Abstract
Activating phosphorylation of cyclin-dependent kinases (Cdks) is mediated by at least two structurally distinct types of Cdk-activating kinases (Caks): the trimeric Cdk7–cyclin H–Mat1 complex in metazoans and the single-subunit Cak1 in budding yeast. Fission yeast has both Cak types: Mcs6 is a Cdk7 ortholog and Csk1 a single-subunit kinase. Both phosphorylate Cdks in vitro and rescue a thermosensitive budding yeast CAK1 strain. However, this apparent redundancy is not observed in fission yeast in vivo. We have identified mutants that exhibit phenotypes attributable to defects in either Mcs6-activating phosphorylation or in Cdc2-activating phosphorylation. Mcs6, human Cdk7 and budding yeast Cak1 were all active as Caks for Cdc2 when expressed in fission yeast. Although Csk1 could activate Mcs6, it was unable to activate Cdc2. Biochemical experiments supported these genetic results: budding yeast Cak1 could bind and phosphorylate Cdc2 from fission yeast lysates, whereas fission yeast Csk1 could not. These results indicate that Mcs6 is the direct activator of Cdc2, and Csk1 only activates Mcs6. This demonstrates in vivo specificity in Cdk activation by Caks.
Keywords: Cak1/Cdk-activating kinases/Csk1/Mcs6
Introduction
Cyclin-dependent kinases (Cdks) are a family of enzymes that initiate and coordinate cell cycle progression. The Cdk alone is inactive and requires both association with a regulatory subunit and an activating phosphorylation on a conserved residue in the ‘T-loop’ of the kinase in order to be fully active (reviewed in Morgan, 1997; Solomon and Kaldis, 1998). The T-loop phosphorylation site is conserved in Cdks from yeast to mammals, and is essential for cell viability in both Schizosaccharomyces pombe Cdc2 (T167; Gould et al., 1991) and Saccharomyces cerevisiae Cdc28 (T169; Lim et al., 1996).
The activating phosphorylation on the T-loop of Cdks is mediated by Cdk-activating kinases (Caks; reviewed in Kaldis, 1999). Biochemical purification of a Cak activity for Cdc2 and Cdk2 (Solomon et al., 1992) subsequently led to the identification of the previously cloned MO15 serine-threonine kinase (Shuttleworth et al., 1990) as the catalytic subunit of the purified Cak (Fesquet et al., 1993; Poon et al., 1993; Solomon et al., 1993). Immunoprecipitation of MO15 revealed stoichiometric binding of 37 and 32 kDa proteins (Tassan et al., 1994) identified as cyclin H (Fisher and Morgan, 1994; Mäkelä et al., 1994) and assembly factor Mat1 (Devault et al., 1995; Fisher et al., 1995; Tassan et al., 1995). As MO15 activity was dependent on the regulatory cyclin subunit, the kinase was renamed Cdk7. The physiological role of the Cdk7–cyclin H–Mat1 complex as a Cak has been addressed by two lines of experimentation. In cycling Xenopus egg extracts, immunodepletion of Cdk7 suppressed Cak activity and inhibited entry into M phase (Fesquet et al., 1997). The Cak activity was restored by injection of Cdk7 and cyclin H mRNA, demonstrating that a Cdk7 complex is necessary for activation of mitotic Cdk–cyclin complexes (Fesquet et al., 1997). In a separate approach, Drosophila Cdk7 was found to be necessary for Cak activity of Cdc2–cyclin B and Cdc2–cyclin A in vivo using both temperature-sensitive and null alleles of the Drosophila CDK7 gene (Larochelle et al., 1998). These results strongly suggest that the Cdk7–cyclin H–Mat1 complex functions as a Cak in vivo, while not excluding the possibility that other Caks exist, as suggested by recent biochemical approaches (Edwards et al., 1998; Kaldis and Solomon, 2000).
The trimeric complex of Cdk7–cyclin H–Mat1 is also part of the general transcription factor TFIIH, where the complex phosphorylates the C-terminal domain (CTD) of the large subunit of RNA polymerase II (Pol II) (Feaver et al., 1994; Roy et al., 1994; Mäkelä et al., 1995; Serizawa et al., 1995; Shiekhattar et al., 1995) and is important for transcription in the early Drosophila embryo (Leclerc et al., 2000). Cdk7–cyclin H–Mat1 binds the core TFIIH through ERCC2/XPD and can also exist as a free complex (Drapkin et al., 1996; Reardon et al., 1996). The budding yeast S.cerevisiae has a complex closely related to Cdk7–cyclin H–Mat1 that consists of the kinase Kin28, the cyclin Ccl1 and the Mat1 homolog Tfb3/Rig2 (Simon et al., 1986; Valay et al., 1993; Faye et al., 1997; Feaver et al., 1997). This complex is also associated with TFIIH and is required for the transcription of most but not all genes (Cismowski et al., 1995; Valay et al., 1995; Hengartner et al., 1998; Lee and Lis, 1998).
In contrast to the Cdk7 complex, the Kin28 complex does not display Cak activity in vitro and is not a Cak in vivo (Cismowski et al., 1995; Valay et al., 1995). Instead, budding yeast S.cerevisiae contains a single Cdk-activating kinase, Cak1/Civ1, discovered by biochemical purification of Cak activity (Espinoza et al., 1996; Kaldis et al., 1996; Thuret et al., 1996). Sequence alignments suggest that Cak1 is distantly related to the Cdk family, but biochemical characterization showed that it is active as a monomer. Both genetic and biochemical evidence indicate that Cak1 is the physiological activating kinase of Cdc28 and is important for both G1–S and G2–M transitions (Kaldis et al., 1996; Thuret et al., 1996; Sutton and Freiman, 1997). Cak1 has been demonstrated to be a physiological Cak of more than one Cdk as it also activates Kin28 (Espinoza et al., 1998; Kimmelman et al., 1999). Thus, it appears that budding yeast has only one Cak.
Cdk activation by Cak thus appears to be mediated by two structurally distinct kinases: a single-subunit kinase in budding yeast and a multi-subunit kinase in metazoans. Interestingly, the fission yeast S.pombe is the only known species expressing both Cak types. Mcs6 is the ortholog of Cdk7 and phosphorylates both Cdks and Pol II CTD (Buck et al., 1995; Damagnez et al., 1995). Mcs6 associates with the cyclin H ortholog Mcs2 (Buck et al., 1995; Damagnez et al., 1995) and with the Mat1 ortholog Pmh1 (our unpublished results; DDBJ/EMBL/GenBank accession No. AF191500). Both mcs2 and mcs6 were originally isolated as potential mitotic inducers in a screen for extragenic suppressors of ‘mitotic catastrophe’ or premature entry into mitosis resulting from elevated Cdc2 activity (Molz et al., 1989). The alleles isolated during the screen (mcs2-75 and mcs6-13) display allele-specific interactions with cdc2, reminiscent of the range of interactions described between cdc2 and cdc13 (Booher and Beach, 1987; Molz et al., 1989). The second fission yeast kinase with Cak activity is the single-subunit Csk1 (Hermand et al., 1998; Lee et al., 1999). The csk1 gene was first identified as a multicopy suppressor of the synthetic lethality of mcs2-75 cdc2-3w cdc25-22 (Molz and Beach, 1993). Subsequently, Csk1 was found to phosphorylate Mcs6 on the T-loop activation site (S165) and activate the Mcs6–Mcs2 complex in vivo (Hermand et al., 1998). A recent report also implicated Csk1 as a direct activator of Cdc2 (Lee et al., 1999), suggesting that Mcs6–Mcs2–Pmh1 and Csk1 function redundantly in Cdc2 activation. In this work, we addressed the in vivo functions of the fission yeast Caks Mcs6 and Csk1. Our results indicate that these kinases have distinct non-overlapping functions: Mcs6 acting as the Cdc2-activating kinase and Csk1 as the Mcs6-activating kinase.
