Abstract
The EBNA1 protein of Epstein–Barr virus (EBV) mediates the partitioning of EBV episomes and EBV-based plasmids during cell division by a mechanism that appears to involve binding to the cellular EBP2 protein on human chromosomes. We have investigated the ability of EBNA1 and the EBV segregation element (FR) to mediate plasmid partitioning in Saccharomyces cerevisiae. EBNA1 expression alone did not enable the stable segregation of FR-containing plasmids in yeast, but segregation was rescued by human EBP2. The reconstituted segregation system required EBNA1, human EBP2 and the FR element, and functionally replaced a CEN element. An EBP2 binding mutant of EBNA1 and an EBNA1 binding mutant of EBP2 each failed to support FR-plasmid partitioning, indicating that an EBNA1–EBP2 interaction is required. The results provide direct evidence of the role of hEBP2 in EBNA1-mediated segregation and demonstrate that heterologous segregation systems can be reconstituted in yeast.
Keywords: CEN/EBNA1/EBP2/Epstein–Barr virus/segregation
Introduction
Epstein–Barr virus (EBV) genomes are maintained in latently infected human cells as low copy number, double-stranded circular DNA episomes (reviewed in Kieff, 1996). These episomes replicate once per cell cycle and are efficiently partitioned during cell division, resulting in a constant copy number per cell (Adams, 1987; Yates and Guan, 1991). The replication and partitioning of the episomes require two viral components: a cis-acting sequence called oriP and the EBNA1 protein (Yates et al., 1984, 1985; Lupton and Levine, 1985). oriP-containing plasmids that express EBNA1 are stably maintained in primate cells, making them useful gene delivery constructs (Yates et al., 1985). oriP is comprised of two functional components termed the dyad symmetry (DS) element and the family of repeats (FR), each of which contains multiple EBNA1 binding sites (Rawlins et al., 1985; Reisman et al., 1985). The DS element is the initiation site for DNA replication (Gahn and Schildkraut, 1989; Wysokenski and Yates, 1989; Harrison et al., 1994; Niller et al., 1995). The FR element enhances both DNA replication and transcription and is the viral segregation element (Lupton and Levine, 1985; Reisman and Sugden, 1986; Krysan et al., 1989; Wysokenski and Yates, 1989; Gahn and Sugden, 1995).
The EBNA1 protein fulfils several roles during EBV latent infection. First, EBNA1 is the origin binding protein that activates DNA replication from the oriP DS element (Yates and Camiolo, 1988; Harrison et al., 1994; Shire et al., 1999; Yates et al., 2000). Secondly, through interactions with the FR element of oriP, EBNA1 governs the segregation of EBV episomes and oriP-containing constructs during cell division (Krysan et al., 1989; Middleton and Sugden, 1994). Thirdly, when bound to the FR element, EBNA1 transactivates the expression of other latent viral gene products (Reisman and Sugden, 1986; Wysokenski and Yates, 1989; Gahn and Sugden, 1995). Finally, EBNA1 can also repress its own transcription by binding to sites downstream of the viral Qp promoter (Sample et al., 1992; Nonkwelo et al., 1996). The mechanisms by which EBNA1 contributes to viral replication and transcription are not yet known, but are likely to involve the recruitment of cellular factors.
EBNA1 is thought to govern the partitioning of EBV episomes by mediating their attachment to the cellular mitotic chromosomes. This segregation model is based on the observations that EBNA1, EBV episomes and oriP-containing constructs localize to the condensed cellular chromosomes during mitosis (Reedman and Klein, 1973; Grogan et al., 1983; Harris et al., 1985; Petti et al., 1990; Delecluse et al., 1993; Simpson et al., 1996). EBNA1 interacts stably with both the FR and DS elements of oriP (Hsieh et al., 1993), but binding to the FR element alone is sufficient for EBNA1-mediated partitioning (Chittenden et al., 1989; Krysan et al., 1989). The FR element contains 20 EBNA1 recognition sites, but only six to eight of these sites are required for episomal persistence (Chittenden et al., 1989; Wysokenski and Yates, 1989). The recognition sites are recognized by amino acids 459–607 of EBNA1; these residues form a complex structure that mediates both the DNA interactions and the dimerization of the protein (Ambinder et al., 1991; Chen et al., 1993; Bochkarev et al., 1995, 1996; Summers et al., 1996). Although the mechanism by which EBNA1 binds oriP has been well defined, the mechanism by which EBNA1 attaches to mitotic chromosomes is just beginning to be revealed.
We recently identified a human cellular protein, hEBP2 (EBNA1 binding protein 2), which interacts specifically with both the DNA-bound and unbound forms of EBNA1 (Shire et al., 1999). hEBP2 binds to a Gly/Arg-rich region of EBNA1 between amino acids 325 and 376, a region that is distinct from the DNA binding and dimerization domain of EBNA1 (Shire et al., 1999). An EBNA1 mutant lacking the EBP2 binding site (Δ325–376) supports the replication of oriP plasmids but is defective in mediating their segregation (Shire et al., 1999). This EBNA1 mutant is nuclear but fails to attach to the cellular chromosomes in mitosis (Wu et al., 2000). The ability of the 325–376 region to interact with mitotic chromosomes was also reported by Marechal et al. (1999). The behaviour of Δ325–376 provided genetic support for the model that EBNA1-mediated segregation involves chromosome attachment and also suggested that EBNA1 binding to EBP2 was important for segregation activity. To explore the relationship between EBNA1 and hEBP2 further, the localization of hEBP2 was examined. In both EBV-positive and -negative cell lines, hEBP2 is nucleolar in interphase and localizes to the condensed cellular chromosomes in mitosis (Chatterjee et al., 1987; Wu et al., 2000). The apparent co-localization of EBNA1 and hEBP2 on mitotic chromosomes suggested that hEBP2 is the component of the mitotic chromosomes with which EBNA1 interacts to mediate DNA segregation.
EBP2 is highly conserved in eukaryotes and the cellular function of this protein has been studied in Saccharomyces cerevisiae (yEBP2). yEBP2 is a nucleolar protein that plays an essential role in ribosome biogenesis; in the absence of functional yEBP2, cell division slows and eventually ceases as ribosome pools are depleted (Huber et al., 2000; Tsujii et al., 2000). Characterization of a temperature-sensitive mutant of yEBP2 revealed a specific role for yEBP2 in the processing of the 27 SA precursor of the 5.8S and 25S rRNA (Huber et al., 2000). Based on the extensive sequence homology between hEBP2 and yEBP2 and on their similar nucleolar localization, it is likely that the human and yeast versions of EBP2 fulfil the same cellular functions.
