Abstract
A clathrin homolog encoded on human chromosome 22 (CHC22) displays distinct biochemistry, distribution and function compared with conventional clathrin heavy chain (CHC17), encoded on chromosome 17. CHC22 protein is upregulated during myoblast differentiation into myotubes and is expressed at high levels in muscle and at low levels in non-muscle cells, relative to CHC17. The trimeric CHC22 protein does not interact with clathrin heavy chain subunits nor bind significantly to clathrin light chains. CHC22 associates with the AP1 and AP3 adaptor complexes but not with AP2. In non-muscle cells, CHC22 localizes to perinuclear vesicular structures, the majority of which are not clathrin coated. Treatments that disrupt the actin–myosin cytoskeleton or affect sorting in the trans-Golgi network (TGN) cause CHC22 redistribution. Overexpression of a subdomain of CHC22 induces altered distribution of TGN markers. Together these results implicate CHC22 in TGN membrane traffic involving the cytoskeleton.
Keywords: clathrin/cytoskeleton/homolog/muscle/trans-Golgi network
Introduction
In multicellular organisms, different cell types utilize specialized pathways of intracellular membrane traffic to facilitate a specific physiological function. Such pathways are presumably mediated or enhanced by various tissue-specific membrane vesicle coat proteins that have been identified. For example, clathrin-coated vesicles (CCVs), which control receptor-mediated endocytosis and organelle biogenesis in eukaryotic cells (Schmid, 1997), have several neuron-specific polymorphic components, including clathrin light chains and adaptor subunits (Pley and Parham, 1993). However, a clear definition of functional differences between most of the coat protein tissue isoforms remains to be established, and it is possible that some isoforms, expressed at high levels in differentiated cells, carry out low level specialized functions in other cells. The recent discovery of a second clathrin heavy chain gene in humans, with elevated levels of mRNA in muscle, was the first suggestion of tissue polymorphism for this highly conserved CCV component (Gong et al., 1996; Kedra et al., 1996; Lindsay et al., 1996; Long et al., 1996; Sirotkin et al., 1996). The novel clathrin heavy chain gene (CHC22) is encoded on human chromosome 22 in a region whose deletion is associated with velo-cardio-facial syndrome (VCFS) and/or DiGeorge syndrome (DGS) (Ryan et al., 1997). The predicted CHC22 polypeptide is composed of 1640 amino acids and shares 85% sequence identity with the conventional clathrin heavy chain (CHC17, 1675 residues) encoded on human chromosome 17 (Dodge et al., 1991). Although RNA transcripts of CHC22 were found to be predominantly expressed in skeletal muscle, they were also detected to a lesser extent in heart muscle and testis and at low levels in a variety of tissues. We report here the identification and characterization of the protein product of CHC22 and demonstrate that it is structurally related to ubiquitous clathrin but displays critical biochemical differences in spite of their high sequence homology. Studies described here implicate CHC22 in muscle-specific functions and in cytoskeleton-associated membrane traffic in the Golgi/TGN (trans-Golgi network) region of non-muscle cells.
The basic structural unit of CCVs is the clathrin triskelion, comprising three 192 kDa heavy chains (CHC17) trimerized near their C-terminus, each of which binds a light chain subunit. In mammals, birds and amphibians, there are two types of light chain, LCa and LCb, which compete with each other for heavy chain binding (Pley and Parham, 1993). Both light chains have a negative regulatory role for clathrin assembly and ensure that cellular clathrin polymerization depends on interaction with adaptor molecules, which interact with cellular membranes and determine what specific cargo will be recruited to CCVs (Ungewickell and Ungewickell, 1991; Liu et al., 1995; reviewed by Hirst and Robinson, 1998). Adaptors are heterotetrameric complexes and the most well-characterized, AP2 and AP1, are responsible for the formation of CCVs on the plasma membrane and TGN, respectively. There are two additional adaptor-related complexes, AP3 and AP4, with a molecular composition similar to AP1 and AP2. However, AP4 lacks clathrin-binding sequences (Dell’Angelica et al., 1999; Hirst et al., 1999) and, although AP3 has been shown to bind clathrin, it can function independently (Cowles et al., 1997; Simpson et al., 1997; Dell’Angelica et al., 1998). The protein domains involved in CHC17 trimerization, light chain binding and adaptor binding are well defined, but the role of individual residues in determining each of these properties has not yet been established. Given that the 15% sequence differences between CHC22 and CHC17 are evenly distributed, it is not possible to determine from sequence analysis whether CHC22 should retain any of these properties. These issues were addressed by biochemical analysis in this study and it was established that CHC22 forms a trimer but does not associate significantly with known clathrin light chains or co-polymerize with CHC17. CHC22 associates only with a subset of adaptor molecules, AP1 and AP3, but not AP2. Consistent with these findings, CHC22 localizes to perinuclear vesicles in non-muscle cells, which are primarily devoid of conventional clathrin.
The potential interaction of CHC22 with elements of the actin cytoskeleton was also addressed in this study, because of the high expression of CHC22 mRNA in skeletal muscle. There are numerous indications of a role for the microfilament network in both the endocytic and secretory pathways (reviewed by Stow and Heimann, 1998; Allan and Schroer, 1999). Endocytosis in yeast is coupled to actin assembly (reviewed by Wendland et al., 1998). Whether actin assembly is absolutely required for clathrin-mediated endocytosis in mammalian cells is still under investigation (Fujimoto et al., 2000), but some data indicate that microfilaments provide the force needed for endocytic vesicle movement and a tethering point for vesicle formation (Gaidarov et al., 1999; Merrifield et al., 1999; Taunton et al., 2000). Several actin-binding proteins are associated with the Golgi membrane, including spectrin, comitin and certain myosins (reviewed by Holleran and Holzbaur, 1998). Non-muscle myosin II has been shown to participate in forming TGN-derived transport vesicles (Ikonen et al., 1997; Müsch et al., 1997), and myosin VI has been localized to the Golgi complex and the leading edge of fibroblasts (Buss et al., 1998). Here we establish that the CHC22 protein is increased in expression during myoblast differentiation, and redistributes with myosin II in non-muscle cells upon treatment of cells with agents that disrupt the actin cytoskeleton. Expression of a fragment of CHC22 in non-muscle cells caused a change in localization of TGN markers, demonstrating a role for CHC22 in TGN membrane traffic, potentially through its interaction with components of the actin cytoskeleton.
Results
Induction of CHC22 during myogenesis
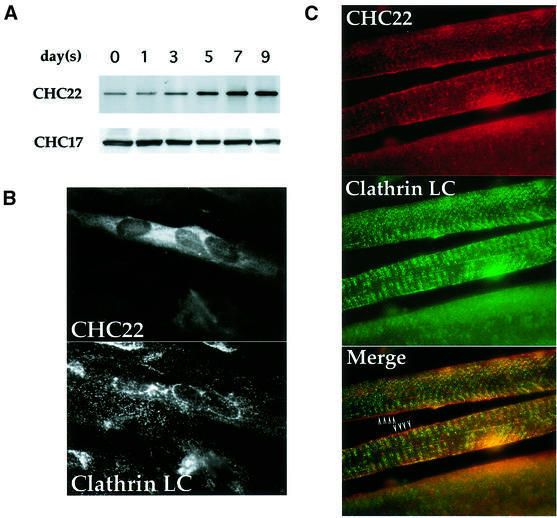
The mRNA for CHC22 was reported to be predominantly expressed in human skeletal muscle. To establish whether this mRNA encodes a viable gene product and whether CHC22 protein expression mirrored its mRNA distribution, two specific antibody reagents recognizing CHC22 were developed. The C-terminal domain of CHC22 (residues 1521–1640) was cloned and expressed in bacteria, and the purified CHC22 fragment was used as an antigen to generate a polyclonal rabbit antiserum and a monoclonal antibody (MAb) against CHC22. A 180 kDa polypeptide was isolated from human skeletal myoblast cell lysate by immunoprecipitation with the CHC22-specific MAb. It was identified by immunoblotting with polyclonal rabbit serum against the same CHC22 C-terminal domain (Figure 1A). The immunoprecipitated protein was not recognized by the TD.1 antibody specific for the N-terminal domain of conventional clathrin heavy chain (CHC17), indicating that the MAb against CHC22 does not immunoprecipitate CHC17 (see Figure 2C) and that TD.1 does not recognize CHC22. A low level of CHC22 protein was detected in undifferentiated human myoblasts (Figure 1A, day 0) and non-muscle cells such as HeLa (Figure 2C), transformed human B cells (JY) and human foreskin fibroblasts (data not shown). This is consistent with the previous observation that CHC22 mRNA can be detected at low levels in all tissues examined (Kedra et al., 1996), suggesting that it may function in a specialized pathway in most cells that becomes amplified in muscle cells. Consistent with this prediction, CHC22 expression was upregulated ∼50-fold upon myoblast differentiation and fusion into myotubes, after 5 days of serum withdrawal (Figure 1A, the percentage of differentiated cells is ∼10–15% in the culture). In contrast, the expression level of CHC17 protein remained unchanged throughout myogenesis. Induction of CHC22 protein in differentiated myotubes is further illustrated by morphological analysis of the myotube culture. CHC22 staining is strongly accentuated in differentiated myotubes containing multiple nuclei (Figure 1B, CHC22), while conventional CCVs are present in all cells, whether differentiated or not (Figure 1B, Clathrin LC). Increase of CHC22 expression in differentiated myotubes also coincides with the appearance of several muscle-specific markers such as skeletal muscle myosin and caveolin-3 (Parton et al., 1997) (data not shown). In addition, prominent staining of CHC22 in human skeletal muscle tissue sections was readily observed (Figure 1C). This staining pattern was different from the pattern seen with antibody to conventional clathrin (Figure 1C). Lack of co-localization is particularly visible at the edge of the muscle fiber in the sites denoted by arrowheads. In addition, the interior core region of the middle fiber, in the segment delineated by the arrowheads, has distinct vertical green lines and horizontal red lines where the respective staining patterns for CHC17 and CHC22 do not merge. The upregulation of CHC22 during muscle differentiation suggests that it has a specialized function, distinct from that of conventional clathrin CHC17.
