Abstract
Complete activation of signal transducer and activator of transcription 1 (STAT1) requires phosphorylation at both Y701 and a conserved PMS727P sequence. S727 phosphorylation of STAT1 in interferon-γ (IFN-γ)-treated mouse fibroblasts occurred without a need for p38 mitogen-activated protein kinase (MAPK), extracellular signal-regulated kinases 1 and 2 or c-Jun kinases, and required both an intact SH2 domain and phosphorylation of Y701. In contrast, UV irradiation-induced STAT1 phosphorylation on S727 required p38MAPK, but no SH2 domain– phosphotyrosine interactions. Mutation of S727 differentially affected IFN-γ target genes, at the level of both basal and induced expression. Particularly strong effects were noted for the GBP1 and TAP1 genes. The PMS727P motif of STAT3 was phosphorylated by stimuli and signaling pathways different from those for STAT1 S727. Transfer of the STAT3 C-terminus to STAT1 changed the stimulus and pathway specificity of STAT1 S727 phosphorylation to that of STAT3. Our data suggest that STAT C-termini contribute to the specificity of cellular responses by linking individual STATs to different serine kinase pathways and through an intrinsically different requirement for serine phosphorylation at different target gene promoters.
Keywords: phosphorylation/signal transduction/STAT1/transcription
Introduction
The JAK–STAT paradigm specifies a signaling pathway employed by most cytokine receptors to reprogram gene expression rapidly (Darnell et al., 1994; Schindler and Darnell, 1995). Receptor-associated JAKs are activated after binding of ligand, whereupon they phosphorylate receptor chains and thus create docking sites for the STAT SH2 domains. Receptor-associated STATs are phosphorylated by JAKs on a single tyrosine residue. They dimerize by reciprocal phosphotyrosine–SH2 domain interactions, traverse the nuclear pore and bind a class of DNA sequences collectively named GAS elements (Decker et al., 1997). The only two firmly established interaction partners for the STAT SH2 domains are cytokine receptors and homo- or heterotypic STATs; in addition, the JAKs may possibly also interact (Gupta et al., 1996; Barahmand-Pour et al., 1998).
The JAK–STAT pathway is intertwined with other signaling routes (Briscoe et al., 1997; Decker and Kovarik, 1999; Ramana et al., 2000a). An important signal convergence point was uncovered with the identification of a STAT serine phosphorylation site (Wen et al., 1995). With the exception of STATs 2 and 6, the C-termini of all family members contain a conserved PMSP or a PSP motif located between amino acids 720 and 730. Its phosphorylation can be caused by a variety of different stimuli and signaling pathways (Decker and Kovarik, 2000). Mutation of the C-terminal serine residue to alanine decreases the transcription factor activity of STAT1 and STAT4 (Wen et al., 1995; Visconti et al., 2000), prolongs tyrosine phosphorylation and DNA binding of STAT5a (Beuvink et al., 2000), or causes both a decrease in transcriptional activity and sustained tyrosine phosphorylation of STAT3 (Wen et al., 1995; Chung et al., 1997; Schuringa et al., 2000). How these seemingly opposing effects on activity are embedded into STAT3-dependent cytokine responses still needs to be solved.
Type I and type II interferons (IFNs) cause phosphorylation of STAT1 on both Y701 and S727 (Wen et al., 1995; Kovarik et al., 1998; Goh et al., 1999; Uddin et al., 2000). Reconstitution of STAT1-deficient U3A fibrosarcoma cells with STAT1-S727A restored biological responses to IFN-α. However, the mutation severely reduced the ability of STAT1 to establish the antiproliferative or antiviral effects after IFN-γ treatment (Bromberg et al., 1996; Horvath and Darnell, 1996). Therefore, phosphorylation of the STAT1 dimer on S727 appears to be critical for IFN-γ responses, but not for IFN-α responses. Moreover, the heterotrimeric ISGF3 (STAT1–STAT2– IRF9) complex that binds to a distinct response element, the ISRE (Levy et al., 1988), appears to dominate the response to type I IFN without requiring S727-phosphorylated STAT1.
IRF1 is the only target gene analyzed so far for its dependence on STAT1 S727 phosphorylation. In STAT1-S727A-reconstituted U3A cells, transcription was reduced by ∼80% (Wen et al., 1995). It is unclear whether this result can be extrapolated to STAT1 target genes in general, or whether the effect of the mutation differs for individual genes.
IFN-γ-stimulated phosphorylation of STAT1 on S727 and Y701 was reported to occur without a mechanistic interdependence because mutation of one residue did not affect phosphorylation of the other (Zhu et al., 1997). However, in IFN-γ-treated murine macrophages, most of the S727-phosphorylated STAT1 was contained in the fraction of molecules also phosphorylated on Y701 (Kovarik et al., 1998). This result suggested some positive influence of one phosphate on the presence of the other. The IFN-γ-stimulated STAT1 serine kinase remains to be identified. IFN-γ stimulates S727 phosphorylation independently of p38 mitogen-activated protein kinase (MAPK; Kovarik et al., 1999). JAK2 or protein kinase R (PKR) deficiency, or expression of a dominant-negative allele of the Pyk2 tyrosine kinase, abrogates IFN-γ-induced STAT1 S727 phosphorylation (Zhu et al., 1997; Takaoka et al., 1999; Ramana et al., 2000b). A link between these transducers, if any, has not been established, and their position with respect to STAT1 is unclear.
Phosphorylation of STAT1 on S727 in response to signals of stress, inflammation or infection (hereafter collectively referred to as stress) occurs in the absence of tyrosine phosphorylation. The biological relevance is to prime STAT1 for an increased transcriptional response once IFN-γ provides the stimulus for tyrosine phosphorylation (Kovarik et al., 1998, 1999; Stoiber et al., 1999). Stress signals to STAT1 through a p38MAPK pathway. p38MAPK specifically phosphorylates STAT1 on S727 in vitro and may be the stress-induced STAT1 kinase, rather than a signaling intermediate. It is an unresolved question whether stress- and IFN-γ-mediated phosphorylation of STAT1 on S727 involve common steps and signal transducers.
The biological activities of STAT1 and STAT3 differ strikingly (Levy, 1999; Akira, 2000; Bromberg and Darnell, 2000). In simple terms, STAT1 activity is correlated with pro-inflammatory and antiproliferative responses, whereas STAT3 activity is linked to proliferation and mostly anti-inflammatory cytokines. This suggests that in spite of the identical position of the PMSP motif in their C-termini, serine phosphorylation of STAT1 and STAT3 might be regulated by different cell surface stimuli and via different signaling pathways. The structural basis for phosphorylation by different kinases in vivo is not known. Compared with the STAT1 C-terminus, the STAT3 C-terminus appears to be a far better substrate for extracellular signal-regulated kinases (ERKs; Chung et al., 1997).
In this study, we pursued three major goals related to open questions outlined above. First, we investigated STAT1 S727 phosphorylation by either IFN-γ or UV irradiation and report that the former requires an intact SH2 domain and tyrosine phosphorylation of STAT1 whereas the latter does not. Secondly, we show that the C-termini of STATs 1 and 3 specify the interaction with different MAPKs. Thirdly, we report striking differences for IFN-inducible genes in their requirement for STAT1 serine phosphorylation.
