly translates into a symmetry in the hydroxyl radical footprints, and reflects an inherent symmetry in the structures of the Ptr1– and Ptr2–operator DNA complexes.
Dimethyl sulfate methylation interference analysis of Ptr1 and Ptr2 binding
To characterize the DNA base- and groove-specific contacts between Ptr1, Ptr2 and their respective operators, we performed in vitro dimethyl sulfate (DMS) methylation interference experiments. DMS methylates N-7 of guanine, in the major groove of the DNA double helix, and N-3 of adenine, in the minor groove. A guanine methylation that interferes with protein binding implies protein contact in the major groove at that site. An operator DNA fragment, labeled at the 5′ end of either the top or bottom strand, was incubated with DMS at a concentration intended to introduce less than one methylation, on average, in each DNA molecule. DMS-treated DNA was then used in standard protein-binding reactions. After separation and isolation from a native gel, approximately equal quantities of total, complexed and free DNA were subjected to chemical cleavage and denaturing PAGE analysis.
On the bottom strand of site 1-7, methylation of either G residue in the 3′ half-site strongly interfered with Ptr1 binding (compare lanes 3 and 4 in Figure 6A). Methylation of G in the 5′ half-site also interfered with Ptr1 binding, albeit to a lesser extent. We conclude that Ptr1 binding involves base-specific contacts in the DNA major groove. We also observed that two minor groove methylations, the AA dinucleotide step at the operator dyad (Figure 6C), strongly inhibit Ptr1 binding. We cannot exclude the possibility that Ptr1 might reach around the DNA backbone to make specific base contacts in the minor groove, but this same AA dinucleotide step retains accessibility to ⋅OH attack through the minor groove in a Ptr1–DNA complex (Figure 4C). We favor the simpler explanation that Ptr1 binding distorts DNA, with a narrowing of the minor groove around the dyad of the binding site. Because the introduction of a methyl group into the minor groove limits the degree of flexibility of the DNA, it hinders the formation of specific Ptr1–DNA contacts on both sides of the dyad. For Ptr2, methylation of G at two sites and of one A on the top strand was found to interfere strongly with Ptr2 binding to site 2-17 (compare lanes 3 and 4 in Figure 6B).
Fig. 6. DMS methylation interference. 1-7 DNA, 5′-end-labeled on the bottom strand (A) and 2-17 DNA, 5′-end-labeled on the top strand (B) were methylated with DMS in vitro, at no more than one position per molecule. Each methylated DNA was incubated with its cognate protein and subjected to an EMSA. Bands corresponding to bound and free DNA were excised, the DNA therein was eluted and analyzed on a sequencing gel. The positions at which methylation strongly interferes with protein binding are indicated by closed circles; weaker interference is indicated by open circles. (C) Summary of the methylation interference data. The consensus half-site hexamers are boxed, and the positions of interference are indicated.
Taken together, the above data suggest that Ptr1 and Ptr2 interact with their respective operators as dimers, with each protein monomer making base-specific contacts in the major groove, most probably through the recognition helix (helix 3) of its putative HTH DNA-binding domain (as indicated below). Effects on the minor groove, either directly by protein contact or indirectly by compression of the minor groove, are also indicated.
Site-directed mutagenesis and deletion analyses of Ptr1 and Ptr2
Ptr1 and Ptr2 are predicted to have an N-terminal HTH DNA-binding domain (program HTH; Dodd and Egan, 1990). The existence of this HTH motif is also inferred from sequence alignments with other putative archaeal as well as known bacterial HTH DNA-binding proteins (Aravind and Koonin, 1999). Minimally, HTH domains consist of a right-handed three-helix bundle, of which the third helix (the recognition helix) makes contacts with the DNA major groove and contains the residues that, in part, determine DNA binding specificity. The Ptr1 and Ptr2 HTH domains, like those from the vast majority of putative HTH proteins identified in archaeal genomes, belong to the winged-helix (wHTH) subfamily (Clark et al., 1993; Aravind and Koonin, 1999), which contain an additional C-terminal β-hairpin called the wing.