The studies to date suggest that the interaction of EBNA1 with hEBP2 is important for EBNA1-mediated partitioning, but proof of the requirement of hEBP2 for segregation was lacking. To provide direct evidence that hEBP2 is responsible for EBNA1-mediated segregation, we sought to identify a eukaryotic system that does not support the partitioning of plasmids containing the EBV segregation element (FR) in the presence of EBNA1, and to determine whether partitioning could be rescued by the expression of hEBP2. Here we show that EBNA1 expression alone was insufficient to support the stable segregation of FR-containing plasmids in budding yeast, but that EBNA1-mediated segregation was reconstituted by the addition of hEBP2.
Results
EBNA1 does not support FR-plasmid segregation in yeast
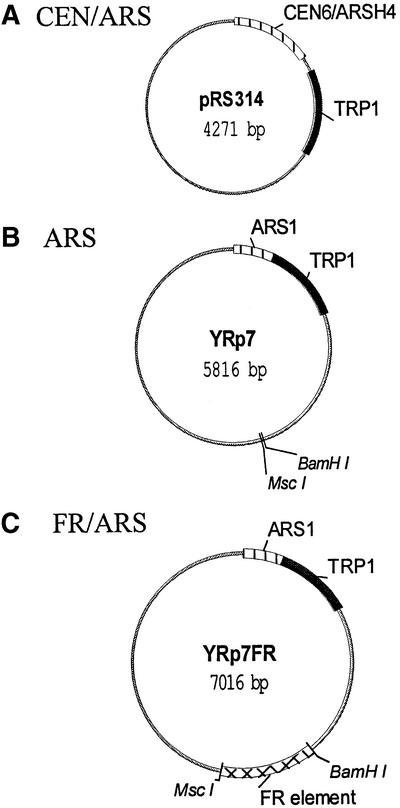
As part of our efforts to understand how EBNA1 mediates EBV segregation, we asked whether EBNA1 could mediate the partitioning of plasmids containing the EBV segregation element (the FR element from oriP) in S.cerevisiae. Yeast was chosen because the cis-acting sequence elements required for the initiation of DNA replication and DNA segregation are well defined and distinct. A segregation test construct (YRp7FR) was generated in which the EBV FR element was inserted into a plasmid (YRp7) that contains a yeast origin of replication (ARS1) and the TRP1 gene but lacks a yeast segregation element (Figure 1). An EBNA1 expression plasmid (p416MET.EBNA1) was also constructed in which EBNA1 was expressed from the MET25 promoter in p416MET25. The plasmid loss rate of YRp7FR, in the presence and absence of EBNA1 expression, was then compared with that of YRp7 and with a positive control segregation construct (pRS314) that contains both an ARS and a CEN element (Figure 1).
Fig. 1. Plasmids used in budding yeast assay for plasmid loss. The stabilities of three different plasmids, all of which contain an ARS element and a TRP1 selectable marker, were examined. (A) Positive control plasmid for stable segregation containing a yeast CEN element (Sikorski and Hieter, 1989). (B) Negative control segregation plasmid lacking a segregation element (Stinchcomb et al., 1979). (C) Experimental plasmid containing the EBV segregation element (FR).
For plasmid loss assays, the KY320 yeast strain was transformed to tryptophan (Trp) prototrophy with either YRp7FR, YRp7 or pRS314 and to uracil (Ura) prototrophy with either p416MET.EBNA1 or p416MET25. The ‘empty’ p416MET25 plasmid, which does not encode a recombinant protein, was used in instances where EBNA1 expression was not desired. The presence of this empty plasmid enabled all yeast samples to be grown under identical conditions (i.e. medium lacking uracil), reducing potential complications caused by growth under different conditions. The stability of the segregation plasmids was monitored by removing the Trp selection for a fixed amount of time and then plating serial dilutions of the cultures on plates that do and do not contain Trp. The Ura selection was maintained at all times to ensure maintenance of the EBNA1-expressing plasmid.
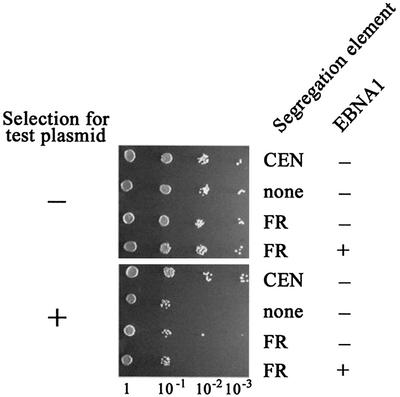
The results from a representative plasmid loss assay are shown in Figure 2. As expected, the CEN-containing plasmid was stably maintained, as indicated by the similar growth of this culture on selective and non-selective plates. In contrast, pronounced loss of YRp7, which lacks a segregation element, was indicated by the decreased growth of this culture on selective plates. The FR-containing segregation plasmid, in the presence and absence of EBNA1 expression, exhibited a plasmid loss profile that was indistinguishable from YRp7, indicating that EBNA1 did not mediate the stable segregation of FR-containing plasmids in yeast.
Fig. 2. Loss of YRp7FR in the presence and absence of EBNA1. Yeast (KY320) were transformed with one of the segregation plasmids shown in Figure 1 and a URA3 expression plasmid that did (+) or did not (–) contain the EBNA1 gene. After 48 h of growth (approximately eight doublings) without selection for the segregation plasmids, 10-fold dilutions of the cultures were plated on SC-Ura (no selection for test plasmid) or SC-Ura,Trp (selection for test plasmid) plates.