Results
Complementation of a temperature-sensitive CAK1 allele by either Csk1 or Mcs6–Mcs2 in budding yeast
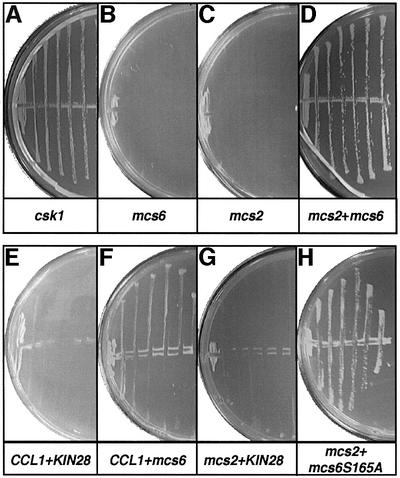
The identification of two kinases in fission yeast with Cak activity in vitro (Buck et al., 1995; Damagnez et al., 1995; Hermand et al., 1998; Lee et al., 1999) prompted us to study whether these kinases could complement a temperature-sensitive (ts) allele of S.cerevisiae CAK1 (civ1-4) (Thuret et al., 1996). To this end we initially used an unbiased approach by screening a S.pombe cDNA library for clones capable of suppressing the civ1-4 cell cycle arrest at the restrictive temperature. The only cDNA identified in this screen was csk1 (Figure 1A). As Mcs6 is a cyclin-dependent kinase, one possibility why it was not identified in this screen is that it requires its cognate cyclin (Mcs2) for activity. To investigate this possibility, plasmids expressing mcs6 and mcs2 were transformed into the thermosensitive CAK1 strain. As shown in Figure 1B–D, overexpression of either mcs6 or mcs2 alone does not suppress the CAK1 thermosensitive phenotype at 35°C, but when both subunits are expressed, the phenotype is fully rescued. This result indicates that the Mcs6–Mcs2 complex has Cak activity in vivo.
Fig. 1. Complementation of a temperature-sensitive CAK1 allele by either Csk1 or Mcs6–Mcs2 in budding yeast. The CAK1 ts strain (civ1-4) was transformed by plasmids overexpressing csk1 (csk1-p426), mcs6 (mcs6-p425), mcs2 (mcs2-p424), KIN28 (KIN28-p425), CCL1 (CCL1-p426), mcs6-S165A (mcs6-S165A-p425), or a combination of these as indicated. Resulting strains were streaked on selective minimal media at 35°C for 4 days and photographed.
As might be expected, co-expression of the budding yeast KIN28 and CCL1 does not rescue the phenotype (Figure 1E) as the complex lacks Cak activity in vitro and is not a Cak in vivo (Cismowski et al., 1995). The experimental set-up allowed us to investigate whether the heterologous Cdk–cyclin pairs would suppress the thermosensitive CAK1 strain. The result indicates that Mcs6 and Ccl1 weakly suppress the strain (Figure 1F), whereas Kin28 and Mcs2 do not (Figure 1G).
Phosphorylation of the activation site of Mcs6 was not required for the rescue as a Cak site mutant (Mcs6-S165A) was indistinguishable from Mcs6 (Figure 1H). The functional complementation of Cak1 by Mcs6–Mcs2 demonstrates that a Cak from the Cdk7 family can perform all essential functions of a Cak in heterologous budding yeast cells.
Activators of Cdc2 suppress the mcs6-13 csk1Δ phenotype but not the csk1Δ phenotype
The complementation results described above show that both Mcs6 and Csk1 have Cak activity when expressed in budding yeast. This result thus suggests that Mcs6 and Csk1 could be functionally redundant for Cdc2 activation in fission yeast cells, as previously suggested (Lee et al., 1999). To address this issue more directly we compared the ability of various Cdc2 regulators to suppress the phenotype of two strains: (i) csk1::sup3-5, a simple csk1 disruption (and referred to as csk1Δ subsequently) displaying a delay of entry into exponential growth (Hermand et al., 1998); and (ii) mcs6-13 csk1Δ (Hermand et al., 1998), in which the csk1 disruption is combined with the mcs6-13 allele (Molz et al., 1989). In combination, these mutations confer a synthetic lethality at 35°C (Hermand et al., 1998), whereas the mcs6-13 mutation alone shows no phenotype (Molz et al., 1989).
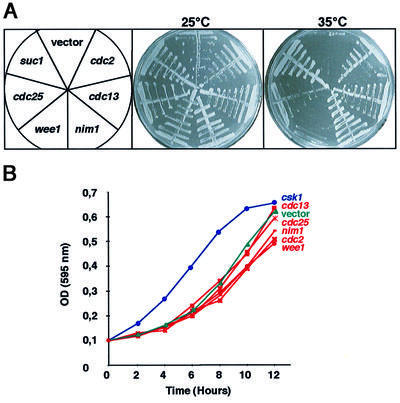
Transformation of plasmids harboring genomic inserts encoding for cdc2, cdc13, nim1, cdc25 and suc1 enabled the growth of mcs6-13 csk1Δ at 35°C, while wee1 did not (Figure 2A). Thus, all of the activators of Cdc2 tested rescued the phenotype, suggesting that impairment of Cdc2 activation is critical for the lethality of mcs6-13 csk1Δ, in agreement with Lee et al. (1999). We also found that the G1- and S-phase cyclins Cig1, Cig2 and Puc1 (a kind gift of Sergio Moreno) were unable to suppress the thermosensitive growth of the mcs6-13 csk1Δ double mutant (data not shown). As only G2 activators of Cdc2 rescue this strain, it is likely that it is mainly affected in the G2–M transition. This would be in agreement with previous observations indicating that higher levels of Cdc2 activity are required at the CG2–M transition compared with the G1–S transition (Stern and Nurse, 1996).
Fig. 2. Activators of Cdc2 suppress the mcs6-13 csk1Δ phenotype but not the csk1Δ phenotype. (A) In order to study the role of the Cdc2–Cdc13 complex in the lethality of the mcs6-13 csk1Δ strain, it was transformed with plasmids containing genomic inserts of cdc2, cdc13, nim1, wee1, cdc25, suc1 in pWH5 or an empty vector. Subsequently, these strains were streaked as sectors on minimal media and incubated at either 25 or 35°C, as indicated, for 4 days. (B) The csk1Δ strain was transformed with the same plasmids as in (A). The resulting strains were inoculated to an optical density (OD) of 0.1 from stationary phase cultures in minimal media. Subsequently, ODs were measured at the indicated time points. A representative experiment is shown.
Remarkably, none of the Cdc2 regulators that suppressed the mcs6-13 csk1Δ phenotype were able to suppress the csk1Δ growth delay phenotype (Figure 2B), suggesting that this phenotype is not directly related to Cdc2 activation.