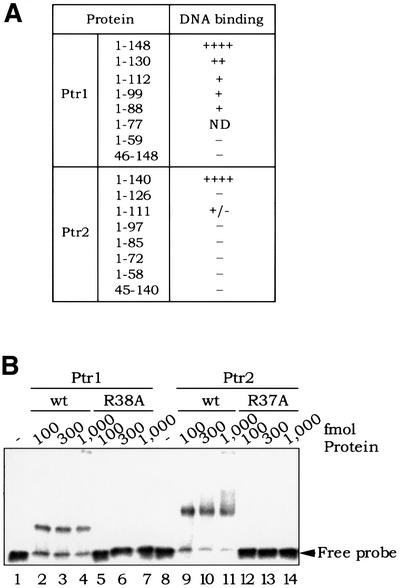
In an attempt to characterize the protein domain, as well as primary sequence requirements for DNA binding by Ptr1 and Ptr2, we have generated a series of derivatives deleted, at their N- or C-terminal regions, of portions of varying length, and tested their ability to bind DNA in standard EMSAs. To varying degrees, all Ptr1- and Ptr2-deleted derivatives lose the ability to bind DNA efficiently and/or stably (Figure 7A).
Fig. 7. (A) Deletion analysis of Ptr1 and Ptr2. Ptr1 and Ptr2 variants, lacking either their predicted HTH domains (N-terminal deletions Ptr1[46–148] and Ptr2[45–140]) or C-terminal segments of varying length, were produced in E.coli and purified as described for the wild-type proteins. The ability of each Ptr1 and Ptr2 variant to bind its cognate DNA (site 1-7 for Ptr1, and site 2-13 for Ptr2) was compared with that of the wild-type proteins Ptr1[1–148] and Ptr2[1–140], respectively. The Ptr1[1–77] variant protein could not be obtained in soluble form from overproducing E.coli cells, and was not tested for DNA binding. (B) Analysis of Ptr1 and Ptr2 alanine substitution mutants. DNA binding by Ptr1-R38A (lanes 5–7) and wild-type Ptr1 (lanes 2–4) was compared in a standard EMSA. Similarly, Ptr2-R37A binding to DNA (lanes 12–14) was compared with that of wild-type Ptr2 (lanes 9–11).
Arginine at positions 38 and 37 of Ptr1 and Ptr2, respectively, is part of their putative recognition helices and is highly conserved among both the archaeal and the bacterial members of the Lrp/AsnC family of proteins (Figure 1). Substitutions of this residue in E.coli Lrp (R48→C) as well as in S.acidocaldarius Sa-Lrp (R44→A) were shown to abolish DNA binding (Platko and Calvo, 1993; Enoru-Eta et al., 2000). Accordingly, alanine substitutions were made at positions 38 and 37 of Ptr1 and Ptr2, respectively. The recombinant mutant proteins, Ptr1-R38A and Ptr2-R37A, were produced in E.coli as fusions with an N-terminal histidine tag, and were purified by metal affinity chromatography (see Materials and methods). Figure 2A shows side-by-side SDS–PAGE analyses of the wild-type and mutant proteins. The ability of these proteins to bind their respective operator DNAs was then compared with that of wild-type Ptr1 and Ptr2. No complex formation was detected with the Ptr1-R38A (Figure 7B, lanes 5–7) or Ptr2-R37A (Figure 7B, lanes 12–14). These results lend credence to the direct involvement of the Ptr1 and Ptr2 HTH motifs in DNA recognition.