Fig. 1. Expression of CHC22 in human myoblasts and muscle tissue. (A) Induction of CHC22 during muscle differentiation. Human skeletal myoblasts SKMC were induced to differentiate in medium with low serum. After serum reduction, cells were harvested on the days indicated and CHC22 proteins were immunoprecipitated from cell lysates (adjusted to equal protein concentrations) using a CHC22-specific MAb. The immunoprecipitates were resolved by SDS–PAGE and CHC22 was detected with purified anti-CHC22 antiserum (CHC22) by immunoblotting. The expression levels of ubiquitous clathrin heavy chain CHC17 in the same lysates were detected by blotting with CHC17-specific MAb TD.1 (CHC17). (B) Immunofluorescence of differentiated human myoblasts. SKMC cells grown on collagen-coated coverslips were induced to differentiate as in (A) for 7 days in low serum medium. Cells were then analyzed by immunofluorescence for the distribution of CHC22, using a specific MAb followed by rhodamine (LRSC)-conjugated goat anti-mouse IgG, and for conventional CCVs (Clathrin LC) using anti-clathrin light chain antiserum α-cons followed by FITC-conjugated goat anti-rabbit IgG. Note that not all cells differentiated and that only multinucleated cells express detectable levels of CHC22. (C) Distribution of CHC22 and clathrin in human skeletal muscle. Tissue sections from normal human muscle were prepared for immunofluorescence and labeled for CHC22 (red) and clathrin light chains (Clathrin LC, green) as in (B). The three panels show the same sample viewed with different filters and the bottom panel shows the merged staining with both antibodies. Arrowheads indicate regions where lack of co-localization (separate red and green staining) is readily observed (see text), while a yellow signal indicates an overlap of staining patterns.
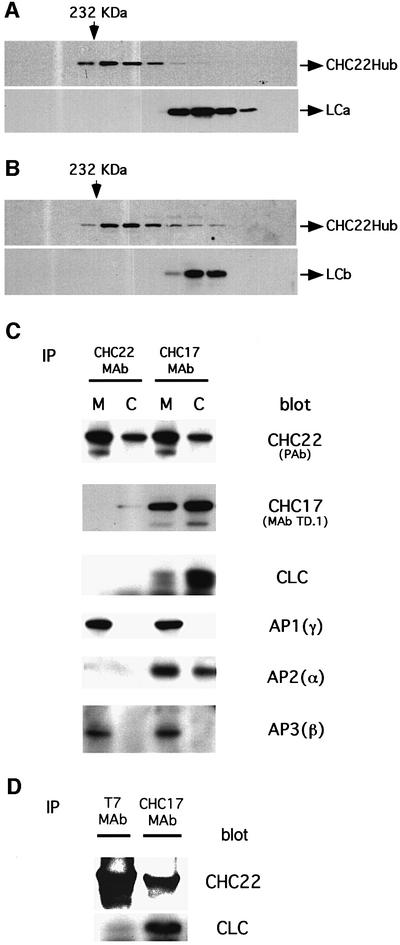
Fig. 2. Differential association of CHC22 with clathrin coat components. (A) Bacterial lysate containing recombinant CHC22Hub and co-expressed bovine neuronal LCa was separated by Superose 6 size exclusion chromatography. The column fractions were collected and resolved in sequence (left to right) by SDS–PAGE. The presence of CHC22Hub polypeptides or LCa was established by immunoblotting using rabbit serum against CHC22 (CHC22Hub) or MAb CON.1, which recognizes a determinant shared by both light chains (LCa and LCb). Arrows on the top indicate the fractions flanking the elution positions of molecular weight standard catalase (232 kDa). (B) Bacterial lysate containing recombinant CHC22Hub and co-expressed bovine neuronal LCb was separated by Superose 6 size exclusion chromatography and analyzed for the elution position of CHC22Hub polypeptides and LCb, as in (A). (C) Cytosolic and membrane-enriched fractions of HeLa 229 cells were prepared. CHC22 was then immunoprecipitated from each fraction using the MAb specific for CHC22. Conventional clathrin heavy chains (CHC17) were isolated with MAb X22 (Blank and Brodsky, 1986). The immunoprecipitates were then subjected to SDS–PAGE and probed with the following antibodies: anti-CHC22 polyclonal antiserum [CHC22 (PAb)], CHC17-specific monoclonal antibody TD.1, anti-clathrin light chain antiserum (CLC), anti-AP1 γ subunit monoclonal antibody 100/3 [AP1(γ)], anti-AP2 α subunit AC1M11 [AP2(α)] and anti-AP3 β3 antiserum [AP3(β)]. The CHC22 signals in CHC17 immunoprecipitates are due to cross-reactivity of X22 with CHC22, causing co-precipitation of CHC22 with CHC17 and possibly explaining the apparent association of AP3 with CHC17. (D) HeLa-tet/on cells were permanently transfected with T7-epitope-tagged full-length CHC22, under the tet operator (pJM601CHC22), and CHC22 expression was induced for 24 h with doxycycline. CHC22 full-length protein was then immunoprecipated using anti-T7 MAb and the sample was analyzed by SDS–PAGE and immunoblotting for CHC22 using a specific polyclonal antiserum, or for clathrin light chains (CLC) using the α-cons polyclonal antibody. Conventional clathrin (CHC17) was immunoprecipitated from the same sample using MAb X22 and similarly analyzed. Note that X22 immunoprecipitates some CHC22 along with CHC17 due to cross-reactivity (see C).
Differential association of CHC22 with clathrin coat components
There is high sequence identity (85%) between CHC22 and CHC17. To begin to understand the cellular function of CHC22, its biochemical properties were compared with those of CHC17. In vivo, the clathrin (CHC17) homotrimer has three associated light chain subunits. We have shown previously that light chains associate with the hub domain of clathrin heavy chain (residues 1074–1675), when co-expressed in bacteria, and co-elute with trimerized hubs during size exclusion chromatography (Liu et al., 1995). To determine whether CHC22 also interacts with clathrin light chain subunits, the equivalent hub domain of CHC22 (residues 1074–1640) was cloned into the bacterial expression vector pET15b in tandem with the cDNA for either light chain LCa or LCb (neuronal isoforms). This fragment of CHC22 also contains a potential trimerization domain (residues 1550–1615 in CHC17). The recombinant CHC22Hub was expressed at a high level after isopropyl-β-d-thiogalactopyranoside (IPTG) induction, but most of the protein was insoluble, in contrast to the hub domain of CHC17, which is >50% soluble under the same expression conditions. Solubility of CHC22 hubs was improved by expression in a bacterial strain carrying the chaperone proteins GroEL and GroES (Caspers et al., 1994). Analysis by size exclusion chromatography then showed that the soluble 64 kDa CHC22Hub polypeptide eluted at fractions equivalent to the 200 kDa marker, indicating that it is a trimer, similar to the CHC17 hub (Figure 2A and B). However, there was no detectable binding of the co-expressed neuronal light chains LCa or LCb, which were recovered in fractions completely segregated from those containing CHC22Hub.
