Abstract
Infection of eukaryotic cells with lytic RNA viruses results in extensive interactions of viral gene products with macromolecular pathways of the host, ultimately leading to death of the infected cells. We show here that infection of cells with poliovirus results in the cytoplasmic accumulation of a variety of shuttling and non-shuttling nuclear proteins that use multiple nuclear import pathways. In vitro nuclear import assays using semi-permeabilized infected cells confirmed that nuclear import was blocked and demonstrated that docking of nuclear import receptor–cargo complexes at the cytoplasmic face of the nuclear pore complex (NPC) was prevented. Analysis of components of the NPC revealed that two proteins, Nup153 and p62, were proteolyzed during poliovirus infection. These results suggest that the cytoplasmic relocalization of numerous cellular proteins is caused by the inhibition of multiple nuclear import pathways via alterations in NPC composition in poliovirus-infected cells. Blocking of nuclear import points to a novel strategy by which cytoplasmic RNA viruses can evade host immune defenses, by preventing signal transduction to the nucleus.
Keywords: hnRNPs/nuclear import/nuclear pore complex/Nup 153/poliovirus
Introduction
Poliovirus is a positive-stranded RNA virus whose replicative cycle is carried out entirely in the cytoplasm of the infected host cell. The 7500 nucleotide genome encodes a 220 kDa polyprotein, which is processed by virus-encoded proteases to yield the final structural and non-structural proteins (reviewed in Rueckert, 1996). Extensive virus–host interactions can be observed in infected cells, such as induction and subsequent inhibition of apoptosis (Tolskaya et al., 1995; Agol et al., 2000), inhibition of host cell mRNA translation (Etchison et al., 1982; Gradi et al., 1998), inactivation of the cellular transcription machinery (Clark et al., 1993; Yalamanchili et al., 1997), inhibition of host protein secretion (Doedens and Kirkegaard, 1995) and subversion of endoplasmic reticulum membranes to generate sites for viral replication complexes (Bienz et al., 1983; Suhy et al., 2000).
Despite the cytoplasmic location of the viral replication complexes, a number of host nuclear proteins have been shown to interact with viral RNA or viral gene products. For example, the La autoantigen enhances translation of the viral mRNA in cell-free translation systems through interaction with the internal ribosome entry site (IRES) (Meerovitch et al., 1989). Additionally, the nuclear factor Sam68 interacts with the viral RNA-dependent RNA polymerase 3D (McBride et al., 1996), and nucleolin can be crosslinked to the 3′ non-coding region (3′ NCR) of replication-competent, but not replication-defective viral mRNAs (Waggoner and Sarnow, 1998). Although clear roles for La, Sam68 and nucleolin in the viral life cycle have yet to be defined, they all relocalize from the nucleus to the cytoplasm during poliovirus infection (Meerovitch et al., 1989; McBride et al., 1996; Waggoner and Sarnow, 1998).
In this study, we have examined the mechanism responsible for the accumulation of host nuclear proteins in the cytoplasm of poliovirus-infected cells. We show that reporter fusion proteins carrying either a classical or a transportin nuclear localization signal (NLS) relocalize from the nucleus to the cytoplasm in poliovirus-infected cells. Furthermore, several endogenous nuclear proteins that utilize either the classical, transportin or other nuclear import pathways were found to accumulate in the cytoplasm of infected cells, suggesting that poliovirus infection causes a general inhibition of nuclear– cytoplasmic trafficking. This notion was substantiated by the finding that NLS-containing cargo was blocked from entering the nucleus in a cell-free nuclear import assay derived from infected cells. Analysis of the nuclear pore complex (NPC) revealed that at least two proteins, Nup153 and p62, were proteolyzed in infected cells. These findings suggest an intriguing mechanism by which lytic cytoplasmic RNA viruses may subvert nuclear import and, likely, cell signaling pathways to prevent onset of antiviral responses.
Results
Reporter proteins that contain either classical or transportin nuclear import signals accumulate in the cytoplasm of poliovirus-infected cells
We have reported that nucleolin relocalizes from the nucleolus to the cytoplasm in poliovirus-infected cells. In contrast, fibrillarin, another resident of the nucleolus, did not exit the nucleolus in infected cells (Waggoner and Sarnow, 1998). Nucleolin contains a bipartite NLS (Schmidt-Zachmann and Nigg, 1993), which serves as cargo for the classical import pathway. Nuclear import by this pathway is mediated by a heterodimer, consisting of the NLS binding adapter importin α and the receptor importin β, which mediates transport of the cargo–receptor complex through the NPC (reviewed in Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Nakielny and Dreyfuss, 1999). To examine the mechanism of cytoplasmic accumulation of nucleolin in infected cells, we tested whether nuclear import and export pathways were functional in poliovirus-infected cells.
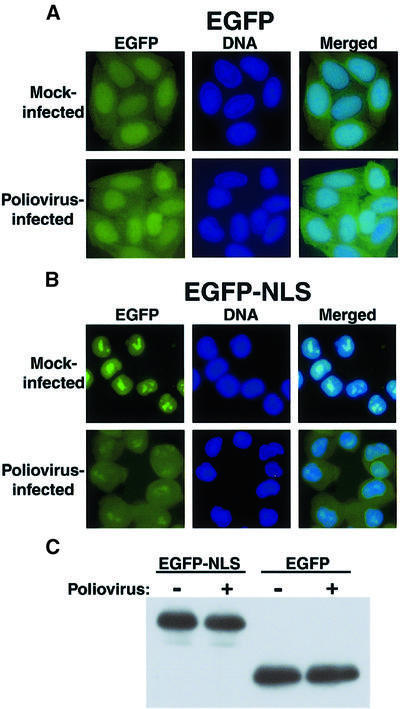
To monitor whether the classical nuclear import pathway was altered in poliovirus-infected cells, cell lines that stably express the enhanced green fluorescent protein (EGFP) or EGFP linked to a NLS derived from the large T antigen of simian virus 40 (EGFP–NLS) were isolated and infected with poliovirus. Cells expressing EGFP displayed fluorescence in both the nucleus and the cytoplasm (Figure 1A, Mock-infected). This finding was expected because the size of EGFP (∼30 kDa) permits diffusion between the nucleus and the cytoplasm. The distribution pattern of EGFP without an NLS was not altered when the cells were infected with poliovirus for a period of 4.5 h (Figure 1A, Poliovirus-infected). Thus, infection with poliovirus had no significant effect on the cellular distribution of a reporter protein lacking an NLS. As expected, EGFP containing a NLS resided entirely in the nucleus of mock-infected cells (Figure 1B, Mock-infected). Infection of EGFP–NLS-expressing cells with poliovirus resulted in significant accumulation of EGFP–NLS in the cytoplasm (Figure 1B, Poliovirus-infected). Because poliovirus encodes several proteases that can degrade non-viral proteins (Clark et al., 1993; Yalamanchili et al., 1997; Gradi et al., 1998; Joachims et al., 1999), it was possible that proteolysis of EGFP–NLS resulted in cytoplasmic relocalization of EGFP molecules that had lost the NLS. Such a mechanism has been postulated for the cytoplasmic relocalization of the La autoantigen in infected cells (Shiroki et al., 1999). However, Figure 1C shows that the EGFP–NLS and EGFP proteins were not proteolyzed in infected cells. Therefore, either poliovirus infection results in the inhibition of nuclear import of EGFP–NLS, or viral infection stimulates enhanced export of NLS-containing EGFP from the nucleus to the cytoplasm. If the latter scenario were the case, enhanced export would be specific to NLS-containing cargo molecules, because EGFP did not relocalize to the cytoplasm in infected cells (Figure 1A).