Putative Ptr1- and Ptr2-binding sites in the M.jannaschii genome
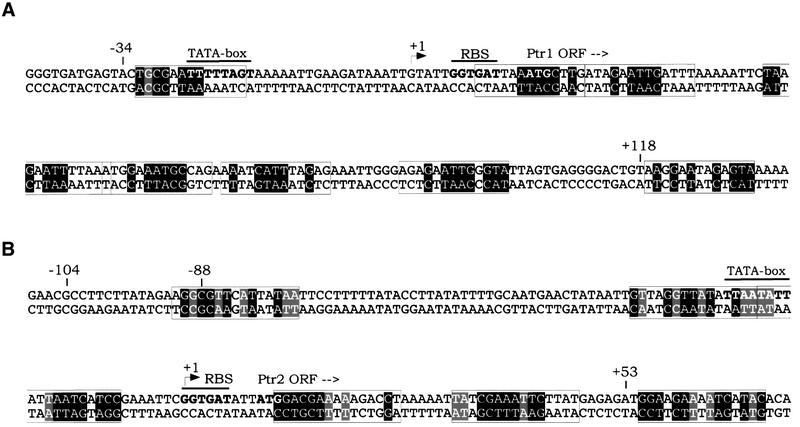
The identification of consensus binding sites for Ptr1 and Ptr2 allows one to re-examine the conjugate promoter regions to which these proteins bind specifically (Figures 2 and 8). A search in these regions for sequences matching the consensus for Ptr1 or Ptr2 binding reveals the presence of clusters of closely spaced sites that are imperfect to varying degrees (Figure 8), consistent with the extended DNase I footprints to which reference has already been made. Ptr1 and Ptr2 binding to clusters of closely spaced, non-consensual (and probably weaker) sites is reminiscent of the properties of E.coli Lrp, whose DNA binding involves some degree of cooperativity (Wang and Calvo, 1993). This is also likely to be the case for Ptr1 and Ptr2, but remains to be analyzed in detail.
Fig. 8. Binding site clusters within the Ptr1 (A) and Ptr2 (B) promoter regions. Sequences resembling the Ptr1 or Ptr2 consensus are boxed, with nucleotide positions matching the consensus shown in black boxes, while those present in at least one of the SELEX clones are shown in gray boxes.
The search for Ptr1- and Ptr2-binding sites has been extended to include the entire M.jannaschii genome. Sites most closely matching the consensus binding sequences for Ptr1 and Ptr2 are shown in Table I. It is impressive that no perfect match to either consensus was found; one site identical to SELEX ligand 1-18, and differing from the Ptr1 consensus only at one position, is located in the intergenic region between the Ptr1 gene and ORF MJ0152, encoding the putative carbon monoxide dehydrogenase α-subunit. Numerous sites containing at least a consensus half-site for either protein were also found. Many such sites lie within intergenic regions or at the ends of ORFs (Table I); some are located within ORFs, often hundreds of base pairs from the nearest putative promoter. Although such sites have been omitted from Table I, neither their functionality nor their physiological relevance can be dismissed at this point. Visual inspection of sequences in the immediate vicinity of the identified sites in Table I reveals additional, more divergent sites. This is precisely the configuration of the Ptr1 and Ptr2 promoters, suggesting that a clustered mode of binding may be a general tendency rather than an exception. Ongoing analysis of the Ptr1- and Ptr2-binding site architecture within these clusters, their positioning relative to transcription factor binding and/or initiation sites, as well as possible differential occupancies and interactions between adjacent sites should provide insights into the underlying modes of regulation.
Table I. Putative Ptr1- and Ptr2-binding sites in the genome of M.jannaschii.