Interaction between CHC22 and non-neuronal clathrin light chains was investigated by analyzing membrane (M) and cytosolic (C) fractions of HeLa cells (Figure 2C). CHC22 proteins were immunoprecipitated from both fractions with anti-CHC22 MAb and resolved by SDS–PAGE. Immunoblots were then probed for clathrin light chains using an antiserum that recognizes the conserved region shared by all known isoforms of mammalian light chains LCa and LCb (Acton and Brodsky, 1990). Again, no clathrin light chains were detected in association with CHC22 isolated from either HeLa cell fraction (Figure 2C, CLC blot). To rule out the possibility that bound light chains were not immunoprecipitated because our antibody might react with a putative light-chain binding region of CHC22, cells were transfected with full-length CHC22 tagged at the N-terminus with the T7 epitope. When these transfected proteins were immunoprecipitated using anti-T7 MAb, they were also not associated with clathrin light chains (Figure 2D). Lastly, in preliminary studies, the potential light-chain binding sequence of CHC22 was tested for interaction with the LCb light chain in a quantitative yeast two-hybrid assay. An interaction signal was detected, but it was considerably weaker than the signal generated by LCb interaction with the equivalent region from CHC17. Together these data demonstrate that the clathrin light chains do not bind to CHC22 with the same affinity as they interact with CHC17. In a co-immunoprecipitation assay their binding to CHC22 is undetectable and only a weak interaction can be detected in the yeast two-hybrid system, which favors low-affinity interactions. Thus, if clathrin light chains bind to CHC22, they readily dissociate, unlike their tight interaction with CHC17 (Figure 2C and D), which can be perturbed only under denaturing conditions. In addition, we did not observe any potential light-chain-like polypeptides, by SDS–PAGE, in CHC22 immunoprecipitates isolated from 35S-labeled cell lysates (data not shown). We can not rule out the possibility that CHC22 binds novel light chains or other subunits that are not amenable to biosynthetic labeling. Nonetheless, our results indicate that there are crucial variant residues between CHC22 and CHC17 that disable the binding of either LCa or LCb to CHC22.
There was also no significant association detected between CHC22 and CHC17 by immunoprecipitation (Figure 2, CHC17 blot). Nor was CHC22 preferentially enriched in CCVs isolated by standard ficoll/sucrose sedimentation (data not shown). It should be noted that MAb X22, used to immunoprecipitate CHC17, cross-reacts to some extent with CHC22, as indicated by blotting with the antiserum specific for CHC22 in the top panels of Figure 2C and D. In contrast, MAb TD.1 specifically recognizes the CHC17 immunoprecipitated by X22, and blotting with TD.1 reveals that a higher proportion of endogenous CHC22 is associated with membranes following subcellular fractionation in comparison with the membrane-associated level of CHC17 (Figure 2C, top two panels). This suggests that CHC22 is more likely to exist in assembled structures than ubiquitous CHC17 inside cells.
To determine whether CHC22 associates with adaptor complexes during its cellular function, the interaction between CHC22 and different adaptors was assessed both biochemically and morphologically. CHC22 was immunoprecipitated from HeLa cell lysates and isolates were probed with antibodies specific for the AP1, AP2 or AP3 adaptors (Figure 2C). All three adaptors were co-precipitated with MAb X22 against CHC17 (Figure 2C, anti-CHC17). In contrast, CHC22 was preferentially associated with AP1 and AP3 but not with AP2 adaptors (Figure 2C, anti-CHC22). The presence of AP3 in immunoprecipitates with either antibody was confirmed by blotting with an antibody to the δ subunit of AP3 (not shown), but its presence in the X22 immunoprecipitate could be due to binding to CHC22 isolated through antibody cross-reaction, rather than to direct CHC17 binding.
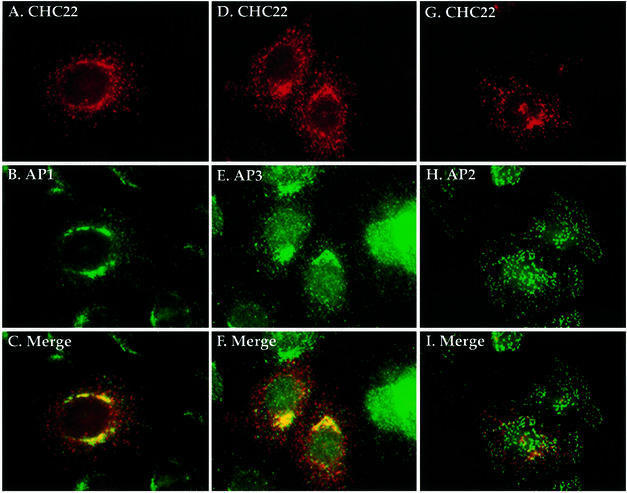
To confirm further the specificity of adaptor binding, the cellular localization of CHC22 and different adaptors was examined by immunofluorescence. To enhance the weak signal for endogenous CHC22 staining, full-length CHC22 cDNA was introduced into HeLa cells by retroviral expression. The transfected CHC22 protein had the same mobility as endogenous CHC22 on SDS–gels (data not shown). Initial CHC22 staining detected by the specific MAb was amorphous, but a distinct punctate morphology, reproducing the weak endogenous staining pattern, was revealed after brief digitonin treatment of cells to remove the cytosol prior to staining (Figure 3A, D and G). Most of the detergent-resistant CHC22 did not co-localize with CCVs, detected by clathrin light chain staining (not shown). The membrane-bound CHC22 partially overlapped with AP1 and AP3 staining, but was largely absent from peripheral AP2-containing vesicles (Figure 3I). A subset of the punctate structures staining for CHC22 did not co-localize with any of the other markers tested. These structures may be associated with novel adaptors or may be induced structures formed from self-assembly of the transfected CHC22 protein. The perinuclear localization of CHC22 was abolished by brefeldin A treatment (data not shown), a fungal metabolite that disrupts binding of AP1 and AP3 to the Golgi/TGN membranes (Simpson et al., 1997; Ooi et al., 1998). Taken together, these results indicate that the membrane-bound portion of cellular CHC22 is specifically localized at the TGN, through its interaction with AP1 and AP3 adaptors, and is generally segregated from clathrin coats.
Fig. 3. Co-localization of CHC22 with AP1 and AP3 but not AP2. HeLa cells infected with pLNCXCHC22 to obtain low-level enhanced expression of CHC22 were treated with 0.004% digitonin, in order to remove cytosolic protein prior to being processed for immunofluorescence. The cells were then stained red with CHC22 MAb (isotype IgG2a) followed by biotinylated goat anti-mouse IgG2a antibody and LRSC-conjugated streptavidin (A and G). AP1 and AP2 were detected in green by monoclonal antibodies 100/3 and AP.6 followed by FITC-conjugated goat anti-IgG2b (B) and IgG1 (H) antibodies, respectively. Double staining of CHC22 (red) and AP3 (green) was achieved by using anti-CHC22 MAb followed by LRSC-conjugated goat anti-mouse IgG (D) and anti-AP3 δ subunit antiserum followed by FITC-conjugated goat anti-rabbit IgG (E). Each column represents the same sample viewed with different filters, and merged images for CHC22 with AP1, AP2 or AP3 are shown in (C), (F) and (I), respectively.
Ultrastructural analysis of intracellular CHC22 distribution
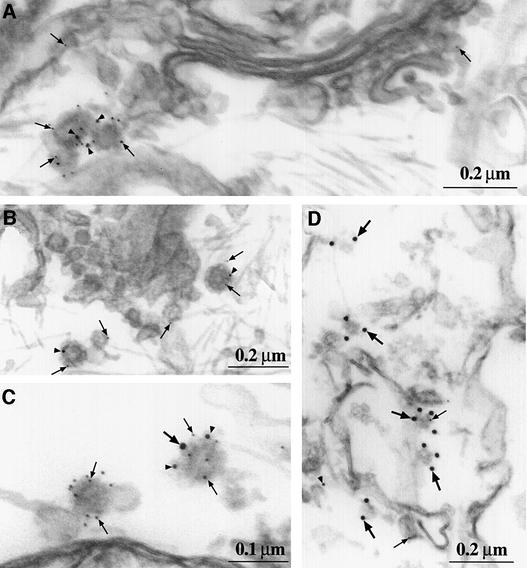
The association of CHC22 with elements of the TGN was confirmed by immunoelectron microscopy (Figure 4) of stably transfected HeLa cells expressing a high level of CHC22 following induction with doxycycline. These cells were sequentially labeled with antibodies to CHC22, the AP1 adaptor molecule or the clathrin light chains, each detected with protein A attached to different sizes of gold particles. Antibodies to CHC22 labeled the cytoplasmic side of vesicular structures, primarily in the Golgi/TGN region of the cell (Figure 4A–C), although some localized to the cell periphery (Figure 4D). In some cases a cytoplasmic coat was observed to surround small (80–100 nm in diameter) CHC22- and AP1-labeled vesicles (Figure 4B). Of the vesicles labeled with antibody to CHC22, 9.4% were double-labeled for AP1 and 18.7% were double-labeled for clathrin light chain. There are several explanations for this unexpected morphological co-localization of CHC22 and clathrin light chains in a subpopulation of CHC22 vesicles. The weak interaction of CHC22 with clathrin light chains in a yeast two-hybrid experiment suggests that there could be a transient in vivo association of clathrin light chain with CHC22 trimers. This transient interaction could potentially be captured by electron microscopy even if it is not obvious by immunofluorescence (where a subpopulation would be hard to detect) or by co-immunoprecipitation (where washing procedures could cause light chain dissociation). In addition, a weak association of CHC22 with clathrin light chains could be enhanced in the cells analyzed, which express relatively higher levels of CHC22 than normal cells after induction by doxycycline. Given that 6% of vesicles labeled for CHC22 were labeled simultaneously for clathrin light chain and for AP1, it is also possible that AP1 could potentially draw the two clathrins into the same coat, promoting an interaction that is not detectable biochemically. It should be noted, however, that the majority of CHC22-labeled vesicles were distinct from CCVs and that these were localized primarily to the TGN region of the cell.