Fig. 1. Intracellular localization of EGFP and EGFP–NLS molecules in uninfected and poliovirus-infected cells. (A) HeLa cells stably expressing EGFP were mock infected or infected with poliovirus as indicated. Cells were processed and examined by fluorescent microscopy at 4.5 h after infection. EGFP fluorescence was visualized using a FITC filter. DNA: Hoechst 33258-stained nuclei were examined with a UV filter. Merged: shows the FITC and Hoechst images merged. (B) HeLa cells stably expressing EGFP–NLS fusion proteins were examined as described in (A). (C) Immunoblot analysis of EGFP–NLS- and EGFP-expressing cell lines. Whole-cell lysates were prepared after 4.5 h of infection from either mock- or poliovirus-infected cells. EGFP was detected by immunoblotting using a polyclonal antibody to GFP (Clontech).
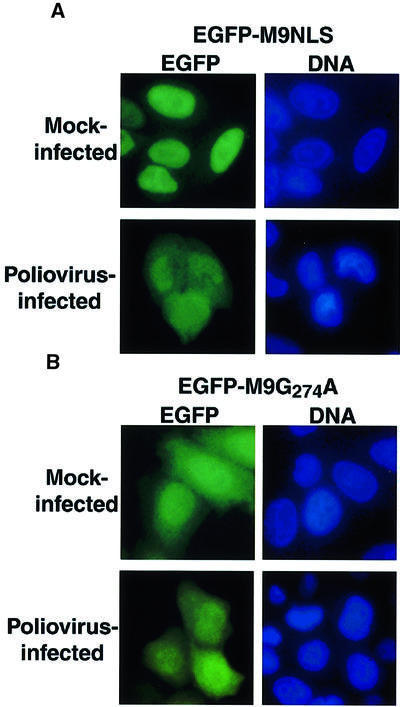
A similar strategy was used to determine whether poliovirus infection affected the transportin import pathway. Cargos that use the transportin pathway consist of a variety of heterogeneous nuclear ribonucleoproteins (hnRNPs) that contain a glycine-rich motif, known as the M9 NLS, which is recognized by the import receptor transportin (reviewed in Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Nakielny and Dreyfuss, 1999). Shuttling is mediated by the bidirectional M9 signal, which targets M9-containing cargo to the nuclear import receptor transportin and to an as yet unidentified nuclear export receptor (Nakielny and Dreyfuss, 1999). The M9 NLS of hnRNP A1 (Pollard et al., 1996) was fused to EGFP and the intracellular distribution of EGFP– M9NLS molecules was examined after transient transfection of EGFP–M9NLS-expressing plasmids into HeLa cells. Again, this molecule is small enough to diffuse across the NPC. Figure 2A shows that the EGFP–M9NLS fusion protein resided predominantly in the nucleus in mock-infected cells. However, infection with poliovirus resulted in a noticeable accumulation of EGFP–M9NLS in the cytoplasm (Figure 2A). This redistribution was not due to proteolytic removal of the M9 NLS, as no difference in the mobility of the EGFP–M9NLS protein was observed in uninfected and infected cell lysates (data not shown). To determine whether the relocalization of EGFP–M9NLS required a functional NLS, we examined the distribution of EGFP–M9NLS molecules in which the glycine at position 274 was changed to alanine (EGFP–M9G274A) (Pollard et al., 1996). This mutation has been shown to abrogate the ability of the M9 sequence to mediate nuclear import and export. EGFP–M9G274A resided both in the cytoplasm and in the nucleus in both mock- and poliovirus-infected cells (Figure 2B), consistent with the expected defect of this molecule in nuclear import. Importantly, the intracellular distribution of EGFP– M9NLS in poliovirus-infected cells was indistinguishable from the distribution of EGFP–M9G274A in mock- or poliovirus-infected cells (Figure 2). These findings show that reporter proteins that contain either classical or M9-type NLS motifs relocalize to the cytoplasm in poliovirus-infected cells; therefore, it is suggested that these two import pathways are disrupted in some way in infected cells.
Fig. 2. Intracellular localization of EGFP–M9NLS and EGFP–M9G274A molecules in mock-infected and poliovirus-infected cells. (A) HeLa cells were transiently transfected with plasmid pEGFP-M9NLS and mock infected or infected with poliovirus at 40 h after transfection and examined 4.5 h after infection. Labeling of panels is as described in Figure 1. (B) Same as in (A), except that cells were transfected with plasmid pEGFP-M9G274A.
Endogenous proteins that utilize several different nuclear import pathways are relocalized to the cytoplasm in poliovirus-infected cells
To determine whether the nuclear localization of endogenous cellular proteins besides La, Sam68 and nucleolin was affected in poliovirus-infected cells, we examined the subcellular distribution of hnRNPs A1, K and C in the presence and absence of poliovirus infection.
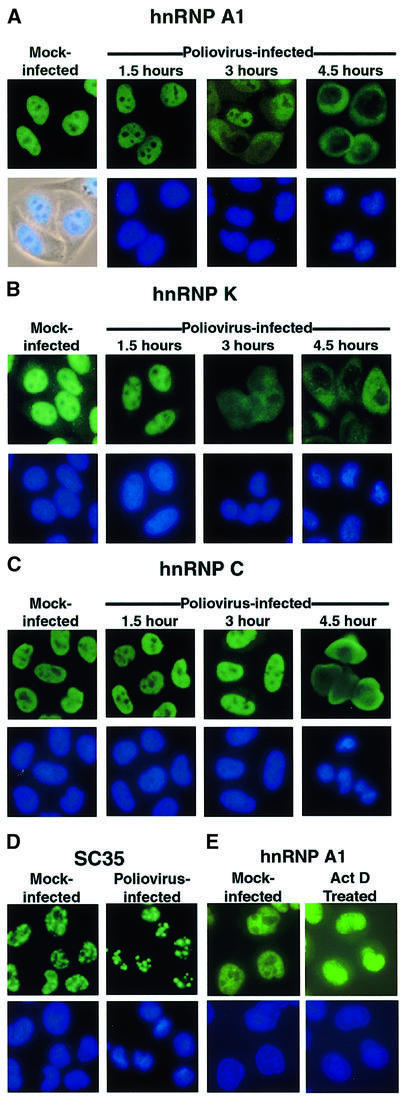
The well-studied hnRNP A1 protein shuttles between the nucleus and the cytoplasm and is involved in both pre-mRNA splicing and nuclear export of mRNA molecules (reviewed in Dreyfuss et al., 1993). Indirect immunofluorescence analysis of cells revealed that hnRNP A1 did relocalize from the nucleus to the cytoplasm during the course of infection with poliovirus (Figure 3A). The kinetics of relocalization were very similar to those reported previously for La (Meerovitch et al., 1993), Sam68 (McBride et al., 1996) and nucleolin (Waggoner and Sarnow, 1998), with cytoplasmic staining becoming detectable at 3 h after infection and essentially complete relocalization occurring by 4.5 h after infection (Figure 3A). Previous work has shown that inhibitors of RNA polymerase II can cause relocalization of hnRNP A1 from the nucleus to the cytoplasm (Pinol-Roma and Dreyfuss, 1991). To examine whether the cytoplasmic accumulation of hnRNP A1 in infected cells was caused by the poliovirus-induced inhibition of cellular transcription (Clark et al., 1993), we examined the cytoplasmic accumulation of hnRNP A1 in cells treated with actinomycin D. Comparison of Figure 3A with E shows that the cytoplasmic accumulation of hnRNP A1 observed in infected cells was more dramatic than in actinomycin D-treated cells. Thus, inhibition of transcription alone is not sufficient to explain the relocalization of hnRNP A1 in poliovirus-infected cells.