| Site | Coordinates | Location | Potentially regulated gene | Function |
|---|---|---|---|---|
| Ptr1 | ||||
| ATCGCATTTGAAT | 145 989–145 977 | MJ0149 | MJ0150 | conserved hypothetical protein |
| TACGCATTGCGAA | 147 231–147 219 | IG | MJ0152 | carbon monoxide dehydrogenase α-subunit (putative) |
| TTTTTATTGCGTA | 159 707–159 719 | IG | MJ0157 | hypothetical protein |
| TTAATTTTGCGTA | 677 806–677 818 | MJ0752 | MJ0751 | transposase-related protein |
| ATGACATTGCGTA | 685 027–685 039 | MJ0762 | MJ0761 | conserved hypothetical protein |
| CGTTCATTGCGTA | 773 274–773 262 | MJ0846 | MJ0847 | N5-methyl-tetrahydromethanopterin:coenzyme Mmethyltransferase, subunit E (mtrE) |
| CAGTGATTGCGTA | 1 030 136–1 030 124 | MJ1092 | MJ1092 | conserved hypothetical protein |
| TACGCATGCATTT | 1 275 882–1 275 870 | IG | MJ1326 | GTP-binding protein, GTP1/OBG family |
| Ptr2 | ||||
| CCACGATTTTCGGTA | 308 286–308 300 | MJ0331 | MJ0330 | hypothetical protein |
| TAAAAATTTTCGTCC | 430 798–430 784 | IG | MJ0486/MJ0487 | conserved hypothetical protein, phenylalanyl-tRNAsynthetase, subunit α (pheS) |
| TTTGGATTTTCGTAT | 555 672–555 658 | MJ0627 | MJ0628 | hypothetical protein |
| AGACGATTATCGTAT | 668 955–668 969 | IG | MJ0740 | rubredoxin |
| AGACGATTTTTAATA | 759 162–759 148 | MJ0834 | MJ0833 | hypothetical protein |
| GGACGATTTTTATGT | 1 277 591–1 277 605 | MJ1327 | MJ1327-2 | adenine-specific DNA methyltransferase |
| AGATGATTTTCGTCA | 1 650 252–1 650 266 | MJ1667 | MJ1666 | hypothetical protein |
A search in the M.jannaschii genome (Bult et al., 1996) for sites closely matching the SELEX-derived Ptr1 and Ptr2 consensus binding sites yields a number of potential candidates, some of which are listed. For each site, the coordinates of the first and last nucleotide, the location [ORF or intergenic region (IG)] and the potentially regulated gene are indicated. The nucleotide positions matching the consensus are shown in bold, and those present in at least one SELEX clone are underlined.
Discussion
Ptr1 and Ptr2, the Lrp-related proteins of M.jannaschii (Figure 1), are site-specific DNA-binding proteins with specificity for their own promoter regions (Figure 2). A high-temperature (65°C) SELEX has been used to identify a consensus DNA site for each protein. Each consensus is a palindrome, clearly reflecting binding by a symmetric protein dimer. The Ptr1 and Ptr2 consensus sites are clearly different in sequence and also in structure, with a single T–A base pair at the Ptr1 dyad and three T–A base pairs (TTT in the top strand) at the Ptr2 dyad. Six base pairs have been specified in each half of these palindromes, but inspection of the sequenced selectants in Figure 3 indicates that the Ptr1 consensus can also be written as (py)TACGCATTGCGTA(pu) and the Ptr2 consensus as (pu)GGACGATTTTCGTCC(py). Pushing the SELEX further, perhaps under conditions that ensure equilibration as the affinity increases, or with Ptr derivatives that bind DNA less well (Figure 7A), could drive additional flanking base pairs into a ‘hyperconsensus’. The two Ptr consensus sequences differ from a sequence previously derived for E.coli Lrp, also by SELEX (Cui et al., 1995), i.e. (c/t)AG(a/c/t)A(a/t)ATT(a/t)T(a/g/t) CT(a/g), but share two general characteristics with it: the E.coli Lrp consensus is also a palindrome (probably imperfect because selection was not driven to completion, and generated sites with a wide range of affinity), and it has a T–A base pair at its dyad.
Finding different consensus sites for Ptr1 and Ptr2 establishes the expectation that paralogous Lrp/AsnC- related proteins in other archaea will also turn out to have separate DNA specificities. This need not mean that they bind exclusively to separate sites; regulatory cross-talk could also be generated at loci consisting of clusters of imperfect sites and half-sites.
Ptr1 and Ptr2 bind their symmetric consensus sites from one side of the DNA helix (protecting ∼22 and 25 bp of DNA, respectively, from DNase I) with evidence for possible DNA distortion at the edge of the protected segment for Ptr2 (Figure 5B) and for possible minor groove compression at the dyad of the Ptr1 site (Figures 5C and 6A). In a similar vein, a T–A base pair at the dyad of a phage 434 cI symmetric operator has been argued to be an important contributor to affinity for the repressor because of its ability to accommodate groove compression that is required at the center of the operator for optimal alignment of protein–DNA interactions on either side (Koudelka and Carlson, 1992). Whether the (predominantly) tetrameric Ptr2 is bivalent for DNA and prone to forming DNA loops, as the Lac repressor does (Oehler et al., 1990), remains to be seen.