Fig. 4. Localization of CHC22 by immunoelectron microscopy. HeLa-tet/on cells transfected with pJM601CHC22 were induced for high level CHC22 expression for 24 h, then permeabilized, lightly fixed and labeled with antibodies to CHC22 (5 nm gold, small arrows), components of AP1 (10 nm gold, arrowheads) and clathrin light chains (15 nm gold, large arrows). Prior to embedding and sectioning, each antibody was sequentially applied and detected with protein A–gold attached to gold particles of the size indicated, followed by blocking with excess protein A. The images were selected to be representative of the intracellular distribution of CHC22 (statistics are provided in the text), showing vesicles of 80–100 nm in the TGN labeled for CHC22 and AP1 (γ subunit) (A) or labeled for CHC22 and AP1 (σ1 subunit) with notable protein coats (B). (C) Similar vesicles, with one labeled exclusively for CHC22 and the other for CHC22, clathrin light chain and AP1. (D) Peripheral vesicles, primarily labeled for clathrin light chain, with one co-labeled for CHC22. The labeling for AP1 (σ1 subunit) at the left suggests that these coated vesicles are near endosomes.
Co-localization of CHC22 and non-muscle myosin II
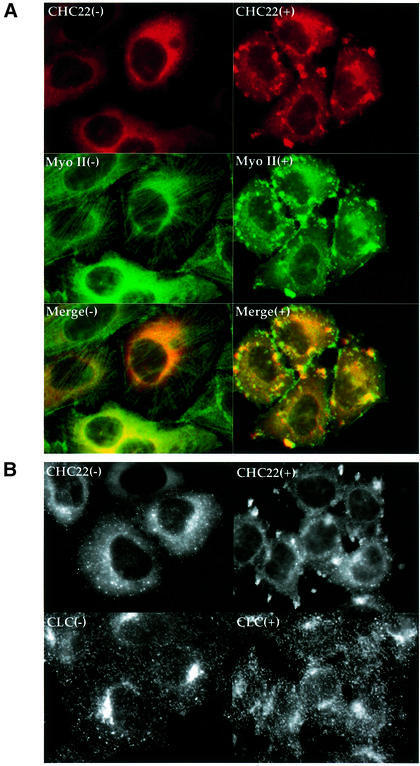
The enhanced expression of CHC22 in human skeletal muscle suggested that the protein might interact with elements of the actin–myosin cytoskeleton. Due to the low resolution of immunofluorescent staining of thick sections of human muscle, localization of CHC22 to specific bands within the muscle tissue could not be determined with any confidence. Therefore, we investigated the interaction between CHC22 and actin–myosin microfilaments in HeLa cells transfected with CHC22. Myosin II is the dominant myosin in human skeletal muscle, and one of the non-muscle myosin II homolog(s), p200, is suggested to have a role in the production of transport vesicles from the TGN (Ikonen et al., 1997; Müsch et al., 1997), where CHC22 has been localized. When we compared the distribution of non-muscle myosin II and CHC22 in HeLa cells transfected with CHC22, it was initially difficult to establish whether there was co-localization of these two proteins, due to extensive cellular staining by antibodies to both proteins (Figure 5A). However, after the cells were treated with cytochalasin D to disrupt the actin cytoskeleton, microfilaments were rapidly depolymerized and both myosin (Figure 5A) and actin (not shown) formed large aggregates in the cell periphery. Most of the intracellular CHC22 also redistributed to these cytoskeletal foci and this behavior was observed for both transfected CHC22 (Figure 5A) and endogenous CHC22 in non-transfected HeLa cells (not shown). After equivalent cytochalasin D treatment, ubiquitous clathrin molecules remained in small vesicles, which dispersed near these structures, but did not co-localize with them (Figure 5B). This lack of clathrin co-localization with CHC22 following cytochalasin D treatment represents a further distinction between these two proteins. It also demonstrates that the re-distribution of CHC22 into the aggregates of cytoskeletal proteins is not due to non-specific effects of cytochalasin D on cytoplasmic proteins in general or to labeling artifacts in the aggregates themselves. Nocodazole treatment, which disassembles microtubules, had no detectable effect on the CHC22 staining pattern (not shown). The redistribution of CHC22 following disruption of microfilaments suggests that this novel clathrin homolog associates with components of the actin–myosin cytoskeleton and may be involved in their function in the TGN.
Fig. 5. Redistribution of CHC22 with non-muscle myosin II following cytochalasin D treatment. HeLa cells, infected with pLNCXCHC22 to obtain low-level enhanced expression of CHC22, were treated with (+) or without (–) 1 µg/ml cytochalasin D for 20 min, then processed for immunofluorescence. Cells were stained with anti-CHC22 MAb followed by LRSC-conjugated goat anti-mouse IgG (red) and anti-non-muscle myosin II antiserum, followed by FITC-conjugated goat anti-rabbit IgG (green) (A). Each column in (A) shows the same sample viewed with different filters, and merged images are shown in the bottom panels. Note that in the left hand panels of (A), some of the cells are only expressing endogenous levels of CHC22, which is faintly detectable by staining with anti-CHC22 (e.g. cells in upper left and lower right corners). HeLa cells, infected with pLNCXCHC22 to obtain low-level enhanced expression of CHC22, treated with (+) or without (–) 1 µg/ml cytochalasin D for 20 min as above, were also stained with anti-CHC22 MAb, detected by LRSC-conjugated goat anti-mouse IgG, and anti-clathrin light chain antiserum, detected with FITC-conjugated goat anti-rabbit IgG (B). Each column represents the same sample viewed with a different filter to detect red (top) or green (bottom) staining.
Cellular localization of CHC22 is sensitive to low temperatures that affect TGN transport
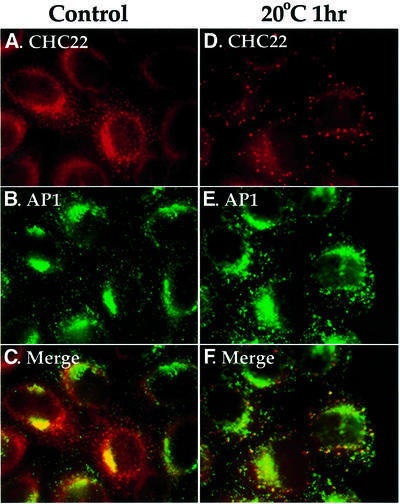
It is a long-standing observation that transport from the TGN to the cell surface is completely inhibited when cells are exposed to mildly low temperatures (16–20°C) (Matlin and Simons, 1983; Saraste and Kuismanen, 1984; Griffiths et al., 1985). Other cellular trafficking steps, such as endocytosis and recycling, continue under these conditions, albeit at slower rates (Sandvig and Olsnes, 1979; Dunn et al., 1980). The exact mechanism of the low temperature effect is still unknown, although it has been observed that low temperatures generally cause accumulation of vesicles in the Golgi region in treated cells (Griffiths et al., 1985). In order to investigate the potential function of CHC22 in TGN transport, the distribution of CHC22 was monitored after 1 h of 20°C treatment (Figure 6). In control cells, transfected CHC22 was observed in association with perinuclear membranes and partially overlapped with AP1 adaptor staining (Figure 6A–C). However, the TGN localization of CHC22 was disrupted when cells were exposed to low temperature, and most of the CHC22 staining was seen in large aggregates dispersed in the cytoplasm (Figure 6D). Conversely, AP1 staining was not substantially altered, although in some cells it appeared slightly increased in the TGN area (Figure 6E), consistent with the accumulation of CCVs in the Golgi/TGN area after 20°C treatment. This temperature-dependent segregation of CHC22 and AP1 suggests either that CHC22 binding to AP1 is temperature sensitive or that CHC22 interacts with additional temperature-sensitive components in the TGN, which stabilize its interaction with AP1.
Fig. 6. Disruption of CHC22 TGN localization by low temperature. HeLa cells, infected with pLNCXCHC22 to obtain low-level enhanced expression of CHC22, were either left untreated (A–C) or incubated at 20°C for 1 h (D–F) before being subjected to digitonin wash to remove cytosol, and then processed for immunofluorescence. CHC22 (red) was detected with anti-CHC22 antiserum followed by LRSC-conjugated goat anti-rabbit IgG (A and D). AP1 (green) was stained with MAb 100/3 followed by FITC-conjugated goat anti-mouse IgG (B and E). The columns represent the same images viewed with different filters and the bottom panels are merged images.