Results
Phosphotyrosine–SH2 domain interactions are required for S727 phosphorylation of STAT1 in response to IFN-γ, but not in response to UV irradiation
To determine whether stress- and IFN-γ-induced phosphorylation of STAT1 on S727 might differ in their requirement for SH2 domain–phosphotyrosine interactions, we transfected 3T3 fibroblasts derived from STAT1-deficient mice (Durbin et al., 1996; Marie et al., 1998) with plasmids expressing either wild-type STAT1 (STAT1-WT) or the R602K mutant with a binding-deficient SH2 domain (Gupta et al., 1996). Pools of stable transfectants were selected and clonal lines expressing comparable amounts of STAT1 were derived by a limiting dilution protocol. As expected, STAT1-R602K was not phosphorylated on tyrosine when isolated from IFN-γ-treated cells (data not shown). Western blots using phosphospecific antiserum demonstrated the expected S727 phosphorylation of STAT1-WT by both IFN-γ and UV irradiation (Figure 1A). In contrast, STAT1-R602K was phosphorylated only in the case of UV irradiation, but not in the case of IFN-γ treatment (Figure 1B). To test whether STAT1 tyrosine phosphorylation is needed for S727 phosphorylation in response to IFN-γ, we reconstituted Stat1-deficient cells with the Y701F mutant of STAT1. Again, S727 phosphorylation was triggered by UV irradiation, but not by IFN-γ treatment (Figure 1C). Therefore, lack of S727 phosphorylation in the case of the STAT1-R602K mutant most probably reflects the SH2 domain requirement for tyrosine phosphorylation.
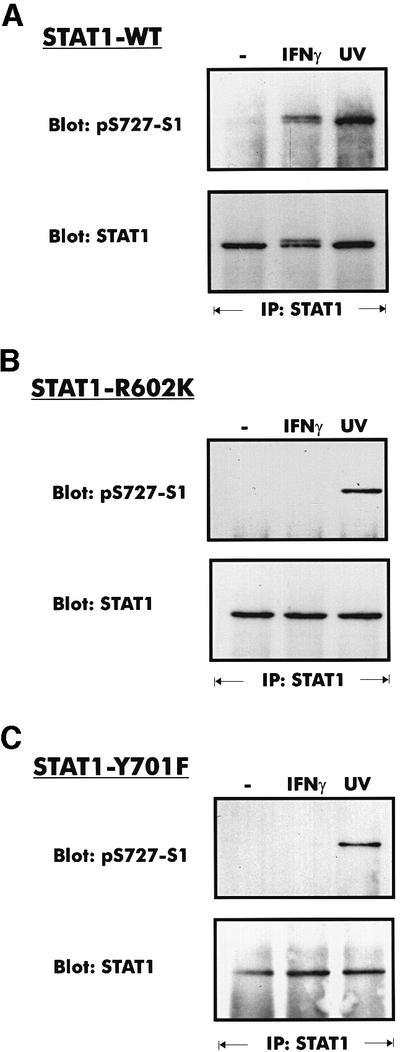
Fig. 1. Phosphorylation of S727 in fibroblast clones from STAT1-deficient mice after stable introduction of STAT1-WT (A), STAT1-R602K (B) or STAT1-Y701F (C). Where indicated, the cells were treated with IFN-γ for 30 min, or irradiated with UV light. STAT1 was precipitated from cellular extracts with a C-terminal antiserum. Western blots of the precipitated proteins were stained with phosphospecific antiserum for S727 (upper panels) and reprobed with a STAT1 C-terminal antiserum (lower panels).
Promoters of IFN-inducible genes can be distinguished by a different requirement for STAT phosphorylation on S727
IFN-inducible promoters contain the GAS or ISRE sequences, or both elements (Darnell et al., 1994; Stark et al., 1998). The IRF1 promoter, whose activation requires STAT1 S727 phosphorylation, contains only the GAS (Pine et al., 1994). No data are available on how the other promoter types are affected by the S727A mutation. To address the importance of STAT1 serine phosphorylation for different target genes, STAT1-deficient 3T3 fibroblasts were reconstituted with the STAT1-S727A mutant as described above. Clonal lines expressing levels of STAT1-S727A comparable to STAT1-WT-reconstituted cells (shown in Figure 2A) were of particular importance, because vast overexpression tended to reduce the penetrance of the S727A mutation.
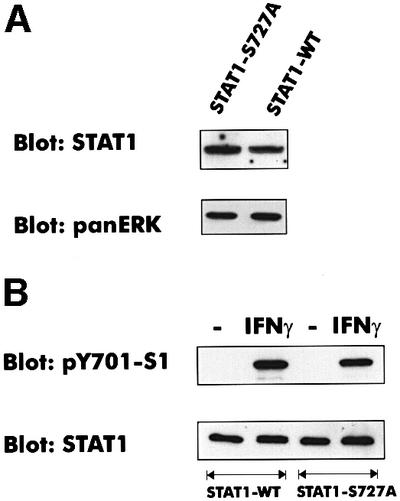
Fig. 2. Expression and tyrosine phosphorylation of STAT1-S727A. (A) Cellular lysates of fibroblasts from STAT1-deficient mice, reconstituted with either STAT1-WT or STAT1-S727A, were analyzed by western blotting with a STAT1 N-terminal monoclonal antibody. For loading control, the blot was reprobed with a panERK monoclonal antibody. (B) Tyrosine phosphorylation in cellular lysates was determined using a phosphotyrosine 701-specific antiserum. Equal loading was confirmed with a STAT1 N-terminal monoclonal antibody.
Following IFN-γ treatment, STAT1-WT and STAT1-S727A were phosphorylated on tyrosine with almost identical efficiency (Figure 2B). This confirms that S727 phosphorylation has no detectable effect on STAT1 tyrosine phosphorylation by the JAKs (Zhu et al., 1997; Kovarik et al., 1998) and excludes the transcriptional differences between STAT1-S727A and STAT1-WT resulting from different levels of tyrosine phosphorylation.
The IFN-inducible IFP53 promoter contains a GAS, but no ISRE (Strehlow et al., 1993). In transient transfections, the S727A mutant caused an average reduction in IFN-γ-stimulated IFP53 reporter gene transcription of 50%, whereas in U3A cells the reduction was ∼80%, consistent with published data obtained with a Ly6 reporter gene (data not shown; Wen et al., 1995).
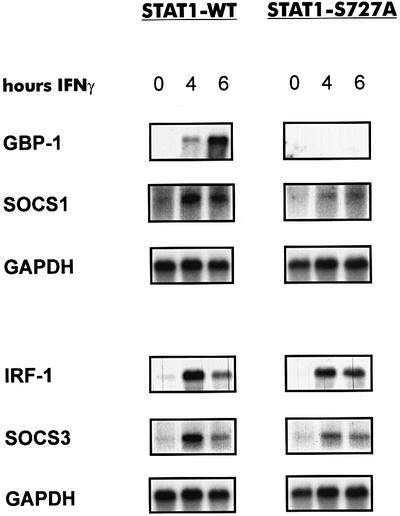
Expression of the genomic IRF1 gene, analyzed by northern blotting (Figure 3), was reduced in STAT1- S727A cells by ∼20–50% in independent experiments. In contrast, GBP1 mRNA was virtually undetectable in IFN-γ-treated STAT1-S727A cells. Since the same RNA preparation was used to analyze IRF1 and GBP1, misinterpretations based on variations in the IFN-γ response can be excluded. SOCS1 and SOCS3 mRNAs encoding IFN-induced feedback inhibitors of cytokine signaling (Hilton, 1999) showed an intermediate dependence on S727 phosphorylation. The reduction in IFN-γ-induced expression was more pronounced than in the case of IRF1, but less than in the case of the GBP gene. Thus, the extent to which serine phosphorylation of STAT1 is required for function appears to be determined largely by the target promoter.
Fig. 3. Northern blot analysis of IFN-inducible mRNAs from STAT1-deficient fibroblasts after reconstitution with STAT1-WT or STAT1-S727A. A 15 µg aliquot of total RNA was blotted to membranes and hybridized to labeled cDNAs corresponding to the indicated mRNAs.