The consensus sites for Ptr1 and Ptr2 are not present in the M.jannaschii genome. A single Ptr1 site that misses its consensus at only one position (Table I), and is actually represented in the final selectant set (Figure 3B), is located in the inter-ORF space downstream of the Ptr1 gene and upstream of an ORF putatively encoding a CO dehydrogenase subunit. There are no sites in the M.jannaschii genome containing just two deviations from either consensus, and only one site that misses the Ptr2 consensus by 3 bp. What we have noted instead are instances of clustering of more imperfect sites around the less imperfect sites shown in Table I. This is also a characteristic of the Ptr1 and Ptr2 sites surrounding their presumptive promoters (Figure 8), where extensive DNase I footprints, covering >100 bp, are generated by the respective proteins. These imperfect sites tend to a half-site-plus-dyad character. Eight potential sites are discerned within a span of ∼150 bp around the putative Ptr1 promoter. It is unlikely that all of these sites can be occupied simultaneously by Ptr1, but full occupancy would generate a distinctive protein array, possibly with a distinctive linear or three-dimensional structure, enveloping the probable promoter.
The properties of E.coli Lrp are strikingly similar. At several operons under its control, extended DNA sites that lack anything close to the Lrp consensus palindrome are occupied by several molecules of dimeric Lrp (Cui et al., 1996). In fact, natural Lrp-binding sites conform to the Lrp consensus palindrome predominantly at the dyad (Shultzaberger and Schneider, 1999).
The preponderance, in archaea, of bacterial-type proteins that could function as regulators of their own eukaryote-like transcription systems has been noted in the Introduction. Repressor activity by proteins with bacterial homology has been demonstrated in vivo (Bell et al., 1999a), and the ability of DNA-binding proteins with sites that overlap promoters to block archaeal transcription in vitro has also been demonstrated (Bell et al., 1999a; Brinkman et al., 2000). The strong parallel between Ptr1, Ptr2 and E.coli Lrp, and the specific properties of the two Ptr proteins that emerge from this analysis, suggest that they also participate in positive as well as negative regulation of archaeal transcription.
The analysis of an Lrp homolog from P.furiosus presents interesting complementarities, but also some contrasts to this work (Brinkman et al., 2000). Pyrococcus furiosus LrpA is reported to be present in alternative, slowly interconverting states of oligomerization (dimer, tetramer and octamer). It binds to a well-defined 40 bp site spanning the start of its transcription unit, but the only palindromic and direct repeat sequence features within this site are not essential for binding, and no similarity between the two halves of the site is discernible. These properties suggest a compact cluster, perhaps a pair, of imperfect binding sites rather than the extended clusters at the Ptr2 and Ptr1 promoters. Nevertheless, P.furiosus LrpA is reported to have an extremely high affinity for this site (apparent equilibrium dissociation constant ∼3 × 10–10 M at room temperature and at an ionic strength of ∼250 mM), an order of magnitude higher affinity than Ptr1 and Ptr2 have for their respective symmetric consensus sites. Pyrococcus furiosus LrpA has been shown to bind to this site from one side of the DNA helix, as individual Ptr1 and Ptr2 molecules also do (Figure 5). It also specifically blocks transcription in vitro by the homologous RNA polymerase at the LrpA promoter at 70°C (and not at two other promoters). Surprisingly, this repression requires LrpA concentrations that exceed its reported (room temperature) Kd by 2–3 orders of magnitude.
The results that are reported here suggest that Ptr1 and Ptr2 also offer strong prospects for analysis of effector interactions with the archaeal transcription apparatus. Effects of Ptr1 and Ptr2 on transcription at their natural promoters are of immediate but not exclusive interest. The ability to define a consensus-type binding sequence for each of these proteins facilitates manipulations of promoter structure for examination of mechanisms of action on the components of the core transcription apparatus at heterologous promoters. Their thermostability (a property that is likely to be shared by other DNA-binding proteins from hyperthermophiles) provides a DNA-binding ‘platform’ that is fully compatible with transcription systems operating at a wide range of temperatures, and should also be compatible with further manipulation at the protein level to construct novel transcription effectors.