Overexpression of a subdomain of CHC22 alters distribution of the mannose 6-phosphate receptor
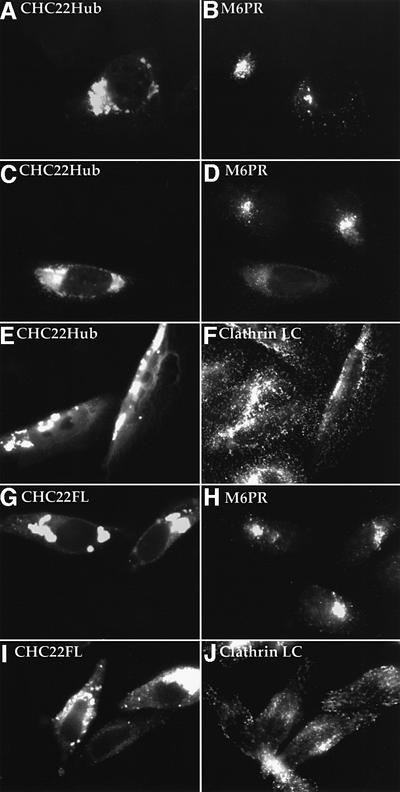
To investigate further the involvement of CHC22 in TGN transport, the C-terminal domain of CHC22 was used to generate a potential CHC22 mutant phenotype. Previously we have shown that cell transfection with CHC17 clathrin hubs (comprising the C-terminal third of clathrin heavy chain) results in the disruption of intracellular clathrin function (Liu et al., 1998). It is conceivable that the CHC22 hub could also interfere with endogenous CHC22 because it contains a potential self-assembly region but lacks the N-terminal domain to interact with adaptor proteins. DNA containing the hub equivalent of CHC22 (residues 1074–1640) was subcloned into a plasmid with the tetracycline-inducible promoter and transfected into HeLa cells harboring the tetracycline-responsive transactivator. After 48 h of doxycycline treatment to induce the tetracycline promoter, CHC22Hub was expressed at high levels in transfected cells. Overexpressed CHC22 hub proteins generally aggregated in the perinuclear region (Figure 7A), although conventional clathrin hubs remain largely soluble at the same expression level (Liu et al., 1998). In CHC22Hub-transfected cells, the distribution of CCVs (illustrated by staining for clathrin LC) was not obviously affected (Figure 7E and F), indicating that CHC22Hub expression does not interfere with conventional CCV formation. Furthermore, the endocytosis of fluorescent transferrin was normal in CHC22Hub-expressing cells (data not shown). These results are consistent with our observation that CHC22 does not interact with conventional clathrin light chains or heavy chains or the AP2 adaptor (Figures 2 and 3). To see whether CHC22Hub could affect TGN function, mannose 6-phosphate receptor (M6PR) was used as a marker to assess transport from the TGN. M6PRs shuttle between TGN and late endosomes to deliver lysosomal hydrolases modified with mannose 6-phosphate. In control HeLa cells (non-transfected within the culture), M6PRs were largely localized at the TGN (Figure 7). However, expression of CHC22Hub caused a dramatic change of M6PR distribution. In cells with a high level of CHC22Hub expression, M6PR staining became more diffuse and was no longer concentrated at the TGN (Figure 7). The distribution of another TGN marker, TGN46, was also affected, but to a lesser degree. This effect was specific to the TGN, as the cellular distribution of Golgi residents such as Golgi p58 and β-cop was not obviously affected (not shown).
Fig. 7. Expression of CHC22 hub domain affects intracellular distribution of M6PR. HeLa-tet/on cells transfected with pJM601CHC22Hub (A–F) or with pJM601CHC22 (G–J) were induced for high level CHC22Hub expression or high level expression of full-length (FL) CHC22 for 24 h. Cells were stained for the expression of CHC22Hub (shown in A, C and E) or CHC22FL (shown in G and I) with anti-CHC22 MAb followed by LRSC-conjugated goat anti-mouse IgG. The distribution of endogenous M6PR (shown in B, D and H) was detected by double staining (A), (C) and (G) with antiserum to the cation-independent M6PR, and the distribution of endogenous clathrin (shown in F and J) was detected by double staining (E) and (I) with anti-clathrin LC antiserum α-cons, both followed by FITC-conjugated goat anti-rabbit IgG. The horizontal rows represent the same images viewed with different filters to see red and green staining on the left and right, respectively. Note that not all cells in the transfected cultures express CHC22Hub or CHC22FL, which was confirmed by staining with anti-T7 MAb, which reacts with the epitope tag on the transfected proteins (not shown). The non-expressing cells serve as negative controls for endogenous staining of M6PR or clathrin LC, and their staining patterns are identical to those of control cells that were either not transfected or not induced for expression of the transfected molecules.
To determine whether the effect of CHC22Hub was due to its mutant structure, we also examined the effects of high expression of the full-length CHC22 molecule in transfected cells (Figure 7G–J). In these cells, the distribution of M6PR and of clathrin light chains was no different from their distributions in non-transfected cells within the same culture (Figure 7H and J) or in control cultures (not shown). These results indicate that CHC22Hub alters membrane traffic by acting as a CHC22 mutant. In addition, they further support the fact that clathrin light chains do not have an avid intracellular association with full-length CHC22. The effect of CHC22Hub on the cellular distribution of M6PR supports a role for CHC22 in TGN transport.
Discussion
We report here the characterization of a novel clathrin-related protein, CHC22. The protein was identified, localized and its biochemistry studied in comparison with conventional clathrin, CHC17. Unlike CHC17, CHC22 is abundant in skeletal muscle relative to its expression in non-muscle cells, and its expression is upregulated during myogenesis. We conclude from a series of biochemical and morphological studies that this protein is structurally related to clathrin, but has a distinct intracellular function and distribution that could contribute to a potential role in tissue-specific membrane traffic.
Comparison between CHC22 and CHC17 peptide sequences reveals no particular domains containing a higher degree of divergence than the other regions. We demonstrate here that, like CHC17, CHC22 forms a trimer. Initial secondary structure analysis suggested that a potential light-chain binding region is also present within the CHC22 hub domain (Sirotkin et al., 1996). However, by biochemical and protein expression analysis, the binding of clathrin light chains (LCa or LCb in neuronal or non-neuronal forms) to CHC22 was not detected, although a weak interaction with neuronal LCb by yeast two-hybrid interaction has been observed. It remains possible that CHC22 could be complexed with an as yet unidentified version of a light chain subunit that is not detected by biosynthetic labeling. On the other hand, lack of light-chain association or weak association could account for differences between CHC22 and CHC17 assembly kinetics, since light chains play a negative regulatory role in clathrin assembly (Ungewickell and Ungewickell, 1991; Liu et al., 1995). Consistent with this speculation, we have observed a ‘ready to assemble’ nature of CHC22 when it is expressed either in bacteria or in mammalian cells. Unlike CHC17Hubs, which remain mostly cytosolic even when expressed at high levels, over-expressed CHC22 or CHC22Hubs usually form big clusters in the perinuclear region. Endogenous CHC22 from HeLa cells also tends to segregate from the soluble cytosolic fractions after ultracentrifugation (Figure 2C).
In mammalian cells, formation of CCVs on specific membranes relies on the association of clathrin with specific adaptors. The primary interaction between clathrin and adaptors is mediated via the N-terminal domain of clathrin heavy chain. We have shown here, by both biochemical and morphological analysis, that CHC22 interacts with adaptor AP1 and AP3 but not AP2 (Figures 2 and 3). Since AP2 is the only adaptor associated with clathrin-mediated endocytosis at the plasma membrane, these observations position CHC22 as a ‘TGN-specific clathrin’. The TGN distribution of CHC22, however, must require multi-component interactions rather than just depend on the adaptor complexes, since 20°C treatment dissociates CHC22 from the TGN membrane without affecting the localization of adaptor complexes or conventional clathrin (Figure 6). The temperature shift could either affect the assembly of CHC22 or disrupt the infrastructure required to maintain CHC22 at the TGN. Low temperatures are known to disrupt transport from the Golgi/TGN through as yet unknown mechanisms. The sensitivity of CHC22 structures to low temperature treatment suggests a role for CHC22 in vesicular trafficking from the TGN and a possible link to the temperature sensitivity of this pathway.
Consistent with a membrane traffic role for CHC22 in the TGN, electron microscopy studies revealed that, in non-muscle cells, CHC22 localizes to coated membrane and vesicular structures in the TGN. Occasional co-localization of clathrin light chains with CHC22 was also observed (18.7% of vesicles). This could be explained by a transient association of clathrin light chains with CHC22 that is disrupted upon biochemical analysis and is not readily detected by immunofluorescent co-localization. Alternatively, the electron microscopy analysis could reflect occasional co-localization of CHC22 with CHC17 bound to clathrin light chains. Our biochemical data indicate that CHC22 and CHC17 do not interact with each other, but both clathrins could be present in the same membrane coat by virtue of their shared interaction with the AP1 adaptor. The majority of CHC22-containing vesicles were not labeled for clathrin light chains, indicating that the two clathrins generally function independently and suggesting that CHC22 is not a component of conventional CCVs in the TGN. This is consistent with the lack of enrichment of CHC22 in ficoll/sucrose preparations of CCVs.