Cak1 suppresses the mcs6-13 csk1Δ but not the csk1Δ phenotype
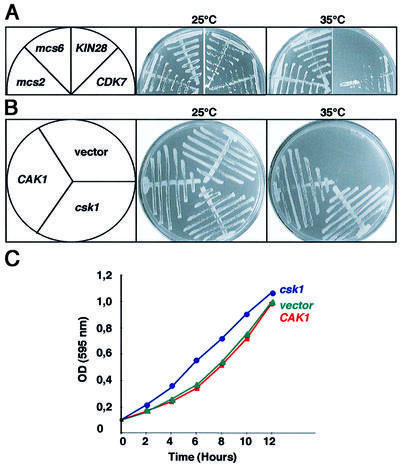
We then extended the suppressor analysis to kinases reported to have Cak activity in other species. For this purpose, initially two kinases of the Cdk7 family (human Cdk7 and budding yeast Kin28) were tested for their ability to suppress the thermosensitive growth defect of the mcs6-13 csk1Δ strain. The results indicate that Cdk7 suppressed the phenotype at 35°C, whereas KIN28 did not (Figure 3A). In addition, the mcs2 cyclin-encoding gene was also found to suppress the lethality when overexpressed (Figure 3A), constituting the first direct genetic interaction between mcs6 and mcs2.
Fig. 3. Cak1 suppresses the mcs6-13 csk1Δ but not the csk1Δ phenotype. (A) The mcs6-13 csk1Δ phenotype is suppressed by heterologous Caks. mcs6-13 csk1Δ was transformed with plasmids overexpressing mcs2 (mcs2-pAHL), mcs6 (mcs6-pREP3), KIN28 (KIN28-pREP3) or CDK7 (CDK7-pREP3). The resulting strains were streaked on minimal media and incubated at either 25 or 35°C, as indicated, for 4 days. (B) As in (A), except that csk1 (csk1-pAHL), CAK1 (CAK1-pAHL) or an empty vector was used. (C) In order to study whether Cak1 could rescue the phenotype of csk1Δ, it was transformed with plasmids overexpressing either csk1 (csk1-pAHL), CAK1 (CAK1-pAHL) or an empty vector and assayed for cell cycle re-entry as in Figure 2B.
The results presented in Figures 2 and 3 suggest that the synthetic lethality of the mcs6-13 csk1Δ strain primarily reflects impaired Cdc2 activation by Cak, and not RNA Pol II large subunit CTD phosphorylation. This was further supported by the ability of budding yeast CAK1 to suppress mcs6-13 csk1Δ at 35°C (Figure 3B). In contrast to the synthetic lethal strain, expression of a CAK1 plasmid did not suppress the csk1Δ growth delay phenotype (Figure 3C), similarly to what was previously observed for the Cdc2 regulators.
Cak1 does not activate Mcs6 unlike Csk1
Based on previous results suggesting that phosphorylation of Mcs6 by Csk1 is required for normal cell cycle entry from stationary phase (Hermand et al., 1998), the inability of CAK1 to suppress the csk1Δ phenotype suggested that Cak1 would not activate Mcs6 in vivo. Accordingly, Mcs2-associated kinase activity was not increased in Cak1-overexpressing S.pombe cells, unlike what has been reported for Csk1-overexpressing cells (Figure 4A; Hermand et al., 1998), although the intrinsic kinase activities of Cak1 and Csk1 in the respective strains toward a glutathione S-transferase (GST)–CDK2 substrate were comparable (Figure 4B). Furthermore, in contrast to Csk1, Cak1 was unable to activate Mcs6 or Mcs6–Mcs2 in vitro (Figure 4C) using baculovirus-expressed GST–Cak1 (or GST–Csk1 as a control) in an activation assay described previously (Hermand et al., 1998). These results demonstrate that Cak1 is unable to activate the Mcs6–Mcs2 complex in S.pombe cells.
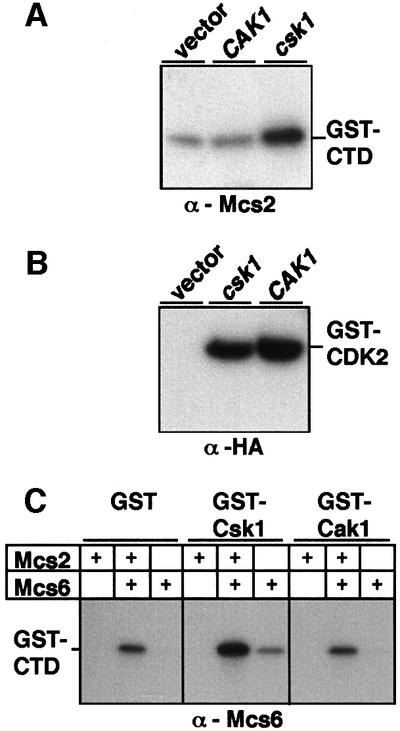
Fig. 4. Cak1 does not activate Mcs6 in vitro or in vivo. (A) In order to analyze the effects of overexpressed Cak1 on Mcs2-associated kinase activity in fission yeast cells, anti-Mcs2 immunoprecipitates made from cells overexpressing HA-Cak1 (CAK1) or HA-Csk1 (csk1) or a control (vector) were subsequently assayed for kinase activity toward GST–CTD, followed by SDS–PAGE and autoradiography. (B) The activity of the overexpressed HA-Cak1 (CAK1) or HA-Csk1 (csk1) from strains used in (A) was controlled by kinase reactions from anti-HA immunoprecipitates using GST–CDK2 as a substrate. (C) Kinase activity of purified Mcs6 or Mcs6–Mcs2 was assessed following a pre-incubation with GST, GST–Csk1 or GST–Cak1 in a cold kinase reaction. The second kinase reaction was performed in the presence of radiolabeled ATP and GST–CTD to assay Mcs6 activity. The basal activity of the Mcs6–Mcs2 complex is shown in lane 2.
Cdc2 associates with overexpressed Cak1 but not Csk1 in fission yeast cells
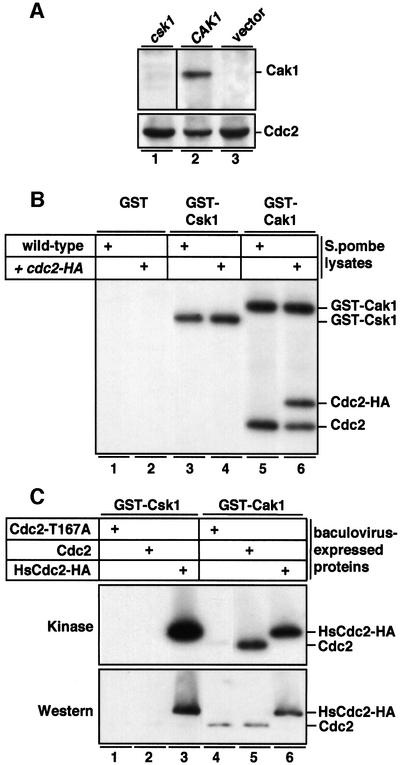
Cak1 has been shown not only to phosphorylate, but also to co-purify with Cdc28 from budding yeast lysates (Thuret et al., 1996); therefore, we were interested in investigating whether Csk1 would associate with its suggested substrate Cdc2 in an analogous manner. Endogenous Csk1 was not detected co-purifying with fission yeast Cdc2 (data not shown), but this could have been due to a detection problem considering the low levels of endogenous Csk1 (our unpublished data). To control this possibility, GST–Suc1 was used to purify Cdc2 (Cdc2-HA) from S.pombe cells also overexpressing either Csk1 or budding yeast Cak1. Subsequent western blotting analysis of the complexes revealed that while overexpressed Cak1 readily associated with Cdc2 in S.pombe lysates, Csk1 did not (Figure 5A).