Materials and methods
Protein production and purification
Ptr1 and Ptr2 were overproduced in E.coli BL21(DE3)pLysS harboring plasmids pLJ-Ptr1 and pLJ-Ptr2, respectively. These plasmids express fusion genes encoding an N-terminal His6 tag, followed by the products of M.jannaschii ORFs MJ0151 and MJ0723, respectively. Similarly, plasmids pLJ-Ptr1-R38A and pLJ-Ptr2-R37A express the N-terminal His6-tagged alanine substitution mutant proteins Ptr1-R38A and Ptr2-R37A, respectively. Complete sequences of these plasmids are available upon request. The His-tagged proteins were affinity purified on Ni-NTA–agarose, and eluted with 250 mM imidazole. These proteins were at least 95% pure, as judged by SDS–PAGE analysis. Protein concentrations were measured using the Micro BCA assay, with bovine serum albumin (BSA) as the standard. Molar concentrations are expressed in terms of protein monomer.
DNA binding and EMSA
32P-labeled DNA fragments were generated by PCR amplification using 5′-end-32P-labeled oligonucleotides and purified by native PAGE. Typically, protein–DNA complexes were formed in 25 µl of binding buffer [20 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 4% (v/v) glycerol] with 10 fmol of DNA probe, at 65°C for 15–20 min, and loaded onto a non-denaturing 4 or 5% polyacrylamide gel. The running buffer and gel contained 25 mM Tris–HCl pH 8.5, 190 mM glycine, 1 mM EDTA; the gel also contained 5 mM MgCl2, 5% (v/v) glycerol and 0.5 mM 2-mercaptoethanol (Bartlett et al., 2000). After electrophoresis at 25–30°C for 2–3 h, gels were dried and analyzed by phosphoimaging.
SELEX
To construct the starting pool of randomized DNA used to select Ptr1- and Ptr2-binding sites, 5′-end-labeled primer B was annealed to the single-stranded template ON61 (Figure 3A), extended with Exo– Klenow DNA polymerase and purified by native PAGE. During the first round of selection, 183 pmol of the resulting 62 bp double-stranded DNA (∼100 copies of each of the 1.1 × 1012 possible variants) were incubated with 3 pmol of Ptr1 or Ptr2, in 200 µl of binding buffer at 65°C for 20 min, and subjected to EMSA. Gel slices corresponding to the location of protein–DNA complexes were excised, and their DNA content was eluted and PCR amplified. In rounds 2, 3 and 4, ∼21, 10.5 and 3 pmol of DNA were incubated with 0.75, 0.4 and 0.4 pmol of protein, respectively. In the fifth round of selection for Ptr1 sites, ∼1 pmol of DNA was incubated with 0.2 pmol of Ptr1. The binding reaction volumes were lowered from 200 µl in the first round, to 100 µl in the second, and to 50 µl in the subsequent rounds of selection. In rounds 2–5, protein–DNA complexes were allowed to form for 15 min only. After each round of selection, the selected DNA mixtures were tested by EMSA for their ability to bind Ptr1 or Ptr2. DNA from Ptr1 and Ptr2 SELEX rounds 5 and 4, respectively, was cloned into pBlueScript(II). Individual clones were isolated and their sequences determined.
DNase I and hydroxyl radical footprinting
Ptr1– and Ptr2–DNA complexes were assembled as described above, on DNA fragments 5′-end-32P-labeled on either the top or bottom strands, in 50 µl of binding buffer (specified above), and subjected to digestion with 1.25 ng of DNase I for 30 s at 37°C. For hydroxyl radical footprinting, protein–DNA complexes were assembled in 50 µl of binding buffer lacking DTT and glycerol, and subjected to hydroxyl radical cleavage for 30 s at 65°C, as described by Tullius et al. (1987). After phenol–chloroform extraction and ethanol precipitation, the DNA fragments were separated on 11.5% polyacrylamide–7 M urea gels and analyzed by phosphoimaging. A+G chemical sequencing ladders were generated according to Sambrook et al. (1989).