The fact that CHC22 appears to assemble more readily than conventional clathrin, and has either no association or a low-affinity transient association with conventional clathrin light chains, indicates that CHC22 and CHC17 have different biochemical properties and that cellular regulation of their assembly will consequently be different. Its apparent stability as an assembled structure suggests that CHC22 could play a role in maintaining cellular structure, such as mediating the interaction between the membranes it associates with and the cytoskeleton. Furthermore, the redistribution of CHC22 in response to cytochalasin D (Figure 5A), which is not a property of CHC17 (Figure 5B), indicates that CHC22 could participate in the function of the cytoskeleton at the TGN. Redistribution of CHC22 with myosin II is particularly interesting in light of the data implicating non-muscle myosin II in TGN transport (Ikonen et al., 1997; Müsch et al., 1997). To investigate the role of CHC22 in TGN transport, the hub domain of CHC22 was expressed in HeLa cells. The CHC22Hub transfectants exhibited no obvious defect in endocytosis measured by FITC-transferrin uptake (data not shown), and CCV distribution in these cells was largely unaffected (Figure 7). However, the cellular distribution of M6PR and TGN46 (not shown) became more widely dispersed and was no longer concentrated at the TGN and late endosomes, as normally seen in control cells (Figure 7). It remains to be determined whether a defect in transport or an overall change of TGN infrastructure contributes to this phenomenon, as CHC22Hub could either affect the formation of transport vesicles containing CHC22 or affect CHC22 interactions with the cytoskeleton framework at the TGN. With regard to the latter, CHC22Hub did not seem to disrupt the cellular distribution of myosin II (data not shown), so its effects on transport are not due to causing a major change in the cytoskeleton. We would also like to point out the different effects on M6PR trafficking of conventional clathrin hub compared with CHC22 hub mutants. While CHC17Hub traps M6PR in the secretory pathway by disabling export out of the TGN by CCVs (Liu et al., 1998), the CHC22Hub mutant seems to prevent M6PR from being concentrated at the TGN. This is an effect specific to the mutant activity of CHC22Hub, as it is not induced by overexpression of full-length CHC22. In summary, the sensitivity of CHC22 to temperatures that affect TGN transport, its affinity for the actin cytoskeleton, and the mutant’s effect on M6PR trafficking support a role for CHC22 in transport at the TGN that is different from that of conventional CCVs.
The high expression of CHC22 in muscle cells suggests that it might have some vital function in muscle-specific membrane traffic pathways or other muscular functions. There is only one gene homologous to clathrin heavy chain in yeast, Dictyostelium and Drosophila. Interestingly, the CHC-deficient mutants of the above organisms exhibit unexpected phenotypes rather than an obvious endocytosis defect. Disruption of the CHC gene in yeast has defects in retention of Golgi enzymes (Seeger and Payne, 1992). Clathrin-minus Dictyostelium cells exhibit a cytokinesis deficiency due to the failure to assemble myosin II into a functional contractile ring (Niswonger and O’Halloran, 1997). An actin-based sperm individualization complex is disrupted in a Drosophila male-sterile clathrin heavy chain mutant (Fabrizio et al., 1998). These unusual phenotypes coincide with possible physiological defects that could be associated with aberrant CHC22 functions in mammalian cells. Sequence comparison reveals similar degrees of homology between CHC22 and CHC17 and the non-mammalian clathrin heavy chain genes (Long et al., 1996). In other words, the CHC gene in each of the above species could represent the common ancestor of both CHC22 and CHC17 in mammalian cells and execute the functions of both. A CHC22-equivalent gene in mouse has not been found (Botta et al., 1997) but there is an indication that CHC22 orthologs exist in dogs, rabbits and primates (Holmes et al., 1997). The evolutionary emergence of CHC22 may reflect an expanded need for membrane traffic pathways interacting with the cytoskeleton in muscle of certain vertebrates. Many patients with DGS and VCFS have a hemizygous deletion of the gene encoding CHC22, as part of a larger genetic deletion in the chromosome 22q11 region (Ryan et al., 1997). These patients suffer from serious developmental defects that affect the heart, thymus, thyroid and parathyroid glands, no doubt due to the haplo-insufficiency of many gene products encoded by the deleted region. Their neonatal problems include hypocalcemic tetany, muscle seizures that occur due to lower than normal levels of calcium. While the hypocalcemia can be attributed to parathyroid problems, it is tempting to speculate that the muscle problems in neonatal DGS/VCFS patients might result from or be exacerbated by a genetic reduction in CHC22 expression. The sarcoplasmic reticulum (SR) is a muscle-specific membranous organelle that sequesters calcium for release in muscle, through calcium channels in the adjacent membranous transverse (T)-tubules. T-tubule development has already been shown to involve caveolin-3, a muscle-specific caveolin (Parton et al., 1997). Some aspects of T-tubule/SR formation or function may also involve CHC22. Alternatively or additionally, CHC22 could play a role in mediating interactions between muscle membrane traffic and its cytoskeletal structures, as has been suggested by our analysis of CHC22 distribution and function in non-muscle cells.
Materials and methods
cDNA isolation and plasmid construction
Partial CHC22 cDNA construct (HS6) was a gift of Raju Kucherlapati, Albert Einstein College of Medicine, NY (Sirotkin et al., 1996). Recombinant CHC22Hub (residues 1074–1640) was produced by PCR amplification from HS6 and insertion into vector pET15b (Novagen), and bovine neuronal light chains LCa and LCb cDNA were each cloned upstream of CHC22Hub as described (Liu et al., 1995). The C-terminal portion of CHC22 (residues 1521–1640) was cloned into the pET15b vector. To generate stable CHC22 transfectants in mammalian cells, full-length CHC22 cDNA was isolated by reverse transcriptase (RT)–PCR using human skeletal muscle Marathon-Ready cDNA (Clontech) as template. A 5 kb DNA fragment corresponding to the entire CHC22 transcript was isolated and cloned into the vector pET23a (Novagen), encoding an N-terminal T7 epitope tag. The CHC22 cDNA insert was then subcloned into the retroviral vector pLNCX (Clontech) for transfection into virus-packaging cells Phoenix (Clonetech). Full-length and the hub portion of CHC22 (both with an N-terminal T7 epitope tag) were also cloned into vector pJM601, a gift from John Morgan, University of Michigan Medical School, who produced it from pCEP4 (Invitrogen) by replacing the CMV promoter with the tetracycline operator sequence.
Cell culture and transfection
Human skeletal muscle myoblasts (SKMC) were obtained from Clonetics and cultured on collagen-coated Petri dishes with SkGM SingleQuots (Clonetics) according to the manufacturer’s instruction. Myotube fusion was induced by changing the confluent SKMC cells to differentiation medium containing Dulbecco’s modified Eagle’s medium (DMEM) and 2% horse serum. HeLa 229 (ATCC) and Phoenix cells were cultured in DMEM with 10% fetal bovine serum. HeLa-tet/on cells (Clontech) were maintained in DMEM with 10% FBS and 0.5 mg/ml G418. To produce CHC22 stable clones with retroviral vector pLNCX, pLNCXCHC22 plasmid was first transfected into Phoenix cells using the standard calcium phosphate method. Thirty-six hours later, the culture supernatant was collected and applied to HeLa cells for 24–48 h. Infected cells were selected by the same culture medium containing 1 mg/ml G418 and used for low-level enhanced expression of CHC22 (Figures 3, 5 and 6). To express high levels of full-length CHC22 protein (Figures 2D, 4 and 7) and CHC22Hub (Figure 7) with the tetracycline-inducible system, pJM601-derived DNA constructs were transfected into HeLa-tet/on cells using Fugen 6 (Boehringer Mannheim). Transfected cells were then selected in culture medium containing 0.5 mg/ml G418 and 0.4 mg/ml hygromycin B and induced for expression using doxycycline.