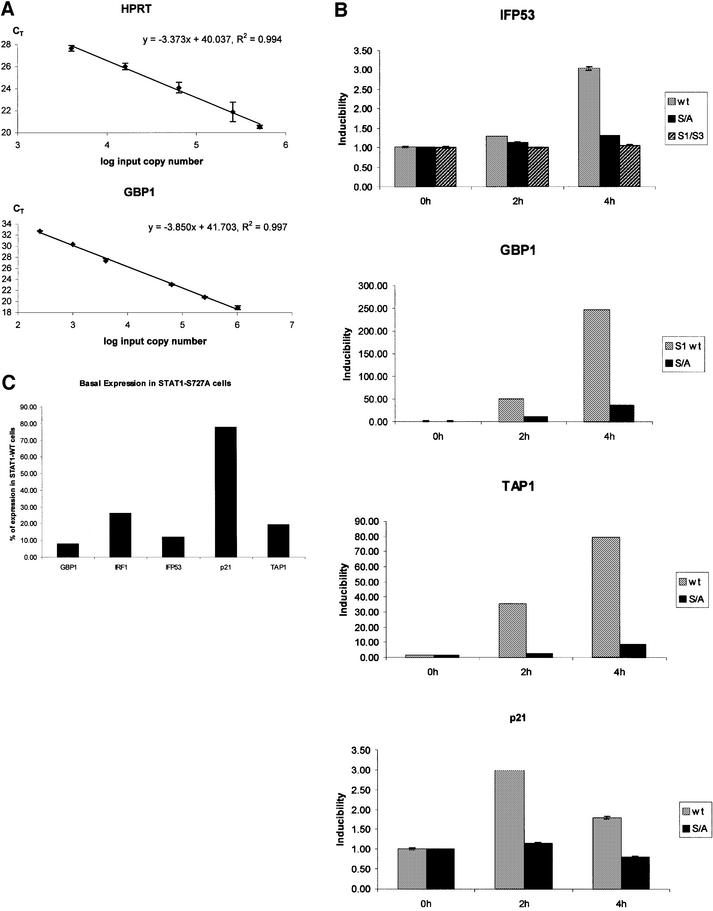
To investigate the effect of the S727A mutation on the GBP promoter beyond the sensitivity of the northern blot experiment, we employed real-time PCR of reverse-transcribed mRNA. Parameters were adjusted to obtain a linear relationship between input mRNA (cDNA) copy number and the number of PCR cycles required to reach a fixed threshold of DNA synthesis (CT value). In this range, the gene of interest can be normalized to the housekeeping HPRT gene product and a quantitative comparison of mRNA amounts in different situations is possible. Figure 4A shows the standard curves obtained for GBP and HPRT; however, similar standard curves were obtained for all genes analyzed below (data not shown).
Fig. 4. Real-time PCR analysis of reverse-transcribed IFN-γ-induced mRNAs. (A) Standard curves for HPRT and GBP1. The input cDNA templates (log input copy number) are plotted against the CT values. The HPRT standard curve was used to normalize the target gene expression for differences in the amount of cDNA added to each reaction. (B) mRNA inducibility of the indicated genes determined in fibroblasts reconstituted with STAT1-WT- (light gray), STAT1-S727A- (dark gray) or STAT1–STAT3 chimera-reconstituted fibroblasts (hatched, IFP53 only). mRNA quantities were determined in samples from untreated cells or after 2 and 4 h of IFN-γ treatment. Inducibility was calculated as the difference of HPRT-normalized CT values in control and IFN-γ-treated cells. (C) Quantitative comparison of the basal expression levels of the indicated genes in STAT1-WT- and STAT1-S727A-reconstituted cells. The numbers of mRNA molecules in each sample were calculated from HPRT-normalized standard curves and the percentage reduction caused by the S727A mutation was calculated.
Real-time PCR analysis of reverse-transcribed, IFN-γ-induced GBP1 mRNA confirmed the strong decrease caused by the lack of STAT1-S727 phosphorylation (Figure 4B). However, a second effect of S727A mutation became apparent: the severe reduction of basal GBP expression to 8% of the amount found in STAT1-WT cells (Figure 4C). Compared with STAT1-WT cells, the combined effect of reduced basal and induced expression caused an ∼85-fold decrease in the number of mRNA molecules found in IFN-γ-treated STAT1-S727A cells. This explains why the low residual GBP inducibility in these cells is below the detection threshold of a northern blot. By comparison, the reduction of basal IRF1 expression in STAT1-S727A cells was less pronounced (28% of wild type; Figure 4C) and a smaller reduction in inducibility was found (data not shown).
A number of IFN-γ-inducible genes not previously analyzed by northern blotting were investigated by real-time PCR (Figure 4B). In agreement with the reporter gene assay, induction of the endogenous IFP53 gene by IFN-γ was reduced by ∼50% and a similar reduction was found for the p21 cyclin-dependent kinase (CDK) inhibitor gene. In contrast, the reduction of IFN-γ-inducibility found for the transporter associated with the antigen presentation 1 (TAP1) gene was even stronger than that found for the GBP1 gene. Basal expression of the TAP1 and IFP53 genes was also strongly affected by the S727A mutation (Figure 4C), whereas very little effect was noted for the p21 gene. The comparison between IFP53 and p21 suggests that effects on basal and induced expression are not necessarily coupled.
The C-terminus determines the specific interaction of STAT1 with serine kinases
The identical position of a PMS727P motif in the C-terminus of STAT3 implies a similar role and regulation of S727 phosphorylation to those in STAT1. Since the PMSP sequence is a potential phosphorylation site for members of the MAPK family, we first addressed the question of if and how STAT1 and STAT3 serine phosphorylation is triggered by different stimuli and different signaling pathways. Further experiments investigated the structural basis of differential responsiveness.
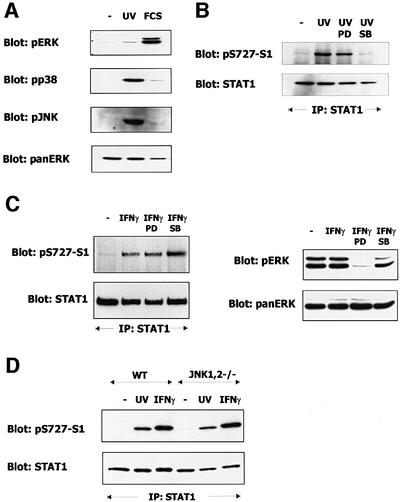
UV irradiation strongly activated c-Jun kinases (JNKs) and p38MAPK. Activation of ERK1/2 was very weak by comparison (Figure 5A). To rule out contributions of the ERK pathway to stress-induced STAT1-S727 phosphorylation, we irradiated the cells with UV light in the presence of PD98059. The MEK inhibitor did not affect STAT1 phosphorylation. In marked contrast, the p38MAPK inhibitor SB203580 completely inhibited the S727 kinase (Figure 5B). In accordance with our previous results, IFN-γ-induced STAT1 phosphorylation on S727 was not affected by either PD98059 or SB203580 (Figure 5C). 3T3 fibroblasts obtained from mice deficient in both JNKs (JNK1,2–/– cells) displayed no impairment in UV-induced STAT1 phosphorylation on S727 (Figure 5D). We conclude that at least in the case of UV irradiation, the STAT1 C-terminus is targeted specifically by the p38MAPK pathway.