Fig. 3. Intracellular localization of endogenous cellular proteins in uninfected and poliovirus-infected cells. (A) Uninfected (Mock-infected) or HeLa cells infected with poliovirus for 1.5, 3 or 4.5 h were fixed and stained with an antibody directed against hnRNP A1. Top panels show cells examined using a FITC filter, bottom panels show Hoechst 33258 staining of the same field using a UV filter or an overlay of the UV image onto a phase image (Mock-infected). (B) At the times indicated cells were fixed and stained with an antibody directed against hnRNP K. Labeling of the panels is as described for (A). (C) At the times indicated, cells were fixed and stained with a monoclonal antibody directed against hnRNP C. (D) Mock- or poliovirus-infected cells were fixed and stained using a monoclonal antibody specific for SC35. (E) Mock-infected HeLa cells were incubated without or with actinomycin D (5 µg/ml) for 4 h (Act D Treated), then fixed and stained with an antibody directed against hnRNP A1.
The shuttling protein hnRNP K can function as an activator or repressor of transcription (reviewed in Krecic and Swanson, 1999) and in translational silencing of the 15-lipoxygenase mRNA (Ostareck et al., 1997). Nuclear–cytoplasmic shuttling is mediated by a 40 amino acid motif, termed KNS, which targets hnRNP K to both import and export receptors that are distinct from the receptors that transport classical NLS- or M9 NLS-containing proteins (Michael et al., 1997). Indirect immunofluorescence analysis showed that hnRNP K was localized predominantly to the nucleus in mock-infected cells. Like hnRNP A1, hnRNP K relocalized to the cytoplasm after 4.5 h of infection with poliovirus (Figure 3B). The kinetics of relocalization were very similar to those described above for hnRNP A1. These results show that proteins containing classical, M9 and KNS NLSs are relocalized in poliovirus-infected HeLa cells.
Nucleolin, Sam68, La, hnRNP A1 and hnRNP K are all known to shuttle between the nucleus and cytoplasm. Thus, it was possible that relocalization to the cytoplasm during poliovirus infection might be limited to shuttling proteins. To explore this possibility, we examined the distribution of hnRNP C in poliovirus-infected cells. hnRNP C is a very abundant nuclear protein, which forms hetero-tetrameric complexes that can bind cooperatively to RNA (Krecic and Swanson, 1999). Although hnRNP C has been implicated in nuclear RNA metabolism (Krecic and Swanson, 1999) and embryonic development in the mouse (Williamson et al., 2000), its exact roles in these processes are unknown. Importantly, hnRNP C resides entirely in the nucleus due to the presence of a 78 amino acid motif, which functions as a nuclear retention signal (Nakielny and Dreyfuss, 1996). Indirect immunofluorescence analysis revealed that hnRNP C resided in the nucleus in mock-infected cells, but relocalized from the nucleus to the cytoplasm by 4.5 h after infection with poliovirus (Figure 3C). Interestingly, hnRNP C seemed to relocalize with slightly delayed kinetics compared with the shuttling proteins (Figure 3, compare C with A and B), suggesting that nuclear retention may affect the redistribution of proteins during poliovirus infection. In any case, these results indicate that nuclear–cytoplasmic shuttling is not required for a nuclear protein to relocalize to the cytoplasm during poliovirus infection.
Nuclear localization of splicing factor SC35 remains unaffected in poliovirus-infected cells
We have shown previously that the nucleolar protein fibrillarin and the nuclear TATA binding protein failed to relocalize during poliovirus infection (McBride et al., 1996; Waggoner and Sarnow, 1998), indicating that not all nuclear proteins relocalize in poliovirus-infected cells. Recently, it has been shown that a group of abundant serine/arginine-rich proteins, known as SR proteins, are imported into the nucleus by a novel receptor, termed transportin-SR (Kataoka et al., 1999). SR proteins, such as SC35, are essential splicing factors that can also regulate alternate splice-site selection (reviewed in Fu, 1995). The SR domain in these proteins functions as an NLS by interacting with transportin-SR, a member of the importin β family of receptor proteins (Kataoka et al., 1999). To determine whether SR proteins are relocalized during poliovirus infection, we examined the subcellular distribution of SC35, a non-shuttling splicing factor, in uninfected and infected cells. Figure 3D shows that SC35 localized to characteristic nuclear speckles (Fu, 1995) both in mock- and poliovirus-infected cells. A similar observation was reported by Meerovitch et al. (1993) who noted that SC35 had not relocalized by 3 h in poliovirus-infected cells. The failure of SC35 to relocalize could indicate that the transportin-SR pathway remains functional in poliovirus-infected cells. Alternatively, pre-existing SC35 may be sequestered in the nucleus and unable to relocalize to the cytoplasm in infected cells, while the levels of newly synthesized SC35 in the cytoplasm may be too low to detect by indirect immunofluorescence. More importantly, these results show that not all nuclear proteins accumulate in the cytoplasm in poliovirus-infected cells, making it unlikely that a poliovirus-induced leakage of the nuclear envelope caused the cytoplasmic accumulation of nucleolin and selected hnRNPs.