Antibodies
Standard methods were used to generate mAbs specific for CHC22 following immunization of mice with recombinant CHC22 C-terminal fragment (residues 1521–1640) with an N-terminal histidine tag, purified by Ni2+-affinity column as described (Liu et al., 1995). Antibody was purified from hybridoma supernatant using a protein G column (Pharmacia). A polyclonal rabbit antiserum against the same fragment of CHC22 was generated by contract laboratory (Covance). The serum was first passed through a CHC17 affinity column [made from the C-terminal residues (1074–1675) of CHC17 coupled to Affi-gel 15 (Bio-Rad)] to remove antibodies that cross-react with conventional clathrin heavy chain. The flowthrough was passed over a second affinity column made from the CHC22 C-terminal fragment. Bound antibodies were eluted with 0.1 M glycine pH 2.5, neutralized and dialyzed into phosphate-buffered saline (PBS) pH 7.4 before use. Polyclonal antibody against clathrin light chains (α-cons), anti-AP2 mAb AP.6 and anti-clathrin heavy chain MAbs TD.1 and X22 were generated in the Brodsky laboratory (Blank and Brodsky, 1986; Chin et al., 1989; Acton and Brodsky, 1990; Näthke et al., 1992). Polyclonal rabbit antibodies against AP3 subunits β3 and δ were gifts of Regis Kelly, University of California, San Francisco, and Margaret Robinson, Cambridge University, UK, respectively. Polyclonal rabbit antibodies to the cation-independent M6PR, γ subunit of AP1 (AE-1) and σ1 subunit of AP1 (DE-1) were gifts of Linton Traub, University of Pittsburgh, Pittsburgh. Purchased antibodies were anti-γ subunit of AP1 (100/3; Sigma), anti-non-muscle myosin II antiserum (Biomedical Technologies Inc.) and anti-α subunit of AP2 (AC1M11; Affinity Bioreagents).
Size exclusion column chromatography
CHC22Hub was co-expressed with either neuronal clathrin light chain LCa or LCb in the BL21 bacterial strain harboring GroES, GroEL and pREP4 GroESL chaperone proteins (Caspers et al., 1994). After 3 h of IPTG induction, the cells were lysed by sonication in 0.5 M Tris pH 7.9 and centrifuged at 100 000 g for 30 min. The resulting supernatant was then loaded onto a Superose 6 size exclusion column (Pharmacia). Column fractions were analyzed by SDS–PAGE and probed with antibodies against either the C-terminal domain of CHC22 or clathrin light chains.
Immunofluorescence
Cells were fixed and processed for immunofluorescence as described (Liu et al., 1998). To remove cytosolic staining, cells were incubated in digitonin buffer [25 mM HEPES–KOH pH 7, 125 mM KOAc, 2.5 mM Mg(OAc)2, 0.004% digitonin] at room temperature for 2 min prior to fixation. Low temperature treatment was achieved by incubating the cells in DMEM plus 20 mM HEPES pH 7.2 at 20°C for 1 h. To disrupt actin–myosin cytoskeleton, 1 µg/ml cytochalasin D (Sigma) was added to the cells for 20 min at 37°C. For human tissue staining, 2–5 µm of human skeletal muscle tissue sections were prepared by the ImmunoPathology Laboratory at University of California, San Francisco. Slides mounted with the formalin-fixed tissue samples were deparaffinized, treated with 0.05% saponin and fixed with 4% paraformaldehyde prior to incubation with 20 µg/ml primary antibodies for at least 3 h. The sections were washed, and staining was detected by incubation with fluorophore-conjugated secondary antibodies.
Electron microscopy
HeLa-tet/on cells transfected with pJM601CHC22 were induced with doxycycline for 24 h, then washed with ice-cold PHEM buffer (60 mM PIPES, 25 mM HEPES pH 6.9, 10 mM EGTA, 2 mM MgCl2), treated with 20–30 µg/ml digitonin for 20 min at 4°C, washed, then fixed [5 min, room temperature (RT)] with 4% paraformaldehyde (80 mM PIPES pH 6.8, 5 mM EGTA and 2 mM MgCl2) followed by 10 min, RT with 4% paraformaldehyde (100 mM sodium borate pH 11.0). Fixation was quenched with 40 mM glycine in PBS and samples were incubated with PBS containing 0.5% (w/v) bovine serum albumin (BSA) (PBS–BSA) and 5% goat serum (15 min, RT). Cells were then incubated with anti-CHC22 polyclonal antibody, diluted in PBS–BSA (45–60 min, RT), washed in PBS–BSA and exposed to protein A–5 nm gold (purchased from Dr Jan Slot, Utrecht University, The Netherlands; diluted in PBS–BSA) (45 min, RT). After washing, cells were incubated with 50 µg/ml protein A in PBS–BSA (15 min, RT), washed, then incubated with the next antibody, diluted in PBS–BSA (45 min, RT) and, after washing, exposed to protein A–10 nm gold, followed by washing. In experiments where a third antibody was used, cells were again incubated with 50 µg/ml protein A, washed, then incubated with the third antibody (45 min, RT), followed by washing, exposure to protein A–15 nm gold, and further washing. Following labeling, cells were fixed with 2.0% (v/v) glutaraldehyde, 100 mM sodium cacodylate pH 7.4, 1 mM CaCl2, 0.5 mM MgCl2 (30 min, RT), rinsed with 100 mM sodium cacodylate pH 7.4 and osmicated with 1.5% OsO4 (w/v), 100 mM sodium cacodylate pH 7.4, 1% (w/v) K4Fe(CN)6 (60 min, 4°C). After several rinses with H2O, samples were treated for 1 min with 0.15% tannic acid (in H2O), then en bloc stained overnight with 0.5% (w/v) uranyl acetate in H2O. Filters were dehydrated in a graded series of ethanol, embedded in the epoxy resin LX-112 (Ladd), and sectioned parallel to the substratum with a diamond knife (Diatome). Sections, gold in color (∼100–120 nm), were mounted on butvar-coated nickel grids and viewed at 100 kV in a JEOL (Japan) 100 CX electron microscope without further contrasting. Changing the order of addition of light chain or AP1-specific antibodies did not affect the results. No staining was observed if specific antibodies were omitted from the staining protocol, and non-specific labeling of mitochondria, Golgi cisternae or the nuclear membrane was rarely observed. For the statistical analysis, 50 random photomicrographs (taken at a magnification of 19 000) were taken from transfectants expressing CHC22. A total of 283 vesicles were counted and categorized according to staining for CHC22, AP1 adaptor complex or light chain.
Cell fractionation, immunoprecipitation and immunoblotting
Membrane and cytosol fractions were prepared from HeLa cells lysed by repeated freeze–thawing in isolation buffer [100 mM 2-(N-morpholino)ethanesulfonic acid (MES), 1 mM EGTA, 1 mM EDTA, 0.5 mM MgCl2, 1 mM dithiothreitol (DTT) pH 6.7]. Lysates were centrifuged at 45 000 r.p.m. (Beckman TLA.45) for 30 min and the cytosol recovered in the supernatant. The membrane pellets were extracted with Triton X-100 lysis buffer (0.5% Triton X-100, 150 mM NaCl, 10 mM imidazole, 1 mM DTT pH 7) (30 min, 4°C), centrifuged, and the resulting supernatant was used as a membrane fraction. For immunoprecipitation, detergent lysates were diluted 5-fold with isolation buffer and Triton X-100 was added to the cytosolic fractions to a final concentration of 0.1%. Both fractions were incubated for 4 h at 4°C with protein G–Sepharose (Pharmacia) prebound with individual antibodies. The immunoprecipitates were then washed with 4:1 isolation buffer:Triton X-100 lysis buffer. Samples were resolved by SDS–PAGE, transferred to nitrocellulose and proteins detected by antibody labeling and chemiluminescence.
Acknowledgments
Acknowledgements
We thank H.Sirotkin and R.Kucherlapati for the CHC22 cDNA fragment, D.A.Riethof for technical assistance in production of the anti-CHC22 MAb, R.Kelly, M.Robinson and L.Traub for antibodies, J.Morgan for vector pJM601 and W.G.Ruiz for outstanding technical support with electron microscopy. This work was supported by NIH grants GM57657 and GM38093 to F.M.B., a Wellcome Prize Travelling Research Fellowship from The Wellcome Trust to M.C.T., an Arthritis Foundation Postdoctoral Fellowship to C.-Y.C., NIH grant AI42093 to W.S. and NIH grant DK51970 to G.A.