Fig. 5. Contribution of MAPK pathways to stress-induced phosphorylation of STAT1 on S727. Where indicated, 3T3 mouse fibroblasts were exposed to UV light, treated with 10% FCS, or with IFN-γ. (A) Activation of MAPK pathways was determined in cell lysates with phosphospecific antibodies to ERK (pERK), JNK (pJNK) or p38MAPK (pp38). The loading control was performed with a monoclonal panERK antibody. (B) The contributions of ERKs and p38MAPK were assessed by adding the MEK inhibitor PD98059 (PD) or the p38MAPK inhibitor SB203580 (SB) 30 min before UV irradiation. STAT1 was precipitated from the cell extracts using a STAT1 C-terminal antiserum. Western blots of the precipitates were stained with phosphospecific antiserum for S727 (upper panel) and reprobed with a STAT1 C-terminal antiserum (lower panel). (C) The contributions of ERKs and p38MAPK to the IFN-γ-induced S727 phosphorylation of STAT1 were assessed as in (B). Aliquots of the cell extracts used for immunoprecipitation of STAT1 were analyzed directly by western blotting with pERK and pan-ERK antibodies to assess ERK activity and confirm equal loading, respectively. (D) The contribution of JNKs was analyzed in fibroblasts deficient for both JNK1 and JNK2. STAT1 was precipitated from control (WT) and JNK/JNK2–/– fibroblasts using STAT1 C-terminal antiserum. STAT1 phosphorylation on S727 (upper panel) and loading (lower panel) were determined with phosphospecific or STAT1 C-terminal antisera.
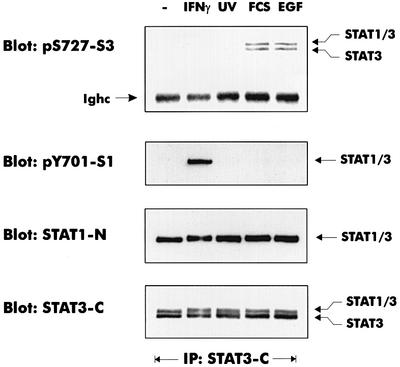
Sensitivity to MEK inhibition and other lines of evidence support the idea that growth factor receptors signal to STAT3 through the ERK pathway (Chung et al., 1997; Ng and Cantrell, 1997; Jain et al., 1998; Stephens et al., 1998). Virtually no phosphorylation of STAT1 on S727 was observed in response to epidermal growth factor (EGF) or serum treatment (Figure 6A), particularly if compared with UV or IFN-γ treatment. In an identical situation, S727 of STAT3 was phosphorylated marginally by IFN-γ treatment or UV irradiation, but strongly phosphorylated by treatment with EGF or fetal calf serum (FCS; Figure 6B). Phosphorylation by EGF was completely abrogated by simultaneous treatment with the MEK inhibitor (Figure 6C). Consistent with the findings of a previous report (Wright et al., 1999), neither STAT3 phosphorylation on S727 nor ERK activation was inhibited by PD98059 if stimulated by FCS (data not shown). Thus, STAT3, but not STAT1, responds to a growth factor-activated ERK1/2 pathway. In conclusion, the STAT1 C-terminus appears to be a good substrate for the p38MAPK pathway, but not the ERK pathway, whereas the opposite is true for STAT3.
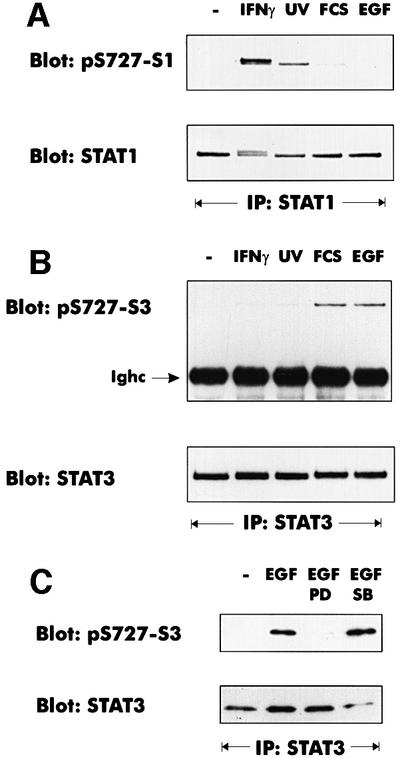
Fig. 6. Different signaling pathways cause S727 phosphorylation of STAT1 and STAT3. Where indicated, mouse 3T3 fibroblasts were treated with IFN-γ for 30 min, irradiated with UV light, or stimulated with 10% FCS (30 min) or EGF (100 ng/ml, 30 min). (A) STAT1 phosphorylation on S727 was analyzed with phosphospecific antiserum. Loading was controlled by reprobing with a monoclonal antibody to the STAT1 N-terminus. (B) STAT3 was precipitated from cellular lysates with an antiserum to the C-terminus. Phosphorylation of S727 was determined with a phosphoserine 727-specific antiserum (Ighc indicates the immunoglobulin heavy chain). The loading control was performed by reprobing the blot with the STAT3 C-terminal antiserum. (C) The pathway-specific inhibitors PD98059 (PD) and SB203580 (SB) were added 30 min prior to EGF treatment. STAT3 phosphorylation on S727 was determined as described for (B).
MAPK substrates contain docking domains that specify the interaction with a particular family member (Tanoue et al., 2000). The portion of the STATs conferring specificity for either a MAPK or a MAPK substrate kinase (MAPKAP) has not been determined. We changed C-terminal amino acids of STAT1 (amino acids 716–750) to those of STAT3 (amino acids 706–770). If the exchanged portion of the molecule contained a specificity element, the chimeric STAT molecule should acquire the substrate properties of STAT3.
STAT1-deficient fibroblasts were stably transfected with the STAT1–STAT3 chimera and clonal lines were selected. Phosphorylation of the chimeric molecule that displayed a slower SDS–PAGE mobility than the endogenous STAT3 protein was analyzed by immunoprecipitation using a STAT3 C-terminal antibody that precipitated both the STAT1–STAT3 chimera and the endogenous STAT3. Tyrosine phosphorylation of STAT1–STAT3 occurred in response to IFN-γ, confirming the structural integrity of the chimera and its ability to interact with the IFN-γ receptor (Figure 7). The IFN-γ-induced expression of the endogenous IFP53 gene was affected similarly to that in the case of the STAT1-S727A mutation (Figure 4B).
Fig. 7. The STAT3 C-terminus determines the link to MAPK pathways. Fibroblasts from STAT1-deficient mice were reconstituted with a STAT1–STAT3 chimera containing STAT3 C-terminal amino acids. Treatment with UV, IFN-γ, FCS or EGF was carried out as described for Figure 66. Endogenous STAT3 and the STAT1–STAT3 chimera were precipitated from cellular lysates using antiserum against the STAT3 C-terminus. Phosphorylation of both proteins on S727 was determined using an antiserum to the phosphoserine 727 of STAT3 (pS727-S3). The position of the immunoglobulin heavy chain is marked by Ighc. Tyrosine phosphorylation of STAT1–STAT3 was determined using an antiserum directed to phosphotyrosine 701 of STAT1 (indicated as pY701-S1). A monoclonal STAT1 N-terminal antibody (indicated as STAT1-N) was used to visualize the STAT1–STAT3 chimeric protein. A STAT3 C-terminal antiserum (indicated as STAT3-C) was employed to detect both the STAT1–STAT3 chimera and the endogenous STAT3.
Following UV irradiation, treatment with IFN-γ, FCS or EGF, S727 phosphorylation of the chimera now followed the pattern of endogenous STAT3 (Figure 7). Thus the specificity for connecting to MAPK pathways appears to reside in the C-terminus of the STATs.