Import of an EGFP–NLS cargo in a cell-free nuclear import assay is impaired in poliovirus-infected cells
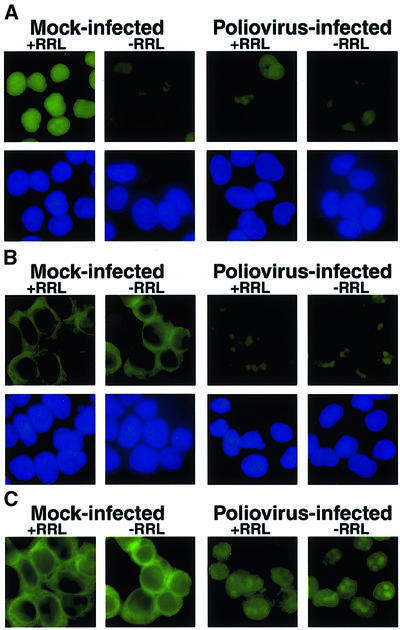
To test directly whether nuclear import is inhibited by poliovirus infection, we monitored the nuclear import of a classical NLS-containing protein in digitonin-permeabilized cells in the presence of cytosol and energy (Adam et al., 1990). When permeabilized mock-infected cells were incubated with GFP–NLS, an energy-regenerating system and rabbit reticulocyte lysate (RRL) as a source of cytosolic factors, nuclear fluorescence was observed (Figure 4A, Mock-infected, +RRL). In contrast, only low levels of the GFP–NLS import substrate accumulated in the nucleus when the RRL was omitted (Figure 4A, Mock-infected, –RRL). Nuclear fluorescence was dependent on an NLS, because substrates lacking an NLS did not exhibit nuclear fluorescence in the presence of RRL (data not shown). When permeabilized poliovirus-infected cells were incubated in the presence or absence of RRL, levels of nuclear fluorescence were indistinguishable from that seen when mock-infected cells were incubated in the absence of RRL (Figure 4A, Poliovirus-infected). These data show that poliovirus infection results in an inability to mediate nuclear import of cargo in a cell-free system, suggesting that components of the nuclear import machinery are altered by poliovirus infection.
Fig. 4. Cell-free nuclear import assays. (A) Uninfected cells (Mock-infected) or cells that had been infected with poliovirus for 4 h (Poliovirus-infected) were permeabilized and used in an in vitro nuclear import assay. Assays were carried out in the presence (+RRL) or absence (–RRL) of rabbit reticulocyte lysate as a source of cytosolic factors. Top panels show GFP using a FITC filter and bottom panels show Hoechst 33258 staining of DNA using a UV filter. All GFP images were acquired using identical exposure times and manipulations in Photoshop 5.0. (B) Same as in (A), except that creatine kinase, creatine phosphate, ATP and GTP were omitted from the reaction. GFP images were acquired using the same exposure time as in (A). (C) Longer exposure of GFP fields in (B).
Receptor–cargo complexes are unable to dock at the NPC in poliovirus-infected cells
For nuclear import to occur, receptor–cargo complexes must first dock at the cytoplasmic face of the NPC, and presumably are then translocated through the nuclear pore before cargo is released in the nucleoplasm. Thus, we tested whether docking of the receptor–cargo complex at the NPC was affected in poliovirus-infected cells. Previous work has shown that in the absence of an energy source, or at non-physiological temperatures, cargo– receptor complexes can not translocate through the pore; instead, they accumulate at the cytoplasmic face of the NPC (Adam et al., 1990). Figure 4B shows that permeabilized mock-infected cells that were incubated in the presence or absence of RRL, but without an energy source (see Materials and methods), accumulated the GFP–NLS import cargo at the nuclear rim, indicating that docking had occurred. In contrast, poliovirus-infected cells that were incubated under identical conditions accumulated very little cargo at the nuclear rim. Figure 4C shows a longer exposure of Figure 4B, displaying more clearly the accumulation of the import cargo at the nuclear rim; at this length of exposure, some nuclear background fluorescence was observed in permeabilized poliovirus-infected cells. Identical results were obtained when the assays were carried out at 0°C (data not shown). The inability of receptor–cargo complexes to dock at the NPC in permeabilized, poliovirus-infected cells indicates that one of the earliest steps in nuclear import is prevented in poliovirus-infected cells.
The CRM1 nuclear export pathway is functional in poliovirus-infected cells
The results presented so far are consistent with poliovirus infection causing an inhibition of multiple import pathways, with the possible exception of the transportin-SR pathway. However, effects on export pathways might also explain the rapid exit of factors from the nucleus and their accumulation in the cytoplasm in infected cells. To examine this possibility, we employed an experimental system that was designed to monitor selectively the export of nuclear molecules (Love et al., 1998). Briefly, the HIV-1 protein Rev, which localizes to the nucleolus, was fused to the hormone binding domain of the glucocorticoid (GC) receptor and GFP (Love et al., 1998). The N-terminal Rev protein contains a classical NLS (Malim et al., 1989) and a leucine-rich nuclear export signal (NES), which is recognized by the Crm1 export receptor (Fischer et al., 1995). The hormone binding domain of the GC receptor harbors a classical NLS (NLS1) adjacent to the hormone binding domain and a second NLS (NLS2) within the hormone binding domain. NLS2, which can not be separated from the hormone binding domain, is dominant over the classical NLS1 and mediates hormone-dependent nuclear import by an unknown importin α-independent pathway (Savory et al., 1999). In the absence of hormone, the Rev–GC fusion protein resides in the cytoplasm; in the presence of hormone, the fusion protein is imported into the nucleus. Because the fusion protein contains the Rev NES, it is rapidly exported to the cytoplasm upon removal of hormone (Love et al., 1998).
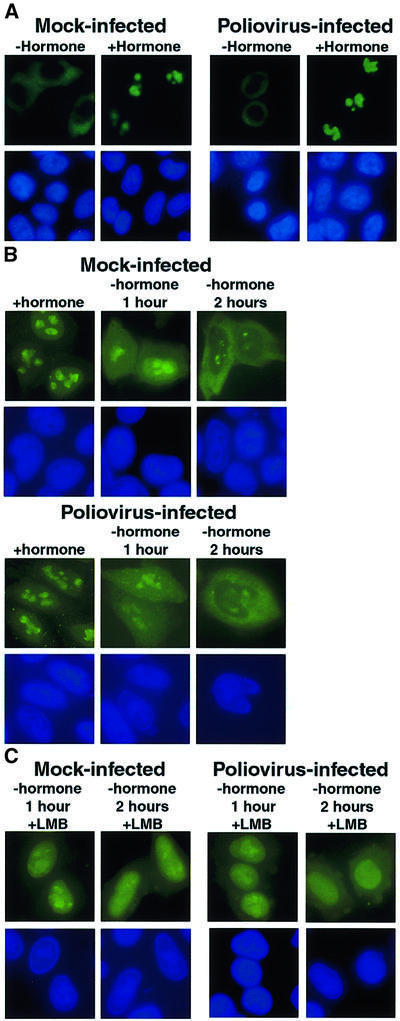
First, we tested whether the fusion protein was imported into the nucleus in poliovirus-infected cells at a time when EGFP–NLS, EGFP–M9 and several cellular proteins had relocalized into the cytoplasm. As expected (Love et al., 1998), in uninfected cells the fusion protein localized to the cytoplasm in the absence of the hormone dexamethasone, but relocalized to the nucleolus 30 min after the addition of hormone (Figure 5A, Mock-infected). Surprisingly, infection with poliovirus did not inhibit nuclear import of the fusion protein (Figure 5A, Poliovirus-infected). This result was not due to poor viral infection efficiency, because nucleolin had relocalized to the cytoplasm in >90% of the cells examined (data not shown). This result demonstrates that not all import pathways are disrupted in poliovirus-infected cells.