Fig. 5. Association and phosphorylation of S.pombe Cdc2 by Cak1, but not Csk1. (A) GST–Suc1 was used to purify Cdc2 from S.pombe cells also overexpressing either Csk1 (csk1-pAAUN) or Cak1 (CAK1-pAHA). Precipitated complexes were analyzed by western blotting using either anti-Csk1 (lane 1) or anti-HA (lanes 2–3) antibodies (upper panel). Subsequently, the blot was stripped and probed using anti-PSTAIRE antibodies (lower panel). (B) Lysates of wild-type S.pombe cells (lanes 1, 3 and 5) or cells moderately overexpressing (see Materials and methods) S.pombe Cdc2-HA (lanes 2, 4 and 6) were incubated for 90 min with GST (lanes 1 and 2), GST–Csk1 (lanes 3 and 4) or GST–Cak1 (lanes 5 and 6). Following glutathione–Sepharose purification, a kinase reaction was performed in the presence of [γ-32P]ATP without additional substrates, followed by SDS–PAGE and autoradiography. Phosphorylated proteins are indicated on the right. (C) Insect cell lysates producing S.pombe Cdc2 (Cdc2), a T167A Cdc2 mutant (Cdc2-T167A) or human HA-tagged Cdc2 (HsCdc2-HA) were incubated with GST–Csk1 or GST–Cak1 as indicated, and subsequently GST proteins were purified and subjected to a kinase reaction (Kinase) as in (B). The kinase gel was subsequently transferred to nitrocellulose and probed with anti-PSTAIRE antibodies.
GST–Cak1, but not GST–Csk1, associates with and subsequently phosphorylates S.pombe Cdc2
In a second approach to address whether Csk1 could associate with S.pombe Cdc2, recombinant GST–Csk1 or GST–Cak1 was added to fission yeast lysates, and following a 30 min incubation on ice GST–Csk1 or GST–Cak1 and associated proteins were purified and subjected to an in vitro kinase reaction with radiolabeled ATP. As no exogenous substrates were added, phosphorylated bands represent potential substrates that had been purified from the fission yeast lysates due to their association with Csk1 or Cak1. This analysis revealed that GST–Cak1 bound and phosphorylated in vitro a protein of 34 kDa from a wild-type fission yeast lysate (Figure 5B, lane 5). Moreover, when a fission yeast lysate expressing Cdc2-HA in addition to endogenous Cdc2 was used in the same assays, an additional band of 36 kDa was labeled (Figure 5B, lane 6). Subsequent western blotting analysis revealed that the 34 and 36 kDa bands co-migrated with Cdc2 and Cdc2-HA, respectively (not shown). When GST–Csk1 was used, no labeled bands were detected (Figure 5B, lane 3–4) nor was Cdc2 or Cdc2-HA detected by western blotting (not shown). As Cdc2 is not autophosphorylated (Gould et al., 1991; Solomon et al., 1992), these results demonstrate that Cak1 can associate with and subsequently phosphorylate fission yeast Cdc2. Furthermore, using a similar approach with baculovirus lysates expressing wild-type S.pombe Cdc2 or a Cdc2 (T167A) activation-site mutant (Gould et al., 1991), we demonstrate that phosphorylation of Cdc2 by GST–Cak1 was directed at Thr167 (Figure 5C).
The inability of Csk1 to associate with and subsequently phosphorylate S.pombe Cdc2 does not reflect a general inability of this kinase to bind to Cdks, as GST–Csk1 readily associated with and subsequently phosphorylated the closely related human Cdc2 (Figure 5C, lane 3) expressed under identical conditions to the S.pombe wild-type and mutant Cdc2. On the other hand, the ability of the single-subunit kinases Cak1 and Csk1 to form stable complexes appears to be substrate specific, as no association of Csk1 with Mcs6 has been detected (data not shown).
A mcs6-S165A mutation mimics the csk1 disruption phenotype in fission yeast
The genetic and biochemical results presented above suggest that Csk1 and Mcs6 are not redundant Caks of S.pombe Cdc2, but instead the data support a linear activation cascade: csk1 → mcs6 → cdc2. In this model, Csk1 phosphorylation of the Mcs6 T-loop Ser165 is not essential (due to an alternative activation mechanism), whereas Mcs6 phosphorylation of Cdc2 on Thr167 is essential.
To test the first part of the model rigorously, we analyzed the effect of replacing mcs6 with a mutant encoding a protein in which the T-loop activation site Ser165 was mutated to Ala, thus blocking Csk1 from activating Mcs6. Comparison of the phenotype of this mcs6::mcs6-S165A strain with that of the csk1 disruption strain (csk1Δ) should reveal to what extent the observed delay of entry into exponential growth reflects an abolished activation of Mcs6 in csk1Δ cells. The results indicate that the mcs6-S165A strain displayed a delay of entry into exponential growth very similar to that observed in the csk1Δ strain (Figure 6, compare mcs6-S165A and csk1Δ). These data strongly suggest that the csk1Δ phenotype is solely due to the absence of T-loop phosphorylation on Mcs6 and are consistent with the observation that overexpression of Mcs6 can rescue the csk1Δ phenotype (Hermand et al., 1998).
Fig. 6. A mcs6-S165A mutation mimics the csk1 disruption phenotype in fission yeast. After integration of the mcs6-S165A allele (see Materials and methods), growth of either wild-type or csk1Δ or mcs6-S165A was compared as in Figure 2.
mcs6-SALR, combining mcs6-S165A and mcs6-13 mutations, arrests at 35°C
The similarity of the mcs6-S165A and csk1Δ phenotypes suggested that it should be possible to generate an mcs6 allele in csk1 wild-type background that would mimic the synthetic lethality observed in the mcs6-13 csk1Δ strain at 35°C. To this end, we sequenced the mcs6-13 allele, and identified a single T → G mutation (nucleotide 832 in DDBJ/EMBL/GenBank accession No. L47353), which results in the replacement of Mcs6 Leu238 with arginine. It is interesting to note that this residue is conserved in Cdk7 and several other Cdks, but not in Kin28. Subsequently, we reproduced this mutation in vitro and combined it with the S165A Cak site mutation to generate mcs6-S165A-L238R (referred to as mcs6-SALR from now on; see Figure 7A for schematic). Following the replacement of the genomic mcs6 with mcs6-SALR, the phenotype of this strain was compared with that of mcs6-13 csk1Δ. As shown in Figure 7B, the mcs6-SALR strain is unable to grow at 35°C, just like mcs6-13 csk1Δ (Figure 7B). Importantly, in contrast to the mcs6-13 csk1Δ strain, the mcs6-SALR strain is not rescued by overexpressed csk1 (Figure 7C), as predicted by the presence of wild-type csk1 in this strain. In all other respects, the mcs6-SALR strain was indistinguishable from mcs6-13 csk1Δ, as shown by suppression of the thermosensitivity by CAK1 (Figure 7C) as well as by all the same Cdc2 regulators and Caks described previously for mcs6-13 csk1Δ and summarized in Figure 7D. These results demonstrate impaired Cdc2 activation in the mcs6-SALR mutant strain at 35°C—a defect that the wild-type Csk1 or even overexpressed Csk1 does not suppress.