Human EBP2 rescues EBNA1 segregation function
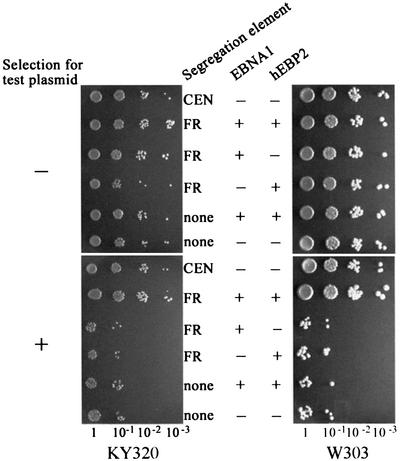
Previously, we have shown that the human cellular protein, hEBP2, interacts specifically with the EBNA1 sequences that mediate DNA partitioning, raising the possibility that hEBP2 binding is important for EBNA1-mediated segregation (Shire et al., 1999). To address the role of hEBP2 in plasmid partitioning further, we tested whether hEBP2 could rescue the segregation function of EBNA1 in yeast. To this end, the plasmid loss assay of the FR-containing plasmid was repeated in the presence of both EBNA1 and hEBP2 in the KY320 yeast strain. The assay was initially performed by expressing hEBP2 on a plasmid containing the LEU2 resistance marker; where hEBP2 expression was not desired, yeast were transformed with the empty LEU2 vector. Cells were grown without selection for the segregation test plasmid for 11 generations, while maintaining selection for the EBNA1 and EBP2 expression vectors. As shown in Figure 3, the YRp7FR construct segregated stably when both EBNA1 and hEBP2 were expressed. The percentage of cells that retained YRp7FR was compared with those for plasmids containing a CEN element or lacking a segregation element (Table I, KY320 column). In the presence of EBNA1 and hEBP2, YRp7FR was slightly more stable than the CEN/ARS plasmid and was ∼50-fold more stable than YRp7 over the assay period.
Fig. 3. Stable segregation of YRp7FR in the presence of EBNA1 and hEBP2. Plasmid loss assays were conducted over 11 generations in KY320 (left panel) or W303-1A (right panel) yeast strains containing a segregation plasmid, a URA3 expression plasmid that contains or lacks the EBNA1 gene, and a LEU2 expression plasmid that contains or lacks the hEBP2 gene. Dilutions of the cultures were then grown on SC-Ura,Leu (no selection for test plasmid) or SC-Ura,Leu,Trp (selection for test plasmid) plates.
Table I. Stability of segregation plasmids in the presence and absence of EBNA1 and hEBP2.
| Segregation plasmid | EBNA1 expression | hEBP2 expressiona | % Plasmid stabilityb |
||
|---|---|---|---|---|---|
| KY320 | W303-1A | KY320.hEBP2 | |||
| pRS314 | – | – | 53 | 74 | NA |
| pRS314 | – | + | NA | NA | 68 |
| YRp7FR | + | + | 62 | 80 | 71 |
| YRp7FR | + | – | 2.1 | 0.9 | NA |
| YRp7FR | – | + | 1.4 | 1.5 | 1.8 |
| YRp7 | + | + | 2.4 | 1.3 | 1.2 |
| YRp7FR | Δ325–376 | + | 0.9 | 0.7 | 2.4 |
| YRp7FR | + | 1–220 | 1.5 | 1.2 | NA |
| YRp7 | – | – | 1.3 | 1.1 | NA |
| YRp7 | – | + | NA | NA | 1.9 |
NA, not applicable.
ahEBP2 was expressed from the p425PGK.hEBP2 plasmid in KY320 and W303.A1, and from the integrated gene in KY320.hEBP2.
bPlasmid stabilities were determined after 11 generations as described in Materials and methods. Values shown are averages from two to four experiments for KY320 and W303.A1, and are the results of the single experiment in Figure 4 for KY320.hEBP2.
Having demonstrated the requirement for hEBP2 for the segregation of YRp7FR, we then assessed the requirements for EBNA1 and the FR element in this system. When the plasmids that express EBNA1 or hEBP2 were replaced by empty vectors, the segregation of YRp7FR was disrupted and loss rates similar to that of YRp7 were observed (Figure 3, left panel; Table I). Identical results were obtained when empty vectors were omitted from samples lacking EBNA1 or EBP2 expression (data not shown). Co-expression of EBNA1 and hEBP2 with YRp7, which lacks the FR element, did not improve the unstable segregation of this plasmid (Figure 3). Thus, efficient segregation of YRp7FR requires EBNA1, hEBP2 and the FR segregation element.
The above experiments were performed in yeast strain KY320, which has a very slow doubling time in minimal medium. To ensure that the observed segregation effect of hEBP2 was not particular to this strain, we repeated the plasmid loss experiments in the faster growing W303-1A strain. The results of plasmid loss assays performed over 11 generations (∼24 h) in W303-1A were essentially the same as those performed over 11 generations (∼72 h) in KY320 (Figure 3; Table I). As in KY320, stable segregation of YRp7FR in W303-1A was observed only when both EBNA1 and hEBP2 were expressed, and depended on the presence of the FR element. The stabilities of both the CEN/ARS plasmid and of YRp7FR in the presence of EBNA1 and hEBP2 were found to be somewhat increased in W303-1A compared with KY320.
The maintenance of YRp7FR was also examined in yeast strains containing an integrated hEBP2 gene. The hEBP2 gene, under control of the PGK promoter, was integrated into KY320 and W303-1A, and expression was confirmed by western blotting (data not shown). In the absence of EBNA1, YRp7FR was not stably maintained in these strains, but in the presence of EBNA1, stable maintenance indistinguishable from that of the CEN/ARS plasmid was observed (Figure 4 and Table I for KY320.hEBP2; data not shown for W303.hEBP2). As in the unintegrated strains, plasmid partitioning by EBNA1 in the hEBP2-integrated strains also required the FR segregation element.
Fig. 4. EBNA1-mediated segregation in KY320.hEBP2. KY320.hEBP2, which contains an integrated hEBP2 gene under control of a constitutively active promoter, was used in plasmid loss assays over 11 generations as described in Figure 2. The assays were conducted in the presence of either wild-type EBNA1 (+) or the EBNAΔ325–376 mutant (Δ325–376), or in the absence of EBNA1 (–).