Discussion
STAT serine phosphorylation is a layer of signaling specificity superimposed on the intrinsic specificity provided by the JAK–STAT paradigm. Here we provide evidence that the implications of STAT1 phosphorylation at S727 for cellular cytokine responses are determined at several distinct stages. First, cytokines such as IFN-γ that cause tyrosine phosphorylation can stimulate a p38MAPK-independent mode of serine phosphorylation, which requires phosphotyrosine binding to SH2 domains. Secondly, when S727 phosphorylation involves MAPK, the connection to individual MAPK family members is determined by C-terminal amino acids, which, at least in the case of STAT3, behave like an independent specificity module. Thirdly, while the effect of S727A mutation is generally to reduce transcription factor activity, striking differences were observed in the magnitude of reduction at individual target genes. Therefore, a specific effect on gene expression is generated by the dependence of individual STAT1 target genes on a phosphorylated S727.
Y701 and S727 phosphorylation are linked in IFN-γ signaling
The strict dependence of IFN-γ-mediated S727 phosphorylation on tyrosine phosphorylation is in line with the previously reported need for JAK2 or the PYK2 PTK (Zhu et al., 1997; Takaoka et al., 1999). In further agreement with our results, tyrosine phosphorylation of STAT1 precedes serine phosphorylation in the IFN-γ response of NIH 3T3 fibroblasts (Zhu et al., 1997). SH2 domain-based interactions involving STAT1 might be necessary for serine phosphorylation at different steps during signaling. The IFN-γ-responsive serine kinase could be part of the receptor complex and phosphorylate STAT1 only after it is bound to the IFNGR1 chain or to one of the JAKs, or the serine kinase may recognize only dimerized STATs. Binding of STAT1 to an unidentified molecular platform might also be necessary or, more simply, its recruitment by an SH2 domain-containing serine kinase. Partial purification of STAT1-S727 kinase produced an enzyme requiring the entire STAT1 for phosphorylation (Zhu et al., 1997). This mode of substrate recognition appears to contrast with that of the major MAPK.
In obvious contrast to our results, S727 phosphorylation of STAT1 by IFN-γ was found to be independent of Y701 phosphorylation in U3A cells. U3A is a human fibrosarcoma line selected for deficiency in IFN signaling after mutagenesis with a frameshift mutagen (Pellegrini et al., 1989; Muller et al., 1993). Thus, there are three major differences from the fibroblasts used in our studies: they are transformed, have been mutagenized and originate from human, not mouse. Wen et al. (1995) reported transfected STAT1α to be constitutively phosphorylated on S727 in U3A cells, even after serum deprivation. Perhaps a mutation in the U3A DNA deregulates a signaling pathway (p38MAPK?), causing phosphorylation of STAT1 on S727 in the absence of tyrosine phosphorylation. The activity of the deregulated kinase might obscure the need for phosphotyrosine-based interactions, as they would occur if phosphorylation involved the proper IFN-γ-stimulated serine kinase. According to our results, the requirement for the JAK2 kinase may derive entirely from the need for tyrosine-phosphorylated STAT1. The role of JAK2 was established in a JAK2-deficient sibling cell line of U3A, γ2A. If our reasoning were correct, the deregulated kinase activity of U3A would not be found in γ2A cells.
The human and mouse origins could also provide an explanation for the differences observed between U3A cells and mouse fibroblasts. In support of this assumption, species differences were noted recently in the activation of STAT4 by type I IFN (Farrar et al., 2000). Moreover, the role of the TYK2 kinase in IFN-α signaling is indispensable in the case of human TYK2-deficient cells, but ancillary in cells from TYK2-deficient mice (Velazquez et al., 1992; Karaghiosoff et al., 2000). Both findings point out major differences in the deployment of JAKs and STATs in mice and men.
Finally, one must bear in mind that the scenario depicted above does not include the PYK2 PTK or PKR. An involvement of these kinases stresses the possibility that the IFN-γ-stimulated signaling network, hence a potential necessity for SH2 domains, extends beyond the JAK–STAT pathway.
STAT1 serine phosphorylation—the MAPK connection
Signaling through MAPK pathways downstream of G-proteins generally proceeds without SH2 domain-based protein interactions (Whitmarsh and Davis, 1998; Burack and Shaw, 2000). During stress responses that do not activate JAKs, but strongly activate MAPK, STAT1 is linked to p38MAPK. However, when IFN induces tyrosine phosphorylation, S727 phosphorylation is not linked to the classical MAPK pathways, even though these pathways may be active, as recently observed for IFN-α (Uddin et al., 1999, 2000).
A role for p38MAPK was postulated recently for STAT1 serine phosphorylation in IFN-γ-treated HeLa cells (Goh et al., 1999). However, our studies and those performed in other laboratories show that most cell types do not employ p38MAPK either in type I or type II IFN-mediated S727 phosphorylation (Kovarik et al., 1999; Uddin et al., 2000). Using JNK-deficient cells, we now provide conclusive evidence that JNKs are not required for stress- or IFN-γ-induced STAT1 phosphorylation on S727. Likewise, and despite some previous support for their potential engagement (David et al., 1995; Wen et al., 1995; Takaoka et al., 1999), a major role for ERKs 1 and 2 in STAT1 phosphorylation can be ruled out. First, MEK inhibition had no influence on stress-induced S727 phosphorylation. Secondly, Zhu et al. (1997) demonstrated that forced ERK activation or disruption of the ERK pathway does not affect the phosphorylation of STAT1. Thirdly, IFN-γ has very limited potential to stimulate ERK, JNK or p38MAPK activation (Kovarik et al., 1998, 1999). The low level of ERK activation by IFN-γ is not sufficient for cross-talk to STATs because phosphorylation of STAT3, a good ERK substrate in vitro and in vivo, was barely detectable upon stimulation with IFN-γ. More importantly, transplanting the STAT3 C-terminus to STAT1 generated a connection to ERKs in growth factor responses, but not in the IFN-γ response. In conclusion, p38MAPK is the only member of the family with a clear link to STAT1, and this link appears to be disconnected once Y701 is phosphorylated.
The striking preferences of the STAT1 and STAT3 C-termini for different MAPK family members are in agreement with current ideas about the cell biology of individual STATs and MAPK family members. STAT1 is linked to the p38MAPK pathway, hence a classical inflammation or stress response, which is likely to occur during TH1-dominated immune responses. In contrast, STAT3 is connected to growth factor responses through the ERK pathway. Surely this simple concept will not apply to all STAT-mediated cellular responses. However, as a rule, the C-terminal amino acids may support the specific biological function of STATs through their influence on serine phosphorylation.
Promoter-specific requirement for STAT1 serine phosphorylation
The results obtained with STAT1-S727A cells confirm that S727 phosphorylation stimulates the function of the transactivating domain. Fairly little is known about the mechanism of the stimulatory activity. Two proteins were recently reported to associate preferentially with S727-phosphorylated STAT1. MCM5, a protein important for DNA replication, causes a fluctuation of STAT1 transcriptional activity during the cell cycle (Zhang et al., 1998). The tumor suppressor BRCA1 enhanced transcription of the p21WAF gene in response to IFN-γ, but had no effect on the induced expression of the IRF1 gene (Ouchi et al., 2000).
Northern blotting and real-time PCR experiments revealed that the STAT1-S727A mutation differentially affects the investigated STAT1 target genes. Inducibility by IFN-γ was significantly affected (∼50% reduction) in the case of the IRF1, IFP53 and p21 genes. These genes are bona fide targets specifically for the STAT1 dimer (Strehlow et al., 1993; Pine et al., 1994; Chin et al., 1996). An 80% reduction in IRF1 gene expression had been noted under similar conditions in U3A cells expressing STAT1-S727A (Wen et al., 1995).