DMS methylation interference
DNA probes methylated at no more than one position per molecule were generated essentially as described by Li and Wrange (1997). Briefly, 5′-end-labeled probes were methylated by incubation in the presence of 50 mM DMS for 5 min at 25°C, in 200 µl of DMS reaction buffer (50 mM Tris–HCl pH 8.0, 1 mM EDTA). The reactions were stopped by addition of 40 µl of ice-cold DMS stop solution (1.5 M NH4OAc, 25 mM EDTA, 2.8 M 2-mercaptoethanol), and precipitated twice with ethanol. Ptr1–DNA and Ptr2–DNA complexes were assembled on the methylated probes and subjected to EMSA, as described above. Bands corresponding to total, unbound and bound DNA were excised from the gel, and the DNA contained therein was eluted and reacted with piperidine. After phenol–chloroform extraction and ethanol precipitation, the resulting DNA fragments were separated on 11.5% polyacrylamide–7 M urea gels and analyzed by phosphoimaging.
DNA and protein sequence analyses
The M.jannaschii genome database was accessed at http://www.tigr.org/tdb/CMR/arg/htmls/SplashPage.html. DNA and protein sequence analyses were performed using the San Diego Supercomputer Center Biology Workbench (http://workbench.sdsc.edu/).
Acknowledgments
Acknowledgements
We gratefully acknowledge valuable advice, as well as helpful comments on this manuscript, from M.S.Bartlett and G.A.Kassavetis, and support of this research by a grant from the NIGMS.
REFERENCES
- Aravind L. and Koonin,E.V. (1999) DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett M.S., Thomm,M. and Geiduschek,E.P. (2000) The orientation of DNA in an archaeal transcription initiation complex. Nature Struct. Biol., 7, 782–785. [DOI] [PubMed] [Google Scholar]
- Bell S.D. and Jackson,S.P. (2000) Mechanism of autoregulation by an archaeal transcriptional repressor. J. Biol. Chem., 275, 31624–31629. [DOI] [PubMed] [Google Scholar]
- Bell S.D., Cairns,S.S., Robson,R.L. and Jackson,S.P. (1999a) Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol. Cell, 4, 971–982. [DOI] [PubMed] [Google Scholar]
- Bell S.D., Kosa,P.L., Sigler,P.B. and Jackson,S.P. (1999b) Orientation of the transcription preinitiation complex in archaea. Proc. Natl Acad. Sci. USA, 96, 13662–13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman A.B. et al. (2000) An Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus is negatively autoregulated. J. Biol. Chem., 275, 38160–38169. [DOI] [PubMed] [Google Scholar]
- Bult C.J. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- Calvo J.M. and Matthews,R.G. (1994) The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev., 58, 466–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier D., Roovers,M., Thia-Toong,T.L., Durbecq,V. and Glansdorff,N. (1997) Cloning and identification of the Sulfolobus solfataricus lrp gene encoding an archaeal homologue of the eubacterial leucine-responsive global transcriptional regulator Lrp. Gene, 201, 63–68. [DOI] [PubMed] [Google Scholar]
- Clark K.L., Halay,E.D., Lai,E. and Burley,S.K. (1993) Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature, 364, 412–420. [DOI] [PubMed] [Google Scholar]
- Cui Y., Wang,Q., Stormo,G.D. and Calvo,J.M. (1995) A consensus sequence for binding of Lrp to DNA. J. Bacteriol., 177, 4872–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Midkiff,M.A., Wang,Q. and Calvo,J.M. (1996) The leucine-responsive regulatory protein (Lrp) from Escherichia coli. Stoichiometry and minimal requirements for binding to DNA. J. Biol. Chem., 271, 6611–6617. [DOI] [PubMed] [Google Scholar]
- Dixon W.J., Hayes,J.J., Levin,J.R., Weidner,M.F., Dombroski,B.A. and Tullius,T.D. (1991) Hydroxyl radical footprinting. Methods Enzymol., 208, 380–413. [DOI] [PubMed] [Google Scholar]
- Dodd I.B. and Egan,J.B. (1990) Improved detection of helix–turn–helix DNA-binding motifs in protein sequences. Nucleic Acids Res., 18, 5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoru-Eta J., Gigot,D., Thia-Toong,T.L., Glansdorff,N. and Charlier,D. (2000) Purification and characterization of Sa-lrp, a DNA-binding protein from the extreme thermoacidophilic archaeon Sulfolobus acidocaldarius homologous to the bacterial global transcriptional regulator Lrp. J. Bacteriol., 182, 3661–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk H.P. et al. (1997) The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature, 390, 364–370. [DOI] [PubMed] [Google Scholar]