EBNA1-mediated segregation in yeast requires EBP2 binding
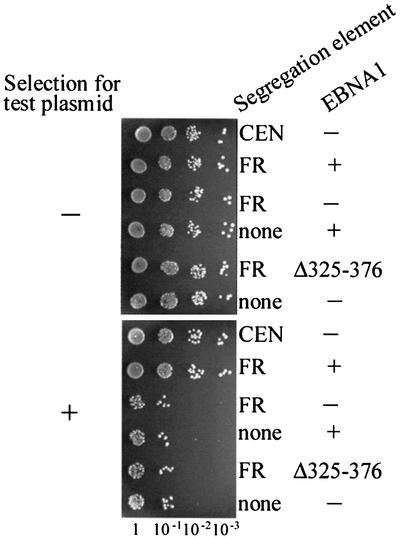
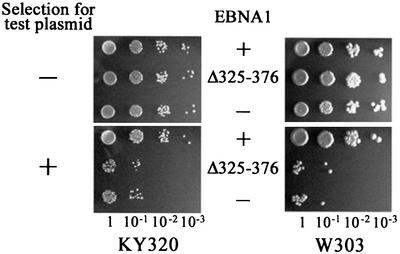
To confirm that EBNA1-mediated segregation in yeast requires the direct interaction of EBNA1 and hEBP2, we tested whether segregation was supported by the EBNAΔ325–376 mutant. We have previously shown that EBNAΔ325–376 supports replication from oriP in human cells but is defective in both EBP2 binding and FR-plasmid segregation in human cells (Shire et al., 1999). EBNAΔ325–376 was expressed from p416MET25 in yeast and its ability to maintain YRp7FR in the presence of hEBP2 was compared with that of wild-type EBNA1. The expression levels of EBNA1 and EBNAΔ325–376 were similar, as determined by western blotting (data not shown). Unlike wild-type EBNA1, EBNAΔ325–376 was unable to mediate the partitioning of YRpFR in either KY320 or W303-1A that contained integrated or plasmid-borne copies of hEBP2 (Figures 4 and 5; Table I). The results suggest that the mechanism by which EBNA1 governs plasmid segregation in yeast is the same as that in human cells and involves EBNA1 binding to EBP2.
Fig. 5. The hEBP2 binding region of EBNA1 is required for EBNA1-mediated segregation in yeast. The loss of YRp7FR was monitored in KY320 (left panel) and W303-1A (right panel) yeast strains in the presence of either wild-type EBNA1 (+) or the EBNAΔ325–376 mutant (Δ325–376), or in the absence of EBNA1 (–). All cultures expressed hEBP2 from p425PGK.hEBP2. Assays were conducted as in Figure 3.
The EBNA1 binding domain of EBP2 is required for EBNA1-mediated segregation
Budding yeast contain an essential gene that encodes a homologue of hEBP2 (referred to as yEBP2). However, the failure of EBNA1 to mediate FR-plasmid segregation in yeast in the absence of hEBP2 indicated that yEBP2 was unable to support the segregation function of EBNA1. Since our data suggested that a direct interaction between EBNA1 and EBP2 was required for segregation, we investigated the possibility that yEBP2 failed to support EBNA1 segregation due to failure to bind EBNA1. The interaction of EBNA1 with yeast and human EBP2 proteins was compared in a yeast two-hybrid assay, where EBNA1 was fused to the GAL4 DNA binding domain and EBP2 was fused to the GAL4 activation domain. While co-expression of EBNA1 and hEBP2 fusion proteins consistently resulted in the activation of both HIS3 and LACZ reporter genes under control of GAL4 binding sites, the co-expression of EBNA1 and yEBP2 fusion proteins did not detectably activate either gene (Figure 6B for HIS3 reporter; data not shown for LACZ reporter). Since western blot analysis confirmed the expression of the yEBP2 fusion protein in the two-hybrid assay (data not shown), the results suggest that EBNA1 cannot attach to yEBP2 in yeast cells.
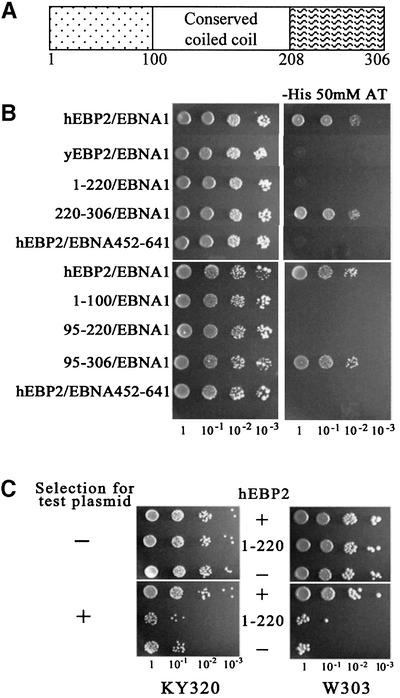
Fig. 6. The EBNA1 binding region of hEBP2 is required for EBNA1-mediated segregation. (A) Schematic representation of hEBP2 showing the most conserved region, which corresponds to a predicted coiled-coil domain. Amino acids numbers are indicated. (B) EBP2 from yeast (yEBP2) and humans (hEBP2), and hEBP2 fragments 1–220, 220–306, 1–100, 95–220 and 95–306 were tested for binding to EBNA1 in a yeast two-hybrid assay, as determined by activation of the HIS3 reporter gene. As a negative control, hEBP2 was tested for binding to EBNA452–641, which lacks the hEBP2 binding sequence. Ten-fold serial dilutions of log-phase cultures were grown on plates containing histidine (left panel) or lacking histidine and containing 50 mM AT (right panel). (C) Plasmid loss assay comparing the ability of hEBP2 (+) and the hEBP2 1–220 mutant (1–220) to support the segregation of YRp7FR in the presence of EBNA1. Assay results in the absence of hEBP2 expression (–) are also shown. Assays were conducted as in Figure 3.
To investigate the EBP2 sequence requirements for EBNA1 binding further, we mapped the EBNA1 binding domain of hEBP2. Clues to the structural organization of hEBP2 were discerned from predictive structural algorithms and from sequence alignments of EBP2 homologues from several eukaryotes (see Shire et al., 1999). These analyses indicated that EBP2 residues corresponding approximately to amino acids 100–208 of the human protein are the most highly conserved and also correspond to a coiled-coil domain (Figure 6A). We reasoned that this central region of EBP2 is probably a structural and functional domain, and divided EBP2 into N-terminal, central and C-terminal domains accordingly. EBP2 fragments spanning one or more of these regions were examined for their ability to bind to EBNA1 in the yeast two-hybrid assay as determined by activation of HIS3 (Figure 6B) and LACZ (data not shown) reporter genes. EBNA1 did not detectably bind to hEBP2 fragments that expressed the N-terminal (amino acids 1–100) and central (amino acids 95–220) domains individually, nor to a C-terminal truncation mutant, hEBP21–220, which spanned both of these domains. However, EBNA1 bound to the remaining C-terminal fragment of hEBP2 (amino acids 220–306) and to a fragment that spanned the central and C-terminal domains (amino acids 95–306), mapping the EBNA1 binding site in hEBP2 between amino acids 220 and 306.