The inducibility of the GBP1 and TAP1 genes by IFN-γ was reduced dramatically to <15% of that found with STAT1-WT. The murine GBP1 promoter contains both a GAS and an ISRE, and the IFN-γ-induced GBP1 gene expression essentially depends on the association of IRF1 with the ISRE (Briken et al., 1995). Similarly, the bidirectional promoter rendering the neighboring TAP1 and LMP2 genes responsive to IFN-γ contains binding sites for both STAT1 and IRF1 (Wright et al., 1995; White et al., 1996; Cramer et al., 2000). The comparatively weak reduction of IRF1 mRNA expression caused by the STAT1-S727A mutation argues against an indirect effect of the mutation on the GBP1 and TAP1 promoters through IRF1. While this cannot be ruled out formally, we favor as one possible explanation that serine phosphorylation might be particularly important when the STAT1 dimer needs to interact with IRF1 for an efficient transcriptional response.
The GAS of the human GBP1 promoter binds a STAT1 dimer associated with the IRF9 (p48) protein (Seegert et al., 1994). The same STAT1–IRF9 complex has also been found to provide for the IFN-γ responses of other genes (Bluyssen et al., 1995; Majumder et al., 1998). Therefore, a second possibility would be that STAT1-S727 phosphorylation is particularly important for inducibility in the context of a STAT1–IRF9 complex.
Surprisingly, the absence of STAT1-S727 phosphorylation affected the basal expression of the GBP1, TAP1 and IFP53 genes and to a lesser extent the IRF1 gene. Again, published results offer at least two potential explanations for this finding. It has been shown that the absence of STAT1 or STAT2 reduces basal expression of IFN-inducible genes (Lee et al., 1999; Park et al., 2000). This effect can be explained at least in part by the autocrine secretion of type I IFN, because a similar reduction in IFN-inducible gene expression is also seen in the absence of the type I IFN receptor. The permanent expression of genes with GAS or ISRE promoter sequences due to autocrine IFN might depend on phosphorylated S727. An alternative explanation for some IFN-inducible genes was suggested recently by Chatterjee-Kishore et al. (2000). This report showed a contribution of non-tyrosine-phosphorylated STAT1 to the constitutive expression of the LMP2 gene, regulated by the bidirectional TAP1/LMP2 promoter. An interaction with IRF1 was important for this constitutive activity of STAT1, thus providing a potential explanation for the strong effect of the S727A mutation on genes containing GAS elements and IRF1-binding sites. In this respect, our analysis would suggest that as in the case of induced GBP1 or TAP1 induction, the ‘constitutive’ STAT1–IRF1 interaction requires serine phosphorylation. This is in agreement with an earlier report by Stark’s group (Kumar et al., 1997).
The differential requirement for S727 phosphorylation might also be explained at the level of association with gene-specific coactivators or chromatin components. The BRCA1 study would be in agreement with this assumption. Thus, while our results support a role for phospho-S727 in promoting the selective transcription of subgroups of STAT1 target genes, it must be the goal of future studies to provide a mechanistic explanation for this finding.
Materials and methods
Cells, cytokines and drugs
3T3 fibroblasts from wild-type mice, STAT1-deficient mice (Durbin et al., 1996), or from JNK1/JNK2–/– embryos (a kind gift from Erwin Wagner, Institute of Molecular Pathology, Vienna) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FCS. Prior to treatment, cells were starved for 16 h in DMEM without FCS. IFN-γ (kindly provided by G.Adolf, Boehringer Ingelheim, Vienna) was used at a concentration of 5 ng/ml for the periods indicated in the figure legends. UV irradiation was with UVC (254 nm, 40 J/m2). Cells were processed for further experimentation 20–30 min after irradiation. EGF was purchased from Calbiochem and used at a concentration of 100 ng/ml. The p38MAPK inhibitor SB203580, kindly provided by Ken Murray (Smith-Kline Beecham), was used at 5 µM. The MEK inhibitor PD98059 was purchased from Calbiochem and used at 50 µM. Both drugs were added 30 min before further treatment.
Plasmids
An expression vector was created by first replacing the cytomegalovirus (CMV) promoter of RcCMV (Invitrogen) with the eIF1α promoter of pEFCX, a pEFBos derivative (Mizushima and Nagata, 1990). Parts of the resulting plasmid, pEF/Rc, were combined with a portion of pcDNA3.1Zeo (Invitrogen) to derive an expression plasmid, pEF-Zeo, containing the eIF1α promoter and a zeocin resistance gene. STAT1-WT cDNA was kindly provided by Chris Schindler (Columbia University, New York), S727A- and Y701-mutated cDNAs were provided by Jim Darnell (Rockefeller Univerity, New York) and R602K-mutated cDNA was obtained from John Krolewski (UC Irvine, Irvine, CA). A chimeric STAT1–STAT3 cDNA was made by substituting the DNA encoding the STAT1 C-terminus beyond amino acid 716 with a PCR-generated fragment encoding STAT3 amino acids 708–770. Correct recombination was confirmed by DNA sequencing. All cDNAs were inserted into pEF-Zeo. The IFP-Luc plasmid containing the IFP53 promoter (Strehlow et al., 1993) fused to the luciferase gene was generated by inserting a 192 bp promoter fragment including the GAS into pGL2basic (Promega).
Cell transfection and selection of transfected clones
A total of 107 STAT1-deficient fibroblasts were transfected using 2 µg of plasmid DNA and Effectene reagent according to the manufacturer’s instructions (Qiagen, Hilden, Germany). Transfected cells were selected in 100 µg/ml zeocin (Invitrogen, Carlsbad, CA). Bulk cultures of zeocin-resistant cells were cloned by limiting dilution in microtiter wells. Individual cell clones were analyzed for STAT1 expression by immunofluorescence with a monoclonal antibody recognizing the STAT1 N-terminus, and by western blotting.
Antibodies
Antisera to the STAT1 C-terminus and to phospho-S727-STAT1 have been described recently (Kovarik et al., 1998). Rabbit antiserum to Y701-phosphorylated STAT1 was purchased from New England Biolabs (Beverly, MA) and used at a dilution of 1:1000. A monoclonal antibody recognizing the STAT1 N-terminus was purchased from Transduction Laboratories (Lexington, KY). Antibodies against the STAT3 C-terminus or the S727-phosphorylated STAT3 C-terminus were obtained, respectively, from Santa Cruz and New England Biolabs. Phosphospecific antibodies against ERKs, p38MAPK and JNKs were bought from New England Biolabs and used for western blotting at dilutions of 1:1000. Monoclonal antibodies recognizing ERKs (panERK) were purchased from Transduction Laboratories (Lexington, KY) and used at a dilution of 1:2000 in western blotting.
Immunoprecipitation and western blotting
A protocol for these procedures has been described recently (Kovarik et al., 1998). Antibodies were used as indicated above or in the figure legends.
Northern blotting
Total RNA was isolated from fibroblasts using the RNeasy reagent kit and the manufacturer’s instructions (Quiagen, Hilden, Germany). A 15 µg aliquot of total RNA was separated on agarose gels and blotted to membranes using standard procedures. The blots were probed with cDNA, labeled with 32P by the random-prime method (Roche, Vienna, Austria). The cDNAs used for labeling encoded the following mouse genes: IRF1 (Miyamoto et al., 1988), GBP1 (Cheng et al., 1991), SOCS1/JAB and SOCS3/CIS3 (Suzuki et al., 1998).