Fig. 7. The mcs6-SALR mutant, combining the mcs6-13 and S165A mutations, is thermosensitive. (A) Schematic comparison of mcs6-13 csk1Δ and mcs6-SALR. (B) The mcs6-13 csk1Δ and mcs6-SALR mutants have an identical thermosensitive phenotype. The mcs6-13 csk1Δ, mcs6-SALR and wild-type control strains were streaked on minimal media and incubated at either 25 or 35°C, as indicated, for 4 days. (C) The mcs6-SALR strain is not rescued by csk1 overexpression. mcs6-SALR strains overexpressing either csk1 (csk1-pAHL), CAK1 (CAK1-pAHL) or an empty vector were streaked on minimal media and incubated at either 25 or 35°C for 4 days. (D) Comparison of suppressors of the mcs6-13 csk1Δ and the mcs6–SALR fission yeast strains by the indicated Cdc2 regulators and Caks.
Discussion
Here we studied the role of Mcs6 and Csk1 in activation of Cdc2 in fission yeast. Previous studies have implicated both kinases in the activation of Cdc2 (Molz et al., 1989; Molz and Beach, 1993; Hermand et al., 1998; Lee et al., 1999). mcs6 and mcs2 encoding for its cyclin partner display strong genetic interactions with cdc2 (Booher and Beach, 1987; Molz et al., 1989).
Csk1 has been implicated in Cdc2 regulation more indirectly by analysis of the mcs6-13 csk1Δ strain, which is synthetically lethal at 35°C (Hermand et al., 1998). Arrested cells from this strain display reduced Cdc2-associated H1 kinase activity not seen in either the mcs6-13 or the csk1Δ single-mutant strains (Lee et al., 1999). These data, together with the fact that the mcs6-13 csk1Δ strain is mutated in two genes encoding proteins with Cak activity in vitro, strongly suggested that the phenotype is due to a Cdc2 activation defect. Results presented here demonstrate this to be the case as both multiple Cdc2 regulators (Figure 2), as well as two heterologous Cdk-activating kinases CDK7 and CAK1 (Figure 3), rescued the thermosensitivity of the strain at 35°C.
The inability of Cdc2 regulators (Figure 2) or CAK1 (Figure 3) to rescue the csk1Δ phenotype suggests that this phenotype is not a Cdc2 activation defect. These data together with the observations that the csk1Δ phenotype is rescued by mcs6 (Hermand et al., 1998) and that the mcs6-S165A strain displays a phenotype indistinguishable from csk1Δ (Figure 6) indicate that T-loop phosphorylation of Mcs6 is not absolutely required for Cdc2 activation. Therefore, the phenotype of csk1Δ and mcs6-S165A strains is likely to reflect a transcription defect relating to the as yet uncharacterized TFIIH-associated function of Mcs6 together with Mcs2 and Pmh1.
The suppressor analyses together with the biochemical data showing that Cak1 is unable to activate Mcs6 and that Csk1 is unable to associate with and subsequently phosphorylate fission yeast Cdc2 argued for a linear activation cascade csk1 → mcs6 → cdc2. Previous reports (Lee et al., 1999) on this subject were based on analysis of the double-mutant mcs6-13 csk1Δ strain, which does not allow a clear distinction between the roles of the two Caks. Encouraged by the phenotype of the mcs6-S165A strain, we therefore attempted to resolve this issue by generating an mcs6 allele that combined the mcs6-13 mutation with the T-loop activation site mutant (S165A). The resulting mcs6-SALR strain was phenotypically indistinguishable from mcs6-13 csk1Δ, and therefore allowed the analysis of the role of Mcs6 in Cdc2 activation. The results demonstrated that Csk1 is not directly involved in Cdc2 activation. Unlike in the report of Lee et al. (1999), in our experimental set-up we did not detect Csk1 phosphorylating fission yeast Cdc2, although we do find that Csk1 can phosphorylate human Cdc2. Our biochemical results are thus in agreement with the genetic results indicating an inability of Csk1 to function as a Cak for Cdc2 in vivo. The functions of Csk1 and Mcs6 are thus distinct and non-redundant.
The linear activation cascade csk1 → mcs6 → cdc2 also explains how Cdc2 activity is unchanged in the csk1Δ strain (Hermand et al., 1998; Lee et al., 1999) and the fact that both mcs6 and mcs2 were isolated as loss-of-function mutants that rescued a hyperactive Cdc2 (Molz et al., 1989) although the strain was wild type with respect to csk1.
When compared with other species, our results on in vivo activation of Cdc2 indicate that fission yeast in this respect is similar to Xenopus (Fesquet et al., 1997) and Drosophila (Larochelle et al., 1998), where Cdk7 has been implicated as the Cak of Cdc2. The role of Cdk7 in Cdc2 activation in Drosophila was questioned by results demonstrating that expression of a dominant-negative Cdk7 mutant during early division cycles did not inhibit Cdc2 phosphorylation (Leclerc et al., 2000). However, this could be due to stable maternal Cdk7 complexes in the early embryonic cycles.
The differences in the Cdc2- and Cdc28-activating kinases in fission yeast and budding yeast, respectively, may also at least partly explain interesting differences noted in the activating phosphorylation of Cdc2 and Cdc28 during a G1 arrest. When fission yeast cells are arrested in G1, Cdc2 is dephosphorylated (Simanis and Nurse, 1986), whereas a G1 arrest in budding yeast cells does not affect Cdc28 phosphorylation (Hadwiger and Reed, 1988). In fission yeast, the Cdc2 dephosphorylation is associated with Rum 1 binding and cyclin B (Cdc13) degradation (Stern and Nurse, 1998), leading to a monomeric kinase, which is not expected to be a substrate of the Cdk7-type Mcs6 kinase (Kaldis et al., 1998). In budding yeast, the G1 arrest is associated with disassembly of the Cdc28 complex (Wittenberg and Reed, 1988), but as Cak1 favors monomers (Kaldis et al., 1998), Cdc28 would continue to be phosphorylated. Interestingly, the mammalian counterpart Cdc2 becomes dephosphorylated upon shifting from exponential growth to quiescence (Lee et al., 1988), which may reflect the inability of Cdk7 to phosphorylate a monomeric Cdc2.
Materials and methods
Yeast strains and techniques
The S.pombe strains used in this study were: h+ ade6-210 ura4D18 leu1-32 his3-D1 (Burke and Gould, 1994), h+ mcs2::mcs2F leu1-32 ura4D18 ade6 (Molz and Beach, 1993); h– csk1Δ mcs6-13 leu1-32 ade6-704 and h+ csk1::ura4+ mcs2::mcs2F leu1-32 ura4D18 ade6 (Hermand et al., 1998). Schizosaccharomyces pombe was transformed using lithium acetate as described previously (Moreno et al., 1991).
The S.cerevisiae strains used were GF2351 MATα civ1-4 ura3 leu2 trp1 lys2 ade2 ade3 (Thuret et al., 1996). A high-efficiency transformation method was used (Gietz and Woods, 1998) in order to screen for S.pombe cDNAs capable of rescuing the GF2351 strain. The S.pombe cDNA library was kindly provided by Dr Michelle Minet and Dr Francois Lacroute.