We then asked whether the EBNA1 binding region of hEBP2 was required for hEBP2 to rescue the segregation function of EBNA1 in yeast. To this end, the stability of YRp7FR in the presence of EBNA1 was compared when hEBP2 and hEBP21–220 were expressed (Figure 6C; Table I). Unlike hEBP2, hEBP21–220 did not enable plasmid partitioning by EBNA1; plasmid loss rates in the presence of hEBP21–220 were indistinguishable from those in the absence of hEBP2. The failure of hEBP21–220 to mediate plasmid partitioning was not due to a low expression level, as this protein was expressed at higher levels than hEBP2 in this assay (data not shown). Thus, the mechanism by which hEBP2 mediates the segregation function of EBNA1 in yeast appears to involve EBNA1 binding. EBNA1 binding is not sufficient for the segregation activity of hEBP2, however, as hEBP2 amino acids 220–306 did not support EBNA1-mediated plasmid partitioning (data not shown).
Discussion
We have shown that plasmids containing the EBV segregation element can be efficiently partitioned in budding yeast in the presence of EBNA1 and human EBP2. This plasmid segregation system requires a version of EBNA1 that can bind hEBP2, a version of hEBP2 that can bind EBNA1, and the FR DNA element. In human cells, EBNA1-dependent plasmid segregation has also been shown to require the EBV FR element as well as the EBP2 binding sequence of EBNA1 (Krysan et al., 1989; Middleton and Sugden, 1994; Shire et al., 1999). Thus, the requirements for EBV-based plasmid segregation in the yeast system, as far as have been determined, are the same as those in human cells.
In EBV-infected human cells, EBNA1 is found throughout the nucleus in interphase and, in mitosis, localizes to the condensed cellular chromosomes (Reedman and Klein, 1973; Grogan et al., 1983; Petti et al., 1990). EBV episomes and oriP-containing constructs have also been observed to associate with the cellular mitotic chromosomes in the presence of EBNA1 (Harris et al., 1985; Delecluse et al., 1993; Simpson et al., 1996). These observations led to the hypothesis that EBNA1 partitions plasmids by mediating their attachment to the cellular mitotic chromosomes. This hypothesis was further supported by the finding that an EBNA1 mutant (Δ325–376), which supports the replication but not the long-term maintenance of oriP plasmids, fails to attach to the cellular chromosomes in mitosis (Shire et al., 1999; Wu et al., 2000). The EBNA1 sequences required for mitotic chromosome attachment are coincident with those required to bind to the cellular EBP2 protein, which localizes with EBNA1 on mitotic chromosomes (Shire et al., 1999; Wu et al., 2000). These findings suggested that EBNA1 attaches to the mitotic chromosomes through an interaction with EBP2. The possible requirement of hEBP2 for EBNA1-mediated segregation in human cells could not be tested, however, due to the essential role of this protein in cell proliferation (Huber et al., 2000). Our discovery that human EBP2 is required for EBNA1 to mediate plasmid segregation in yeast further supports the importance of EBP2 in EBNA1-mediated DNA partitioning.
Although the mechanism by which hEBP2 facilitates EBNA1-mediated plasmid segregation in yeast remains to be determined, the requirement for the EBP2 binding region of EBNA1 and the EBNA1 binding region of EBP2 suggests that the two proteins interact. In human cells, EBNA1 is constitutively bound to its recognition sites in the FR element (Hsieh et al., 1993), and this is also likely to be true in yeast. Based on the localization studies of hEBP2 in human cells, we envisage that, in yeast, the FR-bound EBNA1 would bind to hEBP2, which is or becomes associated with the yeast DNA. The tethering of EBNA1 to the cellular chromosomes probably requires two domains of hEBP2: one that binds EBNA1 (residues 220–306) and a second that associates with the cellular chromosomes (not yet localized). The EBNA1-bound plasmids would remain tethered to the yeast chromosomes throughout mitosis, resulting in approximately half of the plasmids being delivered to the daughter cell. This segregation model implies that in yeast, as in mammalian cells, efficient plasmid partitioning can occur by attachment to cellular chromosomes, a possibility that is also consistent with the segregation behaviour of yeast 2-µm plasmids (Velmurugan et al., 2000).
The failure of EBNA1 to mediate plasmid partitioning in yeast not expressing hEBP2 indicates that yEBP2 does not support EBNA1-mediated segregation. This lack of activity may simply be due to the inability of EBNA1 to bind to yEBP2, as indicated by two-hybrid analysis, but could also reflect differences in the way in which yEBP2 and hEBP2 interact with yeast chromosomes. EBNA1 binds to the C-terminal 86 amino acids of hEBP2, but binding to yEBP2 was not detected. The sequences of the C-terminal domains in the human and yeast EBP2 proteins are 36% identical and 54% similar, with divergent sequences spread throughout the length of the domain. It is not yet clear which of these divergent sequences account for the differential interactions with EBNA1.
Attachment to cellular chromosomes appears to be a common way in which low copy number episomes are partitioned in mammalian cells. Studies on the segregation of bovine papillomavirus (BPV) genomes have shown their association with cellular chromosomes in mitosis (Lehman and Botchan, 1998; Skiadopoulos and McBride, 1998). This association is dependent on the viral E2 protein, which also binds the mitotic chromosomes (Lehman and Botchan, 1998; Skiadopoulos and McBride, 1998), and on the cis-acting minichromosome maintenance element, which contains multiple E2 binding sites (Piirsoo et al., 1996; Ilves et al., 1999). E2 sequences in the N-terminal activation domain as well as phosphorylated serine residues in the central hinge region have been found to be important for chromosome attachment and segregation of BPV genomes (Lehman et al., 1997; Lehman and Botchan, 1998; Skiadopoulos and McBride, 1998). More recently, the episomal genomes of the Kaposi’s sarcoma-associated herpesvirus (KSHV) and the KSHV-encoded LANA protein have been observed to co-localize on mitotic chromosomes (Ballestas et al., 1999; Cotter and Robertson, 1999). LANA was also shown to be the only viral protein required for the persistence of KSHV DNA (Ballestas et al., 1999). Thus, the segregation of BPV and KSHV genomes probably occurs via E2- and LANA-mediated tethering to the cellular chromosomes, respectively. The cellular chromosomal components with which E2 and LANA interact to mediate viral partitioning are not yet clear and it will be interesting to determine whether EBP2 plays a role in these systems. We note, however, that although the E2 and EBNA1 DNA binding domains share a remarkable structural homology (Hegde et al., 1992; Bochkarev et al., 1995), neither E2 nor LANA shares any obvious sequence homology with EBNA1 that would suggest common interacting proteins.