Fig. 5. Intracellular localization of Rev–GC fusion proteins in uninfected and poliovirus-infected cells. (A) HeLa cells were transiently transfected with plasmid pXRGG; after 40 h, cells were either mock infected (Mock-infected) or infected with poliovirus (Poliovirus-infected). Four hours after infection cells were untreated (–hormone) or treated with dexamethasone (+hormone) at 1 µM for 30 min prior to fixation. Top panels show GFP using a FITC filter and bottom panels show Hoechst 33258 staining using a UV filter. (B) Forty hours after transfection with pXRGG, cells were either mock infected or infected with poliovirus and dexamethasone was added. 2.5 h later the cells were either fixed (+hormone), or dexamethasone was removed by washing once with PBS and replacing the medium. Cells were processed for fluorescent microscopy at 1 h (–hormone 1 hour) or 2 h (–hormone 2 hours) following removal of dexamethasone. (C) Same as in (B), except that leptomycin B (5 ng/ml, +LMB) was added following removal of dexamethasone.
Next, we examined the efficiency with which the Rev–GC receptor fusion protein was exported from the nucleus in poliovirus-infected cells. Mock- or poliovirus-infected cells expressing the Rev–GC fusion protein were incubated in the presence of dexamethasone to allow the accumulation of Rev–GC in the nucleus (Figure 5B, +hormone). Dexamethasone was removed 2.5 h after infection and the cells were examined 1 and 2 h later (Figure 5B, –hormone 1 hour and 2 hours). One hour after removal of hormone, the Rev–GC protein was distributed equally between the cytoplasm and nucleus, and by 2 h relocalization was essentially complete in both mock- and poliovirus-infected cells. The compound leptomycin B, which inactivates the Rev NES receptor Crm1 (reviewed in Wolff et al., 1997; Adam, 1999), inhibited the export of Rev–GC in both mock- and poliovirus-infected cells (Figure 5C), suggesting that the fusion protein used the Crm1 pathway in mock- and in poliovirus-infected cells. These results indicate that poliovirus infection does not inactivate the Crm1-mediated export pathway.
Components of the NPC are targeted for degradation during poliovirus infection
Our results indicated that proteins that utilize several distinct nuclear import pathways were relocalized in poliovirus-infected cells. Import receptors use the NPC to transport cargo between the cytoplasm and nucleus. The NPC is a 125 MDa complex that is framed by ∼50 different proteins known as nucleoporins (Nups) (reviewed in Stoffler et al., 1999).
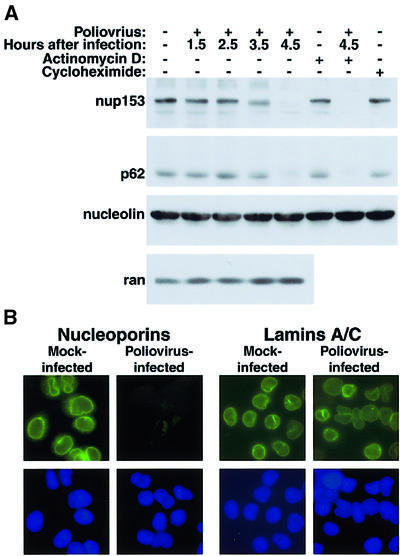
We used monoclonal antibody 414 (Davis and Blobel, 1986), which reacts with a number of nucleoporins in the NPC to examine the status of NPC components in poliovirus-infected cells. In particular, we inspected the integrity of Nup153 which has been reported to interact with importin α/β (Shah et al., 1998), transportin (Nakielny et al., 1999) and various export receptors (Ullman et al., 1999). Immunoblot analysis of whole cell lysates prepared at various times after infection with poliovirus revealed that Nup153 was degraded during the course of infection (Figure 6A). The abundance of Nup153 remained unaltered during the first 2.5 h after infection, began to decline by 3.5 h and was almost undetectable by 4.5 h after infection (Figure 6A). Another nucleoporin, p62 (reviewed in Stoffler et al., 1999), was degraded with similar kinetics to Nup153 in infected cells (Figure 6A). In contrast, the small GTPase Ran (Figure 6A), the nuclear receptor importin β (not shown) and the cargo nucleolin (Figure 6A) were not degraded in infected cells.
Fig. 6. Alterations in the NPC composition in poliovirus-infected HeLa cells. (A) Whole-cell lysates (50 µg) prepared from cells subjected to various treatments were analyzed by immunoblotting with the indicated antibodies. Actinomycin D: cells were treated for 4.5 h with 5 µg/ml actinomycin D. Cycloheximide: cells were treated with 20 µg/ml cycloheximide for 4.5 h. (B) Indirect immunofluorescence using monoclonal antibodies 414 and SC-7292 to detect nucleoporins and lamins, respectively. Cells were either uninfected (Mock-infected) or infected with poliovirus for 4.5 h (Poliovirus-infected). Top panels show cells examined with a FITC filter, bottom panels show the same fields examined with a UV filter to detect Hoechst 33258 staining. FITC images for a given antibody were acquired with identical exposure times and adjustments.
Pulse–chase experiments performed with radiolabeled methionine revealed that the turnover rates of Nup153 and p62 were >5 h in mock-infected cells, but changed to <2 h in poliovirus-infected cells (data not shown). Inhibition of translation by the addition of the elongation inhibitor cycloheximide did not change the relative abundance of Nup153 or p62 (Figure 6A). Inhibition of cellular transcription by actinomycin D did not change the steady-state levels of Nup153 or p62 in uninfected cells, and did not prevent the degradation of both Nup153 and p62 in infected cells (Figure 6A). Because poliovirus replication is insensitive to actinomycin D (reviewed in Schlegel and Kirkegaard, 1995), these results suggest that degradation of Nup153 or p62 was due to ongoing viral gene expression and not due simply to inhibition of cellular transcription or translation.
To examine the status of the NPC in infected cells by another experimental approach, we monitored the NPC in mock-infected and poliovirus-infected cells by indirect immunofluorescence using monoclonal antibody 414 (Davis and Blobel, 1986). Figure 6B shows that uninfected cells stained with antibody 414 displayed the nuclear rim staining characteristic of the NPC (Davis and Blobel, 1986). At higher magnification the fluorescence appears as a speckled staining of the nuclear envelope (data not shown), presumably reflecting staining of individual NPCs. In contrast to what was observed in uninfected cells, a dramatic reduction in staining of the nuclear envelope was observed in infected cells (Figure 6B). This result was not due to a general disruption of proteins associated with the nuclear envelope, as the staining pattern of nuclear lamins A and C was unchanged in poliovirus-infected cells (Figure 6B). These data suggest that the degradation of at least two, perhaps more, components of the NPC resulted in a dramatic perturbation of NPC composition in poliovirus-infected cells. Disruption of the NPC composition could prevent docking, translocation or release of cargo and could have effects that extend to multiple import or export pathways. Importantly, the degradation of Nup153/p62 and the perturbation of NPCs in poliovirus-infected cells occurred at a time that coincided with the relocalization of nuclear proteins.