Fission yeast expression vectors
All expression constructs used in S.pombe are based on two vectors: pREP3 (Maundrell, 1993) and pAAUN (Xu et al., 1990). mcs2-pAHA, csk1-pAHA, myc-mcs6-pREP3 and myc-CDK7-pREP3 have been described (Damagnez et al., 1995; Hermand et al., 1998). mcs2-pAHL and csk1-pAHL result from the exchange of the ura4+ selection marker of pAHA by LEU2 from pREP3. KIN28-pREP3 results from transferring a SalI–BamHI (underlined) PCR fragment from the KIN28 cDNA (a kind gift of Michel Simon) using primers 5′-AGCGGCCGCGTCGACATGAAAGTGAATATGGAG3′ and 5′-GCGGGATCCTCAGTTACGTATTTTTATTG-3′.
The CAK1 open reading frame (ORF) was amplified from S.cerevisiae genomic DNA using primers 5′-CGGAATTCACCATGAAACTGGATAGTATAGAC-3′ and 5′-TATGCGGCCGCTTATGGCTTTTCTAATTCT-3′, and cloned as an EcoRI–NotI (underlined) fragment into pGEX-4T. The ORF was subsequently transferred into pREP3 in order to express wild-type Cak1p, or into pAHA (Hermand et al., 1998) and pAHL (see above) in order to express HA-Cak1.
In the experiment of Figure 2, cdc2, cdc13, nim1, wee1, cdc25 and suc1 were expressed from genomic inserts cloned in pWH5 (Wright et al., 1986). In the experiment in Figure 5B, a cdc2-HA-pREP3 plasmid (a kind gift from B.Ducommun) was used, and the resulting strain was grown in the presence of thiamine to reduce the toxicity of overexpressed Cdc2-HA.
Budding yeast expression vectors
mcs6, mcs2 and csk1 ORFs were cloned under the control of a methionine-repressible promoter in p425, p424 and p426 (Mumberg et al., 1994), respectively. mcs6-S165A (Hermand et al., 1998) was transferred as a ClaI–PstI fragment into mcs6-p425.
KIN28-p425 was made by transfer of the KIN28 cDNA from pREP3 (see above) to p425. The insert for CCL1-p426 was made by PCR from S.cerevisiae genomic DNA using oligonucleotides 5′-ACGCGTCGACGAATCCACCATGACGGATATTCAACTAAATGG-3′ and 5′-GCGGGATCCCTCGAGCGGTAACAGAGCTGTTTCATG-3′, and cloned as SalI–XhoI (underlined) into p426. All plasmids were transformed in GF2351 (Thuret et al., 1996) and streaked out on media lacking methionine and the respective selection nutrients.
Construction and integration of mcs6 mutants
Construction of the myc-mcs6-S165A-pREP3 plasmid was as described previously (Hermand et al., 1998). The S165AL238R mutant of mcs6 was made by replacing the insert in myc-mcs6-pREP3 with a fragment generated by a two-step PCR approach using the following primers: MopS165A1, 5′-AGCGGCCGCGTCGACACCATGGAACAG-3′; MopL238R2, 5′-GGGACGCTGTTGCATACTTTTAATG-3′; MopL238R3, 5′-AAAAGTATGCAACAGCGTCCC-3′; MopS165A4, 5′-TGCGGCCGCGGATCCTTAAACAAATT-3′. The resulting product was verified by direct sequencing.
In order to create the S165E mutant of mcs6, the same strategy was used except that MopS165E2 (5′-TGGTGCTCCATGTGGCTTG-3′) and MopS165E3 (5′-AAGCCACATGGAGCACCAGG-3′) were used. All constructs were verified by sequencing the resulting plasmids. Subsequently, SalI–BamHI inserts containing the SA, SE or SALR mutants were transferred to the pRS306 plasmid (Sikorski and Hieter, 1989) with the kanR cassette of pFA6a-kanMX (Bahler et al., 1998).
An EcoRV fragment of the pSK-mcs6 G, containing a HindIII mcs6 genomic insert, was then replaced by EcoRV inserts harboring the mutants and the kanR cassette. The XhoI–XbaI linear fragments were used to transform the strain: h+ ade6-216 ura4D18 leu1-32 his3-D1. Transformants were plated on YE media for 12 h and replicated on YE media supplemented with G418 in order to select for the presence of integrated mcs6 mutants. Integration at the correct locus was confirmed by Southern blotting.
Antibodies and immunoprecipitations from S.pombe cells
The rabbit polyclonal Csk1, Mcs6 and Mcs2 antisera have been described (Hermand et al., 1998). Monoclonal HA antibody (Boehringer Mannheim) or Anti-FLAG M5 antibody (Eastman Kodak Company) were used as per the manufacturer’s instructions. For immunoprecipitations from S.pombe cells, yeast strains (10 ml) were grown overnight in selective media. Subsequently, cell pellets were washed once in phosphate-buffered saline (PBS), and disrupted in lysis buffer [150 mM NaCl, 50 mM HEPES pH 7.5, 5 mM EDTA, 1% NP-40 with 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM β-glycerophosphate, 5 mM NaF]. Following disruption, lysates were adjusted to 0.5% NP-40 for immunoprecipitation. Immunoprecipitates on protein A–Sepharose (Sigma) beads were washed four times with lysis buffer containing 0.1% NP-40, and once with kinase buffer [20 mM Tris pH 7.5, 50 mM KCl, 5 mM MgCl2, 2.5 mM MnCl2, 1 mM dithiothreitol (DTT)].
Recombinant protein expression and purification
For protein expression, GST–Cak1 (a kind gift from Philipp Kaldis and Mark Solomon; Kaldis et al., 1998), GST–Csk1 (Hermand et al., 1998), Mcs6 (Hermand et al., 1998) and Mcs2 (Hermand et al., 1998) baculoviruses were propagated in Hi5 insect cells (Invitrogen) for 48 h. The GST–Csk1, GST–Cak1 and GST proteins were purified using glutathione–Sepharose and eluted with glutathione. Ten nanograms of purified protein were used per assay. Mcs6, Mcs6–Mcs2 and Mcs6–Mcs2-Pmh1 complexes were purified with Mcs6 antiserum and used bound to protein A–Sepharose. Protein amounts were estimated by silver staining and 100 ng of purified proteins were used for kinase assays. Bacterially expressed GST–CDK2-D155N and GST–CTD proteins were purified as described (Damagnez et al., 1995; Hermand et al., 1998).
Affinity purification with GST–suc1, GST–Cak1 and GST–Csk1
Extracts from fission cells were prepared from 100 ml cultures as above, washed once in PBS, and disrupted in 1× pellet volume of lysis buffer [buffer A (50 mM Tris pH 7.5, 50 mM KCl, 5 mM EDTA, 10 mM β-glycerophosphate, 2 mM PMSF, aprotinin, leupeptin) supplemented with 0.5 M NaCl, 5 mM DTT and 1% Igepal CA-630] and subsequently diluted with 2× volume buffer A.
Baculovirus-infected (48 h) Hi5 cells were washed in PBS, treated 10 min on ice in buffer A with 5 mM DTT and disrupted with a 23G needle. The lysates were diluted with an equal volume of buffer A with 300 mM NaCl and 0.2% Igepal CA-630.