Materials and methods
Plasmid loss assay constructs
Plasmid loss assays were conducted using the centromeric plasmid pRS314 as a positive control (Sikorski and Hieter, 1989) and the replicating plasmid YRp7, which lacks a centromere, as a negative control (Stinchcomb et al., 1979). The segregation test plasmid YRp7FR was constructed by excising the oriP FR element from pGEMoriP (Frappier and O’Donnell, 1991) with BamHI and MscI, and inserting it between the BamHI and MscI sites of YRp7. The EBNA1-expressing vector p416MET.EBNA1 was constructed by excising the EBNA1 gene (lacking most of the Gly–Ala repeat) from pEBNA1 (Shire et al., 1999) with XbaI and BamHI, and inserting it between the XbaI and BamHI sites in p416MET25, downstream of the MET25 promoter. p416MET25 is a low copy expression vector containing the CEN6/ARSH4 element and the URA3 marker (Mumberg et al., 1994). A similar construct, p416MET. EΔ325–376, which expresses the EBNA1 mutant lacking amino acids 325–376 (in addition to most of the Gly–Ala repeat), was also constructed. The gene encoding this EBNA1 mutant was excised from pAS2.EΔ325–376 (Shire et al., 1999) with NdeI and BamHI, filled in with DNA polymerase Klenow and inserted into the SmaI site of p426MET25. To construct the hEBP2 expression vector, p425PGK.hEBP2, the gene for hEBP2 was PCR amplified from pVLEBP2 (Shire et al., 1999), phosphorylated and inserted into the BglII site of pR425/PGK (Marcus et al., 1995), which had been filled in with DNA polymerase Klenow. The resulting construct expresses hEBP2 from the PGK promoter. The pR425/PGK construct that expresses hEBP21–220 was generated by PCR amplification of hEBP2 codons 1–220 from pVLEBP2 using primers containing SmaI sites. After SmaI digestion, the hEBP21–220 fragment was inserted into the blunted BglII site of pR425/PGK.
Yeast two-hybrid constructs
Plasmids pAS2.EBNA1 and pAS2.E452–641 express full-length EBNA1 (lacking most of the Gly–Ala repeat) and EBNA1 amino acids 452–641, respectively, as fusions to the GAL4 DNA binding domain. Their construction has been described previously (Shire et al., 1999). pACT.yEBP2 expresses yEBP2 fused to the GAL4 activation domain from an ADH promoter. It was constructed by PCR amplification of the yEBP2 gene with primers containing SmaI and XhoI sites and insertion of the SmaI/XhoI-digested fragments between the SmaI and XhoI sites in pACTII (Li et al., 1994). pACT63 is a cDNA library construct isolated through its interaction with EBNA1 in a two-hybrid screen (Shire et al., 1999). pACT63 expresses hEBP2 amino acids 21–306 fused to the GAL4 activation domain. Plasmids that express hEBP2 amino acids 1–100, 1–220, 95–220, 95–306 or 220–306 fused to the GAL4 activation domain were generated by PCR amplification of the appropriate hEBP2 fragment from pVLEBP2 using primers containing an XmaI site. The PCR fragments were digested with XmaI and inserted in the XmaI site of pACTII.
Yeast strains
The yeast strains KY320 (MATa leu2-PET56 ura3-52 trp1-Δ1 lys2-801am ade2-101oc his3-Δ200 GAL+) and W303-1A (MATa leu2-3 112 ura3-1 trp1-1 ade2-1 his3-11 can1-100) were used for the plasmid loss assays (Chen and Struhl, 1988; Bailis and Rothstein, 1990). Y190 (MATa leu2-3 112 ura3-52 trp1-901 his3-Δ200 ade2-101 GAL4 Δgal80 ΔURA3 GAL-lacZ LYS GAL-HIS3 Cyhr), which contains integrated HIS3 and LACZ reporter genes under control of GAL4 binding sites, was used for the yeast two-hybrid assays (Harper et al., 1993). Yeast strains containing an integrated copy of the hEBP2 gene were constructed using the plasmid pRS305.hEBP2. To construct pRS305.hEBP2, the fragment containing the hEBP2 gene and the PGK promoter was excised from p425PGK.hEBP2 with HindIII and inserted in the HindIII site of pRS305, a yeast integration vector containing the LEU2 marker (Sikorski and Hieter, 1989). The resulting plasmid contains two EcoRV sites: one in the multiple cloning sequence and one in the LEU2 gene. The EcoRV site in the multiple cloning sequence was eliminated by linearization with BamHI (in the multicloning sequence) and partial digestion with EcoRV, filling in the ends with DNA polymerase Klenow and re-ligating. To generate integrated yeast strains KY320.hEBP2 and W303.hEBP2, pRS305.hEBP2 was linearized within the LEU2 gene with EcoRV and used to transform either KY320 or W303-1A by the lithium acetate method for site-directed integration at the LEU2 locus (Rothstein, 1991; Gietz and Schiestl, 1995). Transformants from SC-Leu plates were then grown in YPD and analysed for hEBP2 expression by western blotting.
Plasmid loss assays
For plasmid loss assays, KY320, W303-1A, KY320.hEBP2 or W303.hEBP2 were transformed to Trp prototrophy with either pRS314 (positive segregation control), YRp7 (negative segregation control) or YRp7FR segregation plasmids, and to Ura prototrophy with a p426MET25 vector that did or did not express EBNA1. Where indicated, KY320 and W303-1A were also transformed to Leu prototrophy with pR425/PGK that did or did not express hEBP2. Positive transformants were grown in selective medium (SC-Trp,Ura,Leu when pR425/PGK vectors were present and SC-Trp,Ura when pR425/PGK vectors were absent) until early to mid-log phase and then diluted 100-fold into medium that did not select for the segregation plasmid (SC-Ura,Leu or SC-Ura) and grown for 11 generations (except where indicated). Ten-fold serial dilutions of the cultures were then generated where the least diluted culture had an OD600 of 0.1. Five microlitres of each dilution were spotted onto selective (SC-Trp,Ura,Leu or SC-Trp,Ura) and non-selective (SC-Ura,Leu or SC-Ura) plates with respect to the segregation plasmid. To quantify the percentage of cells that retained the segregation plasmids, equal amounts of diluted cultures were also spread on the selective and non-selective plates, and the resulting colonies were counted. Plasmid stability was measured as a ratio of the number of colonies on selective versus non-selective plates.