Discussion
In this study we have addressed the mechanism underlying the relocalization of cellular nuclear factors to the cytoplasm in poliovirus-infected cells. Our results indicate that a variety of nuclear–cytoplasmic shuttling and non-shuttling hnRNPs relocalize to the cytoplasm during the course of infection and suggest that several trafficking pathways are altered in infected cells. This effect is most likely due to changes of the NPC structure. However, not all trafficking pathways were disrupted, as the Rev–GC fusion protein was imported to the nucleus in poliovirus-infected cells at a time when many cellular proteins had relocalized to the cytoplasm. Similarly, export of the Rev–GC fusion protein in virus-infected cells indicates that the CRM1 nuclear export pathway was operational under these experimental conditions. One explanation for the cytoplasmic relocalization of several nuclear proteins in infected cells could be virus-induced leakiness of the nuclear envelope. This scenario predicts a uniform distribution of proteins throughout the cell. This was not the case, as nucleolin, La, Sam68, hnRNP A1, K and C all exhibited predominantly cytoplasmic staining. Secondly, nuclear factors such as SC35, TBP and fibrillarin (Meerovitch et al., 1989; McBride et al., 1996; Waggoner and Sarnow, 1998) did not relocalize to the cytoplasm in infected cells. Furthermore, nuclear export of the Rev–GC fusion protein could be inhibited by leptomycin B, arguing that the Crm1 pathway, not nuclear leaking, was facilitating Rev–GC export in infected cells.
The exact mechanism by which poliovirus induces nuclear–cytoplasmic relocalization of cellular proteins is likely to be complex. However, it is striking that relocalization strongly correlated with an inability of infected semi-permeabilized cells to dock cargo in a cell-free nuclear import assay and with proteolysis of Nup153, p62 and possibly other proteins in the NPC. Several reports have suggested that Nup153 may play an important role in a number of different transport pathways. For example, overexpression of Nup153 has been shown to inhibit mRNA export in BHK cells (Bastos et al., 1996), and antibodies directed against Nup153 also block the export of snRNA, mRNA and 5S rRNA in Xenopus oocytes (Ullman et al., 1999). More recently, it has been shown that Nup153 contains an M9 shuttling domain and a RanGDP binding domain that can interact with a variety of import and export receptors (Shah et al., 1998; Nakielny et al., 1999). Proteolysis of Nup153/p62 in poliovirus-infected cells correlated with the inability of a cargo containing a classical NLS to dock at the NPC and to be transported to the nucleus. It is not clear whether proteolysis of Nup153, p62 or both resulted in the generation of non-functional NPCs or reduced the numbers of NPCs in infected cells. Either scenario would result in an overall decrease in docking of NLS-containing cargo at the NPC. The fact that both nuclear import and export of Rev–GC cargo were functional in infected cells suggests that transport of the Rev–GC fusion protein through the NPC does not require intact Nup153/p62. A more extensive analysis of the NPC composition/structure using a large panel of antibodies directed against distinct Nups should distinguish between these possibilities.
There is ample evidence demonstrating that viruses use components of the nuclear import/export machinery for the shuttling of viral macromolecules; however, only a few examples of viral infections that perturb normal nuclear–cytoplasmic trafficking pathways have been reported. For example, at late times during adenovirus infection, host mRNAs accumulate in the nucleus while viral mRNAs are exported efficiently (reviewed in Schneider and Shenk, 1987). Although viral elements that facilitate the export of adenoviral late mRNAs have been identified, the mechanism responsible for inhibiting the export of host mRNAs is not known. More recently, the vesicular stomatitis virus (VSV) matrix (M) protein was shown to inhibit a variety of different import and export pathways in Xenopus laevis oocytes (Her et al., 1997). In this study, the M protein was shown to inhibit the export of small nuclear RNAs, mRNAs and ribosomal RNAs while export of transfer RNAs was unaffected. The M protein was also shown to inhibit the nuclear import of snRNAs and some proteins. A very recent report has shown that the inhibitory effect of the M protein involves targeting nucleoporin Nup98 (von Kobbe et al., 2000). Very recently, Belov et al. (2000) have reported that nuclear-located EGFP–NLS relocalizes to the cytoplasm in cells infected with either poliovirus, coxsackievirus B3 or VSV. These findings together with the results presented here, raise the possibility that targeting components of the NPC may be a more common mechanism by which viruses disrupt normal nuclear/cytoplasmic trafficking pathways.
The reasons why poliovirus inhibits nuclear import pathways are unknown, yet there are intriguing possibilities. A block in nuclear import is predicted to result in the cytoplasmic accumulation of cellular proteins that could be recruited to help in the translation, amplification and packaging steps of the viral genome (see Introduction). Alternatively, poliovirus-induced inhibition of nuclear import might circumvent antiviral responses of the host cell. For example, it is known that viral infection can induce the translocation of latent transcription factors from the cytoplasm to the nucleus, where they can then activate interferon genes (reviewed in Stark et al., 1998). Sequestration of transcription factors in the cytoplasm would greatly reduce the antiviral response of the infected cell.
Finally, it is not known as yet whether inhibition of nuclear import is caused by a specific viral protein or whether it is a consequence of virus-induced alterations in the infected host cell. Studies with translation inhibitors showed that viral proteins were required for the inhibition of nuclear import (our unpublished data). It is unknown at this point whether viral proteases are responsible for the degradation of Nup153/p62, or whether viral proteins play an indirect role. It is known that poliovirus infection can induce apoptosis in cells infected under certain conditions (Tolskaya et al., 1995). Furthermore, previous work has shown that Nup153, but not p62, can be cleaved in a caspase-dependent manner in cells undergoing apoptosis (Buendia et al., 1999). However, treatment of poliovirus-infected cells with caspase inhibitors does not prevent the degradation of Nup153 or p62 shown here (our unpublished data). In poliovirus-infected cells, the host cell secretory pathway is subverted to provide membranes on which RNA replication complexes are assembled and host secretory traffic is inhibited (reviewed in Schlegel and Kirkegaard, 1995). Curiously, a recent study in yeast has provided a link between disruption of the secretory pathway and a block in nuclear import. Specifically, Nanduri et al. (1999) have shown that various yeast sec mutants accumulated import cargo at the NPC. If a similar scenario occurs in mammalian cells, breakdown of the host secretory pathway may have consequences on the nuclear import pathway in virus-infected cells.
Materials and methods
Cell lines and viruses
HeLa cells were maintained as described previously (Waggoner and Sarnow, 1998). Mahoney type 1 poliovirus stocks were prepared and infections were initiated as described (Waggoner and Sarnow, 1998). DNA transfections were performed using the Lipofectin reagent (Gibco/BRL) following the manufacturer’s recommendations.