REFERENCES
- Acton S. and Brodsky,F.M. (1990) Predominance of clathrin light chain LCb correlates with the presence of a regulated secretory pathway. J. Cell Biol., 111, 1419–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan V.J. and Schroer,T.A. (1999) Membrane motors. Curr. Opin. Cell Biol., 11, 476–482. [DOI] [PubMed] [Google Scholar]
- Blank G.S. and Brodsky,F.M. (1986) Site-specific disruption of clathrin assembly produces novel structures. EMBO J., 5, 2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta A., Lindsay,E.A., Jurecic,V. and Baldini,A. (1997) Comparative mapping of the DiGeorge syndrome region in mouse shows inconsistent gene order and differential degree of gene conservation. Mamm. Genome, 8, 890–895. [DOI] [PubMed] [Google Scholar]
- Buss F., Kendrick-Jones,J., Lionne,C., Knight,A.E., Côté,G.P. and Luzio,J.P. (1998) The localization of myosin VI at the Golgi complex and leading edge of fibroblasts and its phosphorylation and recruitment into membrane ruffles of A431 cells after growth factor stimulation. J. Cell Biol., 143, 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers P., Stieger,M. and Burn,P. (1994) Overproduction of bacterial chaperones improves the solubility of recombinant protein tyrosine kinases in Escherichia coli. Cell. Mol. Biol. (Noisy-le-grand), 40, 635–644. [PubMed] [Google Scholar]
- Chin D.J., Straubinger,R.M., Acton,S., Näthke,I. and Brodsky,F.M. (1989) 100-kDa polypeptides in peripheral clathrin-coated vesicles are required for receptor-mediated endocytosis. Proc. Natl Acad. Sci. USA, 86, 9289–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C.R., Odorizzi,G., Payne,G.S. and Emr,S.D. (1997) The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell, 91, 109–118. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E.C., Klumperman,J., Stoorvogel,W. and Bonifacino,J.S. (1998) Association of the AP-3 complex with clathrin. Science, 280, 431–434. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E.C., Mullins,C. and Bonifacino,J.S. (1999) AP-4, a novel protein complex related to clathrin adaptors. J. Biol. Chem., 274, 7278–7285. [DOI] [PubMed] [Google Scholar]
- Dodge G.R., Kovalszky,I., McBride,O.W., Yi,H.J., Chu,M., Saitta,B., Stokes,D.G. and Ioozo,R.V. (1991) Human clathrin heavy chain (CLTC): partial molecular cloning, expression and mapping of the gene to human chromosome 17q11-qter. Genomics, 11, 174–178. [DOI] [PubMed] [Google Scholar]
- Dunn W.A., Hubbard,A.L. and Aronson,N.N.J. (1980) Low temperature selectively inhibits fusion between pinocytic vesicles and lysosomes during heterophagy of 125I-asialofetuin by the perfused rat liver. J. Biol. Chem., 255, 5971–5978. [PubMed] [Google Scholar]
- Fabrizio J.J., Hime,G., Lemmon,S.K. and Bazinet,C. (1998) Genetic dissection of sperm individualization in Drosophila melanogaster. Development, 125, 1833–1843. [DOI] [PubMed] [Google Scholar]
- Fujimoto L.M., Roth,R., Heuser,J.E. and Schmid,S.L. (2000) Actin assembly plays a variable, but not obligatory role in receptor-mediated endocytosis in mammalian cells. Traffic, 1, 161–171. [DOI] [PubMed] [Google Scholar]
- Gaidarov I., Santini,F., Warren,R.A. and Keen,J.H. (1999) Spatial control of coated-pit dynamics in living cells. Nature Cell Biol., 1, 1–7. [DOI] [PubMed] [Google Scholar]
- Gong W., Emanuel,B.S., Collins,J., Kim,D.H., Wang,Z., Chen,F., Zhang,G., Roe,B. and Budarf,M.L. (1996) A transcription map of the DiGeorge and velo-cardio-facial syndrome minimal critical region on 22q11. Hum. Mol. Genet., 5, 789–800. [DOI] [PubMed] [Google Scholar]
- Griffiths G., Pfeiffer,S., Simons,K. and Matlin,K. (1985) Exit of newly synthesized membrane proteins from the trans cisterna of the Golgi complex to the plasma membrane. J. Cell Biol., 101, 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J. and Robinson,M.S. (1998) Clathrin and adaptors. Biochim. Biophys. Acta, 1404, 173–193. [DOI] [PubMed] [Google Scholar]
- Hirst J., Bright,N.A., Rous,B. and Robinson,M.S. (1999) Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell, 10, 2787–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran E.A. and Holzbaur,E.L. (1998) Speculating about spectrin: new insights into the Golgi-associated cytoskeleton. Trends Cell Biol., 8, 26–29. [DOI] [PubMed] [Google Scholar]
- Holmes S.E. et al. (1997) Disruption of the clathrin heavy chain-like gene (CLTCL) associated with features of DGS/VCFS: a balanced (21;22)(p12;q11) translocation. Hum. Mol. Genet., 6, 357–367. [DOI] [PubMed] [Google Scholar]
- Ikonen E., de Almeid,J.B., Fath,K.R., Burgess,D.R., Ashman,K., Simons,K. and Stow,J.L. (1997) Myosin II associated with Golgi membranes: identification of p200 as nonmuscle myosin II on Golgi-derived vesicles. J. Cell Sci., 110, 2155–2164. [DOI] [PubMed] [Google Scholar]
- Kedra D., Peyrard,M., Fransson,I., Collins,J.E., Dunham,I., Roe,B.A. and Dumanski,J.P. (1996) Characterization of a second human clathrin heavy chain polypeptide gene (CLH-22) from chromosome 22q11. Hum. Mol. Genet., 5, 625–631. [DOI] [PubMed] [Google Scholar]
- Lindsay E.A., Rizzu,P., Antonacci,R., Jurecic,V., Delmas-Mata,J., Lee,C.-C., Kim,U.-J., Scambler,P.J. and Baldini,A. (1996) A transcription map in the CATCH22 critical region: identification, mapping and ordering of four novel transcripts expressed in heart. Genomics, 32, 104–112. [DOI] [PubMed] [Google Scholar]
- Liu S.-H., Wong,M.L., Craik,C.S. and Brodsky,F.M. (1995) Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell, 83, 257–267. [DOI] [PubMed] [Google Scholar]
- Liu S.-H., Marks,M.S. and Brodsky,F.M. (1998) A dominant negative clathrin mutant differentially affects trafficking of molecules with distinct sorting motifs in the class II MHC pathway. J. Cell Biol., 140, 1023–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long K.R., Trofatter,J.A., Ramesh,V., McCormick,M.K. and Buckler,A.J. (1996) Cloning and characterization of a novel human clathrin heavy chain gene (CLTCL). Genomics, 35, 466–472. [DOI] [PubMed] [Google Scholar]
- Matlin K.S. and Simons,K. (1983) Reduced temperature prevents transfer of a membrane glycoprotein to the cell surface but does not prevent terminal glycosylation. Cell, 34, 233–243. [DOI] [PubMed] [Google Scholar]
- Merrifield C.J., Moss,S.E., Ballestrem,C., Imhof,B.A., Giese,G., Wunderlich,I. and Almers,W. (1999) Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nature Cell Biol., 1, 72–74. [DOI] [PubMed] [Google Scholar]
- Müsch A., Cohen,D. and Rodriguez-Boulan,E. (1997) Myosin II is involved in the production of constitutive transport vesicles from the TGN. J. Cell Biol., 138, 291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näthke I.S., Heuser,J., Lupas,A., Stock,J., Turck,C.W. and Brodsky,F.M. (1992) Folding and trimerization of clathrin subunits at the triskelion hub. Cell, 68, 899–910. [DOI] [PubMed] [Google Scholar]
- Niswonger M.L. and O’Halloran,T.J. (1997) A novel role of clathrin in cytokinesis. Proc. Natl Acad. Sci. USA, 94, 8575–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi C.E., Dell’Angelica,E.C. and Bonifacino,J.S. (1998) ADP-ribosylation factor (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J. Cell Biol., 142, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Way,M., Zorzi,N. and Stang,E. (1997) Caveolin-3 associates with developing T-tubules during muscle differentiation. J. Cell Biol., 136, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley U. and Parham,P. (1993) Clathrin: its role in receptor-mediated vesicular transport and specialized functions in neurons. Crit. Rev. Biochem. Mol. Biol., 28, 431–464. [DOI] [PubMed] [Google Scholar]
- Ryan A.K. et al. (1997) Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J. Med. Genet., 34, 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K. and Olsnes,S. (1979) Effect of temperature on the uptake, excretion and degradation of abrin and ricin by HeLa cells. Exp. Cell Res., 121, 15–25. [DOI] [PubMed] [Google Scholar]
- Saraste J. and Kuismanen,E. (1984) Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell, 38, 535–549. [DOI] [PubMed] [Google Scholar]
- Schmid S.L. (1997) Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu. Rev. Biochem., 66, 511–548. [DOI] [PubMed] [Google Scholar]
- Seeger M. and Payne,G.S. (1992) Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J. Cell Biol., 118, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F., Peden,A.A., Christopoulou,L. and Robinson,M.S. (1997) Characterization of the adaptor-related protein complex, AP3. J. Cell Biol., 137, 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin H. et al. (1996) Isolation of a new clathrin heavy chain gene with muscle-specific expression from the region commonly deleted in velo-cardio-facial syndrome. Hum. Mol. Genet., 5, 617–624. [DOI] [PubMed] [Google Scholar]
- Stow J.L. and Heimann,K. (1998) Vesicle budding on Golgi membranes: regulation by G proteins and myosin motors. Biochim. Biophys. Acta, 1404, 161–171. [DOI] [PubMed] [Google Scholar]
- Taunton J., Rowning,B.A., Coughlin,M.L., Wu,M., Moon,R.T., Mitchison,T.J. and Larabell,C.A. (2000) Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol., 148, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E. and Ungewickell,H. (1991) Bovine brain clathrin light chains impede heavy chain assembly in vitro. J. Biol. Chem., 266, 12710–12714. [PubMed] [Google Scholar]
- Wendland B., Emr,S.D. and Riezman,H. (1998) Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr. Opin. Cell Biol., 10, 513–522. [DOI] [PubMed] [Google Scholar]