To purify Cdc2 from S.pombe lysates, 1 µl of GST–Suc1 agarose (Upstate Biotechnology) was incubated with 200 µg of lysate on ice for 30 min. Alternatively, yeast or insect cell lysates (1 mg in 400 µl) were supplemented with 2–4 µg of purified GST, GST–Csk1 or GST–Cak1 and incubated on ice for 30 min followed by purification with glutathione–Sepharose as above. Beads were washed four times with washing buffer (50 mM Tris pH 7.5, 50 mM KCl, 0.1% Igepal CA-630) and used in kinase assay (see below).
Kinase assays
Kinase assays on immunoprecipitates from S.pombe cells were performed essentially as described (Hermand et al., 1998). Briefly, reactions were performed in 30 µl of kinase buffer supplemented with 10 µCi of [γ-32P]ATP and indicated substrates (4 µg of GST–CDK2-D155N, 4 µg of GST–CTD) for 30 min at 30°C. When using baculovirus proteins, 10 ng of GST–Csk1, GST–Cak1, or 100 ng of Mcs6 (and complexed proteins as indicated) in immunoprecipitates were used as kinases. In the Cdk-activation experiments, GST–Csk1 or GST–Cak1 was incubated with Mcs6, Mcs2 or Mcs6–Mcs2 bound to beads for 6 min at 30°C in kinase buffer supplemented with 1 mM ATP. Subsequently, the beads were washed three times with kinase buffer before a kinase reaction with [γ-32P]ATP with GST–CTD substrate. Phosphorylated substrates were analyzed by 10% SDS–PAGE followed by autoradiography.
Acknowledgments
Acknowledgements
We thank Philipp Kaldis, Mark Solomon, Paul Nurse, Sergio Moreno, Gerard Faye, Michel Simon, Michelle Minet, Francois Lacroute, Eberhard Schneider, Bernard Ducommun, David Beach, Peter Wagner and Nina Korsisaari for providing reagents. We are grateful to Beata Grallert and Eric Boye for fruitful discussions and help with fission yeast genetics. This study was supported by grants from Academy of Finland, University of Helsinki, Finnish Cancer Organization, Finnish Cancer Institute and Sigrid Juselius Foundation. D.H. is a FNRS Postdoctoral Researcher; A.P. is a graduate student of the Helsinki Graduate School in Biotechnology and Molecular Biology; T.W. and T.V. are graduate students of Helsinki Biomedical Graduate School.
REFERENCES
- Bahler J., Wu,J.Q., Longtine,M.S., Shah,N.G., McKenzie,A.,III, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Booher R. and Beach,D. (1987) Interaction between cdc13+ and cdc2+ in the control of mitosis in fission yeast; dissociation of the G1 and G2 roles of the cdc2+ protein kinase. EMBO J., 6, 3441–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck V., Russell,P. and Millar,J.B. (1995) Identification of a cdk-activating kinase in fission yeast. EMBO J., 14, 6173–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J.D. and Gould,K.L. (1994) Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol. Gen. Genet., 242, 169–176. [DOI] [PubMed] [Google Scholar]
- Cismowski M.J., Laff,G.M., Solomon,M.J. and Reed,S.I. (1995) KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol., 15, 2983–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damagnez V., Makela,T.P. and Cottarel,G. (1995) Schizosaccharomyces pombe Mop1–Mcs2 is related to mammalian CAK. EMBO J., 14, 6164–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devault A., Martinez,A.M., Fesquet,D., Labbe,J.C., Morin,N., Tassan,J.P., Nigg,E.A., Cavadore,J.C. and Doree,M. (1995) MAT1 (‘menage a trois’) a new RING finger protein subunit stabilizing cyclin H–cdk7 complexes in starfish and Xenopus CAK. EMBO J., 14, 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R., Le Roy,G., Cho,H., Akoulitchev,S. and Reinberg,D. (1996) Human cyclin-dependent kinase-activating kinase exists in three distinct complexes. Proc. Natl Acad. Sci. USA, 93, 6488–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.C., Wong,C. and Elledge,S.J. (1998) Human cyclin K, a novel RNA polymerase II-associated cyclin possessing both carboxy-terminal domain kinase and Cdk-activating kinase activity. Mol. Cell. Biol., 18, 4291–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza F.H., Farrell,A., Erdjument-Bromage,H., Tempst,P. and Morgan,D.O. (1996) A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science, 273, 1714–1717. [DOI] [PubMed] [Google Scholar]
- Espinoza F.H., Farrell,A., Nourse,J.L., Chamberlin,H.M., Gileadi,O. and Morgan,D.O. (1998) Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol., 18, 6365–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G., Simon,M., Valay,J.G., Fesquet,D. and Facca,C. (1997) Rig2, a RING finger protein that interacts with the Kin28/Ccl1 CTD kinase in yeast. Mol. Gen. Genet., 255, 460–466. [DOI] [PubMed] [Google Scholar]
- Feaver W.J., Svejstrup,J.Q., Henry,N.L. and Kornberg,R.D. (1994) Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell, 79, 1103–1109. [DOI] [PubMed] [Google Scholar]
- Feaver W.J., Henry,N.L., Wang,Z., Wu,X., Svejstrup,J.Q., Bushnell,D.A., Friedberg,E.C. and Kornberg,R.D. (1997) Genes for Tfb2, Tfb3 and Tfb4 subunits of yeast transcription/repair factor IIH. Homology to human cyclin-dependent kinase activating kinase and IIH subunits. J. Biol. Chem., 272, 19319–19327. [DOI] [PubMed] [Google Scholar]
- Fesquet D. et al. (1993) The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J., 12, 3111–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesquet D., Morin,N., Doree,M. and Devault,A. (1997) Is Cdk7/cyclin H/MAT1 the genuine cdk activating kinase in cycling Xenopus egg extracts? Oncogene, 15, 1303–1307. [DOI] [PubMed] [Google Scholar]
- Fisher R.P. and Morgan,D.O. (1994) A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell, 78, 713–724. [DOI] [PubMed] [Google Scholar]
- Fisher R.P., Jin,P., Chamberlin,H.M. and Morgan,D.O. (1995) Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell, 83, 47–57. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Woods,R.A. (1998) Transformation of yeast by lithium acetate-single stranded carrier DNA/PEG method. In Brown,A.J.P. and Tuite,M.F. (eds), Methods in Microbiology. Vol. 26. Academic Press, New York, NY.