Plasmids
To create pEGFP, nucleotides 941–2621 of pIRES-EGFP (Clonetech) were amplified by PCR (5′ primer: 5′-GCGTCGACGTGGATCCACTAGTAACG-3′, 3′ primer: 5′-GCGAATTCGATATCCTTGTACAGCTCGTC-3′) and cloned into pCR2.1 using the TA cloning kit (Invitrogen). The amplified EGFP coding region was isolated as a HindIII–EcoRI fragment and inserted into the corresponding sites of pCDNA3 (Invitrogen) to yield pEGFP. pEGFP-M9 and M9G274A were created by isolating the respective coding regions from pGST-M9 or pGST-M9G274A (Pollard et al., 1996) as EcoRI–XhoI fragments and inserting into the corresponding sites of pEGFP. To link the SV40 TAg NLS to EGFP, the TAg NLS coding region was isolated as an EcoRI–BamHI fragment from pGST-T NLS (Pollard et al., 1996) and inserted into the corresponding sites of pGEM4. This vector was then digested with EcoRI, overlapping ends were repaired using the large fragment of DNA polymerase I, and digested with HincII. The TAg NLS was then ligated into pEGFP that had been digested with EcoRV. The resulting plasmid, pEGFP-NLS, contains four tandem repeats of the TAg NLS (PKKKRKV). pGEX-NLS-GFP encodes a GST–NLS–GFP fusion protein (Rosorius et al., 1999). pXRGG encodes a fusion protein consisting of the full-length HIV-1 Rev protein, linked to the rat glucocorticoid hormone binding domain and GFP (Love et al., 1998).
Indirect immunofluorescence
Mouse monoclonal antibodies used for indirect immunofluorescence analysis included: 9H10 to detect hnRNP A1, 12G4 to detect hnRNP K/J, 4F4 to detect hnRNP C, #S-4045 (Sigma) to detect SC35, 414 (Convance) to detect nucleoporins and #SC-7292 (Santa Cruz Biotechnology) to detect lamins A and C. Before fixation, cells were washed three times with phosphate-buffered saline (PBS). Immunostaining for hnRNP A1, hnRNP K/J, hnRNP C and SC35 was accomplished by fixing cells in 2% formaldehyde for 30 min at 25°C, washing three times with PBS and permeabilizing in acetone at –20°C for 3 min. Immunostaining using antibodies 414 and #SC-7292 was carried out by fixing cells in 3% formaldehyde for 20 min at 25°C, washing three times with PBS and permeabilizing in methanol at –20°C for 5 min.
Following fixation/permeabilization, cells were washed three times with PBS and then blocked in base solution (PBS containing 0.1% fish skin gelatin and 0.05% Triton X-100) for 40 min at 25°C. Coverslips were inverted into 100 µl of base solution containing diluted primary antibody and incubated overnight at 4°C. Coverslips where washed three times in base solution at 25°C and then inverted into 100 µl of base solution containing a 1:100 dilution of FITC-conjugated goat anti-mouse immunoglobulin (Zymed) and incubated for 1 h at 25°C. Coverslips were washed twice in base solution, once in PBS containing 0.2 µg/ml Hoechst 33258, drained and mounted in Vectashield mounting medium (Vector Labs) onto glass slides. Cells were viewed using an Olympus BX-60 fluorescent microscope with a 40× objective and photographed using a 35 mm camera (Figures 2, 3A–D and 5A and B). Alternatively, images were acquired using a Hamamatsu Orca digital camera and Image Pro Plus software with the 40× (Figures 1, 3E and 5C and D), or 60× objective (Figure 6).
In vitro import assay
RRLs (Promega) were prepared by centrifugation at 100 000 g for 30 min at 4°C, followed by extensive dialysis at 4°C against transport buffer TB (20 mM HEPES pH 7.3, 110 mM KOAc, 5 mM NaOAc, 1 mM MgOAc, 1 mM EGTA, 2 mM dithiothreitol and 1 µg/ml each of chymotrypsin, leupeptin, antipain and pepstatin). Dialyzed RRL was aliquoted, frozen in liquid nitrogen and stored at –80°C. The GST–NLS–EGFP fusion protein used as an import substrate in this assay was purified from bacteria transformed with pGEX-NLS-EGFP (Rosorius et al., 1999) using glutathione–Sepharose as described by the manufacturer (Pharmacia). Purified GST–NLS–EGFP fusion protein was dialyzed against TB as described above, before storage at –80°C.
HeLa cells were seeded onto 12 mm glass coverslips 2 days before use in import assays. Mock- or poliovirus-infected cells were prepared at 4 h post-infection by washing once in PBS and once with ice-cold TB, followed by permeabilization for 5 min at 0°C in TB containing 40 µg/ml digitonin. Permeabilized cells were then washed once in TB and inverted into a 20 µl import reaction consisting of TB supplemented with 50% RRL, 0.4 µM GST–NLS–EGFP, 1 mM ATP, 1 mM GTP, 5 mM creatine phosphate and 20 U/ml creatine kinase. Reactions were incubated at 25°C for 30 min, washed once in TB and fixed in 3% formaldehyde for 20 min at 25°C. Following fixation, cells were washed, stained with Hoechst 33258 and mounted onto glass slides as described above. Images were acquired on an Olympus BX-60 fluorescent microscope equipped with a 60× objective using a Hamamatsu Orca digital camera and Image Pro Plus software.
Immunoblotting
HeLa cell lysates were prepared by washing cells once in PBS, followed by resuspension in lysis buffer (Pante et al., 1994). Samples were prepared and analyzed as described previously (Gustin and Imperiale, 1998). Nup153 and p62 were detected using mouse monoclonal antibody 414 (Convance). Mouse monoclonal antibody MS3 was used to detect nucleolin. Ran was detected using rabbit polyclonal sera (Convance).
Acknowledgments
Acknowledgements
We are very grateful to Karla Kirkegaard and Michael Rexach for stimulating discussions and critical reading of the manuscript. We would also like to thank Harris Busch for anti-nucleolin antibodies, Gideon Dreyfuss for the GST-T NLS and M9-NLS plasmids and antibodies to hnRNPs A, K and C, John Hanover for plasmid pXRGG and Brian Burke for antibody to Nup153. This work was supported by a grant from the National Institutes of Health (AI25105). K.E.G. is a recipient of a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research.