- Koudelka G.B. and Carlson,P. (1992) DNA twisting and the effects of non-contacted bases on affinity of 434 operator for 434 repressor. Nature, 355, 89–91. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C. and Ouzounis,C.A. (1999) Transcription in archaea. Proc. Natl Acad. Sci. USA, 96, 8545–8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer D., Hain,J., Thuriaux,P. and Zillig,W. (1995) Transcription in archaea: similarity to that in eucarya. Proc. Natl Acad. Sci. USA, 92, 5768–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. and Wrange,O. (1997) Assays for transcription factors access to nucleosomal DNA. Methods, 12, 96–104. [DOI] [PubMed] [Google Scholar]
- Napoli A., van der Oost,J., Sensen,C.W., Charlebois,R.L., Rossi,M. and Ciaramella,M. (1999) An Lrp-like protein of the hyperthermophilic archaeon Sulfolobus solfataricus which binds to its own promoter. J. Bacteriol., 181, 1474–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman E.B. and Lin,R. (1995) Leucine-responsive regulatory protein: a global regulator of gene expression in E.coli. Annu. Rev. Microbiol., 49, 747–775. [DOI] [PubMed] [Google Scholar]
- Newman E.B., Lin,R.T. and D’Ari,R. (1996) The leucine/Lrp regulon. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella typhimurium. ASM Press, Washington, DC, pp. 1513–1525.
- Oehler S., Eismann,E.R., Krämer,H. and Müller-Hill,B. (1990) The three operators of the lac operon cooperate in repression. EMBO J., 9, 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavassiliou A.G. (1995) Chemical nucleases as probes for studying DNA–protein interactions. Biochem. J., 305, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platko J.V. and Calvo,J.M. (1993) Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J. Bacteriol., 175, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi S.A. and Jackson,S.P. (1998) Sequence-specific DNA binding by the S.shibatae TFIIB homolog, TFB and its effect on promoter strength. Mol. Cell, 1, 389–400. [DOI] [PubMed] [Google Scholar]
- Rost B. and Sander,C. (1993) Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl Acad. Sci. USA, 90, 7558–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Maniatis,T. and Fritsch,E.F. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shultzaberger R.K. and Schneider,T.D. (1999) Using sequence logos and information analysis of Lrp DNA binding sites to investigate discrepancies between natural selection and SELEX. Nucleic Acids Res., 27, 882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.R. et al. (1997) Complete genome sequence of Methanobacterium thermoautotrophicumδH: functional analysis and comparative genomics. J. Bacteriol., 179, 7135–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J. (1999) Transcription initiation in Archaea: facts, factors and future aspects. Mol. Microbiol., 31, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Thomm M. (1996) Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol. Rev., 18, 159–171. [DOI] [PubMed] [Google Scholar]
- Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- Tullius T.D., Dombroski,B.A., Churchill,M.E. and Kam,L. (1987) Hydroxyl radical footprinting: a high-resolution method for mapping protein–DNA contacts. Methods Enzymol., 155, 537–558. [DOI] [PubMed] [Google Scholar]
- Wang Q. and Calvo,J.M. (1993) Lrp, a global regulatory protein of Escherichia coli, binds co-operatively to multiple sites and activates transcription of ilvIH. J. Mol. Biol., 229, 306–318. [DOI] [PubMed] [Google Scholar]
- Wang Q., Wu,J., Friedberg,D., Plakto,J. and Calvo,J.M. (1994) Regulation of the Escherichia coli lrp gene. J. Bacteriol., 176, 1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins D.A., Ryan,C.W., Platko,J.V. and Calvo,J.M. (1991) Characterization of Lrp, an Escherichia coli regulatory protein that mediates a global response to leucine. J. Biol. Chem., 266, 10768–10774. [PubMed] [Google Scholar]