- Gould K.L., Moreno,S., Owen,D.J., Sazer,S. and Nurse,P. (1991) Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J., 10, 3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger J.A. and Reed,S.I. (1988) Invariant phosphorylation of the Saccharomyces cerevisiae Cdc28 protein kinase. Mol. Cell. Biol., 8, 2976–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner C.J., Myer,V.E., Liao,S.M., Wilson,C.J., Koh,S.S. and Young,R.A. (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin- dependent kinases Mol. Cell, 2, 43–53. [DOI] [PubMed] [Google Scholar]
- Hermand D., Pihlak,A., Westerling,T., Damagnez,V., Vandenhaute,J., Cottarel,G. and Makela,T.P. (1998) Fission yeast Csk1 is a CAK-activating kinase (CAKAK). EMBO J., 17, 7230–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis P. (1999) The cdk-activating kinase (CAK): from yeast to mammals. Cell. Mol. Life Sci., 55, 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis P. and Solomon,M.J. (2000) Analysis of CAK activities from human cells. Eur. J. Biochem., 267, 4213–4221. [DOI] [PubMed] [Google Scholar]
- Kaldis P., Sutton,A. and Solomon,M.J. (1996) The Cdk-activating kinase (CAK) from budding yeast. Cell, 86, 553–564. [DOI] [PubMed] [Google Scholar]
- Kaldis P., Russo,A.P., Chou,H.S., Pavletich,N.P. and Solomon,M.J. (1998) Human and yeast CDK-activating kinases (CAKs) display distinct substrate specificities. Mol. Biol. Cell, 9, 2545–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman J., Kaldis,P., Hengartner,C.J., Laff,G.M., Koh,S.S., Young,R.A. and Solomon,M.J. (1999) Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol., 19, 4774–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle S., Pandur,J., Fisher,R.P., Salz,H.K. and Suter,B. (1998) Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev., 12, 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc V., Raisin,S. and Leopold,P. (2000) Dominant-negative mutants reveal a role for the Cdk7 kinase at the mid-blastula transition in Drosophila embryos. EMBO J., 19, 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. and Lis,J.T. (1998) Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature, 393, 389–392. [DOI] [PubMed] [Google Scholar]
- Lee K.M., Saiz,J.E., Barton,W.A. and Fisher,R.P. (1999) Cdc2 activation in fission yeast depends on Mcs6 and Csk1, two partially redundant Cdk-activating kinases (CAKs). Curr. Biol., 9, 441–444. [DOI] [PubMed] [Google Scholar]
- Lee M.G., Norbury,C.J., Spurr,N.K. and Nurse,P. (1988) Regulated expression and phosphorylation of a possible mammalian cell-cycle control protein. Nature, 333, 676–679. [DOI] [PubMed] [Google Scholar]
- Lim H.H., Loy,C.J., Zaman,S. and Surana,U. (1996) Dephosphorylation of threonine 169 of Cdc28 is not required for exit from mitosis but may be necessary for start in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 4573–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä T.P., Tassan,J.P., Nigg,E.A., Frutiger,S., Hughes,G.J. and Weinberg,R.A. (1994) A cyclin associated with the CDK-activating kinase MO15. Nature, 371, 254–257. [DOI] [PubMed] [Google Scholar]
- Mäkelä T.P., Parvin,J.D., Kim,J., Huber,L.J., Sharp,P.A. and Weinberg,R.A. (1995) A kinase-deficient transcription factor IIH is functional in basal and activated transcription. Proc. Natl Acad. Sci. USA, 92, 5174–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 123, 127–130. [DOI] [PubMed] [Google Scholar]
- Molz L. and Beach,D. (1993) Characterization of the fission yeast mcs2 cyclin and its associated protein kinase activity. EMBO J., 12, 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molz L., Booher,R., Young,P. and Beach,D. (1989) cdc2 and the regulation of mitosis: six interacting mcs genes. Genetics, 122, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Morgan D.O. (1997) Cyclin-dependent kinases: engines, clocks and microprocessors. Annu. Rev. Cell Dev. Biol., 13, 261–291. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon R.Y., Yamashita,K., Adamczewski,J.P., Hunt,T. and Shuttleworth,J. (1993) The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J., 12, 3123–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon J.T., Ge,H., Gibbs,E., Sancar,A., Hurwitz,J. and Pan,Z.Q. (1996) Isolation and characterization of two human transcription factor IIH (TFIIH)-related complexes: ERCC2/CAK and TFIIH. Proc. Natl Acad. Sci. USA, 93, 6482–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Adamczewski,J.P., Seroz,T., Vermeulen,W., Tassan,J.P., Schaeffer,L., Nigg,E.A., Hoeijmakers,J.H. and Egly,J.M. (1994) The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell, 79, 1093–1101. [DOI] [PubMed] [Google Scholar]
- Serizawa H., Makela,T.P., Conaway,J.W., Conaway,R.C., Weinberg,R.A. and Young,R.A. (1995) Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature, 374, 280–282. [DOI] [PubMed] [Google Scholar]
- Shiekhattar R., Mermelstein,F., Fisher,R.P., Drapkin,R., Dynlacht,B., Wessling,H.C., Morgan,D.O. and Reinberg,D. (1995) Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature, 374, 283–287. [DOI] [PubMed] [Google Scholar]
- Shuttleworth J., Godfrey,R. and Colman,A. (1990) p40MO15, a cdc2-related protein kinase involved in negative regulation of meiotic maturation of Xenopus oocytes. EMBO J., 9, 3233–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V. and Nurse,P. (1986) The cell cycle control gene cdc2+ of fission yeast encodes a protein kinase potentially regulated by phosphorylation. Cell, 45, 261–268. [DOI] [PubMed] [Google Scholar]
- Simon M., Seraphin,B. and Faye,G. (1986) KIN28, a yeast split gene coding for a putative protein kinase homologous to CDC28. EMBO J., 5, 2697–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M.J. and Kaldis,P. (1998) Regulation of CDKs by phosphorylation. Results Probl. Cell Differ., 22, 79–109. [DOI] [PubMed] [Google Scholar]
- Solomon M.J., Lee,T. and Kirschner,M.W. (1992) Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol. Biol. Cell, 3, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M.J., Harper,J.W. and Shuttleworth,J. (1993) CAK, the p34cdc2 activating kinase, contains a protein identical or closely related to p40MO15. EMBO J., 12, 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B. and Nurse,P. (1996) A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet., 12, 345–350. [PubMed] [Google Scholar]
- Stern B. and Nurse,P. (1998) Cyclin B proteolysis and the cyclin-dependent kinase inhibitor rum1p are required for pheromone-induced G1 arrest in fission yeast. Mol. Biol. Cell, 9, 1309–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A. and Freiman,R. (1997) The Cak1p protein kinase is required at G1/S and G2/M in the budding yeast cell cycle. Genetics, 147, 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassan J.P., Schultz,S.J., Bartek,J. and Nigg,E.A. (1994) Cell cycle analysis of the activity, subcellular localization and subunit composition of human CAK (CDK-activating kinase). J. Cell Biol., 127, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassan J.P., Jaquenoud,M., Fry,A.M., Frutiger,S., Hughes,G.J. and Nigg,E.A. (1995) In vitro assembly of a functional human CDK7–cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J., 14, 5608–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuret J.Y., Valay,J.G., Faye,G. and Mann,C. (1996) Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell, 86, 565–576. [DOI] [PubMed] [Google Scholar]
- Valay J.G., Simon,M. and Faye,G. (1993) The kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J. Mol. Biol., 234, 307–310. [DOI] [PubMed] [Google Scholar]
- Valay J.G., Simon,M., Dubois,M.F., Bensaude,O., Facca,C. and Faye,G. (1995) The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol., 249, 535–544. [DOI] [PubMed] [Google Scholar]
- Wittenberg C. and Reed,S.I. (1988) Control of the yeast cell cycle is associated with assembly/disassembly of the Cdc28 protein kinase complex. Cell, 54, 1061–1072. [DOI] [PubMed] [Google Scholar]
- Wright A., Maundrell,K., Heyer,W.D., Beach,D. and Nurse,P. (1986) Vectors for the construction of gene banks and the integration of cloned genes in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Plasmid, 15, 156–158. [DOI] [PubMed] [Google Scholar]
- Xu H.P., Wang,Y., Riggs,M., Rodgers,L. and Wigler,M. (1990) Biological activity of the mammalian RAP genes in yeast. Cell Regul., 1, 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]