REFERENCES
- Adam S.A. (1999) Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr. Opin. Cell Biol., 11, 402–406. [DOI] [PubMed] [Google Scholar]
- Adam S.A., Marr,R.S. and Gerace,L. (1990) Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol., 111, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agol V.I., Belov,G.A., Bienz,K., Egger,D., Kolesnikova,M.S., Romanova, L.I., Sladkova,L.V. and Tolskaya,E.A. (2000) Competing death programs in poliovirus-infected cells: commitment switch in the middle of the infectious cycle. J. Virol., 74, 5534–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R., Lin,A., Enarson,M. and Burke,B. (1996) Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol., 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G.A., Evstafieva,A.G., Rubtsov,Y.P., Mikitas,O.V., Vartapetian, A.B. and Agol,V.I. (2000) Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology, 275, 244–248. [DOI] [PubMed] [Google Scholar]
- Bienz K., Egger,D., Rasser,Y. and Bossart,W. (1983) Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology, 131, 39–48. [DOI] [PubMed] [Google Scholar]
- Buendia B., Santa-Maria,A. and Courvalin,J.C. (1999) Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J. Cell Sci., 112, 1743–1753. [DOI] [PubMed] [Google Scholar]
- Clark M.E., Lieberman,P.M., Berk,A.J. and Dasgupta,A. (1993) Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol., 13, 1232–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L.I. and Blobel,G. (1986) Identification and characterization of a nuclear pore complex protein. Cell, 45, 699–709. [DOI] [PubMed] [Google Scholar]
- Doedens J.R. and Kirkegaard,K. (1995) Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J., 14, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- Etchison D., Milburn,S.C., Edery,I., Sonenberg,N. and Hershey,J.W.B. (1982) Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220 000 dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem., 257, 14806–14810. [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Luhrmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Fu X.D. (1995) The superfamily of arginine/serine-rich splicing factors. RNA, 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- Gradi A., Svitkin,Y.V., Imataka,H. and Sonenberg,N. (1998) Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl Acad. Sci. USA, 95, 11089–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.E. and Imperiale,M.J. (1998) Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol., 72, 7860–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her L.S., Lund,E. and Dahlberg,J.E. (1997) Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science, 276, 1845–1848. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. and Adam,S. (1998) Transport of macromolecules between the nucleus and the cytoplasm. RNA, 4, 351–364. [PMC free article] [PubMed] [Google Scholar]
- Joachims M., Van Breugel,P.C. and Lloyd,R.E. (1999) Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol., 73, 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Bachorik,J.L. and Dreyfuss,G. (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol., 145, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Love D.C., Sweitzer,T.D. and Hanover,J.A. (1998) Reconstitution of HIV-1 rev nuclear export: independent requirements for nuclear import and export. Proc. Natl Acad. Sci. USA, 95, 10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M.H., Bohnlein,S., Hauber,J. and Cullen,B.R. (1989) Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell, 58, 205–214. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- McBride A.E., Schlegel,A. and Kirkegaard,K. (1996) Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl Acad. Sci. USA, 93, 2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K., Pelletier,J. and Sonenberg,N. (1989) A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev., 3, 1026–1043. [DOI] [PubMed] [Google Scholar]
- Meerovitch K., Svitkin,Y.V., Lee,H.S., Lejbkowicz,F., Kenan,D.J., Chan,E.K., Agol,V.I., Keene,J.D. and Sonenberg,N. (1993) La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol., 67, 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W.M., Eder,P.S. and Dreyfuss,G. (1997) The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J., 16, 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1996) The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol., 134, 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Shaikh,S., Burke,B. and Dreyfuss,G. (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J., 18, 1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri J., Mitra,S., Andrei,C., Liu,Y., Yu,Y., Hitomi,M. and Tartakoff,A.M. (1999) An unexpected link between the secretory path and the organization of the nucleus. J. Biol. Chem., 274, 33785–33789. [DOI] [PubMed] [Google Scholar]
- Ostareck D.H., Ostareck-Lederer,A., Wilm,M., Thiele,B.J., Mann,M. and Hentze,M.W. (1997) mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89, 597–606. [DOI] [PubMed] [Google Scholar]
- Pante N., Bastos,R., McMorrow,I., Burke,B. and Aebi,U. (1994) Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol., 126, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S. and Dreyfuss,G. (1991) Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science, 253, 312–314. [DOI] [PubMed] [Google Scholar]
- Pollard V.W., Michael,W.M., Nakielny,S., Siomi,M.C., Wang,F. and Dreyfuss,G. (1996) A novel receptor-mediated nuclear protein import pathway. Cell, 86, 985–994. [DOI] [PubMed] [Google Scholar]
- Rosorius O., Heger,P., Stelz,G., Hirschmann,N., Hauber,J. and Stauber,R.H. (1999) Direct observation of nucleocytoplasmic transport by microinjection of GFP-tagged proteins in living cells. Biotechniques, 27, 350–355. [DOI] [PubMed] [Google Scholar]
- Rueckert R.R. (ed.) (1996) Picornaviridae: the Viruses and their Replication. Lippincott-Raven, Philadelphia, PA.
- Savory J.G., Hsu,B., Laquian,I.R., Giffin,W., Reich,T., Hache,R.J. and Lefebvre,Y.A. (1999) Discrimination between NL1- and NL2-mediated nuclear localization of the glucocorticoid receptor. Mol. Cell. Biol., 19, 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A. and Kirkegaard,K. (1995) Cell biology of enterovirus infection. In Rotbart,H.A. (ed.), Human Enterovirus Infections. American Society for Microbiology, Washington, DC, pp. 135–153.
- Schmidt-Zachmann M.S. and Nigg,E.A. (1993) Protein localization to the nucleolus: a search for targeting domains in nucleolin. J. Cell Sci., 105, 799–806. [DOI] [PubMed] [Google Scholar]
- Schneider R.J. and Shenk,T. (1987) Impact of virus infection on host cell protein synthesis. Annu. Rev. Biochem., 56, 317–332. [DOI] [PubMed] [Google Scholar]
- Shah S., Tugendreich,S. and Forbes,D. (1998) Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol., 141, 31–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroki K., Isoyama,T., Kuge,S., Ishii,T., Ohmi,S., Hata,S., Suzuki,K., Takasaki,Y. and Nomoto,A. (1999) Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol., 73, 2193–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber, R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Stoffler D., Fahrenkrog,B. and Aebi,U. (1999) The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol., 11, 391–401. [DOI] [PubMed] [Google Scholar]
- Suhy D.A., Giddings,T.H.,Jr and Kirkegaard,K. (2000) Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol., 74, 8953–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolskaya E.A., Romanova,L.I., Kolesnikova,M.S., Ivannikova,T.A., Smirnova,E.A., Raikhlin,N.T. and Agol,V.I. (1995) Apoptosis-inducing and apoptosis-preventing functions of poliovirus. J. Virol., 69, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman K.S., Shah,S., Powers,M.A. and Forbes,D.J. (1999) The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell, 10, 649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C., van Deursen,J.M.A., Rodrigues,J.P., Sitterlin,D., Bachi,A., Wu,X., Wilm,M., Carno-Fonseca,M. and Izaurralde,E. (2000) Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell, 6, 1243–1252. [DOI] [PubMed] [Google Scholar]
- Waggoner S. and Sarnow,P. (1998) Viral ribonucleoprotein complex formation and nucleolar–cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol., 72, 6699–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D.J., Banik-Maiti,S., DeGregori,J. and Ruley,H.E. (2000) hnRNP C is required for postimplantation mouse development but is dispensable for cell viability. Mol. Cell. Biol., 20, 4094–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff B., Sanglier,J.J. and Wang,Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol., 4, 139–147. [DOI] [PubMed] [Google Scholar]
- Yalamanchili P., Datta,U. and Dasgupta,A. (1997) Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol., 71, 1220–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]