Quantitation of gene expression using real-time PCR
The cDNAs used in the real-time PCR assays were reverse transcribed from 5 µg of total RNA using the Oligo(dT)12–18 Primer and the SuperScript Preamplification System for first strand cDNA synthesis (Life Technologies) according to the manufacturer’s recommendations. Real-time PCR was performed on an ABI PRISMTM7700 Sequence Detection System (Applied Biosystems). Detailed information about real-time PCR conditions, primers and quantification (Heid et al., 1996) is provided as Supplementary data.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
Help by Andreas Pilz in the identification of fibroblast clones expressing mutant STAT1 is gratefully acknowledged. We thank Erwin Wagner for JNK-deficient fibroblasts. Jim Darnell, Chris Schindler and John Krolewski kindly provided STAT cDNAs. Manuela Baccarini is thanked for reading and valuable comments on the manuscript.
REFERENCES
- Akira S. (2000) Roles of STAT3 defined by tissue-specific gene targeting. Oncogene, 19, 2607–2611. [DOI] [PubMed] [Google Scholar]
- Barahmand-Pour F., Meinke,A., Groner,B. and Decker,T. (1998) Jak2–Stat5 interactions analyzed in yeast. J. Biol. Chem., 273, 12567–12575. [DOI] [PubMed] [Google Scholar]
- Beuvink I., Hess,D., Flotow,H., Hofsteenge,J., Groner,B. and Hynes,N.E. (2000) Stat5a serine phosphorylation. Serine 779 is constitutively phosphorylated in the mammary gland and serine 725 phosphorylation influences prolactin-stimulated in vitro DNA binding activity. J. Biol. Chem., 275, 10247–10255. [DOI] [PubMed] [Google Scholar]
- Bluyssen H.A., Muzaffar,R., Vlieststra,R.J., van der Made,A.C., Leung,S., Stark,G.R., Kerr,I.M., Trapman,J. and Levy,D.E. (1995) Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc. Natl Acad. Sci. USA, 92, 5645–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briken V., Ruffner,H., Schultz,U., Schwarz,A., Reis,L.F., Strehlow,I., Decker,T. and Staeheli,P. (1995) Interferon regulatory factor 1 is required for mouse Gbp gene activation by γ interferon. Mol. Cell. Biol., 15, 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., Kohlhuber,F. and Muller,M. (1997) JAKs and STATs get connected. Trends Cell Biol., 6, 336–340. [DOI] [PubMed] [Google Scholar]
- Bromberg J. and Darnell,J.E.,Jr (2000) The role of STATs in transcriptional control and their impact on cellular function. Oncogene, 19, 2468–2473. [DOI] [PubMed] [Google Scholar]
- Bromberg J.F., Horvath,C.M., Wen,Z., Schreiber,R.D. and Darnell,J.E.,Jr (1996) Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon α and interferon γ. Proc. Natl Acad. Sci. USA, 93, 7673–7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack W.R. and Shaw,A.S. (2000) Signal transduction: hanging on a scaffold. Curr. Opin. Cell Biol., 12, 211–216. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M., Wright,K.L., Ting,J.P. and Stark,G.R. (2000) How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J., 19, 4111–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.S., Patterson,C.E. and Staeheli,P. (1991) Interferon-induced guanylate-binding proteins lack an N(T)KXD consensus motif and bind GMP in addition to GDP and GTP. Mol. Cell. Biol., 11, 4717–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin Y.E., Kitagawa,M., Su,W.C., You,Z.H., Iwamoto,Y. and Fu,X.Y. (1996) Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science, 272, 719–722. [DOI] [PubMed] [Google Scholar]
- Chung J., Uchida,E., Grammer,T.C. and Blenis,J. (1997) Stat3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol. Cell. Biol., 17, 6508–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer L.A., Nelson,S.L. and Klemsz,M.J. (2000) Synergistic induction of the Tap-1 gene by IFN-γ and lipopolysaccharide in macrophages is regulated by STAT1. J. Immunol., 165, 3190–3197. [DOI] [PubMed] [Google Scholar]
- Darnell J.E. Jr, Kerr,I.M. and Stark,G.R. (1994) Jak–STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science, 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- David M., Petricoin,E.,III, Benjamin,C., Pine,R., Weber,M.J. and Larner,A.C. (1995) Requirement for MAP kinase (ERK2) activity in interferon α- and interferon β-stimulated gene expression through STAT proteins. Science, 269, 1721–1723. [DOI] [PubMed] [Google Scholar]
- Decker T. and Kovarik,P. (1999) Transcription factor activity of STAT proteins: structural requirements and regulation by phosphorylation and interacting proteins. Cell. Mol. Life Sci., 55, 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T. and Kovarik,P. (2000) Serine phosphorylation of STATs. Oncogene, 19, 2628–2637. [DOI] [PubMed] [Google Scholar]
- Decker T., Kovarik,P. and Meinke,A. (1997) GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res., 17, 121–134. [DOI] [PubMed] [Google Scholar]
- Durbin J.E., Hackenmiller,R., Simon,M.C. and Levy,D.E. (1996) Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell, 84, 443–450. [DOI] [PubMed] [Google Scholar]
- Farrar D.J., Smith,J.D., Murphy,T.L., Leung,S., Stark,G.R. and Murphy,K.M. (2000) Selective loss of type I interferon-induced STAT4 activation caused by a minisatellite insertion in mouse Stat2. Nature Immunol., 1, 65–69. [DOI] [PubMed] [Google Scholar]
- Goh K.C., Haque,S.J. and Williams,B.R.G. (1999) p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J., 18, 5601–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Yan,H., Wong,L.H., Ralph,S., Krolewski,J. and Schindler,C. (1996) The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-α signals. EMBO J., 15, 1075–1084. [PMC free article] [PubMed] [Google Scholar]
- Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- Hilton D.J. (1999) Negative regulators of cytokine signal transduction. Cell. Mol. Life Sci., 55, 1568–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C.M. and Darnell,J.E.,Jr (1996) The antiviral state induced by α interferon and γ interferon requires transcriptionally active Stat1 protein. J. Virol., 70, 647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N., Zhang,T., Fong,S.L., Lim,C.P. and Cao,X. (1998) Repression of Stat3 activity by activation of mitogen-activated protein kinase (MAPK). Oncogene, 17, 3157–3167. [DOI] [PubMed] [Google Scholar]
- Karaghiosoff M. et al. (2000) Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity, 13, 549–560. [DOI] [PubMed] [Google Scholar]
- Kovarik P., Stoiber,D., Novy,M. and Decker,T. (1998) Stat1 combines signals derived from IFN-γ and LPS receptors during macrophage activation. EMBO J., 17, 3660–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik P., Stoiber,D., Eyers,P.A., Menghini,R., Neininger,A., Gaestel,M., Cohen,P. and Decker,T. (1999) Stress-induced phosphorylation of STAT1 at Ser727 requires p38 mitogen-activated protein kinase whereas IFN-γ uses a different signaling pathway. Proc. Natl Acad. Sci. USA, 96, 13956–13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Commane,M., Flickinger,T.W., Horvath,C.M. and Stark,G.R. (1997) Defective TNF-α-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science, 278, 1630–1632. [DOI] [PubMed] [Google Scholar]
- Lee C.K., Gimeno,R. and Levy,D.E. (1999) Differential regulation of constitutive major histocompatibility complex class I expression in T and B lymphocytes. J. Exp. Med., 190, 1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E. (1999) Physiological significance of STAT proteins: investigations through gene disruption in vivo. Cell. Mol. Life Sci., 55, 1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E., Kessler,D.S., Pine,R., Reich,N. and Darnell,J.E.,Jr (1988) Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev., 2, 383–393. [DOI] [PubMed] [Google Scholar]