Yeast two-hybrid assays
Y190 was transformed to Trp and Leu prototrophy by the lithium acetate transformation method with either pAS1.EBNA1 and pACT63 (positive control), pAS1.E452–641 and pACT63 (negative control) or pAS1.EBNA1 and a pACTII vector expressing a hEBP2 fragment. Positive transformants were grown overnight in SC-Trp,Leu to saturation. The saturated cultures were diluted into SC-Trp,Leu,His medium and further grown until mid-log phase. Interactions between EBNA1 and the hEBP2 fragments were determined by activation of the HIS3 reporter gene. Activation of the HIS3 gene was measured by spotting 5 µl of 10-fold serial dilutions of the cultures at an OD600 of 0.5 on SC-Trp,Leu plates and on SC-Trp,Leu,His plates that contained 50 mM 3-aminotriazol (AT). A positive interaction was indicated by the growth of cultures on both types of plates.
Acknowledgments
Acknowledgements
We thank Dr Brenda Andrews and members of the Andrews laboratory for W303-1A, yeast expression plasmids and helpful advice. We also thank Dr Chris Brandl for KY320, Dr Vicki Athanasopoulos for help in construction of the figures and Dr Jim Smiley for the suggestion to study EBNA1-mediated segregation in yeast. This work was supported by a grant from the Medical Research Council of Canada (MRC). L.F. is an MRC scientist.
REFERENCES
- Adams A. (1987) Replication of latent Epstein–Barr virus genomes. J. Virol., 61, 1743–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambinder R.F., Mullen,M., Chang,Y., Hayward,G.S. and Hayward,S.D. (1991) Functional domains of Epstein–Barr nuclear antigen EBNA-1. J. Virol., 65, 1466–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis A.M. and Rothstein,R. (1990) A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination. Genetics, 126, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestas M.E., Chatis,P.A. and Kaye,K.M. (1999) Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science, 284, 641–644. [DOI] [PubMed] [Google Scholar]
- Bochkarev A., Barwell,J., Pfuetzner,R., Furey,W., Edwards,A. and Frappier,L. (1995) Crystal structure of the DNA binding domain of the Epstein–Barr virus origin binding protein EBNA1. Cell, 83, 39–46. [DOI] [PubMed] [Google Scholar]
- Bochkarev A., Barwell,J., Pfuetzner,R., Bochkareva,E., Frappier,L. and Edwards,A.M. (1996) Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin binding protein, EBNA1, bound to DNA. Cell, 84, 791–800. [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Freeman,J.W. and Busch,H. (1987) Identification and partial characterization of a Mr 40 000 nucleolar antigen associated with cell proliferation. Cancer Res., 47, 1123–1129. [PubMed] [Google Scholar]
- Chen M.-R., Middeldorp,J.M. and Hayward,S.D. (1993) Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J. Virol., 67, 4875–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. and Struhl,K. (1988) Saturation mutagenesis of a yeast his3 ‘TATA element’: genetic evidence for a specific TATA-binding protein. Proc. Natl Acad. Sci. USA, 85, 2691–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden T., Lupton,S. and Levine,A.J. (1989) Functional limits of oriP, the Epstein–Barr virus plasmid origin of replication. J. Virol., 63, 3016–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter M.A. and Robertson,E.S. (1999) The latency-associated nuclear antigen tethers the Kaposi’s sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology, 264, 254–264. [DOI] [PubMed] [Google Scholar]
- Delecluse H.-J., Bartnizke,S., Hammerschmidt,W., Bullerdiek,J. and Bornkamm,G.W. (1993) Episomal and integrated copies of Epstein–Barr virus coexist in Burkitt’s lymphoma cell lines. J. Virol., 67, 1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L. and O’Donnell,M. (1991) Overproduction, purification and characterization of EBNA1, the origin binding protein of Epstein–Barr virus. J. Biol. Chem., 266, 7819–7826. [PubMed] [Google Scholar]
- Gahn T.A. and Schildkraut,C.L. (1989) The Epstein–Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell, 58, 527–535. [DOI] [PubMed] [Google Scholar]
- Gahn T. and Sugden,B. (1995) An EBNA1 dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein–Barr virus LMP gene. J. Virol., 69, 2633–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D. and Schiestl,R.H. (1995) Transforming yeast with DNA. Methods Mol. Cell. Biol., 5, 255–269. [Google Scholar]
- Grogan E.A., Summers,W.P., Dowling,S., Shedd,D., Gradoville,L. and Miller,G. (1983) Two Epstein–Barr viral nuclear neoantigens distinguished by gene transfer, serology and chromosome binding. Proc. Natl Acad. Sci. USA, 80, 7650–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Adami,G.R., Wei,N., Keyomarsi,K. and Elledge,S.J. (1993) The p21 Cdk-interaction protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Harris A., Young,B.D. and Griffin,B.E. (1985) Random association of Epstein–Barr virus genomes with host cell metaphase chromosomes in Burkitt’s lymphoma-derived cell lines. J. Virol., 56, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S., Fisenne,K. and Hearing,J. (1994) Sequence requirements of the Epstein–Barr virus latent origin of DNA replication. J. Virol., 68, 1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R.S., Grossman,S.R., Laimins,L.A. and Sigler,P.B. (1992) Crystal structure at 1.7Å of the bovine papillomavirus-1 E2 DNA-binding protein bound to its DNA target. Nature, 359, 505–512. [DOI] [PubMed] [Google Scholar]
- Hsieh D.-J., Camiolo,S.M. and Yates,J.L. (1993) Constitutive binding of EBNA1 protein to the Epstein–Barr virus replication origin, oriP, with distortion of DNA structure during latent infection. EMBO J., 12, 4933–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M., Dworet,J., Shire,K., Frappier,L. and McAlear,M. (2000) The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem., 275, 28764–28773. [DOI] [PubMed] [Google Scholar]
- Ilves I., Kivi,S. and Ustav,M. (1999) Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol., 73, 4404–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. (1996) Epstein–Barr virus and its replication. In Fields,D.M.K.B.N., Knipe,P.M. and Howley,P.M. (eds), Epstein–Barr Virus and its Replication. Lippincott-Raven, Philadelphia, PA, pp. 2343–2396.