- Majumder S., Zhou,L.Z., Chaturvedi,P., Babcock,G., Aras,S. and Ransohoff,R.M. (1998) p48/STAT-1α-containing complexes play a predominant role in induction of IFN-γ-inducible protein, 10 kDa (IP-10) by IFN-γ alone or in synergy with TNF-α. J. Immunol., 161, 4736–4744. [PubMed] [Google Scholar]
- Marie I., Durbin,J.E. and Levy,D.E. (1998) Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J., 17, 6660–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto M., Fujita,T., Kimura,Y., Maruyama,M., Harada,H., Sudo,Y., Miyata,T. and Taniguchi,T. (1988) Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell, 54, 903–913. [DOI] [PubMed] [Google Scholar]
- Mizushima S. and Nagata,S. (1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res., 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Laxton,C., Briscoe,J., Schindler,C., Improta,T., Darnell,J.E.,Jr, Stark,G.R. and Kerr,I.M. (1993) Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J., 12, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J. and Cantrell,D. (1997) STAT3 is a serine kinase target in T lymphocytes. Interleukin 2 and T cell antigen receptor signals converge upon serine 727. J. Biol. Chem., 272, 24542–24549. [DOI] [PubMed] [Google Scholar]
- Ouchi T., Lee,S.W., Ouchi,M., Aaronson,S.A. and Horvath,C.M. (2000) Collaboration of signal transducer and activator of transcription 1 (STAT1) and BRCA1 in differential regulation of IFN-γ target genes. Proc. Natl Acad. Sci. USA, 97, 5208–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Li,S., Cha,E. and Schindler,C. (2000) Immune responses in Stat2 knock-out mice. Immunity, in press. [DOI] [PubMed] [Google Scholar]
- Pellegrini S., John,J., Shearer,M., Kerr,I.M. and Stark,G.R. (1989) Use of a selectable marker regulated by α interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol., 9, 4605–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine R., Canova,A. and Schindler,C. (1994) Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN α and IFN γ and is likely to autoregulate the p91 gene. EMBO J., 13, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana C.V., Chatterjee-Kishore,M., Nguyen,H. and Stark,G.R. (2000a) Complex roles of Stat1 in regulating gene expression. Oncogene, 19, 2619–2627. [DOI] [PubMed] [Google Scholar]
- Ramana C.V., Grammatikakis,N., Chernov,M., Nguyen,H., Goh,K.C., Williams,B.R. and Stark,G.R. (2000b) Regulation of c-myc expression by IFN-γ through Stat1-dependent and -independent pathways. EMBO J., 19, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C. and Darnell,J.E.,Jr (1995) Transcriptional responses to polypeptide ligands: the JAK–STAT pathway. Annu. Rev. Biochem., 64, 621–651. [DOI] [PubMed] [Google Scholar]
- Schuringa J.-J., Jonk,L.J.C., Dokter,W.H.A., Vellenga,E. and Kruijer,W. (2000) IL-6-induced STAT3 transactivation and serine 727 phosphorylation involves Vav, Rac-1 and SEK-1/MKK-4 as signal transduction components. Biochem. J., 347, 89–96. [PMC free article] [PubMed] [Google Scholar]
- Seegert D., Strehlow,I., Klose,B., Levy,D.E., Schindler,C. and Decker,T. (1994) A novel interferon-α-regulated, DNA-binding protein participates in the regulation of the IFP53/tryptophanyl-tRNA synthetase gene. J. Biol. Chem., 269, 8590–8595. [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Stephens J.M., Lumpkin,S.J. and Fishman,J.B. (1998) Activation of signal transducers and activators of transcription 1 and 3 by leukemia inhibitory factor, oncostatin-M and interferon-γ in adipocytes. J. Biol. Chem., 273, 31408–31416. [DOI] [PubMed] [Google Scholar]
- Stoiber D., Kovarik,P., Cohney,S., Johnston,J.A., Steinlein,P. and Decker,T. (1999) Lipopolysaccharide induces in macrophages the synthesis of the suppressor of cytokine signaling 3 and suppresses signal transduction in response to the activating factor IFN-γ. J. Immunol., 163, 2640–2647. [PubMed] [Google Scholar]
- Strehlow I., Seegert,D., Frick,C., Bange,F.C., Schindler,C., Bottger,E.C. and Decker,T. (1993) The gene encoding IFP 53/tryptophanyl-tRNA synthetase is regulated by the γ-interferon activation factor. J. Biol. Chem., 268, 16590–16595. [PubMed] [Google Scholar]
- Suzuki R. et al. (1998) CIS3 and JAB have different regulatory roles in interleukin-6 mediated differentiation and STAT3 activation in M1 leukemia cells. Oncogene, 17, 2271–2278. [DOI] [PubMed] [Google Scholar]
- Takaoka A. et al. (1999) Protein tyrosine kinase Pyk2 mediates the Jak-dependent activation of MAPK and Stat1 in IFN-γ, but not IFN-α, signaling. EMBO J., 18, 2480–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T., Adachi,M., Moriguchi,T. and Nishida,E. (2000) A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nature Cell Biol., 2, 110–116. [DOI] [PubMed] [Google Scholar]
- Uddin S., Majchrzak,B., Woodson,J., Arunkumar,P., Alsayed,Y., Pine,R., Young,P.R., Fish,E.N. and Platanias,L.C. (1999) Activation of the p38 mitogen-activated protein kinase by type I interferons. J. Biol. Chem., 274, 30127–30131. [DOI] [PubMed] [Google Scholar]
- Uddin S., Lekmine,F., Sharma,N., Majchrzak,B., Mayer,I., Young,P.R., Bokoch,G.M., Fish,E.N. and Platanias,L.C. (2000) The Rac1/p38 Map kinase pathway is required for IFNα-dependent transcriptional activation but not serine phosphorylation of Stat-proteins. J. Biol. Chem., 275, 27634–27640. [DOI] [PubMed] [Google Scholar]
- Velazquez L., Fellous,M., Stark,G.R. and Pellegrini,S. (1992) A protein tyrosine kinase in the interferon α/β signaling pathway. Cell, 70, 313–322. [DOI] [PubMed] [Google Scholar]
- Visconti R., Gadina,M., Chiariello,M., Chen,E.H., Stancato,L.F., Gutkind,J.S. and O’Shea,J.J. (2000) Importance of the MKK6/p38 pathway for interleukin-12-induced STAT4 serine phosphorylation and transcriptional activity. Blood, 96, 1844–1852. [PubMed] [Google Scholar]
- Wen Z., Zhong,Z. and Darnell,J.E.,Jr (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell, 82, 241–250. [DOI] [PubMed] [Google Scholar]
- White L.C., Wright,K.L., Felix,N.J., Ruffner,H., Reis,L.F., Pine,R. and Ting,J.P. (1996) Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1–/– mice. Immunity, 5, 365–376. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J. and Davis,R.J. (1998) Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci., 23, 481–485. [DOI] [PubMed] [Google Scholar]
- Wright J.H., Munar,E., Jameson,D.R., Andreassen,P.R., Margolis,R.L., Seger,R. and Krebs,E.G. (1999) Mitogen-activated protein kinase kinase activity is required for the G(2)/M transition of the cell cycle in mammalian fibroblasts. Proc. Natl Acad. Sci. USA, 96, 11335–11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K.L., White,L.C., Kelly,A., Beck,S., Trowsdale,J. and Ting,J.P. (1995) Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med., 181, 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J., Zhao,Y., Chait,B.T., Lathem,W.W., Ritzi,M., Knippers,R. and Darnell,J.E.,Jr (1998) Ser727-dependent recruitment of MCM5 by Stat1α in IFN-γ-induced transcriptional activation. EMBO J., 17, 6963–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.J., Wen,Z.L., Xu,L.Z. and Darnell,J.E. (1997) Stat1 serine phosphorylation occurs independently of tyrosine phosphorylation and requires an activated Jak2 kinase. Mol. Cell. Biol., 17, 6618–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]