- Krysan P.J., Haase,S.B. and Calos,M.P. (1989) Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol., 9, 1026–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman C.W. and Botchan,M.R. (1998) Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl Acad. Sci. USA, 95, 4338–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman C.W., King,D.S. and Botchan,M.R. (1997) A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J. Virol., 71, 3652–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Elledge,S.J., Peterson,C.A., Bales,E.S. and Legerski,R.J. (1994) Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl Acad. Sci. USA, 91, 5012–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton S. and Levine,A.J. (1985) Mapping of genetic elements of Epstein–Barr virus that facilitate extrachromosomal persistence of Epstein–Barr virus-derived plasmids in human cells. Mol. Cell. Biol., 5, 2533–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S.L., Miyata,K.S., Rachubinski,R.A. and Capone,J.P. (1995) Transactivation by PPAR/RXR heterodimers in yeast is potentiated by exogenous fatty acid via a pathway requiring intact peroxisomes. Gene Expr., 4, 227–239. [PMC free article] [PubMed] [Google Scholar]
- Marechal V., Dehee,A., Chikhi-Brachet,R., Piolot,T., Coppey-Moisan,M. and Nicolas,J. (1999) Mapping EBNA1 domains involved in binding to metaphase chromosomes. J. Virol., 73, 4385–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton T. and Sugden,B. (1994) Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein–Barr virus replication protein EBNA1. J. Virol., 68, 4067–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Muller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niller H.H., Glaser,G., Knuchel,R. and Wolf,H. (1995) Nucleoprotein complexes and DNA 5′-ends at oriP of Epstein–Barr virus. J. Biol. Chem., 270, 12864–12868. [DOI] [PubMed] [Google Scholar]
- Nonkwelo C., Skinner,J., Bell,A., Rickinson,A. and Sample,J. (1996) Transcription start site downstream of the Epstein–Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells defines a fourth promoter for expression of the EBV EBNA1 protein. J. Virol., 70, 623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti L., Sample,C. and Kieff,E. (1990) Subnuclear localization and phosphorylation or Epstein–Barr virus latent infection nuclear proteins. Virology, 176, 563–574. [DOI] [PubMed] [Google Scholar]
- Piirsoo M., Ustav,E., Mandel,T., Stenlund,A. and Ustav,M. (1996) cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J., 15, 1–11. [PMC free article] [PubMed] [Google Scholar]
- Rawlins D.R., Milman,G., Hayward,S.D. and Hayward,G.S. (1985) Sequence-specific DNA binding of the Epstein–Barr virus nuclear antigen (EBNA1) to clustered sites in the plasmid maintenance region. Cell, 42, 859–868. [DOI] [PubMed] [Google Scholar]
- Reedman B.M. and Klein,G. (1973) Cellular localization of an Epstein–Barr virus (EBV)-associated complement-fixing antigen in producer and non-producer lymphoblastoid cell lines. Int. J. Cancer, 11, 499–520. [DOI] [PubMed] [Google Scholar]
- Reisman D. and Sugden,B. (1986) Trans activation of an Epstein–Barr viral transcripitonal enhancer by the Epstein–Barr viral nuclear antigen 1. Mol. Cell. Biol., 6, 3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D., Yates,J. and Sugden,B. (1985) A putative origin of replication of plasmids derived from Epstein–Barr virus is composed of two cis-acting components. Mol. Cell. Biol., 5, 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. (1991) Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–301. [DOI] [PubMed] [Google Scholar]
- Sample J., Henson,E.B.D. and Sample,C. (1992) The Epstein–Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J. Virol., 66, 4654–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shire K., Ceccarelli,D.F.J., Avolio-Hunter,T.M. and Frappier,L. (1999) EBP2, a human protein that interacts with sequences of the Epstein–Barr nuclear antigen 1 important for plasmid maintenance. J. Virol., 73, 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson K., McGuigan,A. and Huxley,C. (1996) Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol. Cell. Biol., 16, 5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiadopoulos M.H. and McBride,A.A. (1998) Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol., 72, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb D.T., Struhl,K. and Davis,R.W. (1979) Isolation and characterization of a yeast chromosomal replicator. Nature, 282, 39–43. [DOI] [PubMed] [Google Scholar]
- Summers H., Barwell,J.A., Pfuetzner,R.A., Edwards,A.M. and Frappier,L. (1996) Cooperative assembly of EBNA1 on the Epstein–Barr virus latent origin of replication. J. Virol., 70, 1228–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujii R., Miyoshi,K., Tsuno,A., Matsui,Y., Toh-e,A., Miyakawa,T. and Mizuta,K. (2000) Ebp2p, yeast homologue of a human protein that interacts with Epstein–Barr virus nuclear antigen 1, is required for pre-RNA processing and ribosomal subunit assembly. Genes Cells, 5, 543–553. [DOI] [PubMed] [Google Scholar]
- Velmurugan S., Yang,X.-M., Chan,A.S.-M., Dobson,M. and Jayaram,M. (2000) Partitioning of the 2-µm circle plasmid of Saccharomyces cerevisiae: Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J. Cell Biol., 149, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Ceccarelli,D.F.J. and Frappier,L. (2000) The DNA segregation mechanism of the Epstein–Barr virus EBNA1 protein. EMBO rep., 1, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysokenski D.A. and Yates,J.L. (1989) Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein–Barr virus. J. Virol., 63, 2657–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J.L. and Camiolo,S.M. (1988) Dissection of DNA replication and enhancer activation functions of Epstein–Barr virus nuclear antigen 1. Cancer Cells, 6, 197–205. [Google Scholar]
- Yates J.L. and Guan,N. (1991) Epstein–Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol., 65, 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J.L., Warren,N., Reisman,D. and Sugden,B. (1984) A cis-acting element from the Epstein–Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl Acad. Sci. USA, 81, 3806–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J.L., Warren,N. and Sugden,B. (1985) Stable replication of plasmids derived from Epstein–Barr virus in various mammalian cells. Nature, 313, 812–815. [DOI] [PubMed] [Google Scholar]
- Yates J.L., Camiolo,S.M. and Bashaw,J.M. (2000) The minimal replicator of Epstein–Barr virus oriP. J. Virol., 74, 4512–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]